Abstract
Cell proliferation and survival require the faithful maintenance and propagation of genetic information, which are threatened by the ubiquitous sources of DNA damage present intracellularly and in the external environment. A system of DNA repair, called the DNA damage response (DDR), detects and repairs damaged DNA and prevents cell division until the repair is complete. Here we report that methylation at the 6 position of adenosine (m6A) in RNA is rapidly (within 2 minutes) and transiently induced at DNA damage sites in response to UV. This modification occurs on numerous poly(A)+ transcripts and is regulated by the methyltransferase METTL31 and the demethylase FTO2. In the absence of METTL3 catalytic activity, cells showed delayed repair of UV-induced cyclobutane pyrimidine (CPD) adducts and elevated sensitivity to UV, demonstrating the importance of m6A in the UV-responsive DDR. Multiple DNA polymerases are involved in the UV response, some of which resynthesize DNA after the lesion has been excised by the nucleotide excision repair (NER) pathway3, while others participate in trans-lesion synthesis (TLS) to allow replication past damaged lesions in S phase4. DNA polymerase κ (Pol κ), which has been implicated in both NER and TLS5,6, required the catalytic activity of METTL3 for immediate localization to UV-induced DNA damage sites. Importantly, Pol κ over-expression qualitatively suppressed the CPD removal defect associated with METTL3 loss. Taken together, we have uncovered a novel function for RNA m6A modification in the UV-induced DDR, and our findings collectively support a model whereby m6A RNA serves as a beacon for the selective, rapid recruitment of Pol κ to damage sites to facilitate repair and cell survival.
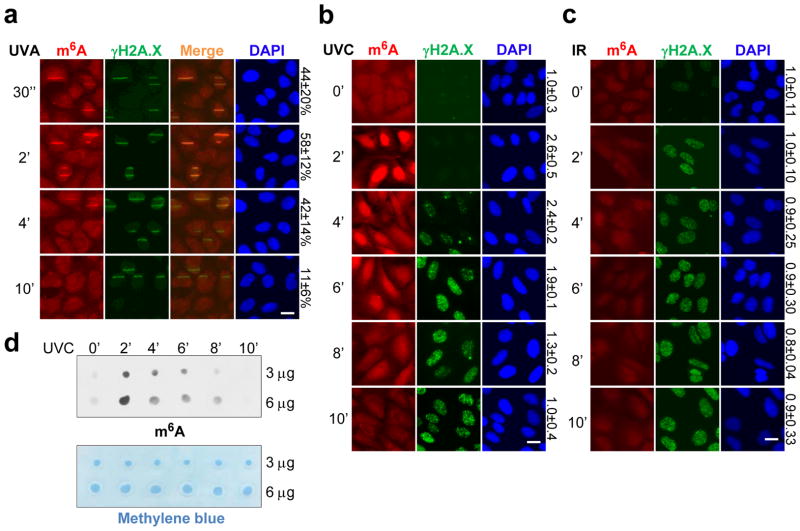
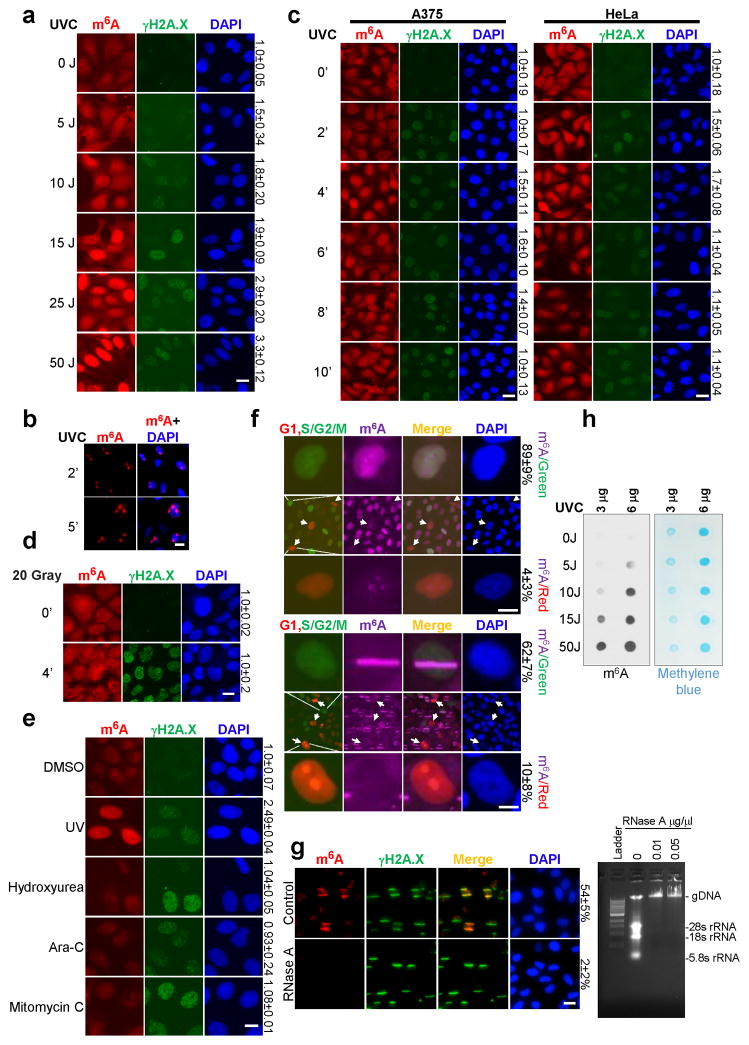
An early step of the DDR includes chemical modifications to chromatin7 that make the region accessible to repair factors and prevent transcription off a damaged template8. To identify novel chromatin regulatory events involved in the DDR, we screened for chromatin factors and modifications localized to damage sites. Surprisingly, we found that an antibody recognizing m6A-modified nucleic acid strongly stained DNA damage sites in U2OS cells generated by UV laser micro-irradiation (Fig. 1a). The signal accumulated in nuclei upon global UVC irradiation in a dose-dependent manner (Fig. 1b, Extended Data Fig. 1a), and localized to damage sites upon focused irradiation (Extended Data Fig. 1b). The staining intensity following laser microirradiation or global UVC irradiation exceeded cytoplasmic levels, peaking at 2 minutes after irradiation, and diminishing over the following 8 minutes (Fig. 1a–b). A375 melanoma and HeLa cells exhibited a similar response (Extended Data Fig. 1c). The response appeared specific to UV damage, as induction of damage by γ-irradiation (Fig. 1c, Extended Data Fig. 1d) or DNA damaging chemicals (Extended Data Fig. 1e) did not induce m6A. Analysis of cell-cycle reporter lines9 suggested that the m6A response was excluded from G1 cells (Extended Data Fig. 1f), providing a possible explanation for the incomplete penetrance of the effect (Fig. 1a). These results suggest that induction of m6A occurs generally in response to UV.
Figure 1. m6A modification on RNA accumulates at sites of DNA damage after UV exposure.
a–c, U2OS cells were subjected or not (0′) to UVA laser (a), 15 J UVC irradiation (b), or 10 Gray γ-irradiation, incubated at 37 °C for the indicated time, and costained for m6A and γH2A.X. The percentage of γH2A.X-positive cells displaying colocalizing m6A signal (a) and relative m6A intensity (b–c) are indicated. d, Poly(A)+ RNA from samples in (b) was subjected to dot-blot analysis with an antibody recognizing m6A. Loading control: methylene blue.
As the m6A antibody recognizes both modified DNA and RNA10, we investigated which type of nucleic acid was modified following UV irradiation. RNase A treatment of cells abrogated m6A accumulation at damage sites (Extended Data Fig. 1g). The poly(A)+ RNA pool (but not total RNA (data not shown)) displayed a UV dose-dependent peak of m6A 2 minutes after irradiation (Fig. 1d, Extended Data Fig. 1h), suggesting that the majority of the signal was derived from poly(A)+ RNA. The reported roles for m6A methylation involve regulation of RNA fate-- such as stability11, translation12–14, splicing15–17, and miRNA processing18,19, and promoting differentiation20, pluripotency21, X chromosome inactivation22, and responses to cellular stresses12,14,15. Our results now demonstrate that methylation of poly(A)+ RNA occurs in response to UV irradiation, suggesting an unprecedented RNA-mediated response to DNA damage.
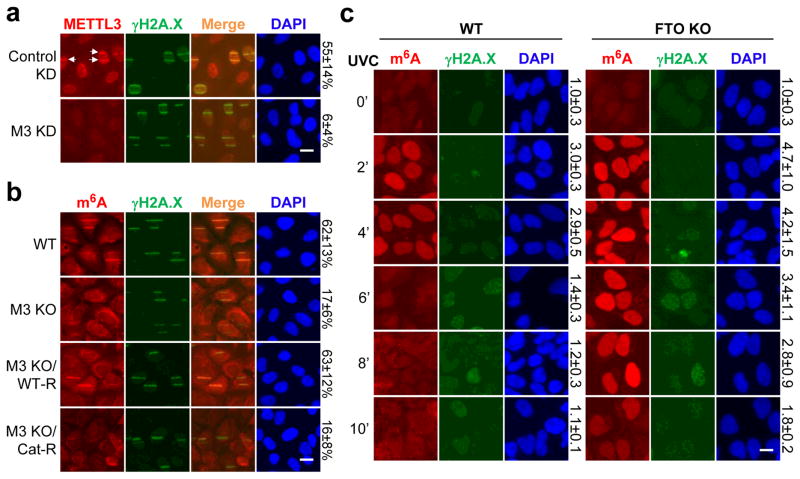
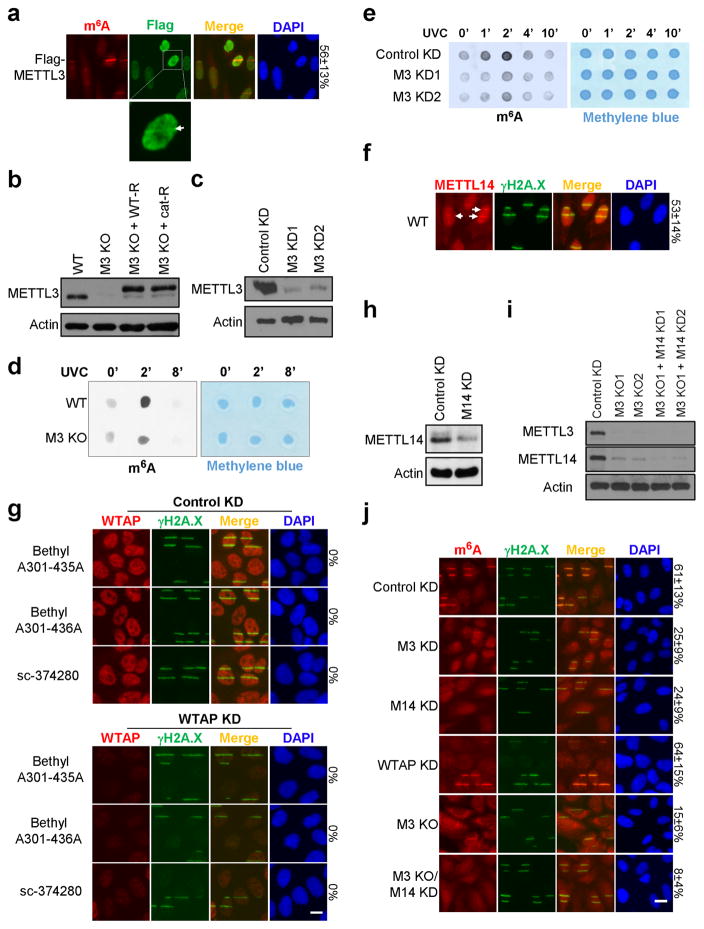
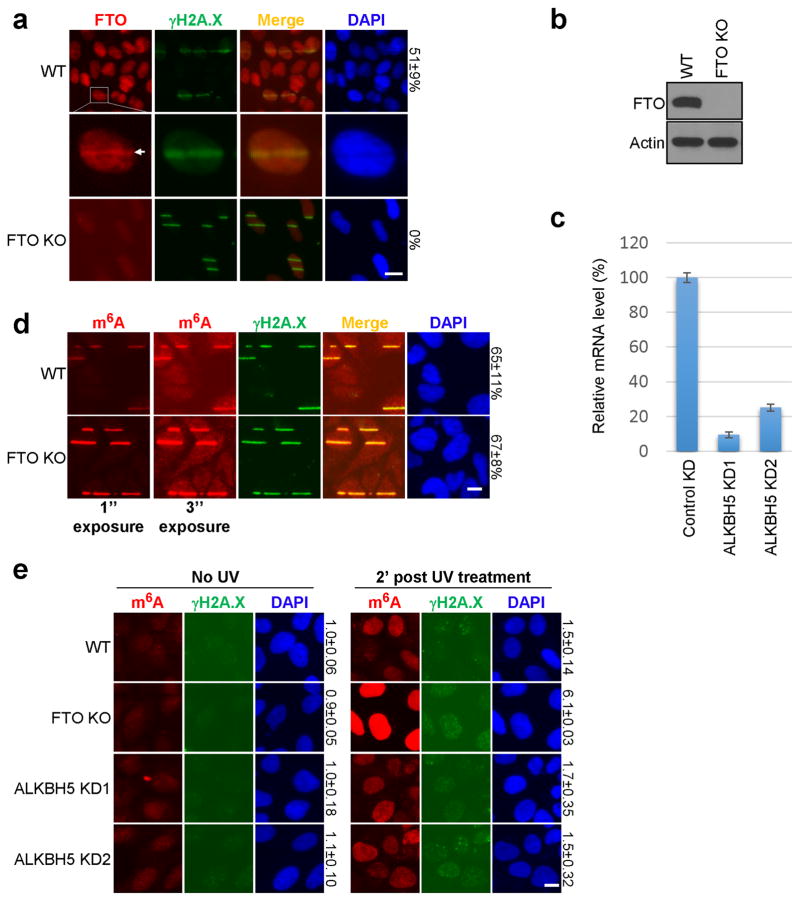
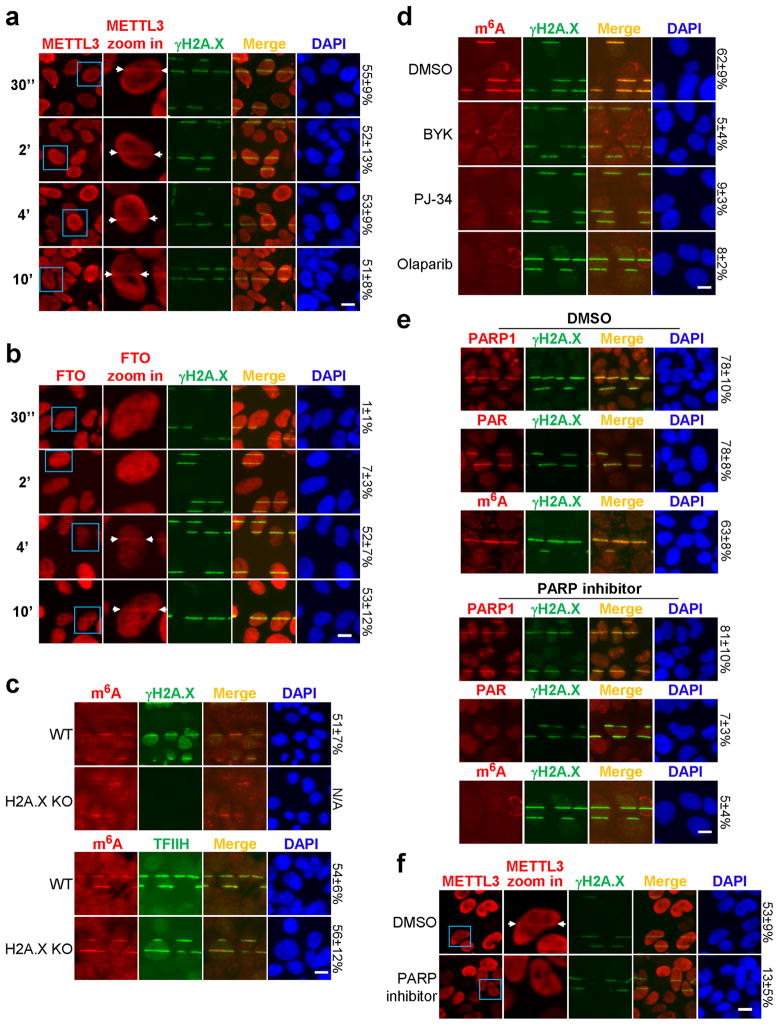
We next identified the enzymes responsible for regulating RNA methylation. The known m6A methyltransferase METTL323 localized within 2 minutes to sites of UV-induced damage (Fig. 2a, Extended Data Fig. 2a), and METTL3 catalytic activity was required for the m6A RNA response (Fig. 2b, Extended Data Fig. 2b–e). Of the two METTL3 cofactors, METTL14 and WTAP1,16, only METTL14 localized to damage sites (Extended Data Fig. 2f–g) and was needed for full m6A RNA induction (Extended Data Fig. 2h–j). Cells lacking both METTL3 and METTL14 (Extended Data Fig. 2i) exhibited nearly background levels of m6A RNA (Extended Data Fig. 2j) and displayed reduced viability compared to METTL3 KO cells (data not shown), consistent with m6A RNA suppressing apoptotic pathways15. Because the m6A signal rapidly disappeared from damage sites (Fig. 1a), we next investigated whether the RNA was actively demethylated. The known demethylase FTO2, localized to damage sites (Extended Data Fig. 3a) and its loss (Extended Data Fig. 3b) (but not that of the known demethylase ALKBH524 (Extended Data Fig. 3c)) increased the intensity of m6A RNA at laser-induced damage sites (Extended Data Fig. 3d), and increased both the nuclear intensity and duration of the m6A signal following UVC irradiation (Fig. 2c, Extended Data Fig. 3e). Although BrdU was incorporated into cellular DNA before UV irradiation to enhance the DDR, as described25, localized m6A RNA also occurred in UV-irradiated FTO KO cells lacking BrdU (data not shown), and no m6A RNA was detected in γ-irradiated cells containing BrdU (Fig. 1c). These results suggest that the m6A response is UV-specific and triggered by DNA damage, as it is enhanced by BrdU incorporation in DNA. Notably, METTL3 localization preceded that of FTO at damage sites, possibly allowing for a brief window of m6A RNA accumulation (Extended Data Fig. 4a–b). Of two critical early regulators of the DDR— PARP and γH2A.X7—only PARP was required for m6A RNA accumulation following UV irradiation (Extended Data Fig. 4c–e), possibly by recruiting METTL3 to damage sites (Extended Data Fig. 4f). These results demonstrate that METTL3, METTL14, and FTO dynamically regulate m6A RNA at DNA damage sites, and that PARP is required for m6A RNA induction in response to UV.
Figure 2. METTL3/14 and FTO oppositely regulate m6A RNA at DNA damage sites.
a, Control or METTL3 (M3 KD) U2OS cells were microirradiated, incubated at 37°C for 2 min, and stained as indicated. Arrows: representative γH2A.X-positive damage sites with METTL3 colocalization. b, WT or METTL3 KO U2OS cells expressing or not WT (WT-R) or catalytically inactive (Cat-R) METTL3, treated as in (a). c, WT or FTO KO U2OS cells subjected or not (0′) to 50 J UVC, treated as in (a). The percentage of γH2A.X-positive damage sites displaying colocalizing METTL3 (a), m6A (b), or relative m6A intensity (c), are indicated. n=3, 50 cells per replicate. Scale bar, 20 μm.
To identify UVC-dependent transcriptome-wide methylation patterns, we performed immunoprecipitations of m6A methylated RNA, followed by sequencing (MeRIP-seq)15. Methylation peaks in poly(A)+ RNAs displayed reproducible methylation patterns consistent with previous studies15 (Extended Data Fig. 5a–c), with UV exposure leading to an increase in 5′ UTR methylation (Extended Data Fig. 5c–d), as described in response to cellular stresses14. Sequence analysis of m6A RNAs revealed the known METTL3 target site GGACU15 (Extended Data Fig. 5e), and additional degenerate motifs (Extended Data Fig. 5f). Gene ontology (GO) analysis of METTL3-dependent, UV-induced methylated transcripts (Extended Data Fig. 5g–i) identified several weakly enriched categories (Enrichment Scores ≤ 2.35), including “DNA damage” (data not shown). As detailed below, a primary role for the m6A-marked transcripts is to recruit a DNA polymerase to sites of damage, but we cannot rule out that these transcripts may themselves play a role in the UV response as reported for other types of cellular stresses12,14,15.
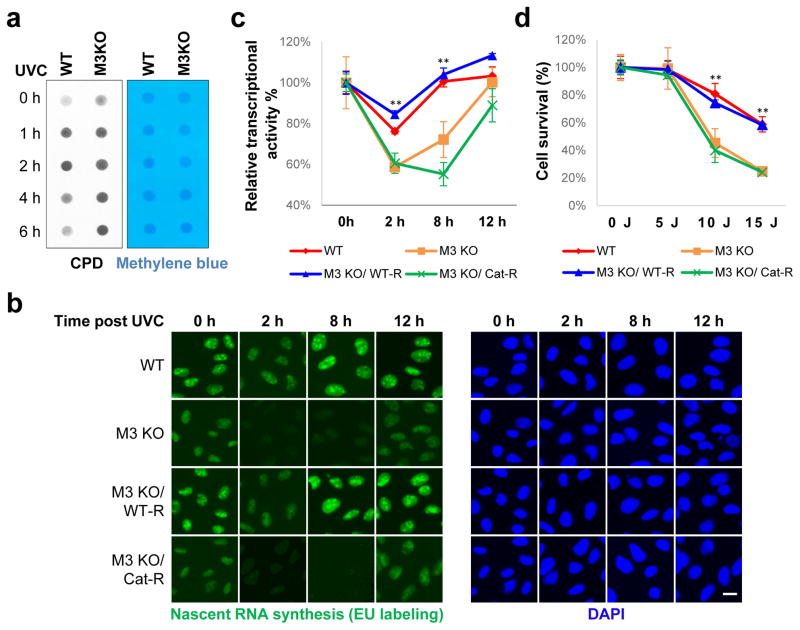
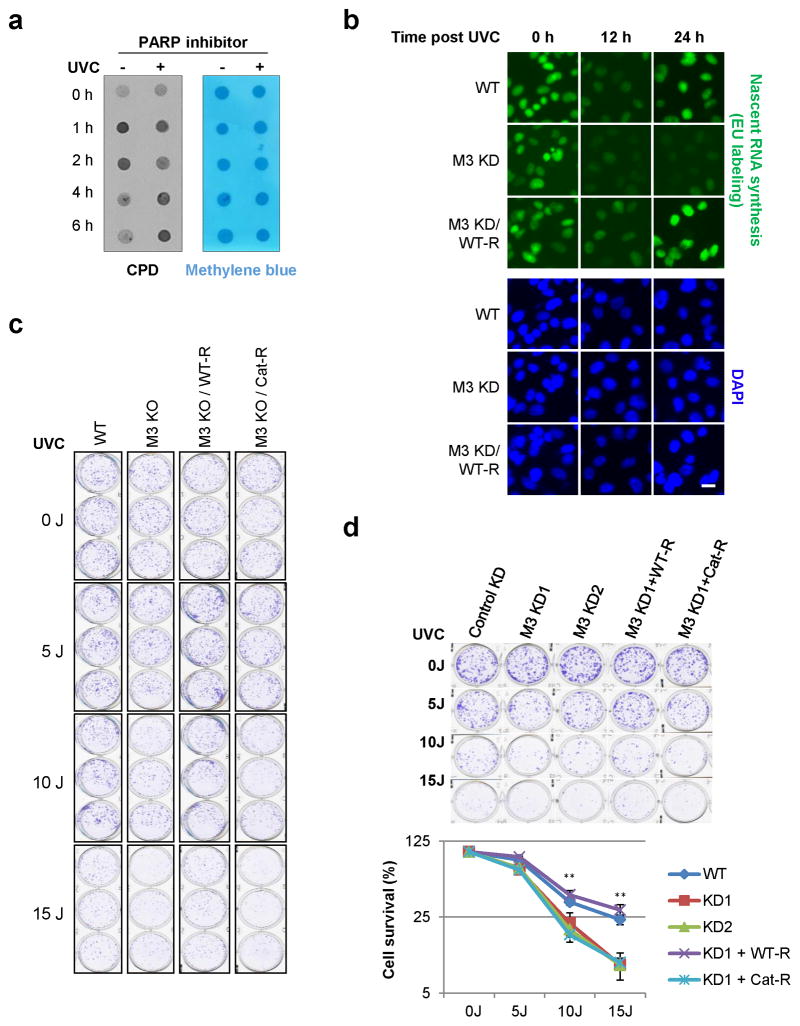
We next investigated whether RNA m6A modification affected the repair process. By performing dot-blots of genomic DNA using an antibody recognizing CPDs, the primary DNA lesion induced by UV exposure26, we found that METTL3 KO cells exhibited a delay in CPD removal (Fig. 3a), which was recapitulated by PARP inhibition (Extended Data Fig. 6a). Consistently, METTL3 catalytic activity was required for timely transcription re-initiation following DNA damage (Fig. 3b–c, Extended Data Fig. 6b). Although the signal intensity of newly-synthesized transcripts varied between cells in METTL3 KD cultures, the differences between individual KD cells were much smaller than the global differences observed between control and KD populations. Finally, we found that METTL3 catalytic activity was required for robust cell survival following 10–15 J of UV irradiation (Fig. 3d, Extended Data Fig. 6c–d). Although we cannot exclude the possibility that transcripts regulated by METTL3 in the absence of UV exposure may also contribute to the DDR, our results have collectively identified a novel role for METTL3-mediated, rapid RNA methylation in promoting the repair of UV-induced lesions and cellular resistance to UV.
Figure 3. METTL3 is important for UV-induced DNA damage repair and cell survival.
a, Dot-blot detection of CPDs in genomic DNA of WT or METTL3 KO (M3 KO) U2OS cells, subjected or not (0 h) to 15 J UVC. Loading control: Methylene blue. b, Nascent RNA in cells from (a) expressing or not WT (WT-R) or catalytically inactive (Cat-R) METTL3, subjected or not (0 h) to 25 J UVC and incubated at 37°C as indicated. (a–b) n=3, 50 cells per replicate. Scale bar, 20 μm. c, Quantification of (b) with each time-point normalized to its unirradiated control. d, Normalized colony formation of UVC-irradiated cell lines from (b). n≥3, results shown as mean ± SEM. (**) P ≤ 0.01.
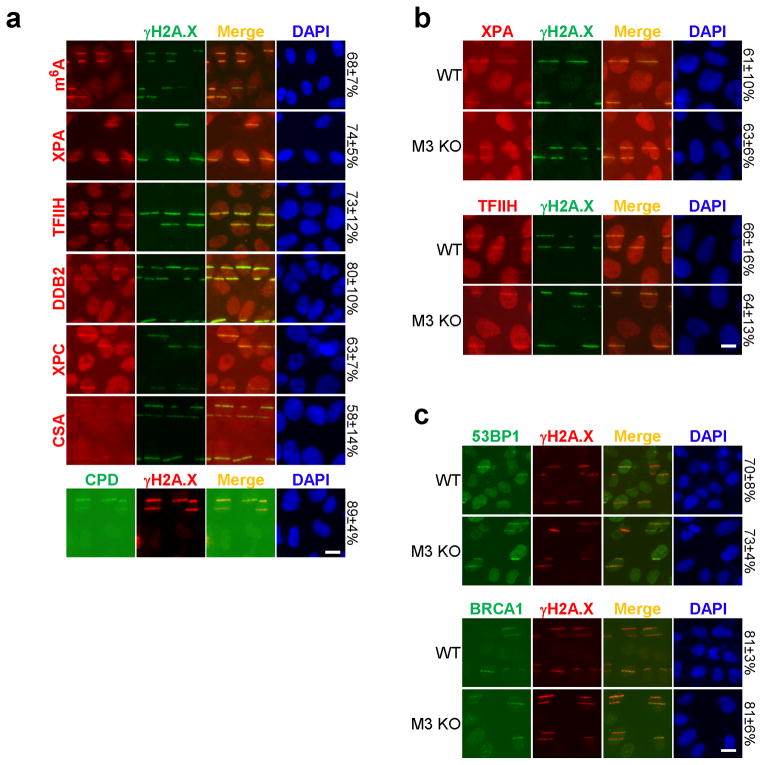
We next investigated how m6A RNA contributes to DNA repair and cell survival following UV damage. UV-induced lesions are repaired by the nucleotide excision repair (NER) pathway, which employs the sequential action of proteins that recognize the damaged lesion (DDB2, XPC, and CSA and CSB), proteins that verify the lesion (XPA and RPA), proteins that remove nucleotides encompassing the damage site (TFIIH, XPG, XPF), and DNA polymerases (δ and ε) that resynthesize the excised region (reviewed in3). Trans-lesion synthesis (TLS), conducted by multiple polymerases4, promotes replication across damage sites during S phase. The core NER factors were recruited to CPD-containing damage sites, and although their recruitment coincided with m6A RNA localization (Fig. 1a, Extended Data Fig. 7a), METTL3 was not required for recruiting the NER factors XPA or TFIIH (Extended Data Fig. 7b), nor the double-strand break (DSB) repair proteins 53BP1 or BRCA1 (Extended Data Fig. 7c), suggesting that RNA methylation is not necessary to initiate canonical NER or DSB repair.
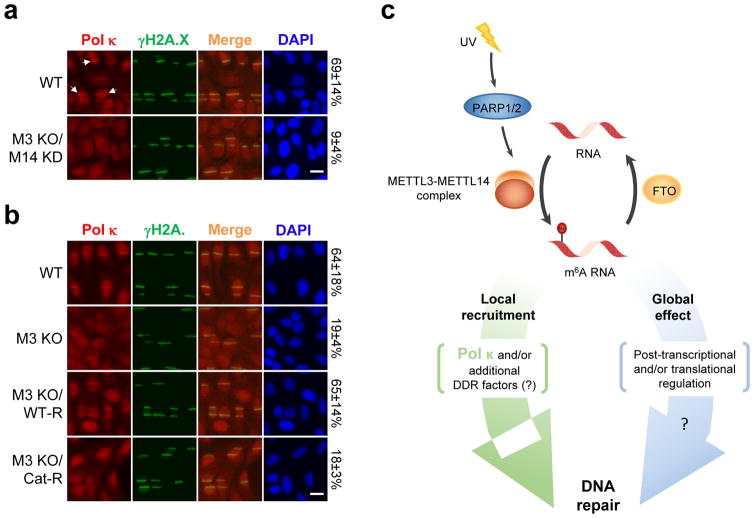
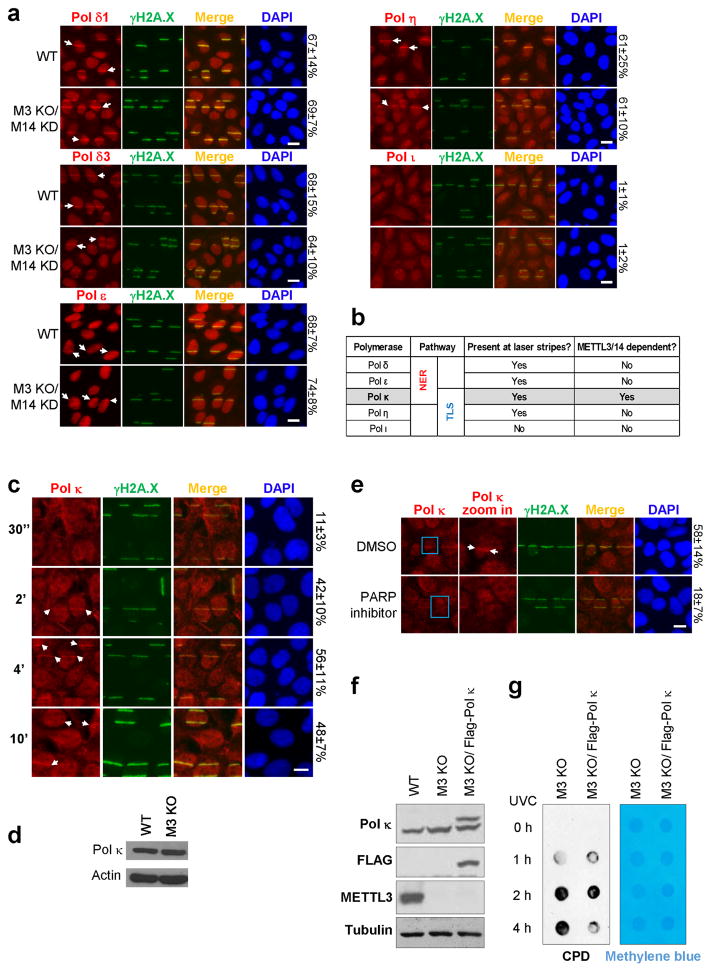
As DNA polymerases are the downstream effectors of the repair pathways responsible for UV resistance, we next investigated whether UV-induced m6A RNA methylation regulated their localization to DNA damage sites. We found that Pol κ, which operates in both NER and TLS pathways5,6, localized to damage sites simultaneously with m6A RNA, and was the only tested polymerase requiring METTL3 and METTL14 for its recruitment (but not expression) (Fig. 4a, Extended Data Fig. 8a–d), in a manner dependent on the catalytic activity of METTL3 (Fig. 4b). Consistent with our observation that PARP functions upstream of m6A RNA methylation, PARP inhibition also diminished Pol κ recruitment (Extended Data Fig. 8e). Importantly, Pol κ over-expression qualitatively rescued the CPD removal defect (Extended Data Fig. 8f–g), suggesting that Pol κ is a key effector of METTL3 in CPD repair, and that its role is distinct from those of polymerases δ, ε, and η, which localize to damage sites independently of METTL3 (Extended Data Fig. 8a–b). We did not find evidence of known m6A RNA-binding proteins localizing to damage sites (Extended Data Fig. 9a) or mediating Pol κ localization (Extended Data Figs. 9b, 10a–d), nor of Pol κ binding directly and specifically to m6A RNA (data not shown). It remains possible that Pol κ might recognize sequence/structural RNA features not represented in our binding assays, as described for other RNA-protein interactions17,27. These results raise the possibility that a novel reader may mediate Pol κ recruitment to DNA damage sites by m6A RNA to ensure efficient repair.
Figure 4. Pol κ localization to damage sites is METTL3-dependent.
a, WT or METTL3 KO/METTL14 KD (M3 KO/M14 KD) U2OS cells were microirradiated, incubated at 37°C for 4 min, and stained as indicated. Arrows: representative γH2A.X-positive damage sites with colocalizing Pol κ. b, WT or METTL3 KO (M3 KO) U2OS cells expressing or not WT (WT-R) or catalytically inactive (Cat-R) METTL3 treated as in (a). (a–b) The percentage of cells showing Pol κ and γH2A.X colocalization is indicated. n=3, 50 cells per replicate. Scale bar, 20 μm. c, Proposed model of m6A RNA regulation and role in DNA repair.
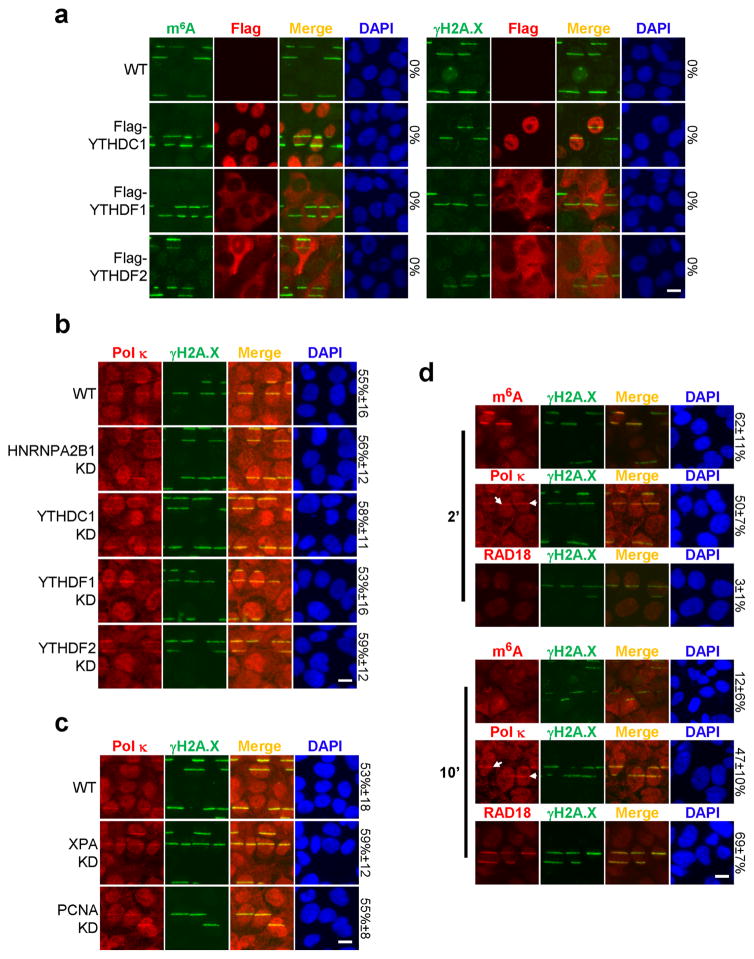
Multiple factors have been implicated in recruiting Pol κ to damage sites in different contexts, and we investigated their involvement in recruiting Pol κ to UV-induced damage sites immediately after irradiation. XPA, which is required for Pol κ recruitment at 30 minutes28, was dispensable at the 2 minute mark (Extended Data Figs. 9c, 10e). Although Pol κ recruitment to DNA lesions 30 minutes or later following damage requires mono-ubiquitination of PCNA by Rad1829, the early localization of Pol κ (at 2 minutes) occurred prior to Rad18 localization (Extended Data Fig. 9d) and even in PCNA KD cells (Extended Data Figs. 9c, 10f). These results suggest that the canonical NER and Rad18 pathways are not required for early METTL3-mediated recruitment of Pol κ to damage sites. At 10 minutes following irradiation, when m6A RNA had decreased nearly to background levels (Fig. 1a–b), Pol κ and Rad18 were simultaneously detectable at damage sites (Extended Data Fig. 9d), suggesting that Rad18 may be responsible for late-stage Pol κ localization. The distinct mechanisms of early and late Pol κ recruitment may operate in different cell-cycle phases, which could possibly account for the published observations that Pol κ was not detected at UV damage sites at 30 minutes in S-phase and XPA mutant cells28, whereas METTL3-dependent recruitment occurred at 2 minutes in the absence of XPA (Extended Data Fig. 9c) and presumably in S/G2 cells (Extended Data Fig. 1f). Taken together, our results suggest that PARP, METTL3, methylated RNA, and Pol κ might constitute a repair pathway operating separately from the canonical NER and TLS systems.
We have reported the identification of RNA m6A methylation as a novel and essential component of the DNA repair system responding to UV-induced lesions (Fig. 4c) that is critical for localizing Pol κ to damage sites for DNA repair (Fig. 4a). Globally, UV-induced RNA methylation could conceivably also affect the expression of genes participating in DNA repair. Although DNA damage is random, some transcripts were reproducibly methylated, and we speculate that these transcripts may be regulated separately from those at sites of damage.
Our results suggest that the m6A RNA- Pol κ pathway operates separately from the canonical NER pathway and Rad18/PCNA-regulated TLS pathway, but the precise role of Pol κ in UV damage repair remains unknown. A canonical role for Pol κ in repair synthesis (i.e. filling in DNA gaps after strand excision) would not be expected to elevate CPDs in the genome when its function is compromised, as these lesions would have been removed in the prior excision step. Pol κ might act as a TLS polymerase to facilitate lesion bypass during S phase (preventing lethal replication fork stalling and/or collapse, and allowing the NER or another system to repair damage subsequently4), and it may additionally participate in gap-filling after nucleotide excision, and/or perform a novel function in NER. The TLS model is consistent with Pol κ loss leading to the accumulation of UV-induced 6-4 pyrimidine-pyrimidone dimers5, Pol κ-deficient cells being uniquely sensitive to UV irradiation in S phase30, and our observation (Extended Data Fig. 9c) that Pol κ localizes to lesion-containing sites in XPA KD cells (which do not excise damaged DNA). Taken together, our results demonstrate a novel role for RNA methylation in promoting cellular resistance to UV damage, and identify a potential new pathway involving METTL3, m6A RNA, and Pol κ in the early UV-induced DNA damage response, with a key role for methylated RNA in recruiting Pol κ to damage sites for efficient DNA repair.
Methods
Plasmids
cDNAs coding METTL3, YTHDC1, YTHDF1, YTHDF2, or Pol κ were cloned into pENTR-D-TOPO (Invitrogen) and recombined into the pHAGE-CMV-Flag-HA Gateway destination vector for lentiviral expression. Lentiviral shRNA constructs for METTL3, METTL14, WTAP, ALKBH5, PCNA, XPA, YTHDC1, YTHDF1, YTHDF2, or HNRNPA2B1 were obtained by cloning into the pLKO.1 vector. shRNA sequences used are described in the Oligonucleotides section. The catalytically inactive METTL3 was produced by site-directed mutagenesis to convert the “DPPW” motif to “APPW” or “APPA” and confirmed by sequencing. Both mutants are expected to lack catalytic activity based on the literature31, and both performed equivalently in our assays. To generate a METTL3 clone resistant to shRNA#3 (see below), the WT sequence starting at nucleotide 1144 (counting the A of the start codon as +1), GACGTCAGTATCTTGGGCAAGTT was mutated to the synonymous GATGTTTCCATTCTAGGAAAATT.
Cell culture and viral transduction
HEK293T, U2OS, HeLa, A375, MEFs and FUCCI cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 4,500 mg/L glucose, 10% heat-inactivated fetal bovine serum, 110 mg/L sodium pyruvate, 2 mM L-glutamine and 100 U/ml penicillin plus 100 μg/ml streptomycin. H2A.X−/− MEFs were provided by Dr A. Nussenzweig (National Cancer Institute, Bethesda, MD)32. FUCCI-U2OS cells were provided by Dr S. Elledge (Harvard Medical School, Boston, MA)9. The PARP inhibitors Olaparib (MedKoo), BYK (MedKoo), or PJ34 (MedKoo) were added to cell culture media at a final concentration of 10 μM, 2 to 8 h prior to irradiation. U2OS cells were treated with hydroxyurea (1 mM, Sigma-Aldrich), cytarabine (Ara-C, 50 nM, Sigma-Aldrich), or mitomycin C (300 ng/ml, Sigma-Aldrich) for 12 h before detection of m6A and H2A.X levels by immunofluorescence microscopy analysis.
Lentiviruses were produced by cotransfection of the lentiviral plasmid with helper vectors (pHDM-VSV-G, pHDM-tat1b, pHDM-HgPM2, and pRC-CMV-RaII) into HEK293T cells, and viral supernatants were collected after 60–72 h. Infected cells were selected with 1 μg/ml puromycin after transduction.
For shRNA-mediated knock-down, cells were infected with virus encoding the indicated shRNA, and selected with 1 μg/ml puromycin for 48 h. Knock-down efficiency was assessed by western blot. To generate CRISPR knock-outs, cells were transiently transfected with the desired Lenti-CrisprV2 plasmids and, beginning on the following day, were selected with puromycin (1 μg/ml) for 48 h. Cells were then transferred to media lacking puromycin, serially diluted, and single-cell cloned in 96-well plates. Clones were screened for knock-out by western blot and immunofluorescence, and verified by sequencing.
UV irradiation and immunofluorescence microscopy
Cells were cultured for 24–48 h prior to irradiation with 10 μM BrdU25 (Sigma-Aldrich cat. #B9285). UVA laser (50 mW) irradiation was conducted using an inverted microscope (Eclipse Ti; Nikon) with a Palm microbeam laser microdissection workstation. UVC irradiation was conducted in a Stratalinker 2400 (Stratagene) at 50 J, or at the indicated doses. Following irradiation, cells were incubated at 37 °C for the indicated times, washed once with cold PBS, and then fixed with PBS containing 4% paraformaldehyde (PFA) for 10 min at room temperature. Cells were then washed three times with cold PBS and permeabilized with PBS containing 0.25% Triton-X100 for 30 min. After permeabilization, cells were washed extensively with IF blocking buffer (PBS containing 1% BSA, 10% FBS, 0.25% Triton-X100, 0.02% NaN3), and then incubated with the indicated antibodies diluted in blocking buffer at room temperature for 1–2 h or at 4°C overnight. Staining was conducted with fluorescent secondary antibodies (conjugated with Alexa Fluor 350, 488, 594, or 647; Invitrogen, cat. #s A-21244, A-11037, A11032, A11034, A11029) and 1 μg/ml DAPI (Invitrogen, cat. #D1306) where indicated. For CPD staining, permeabilized cells were denatured with 2N HCl for 15 min, then neutralized with 100 mM Tris-HCl (pH 8.5) for 5 min before blocking. Images were captured on Nikon Eclipse Ti microscope. Raw images were exported into Adobe Photoshop, and any linear adjustments were applied to all images in a given experiment. For quantification of laser stripes or any fluorescent signals, images of 50–100 irradiated cells were analyzed in triplicate, or as otherwise indicated. EU signal was quantified using ImageJ software.
For RNase A treatment, cells were subjected to UV laser microirradiation, incubated 2 min at 37°C, washed twice with cold PBS, and then permeabilized with PBS containing 1% Triton-X100 for 1 min on ice. Cells were then washed three times with cold PBS and incubated with RNase A (Sigma-Aldrich, cat. #R6513) at the indicated concentrations in 1X RNase H buffer (NEB, cat. #M0297S) for 5 min at 37°C. Cells were then washed twice with cold PBS, fixed with PBS containing 4% PFA for 10 min at room temperature, washed again with cold PBS, and then stained as previously described.
Transcriptional recovery assay
U2OS cells were grown and irradiated with UVC as described above, and then incubated at 37°C for 2, 8, 12 or 24 hours. For each sample of unirradiated and irradiated cells, 5-ethynyl uridine (EU) (1 mM final) was added to the culture medium for 3 hours to label nascent RNA, and then the cells were fixed and permeabilized as previously described. EU labeling of RNAs was detected using the Click-iT RNA Imaging Kit (Life Technologies, cat. #C10329), following the manufacturer’s protocol. The original data used to generate the graph in Fig. 3c is provided in Source Data for Fig. 3.
Colony formation assay
Cells were plated at a density of 50–350 cells per cm2, irradiated with the indicated doses of UVC, cultured for 10–14 days, fixed, and stained with crystal violet solution. The original data used to generate the graphs in Fig. 3d and Extended Data Fig. 6d are provided in Source Data for Fig. 3 and Source Data ED Fig. 6, respectively.
Poly(A)+ RNA and genomic DNA extraction
For genomic DNA extraction, the cells were washed once with PBS, resuspended in lysis buffer (10 mM Tris pH8, 1% SDS, 2 mM EDTA, 100 mM NaCl), and digested with 1 μg/ml proteinase K (Ambion, cat. #AM2542) at 60°C overnight. The next day, genomic DNA was purified by phenol/chloroform (Sigma-Aldrich, cat. #P3803) extraction, resuspended in water, boiled 5 min at 95°C and then quickly put on ice to denature the DNA. To isolate poly(A)+ RNA, total RNA was first extracted using TRIzol reagent (Life Technologies, cat. #15596018) according to the manufacturer’s instructions. Poly(A)+ RNA was then isolated using a magnetic mRNA isolation kit (New England BioLabs, cat. #S1550S) following the manufacturer’s instructions.
RNA extraction and qPCR analysis
Total RNAs were extracted from cells using TRIzol reagent (Life Technologies, cat. #15596018) according to the manufacturer’s instructions. Reverse transcription was carried out on 1 μg of total RNA by using PrimeScript RT reagent kit (Takara; Clontech). cDNAs obtained were diluted and used for quantitative PCR (qPCR). Each qPCR assay was performed with a standard dilution curve of a calibrator, which was a mixture of different cDNA, to precisely quantify relative transcript levels. Gene-specific primers were used together with SYBR green (Roche) for detection on a LightCycler 480 system (Roche). The primer sequences used are described in the Oligonucleotides section.
Protein extraction, western-blot and RNA/DNA dot-blot
Proteins extracts were obtained by washing the cells once with PBS and resuspending in lysis buffer (50 mM Tris pH6.8, 2% SDS, 10% glycerol, bromophenol blue, β-mercaptoethanol). Samples were then boiled 5 min at 95°C, subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. Membranes were blocked overnight in TBS-T (Tris-Buffered Saline (TBS), 0.05% Tween-20) containing 5% skim milk, and incubated with the appropriate antibody. Membranes were washed four times in TBS–T, then incubated for 1 hour with the appropriate peroxidase-conjugated anti-goat or anti-rabbit secondary antibody at 1/5,000 (EMD Millipore). Membranes were washed again as described, incubated with Western Lighting Plus ECL reagents (PerkinElmer) according to manufacturer’s instructions, and exposed to film. Poly(A)+ RNA or genomic DNA was spotted onto Hybond N+ membranes (GE Healthcare, cat. #RPN119B), and subjected to dot blotting with the indicated antibodies as described33. Original western blot and dot blot scans are provided in Supplementary Figure.
Antibodies
αm6A antibody (Synaptic Systems, 202 003) was used at 1 μg/ml for IF and 2 μg/ml for western and dot blots. αMETTL3 (ABclonal, A8370) and αMETTL14 (ABclonal, A8530) antibodies were used at 2 μg/ml for IF and 0.5 μg/ml for western blot. Antibody to Pol κ (ABclonal, P1619) was used at 5 μg/ml for IF or 0.5 μg/ml for western blot. αXPA antibody (Santa Cruz, sc-56831) was used at 3 μg/ml for IF or 0.5 μg/ml for western blot. αFlag antibody (Sigma-Aldrich, F3165) was used at 1 μg/ml for IF or 0.1 μg/ml for western blot. Antibodies to γH2A.X (Abcam, ab26350 (mouse), or ABclonal, AP0099 (rabbit)) were used at 2 μg/ml or 1 μg/ml, respectively, for IF. αRAD18 antibody (Cell Signaling, 9040) was used at 1 μg/ml for IF. Antibodies to POL δ1 (ABclonal A5323), POL δ3 (ABclonal, A7243), POL ε3 (ABclonal, A6469), POL η (Abcam, ab17725), POL ι (Sigma-Aldrich, HPA012000 or ABclonal, A1942), PARP1 (Trevigen, 4338-MC-50), PAR (TULIP BIOLABS, 1023), XPC (Santa Cruz, sc-74410), DDB2 (ABclonal, A1848), CSA (Santa Cruz, sc-10997), TFIIH/p89 (Santa Cruz, sc-293 (rabbit) or sc-377301 (mouse)), WTAP (Bethyl, A301-435A or A301-436A, or Santa Cruz, sc-374280), or FTO (Abcam, ab126605) were used at 3 μg/ml for IF. αYTHDC1 antibody (OriGene Technologies, TA337926) was used at 2 μg/ml for western blot. αYTHDF1 antibody (Abcam, ab99080) was used at 1 μg/ml for western blot. Antibodies to CPD (Kamiya Biomedical Company, MC-062), HNRNPA2B1 (Abcam, ab6102), or PCNA (Santa Cruz, sc-56) were used at 0.5 μg/ml for western blot. Antibodies to β-actin (Sigma-Aldrich, A3854) or α-tubulin (Santa Cruz, sc-23948) were used at 0.1 μg/ml for western blot. Where two different antibodies are listed for a single antigen, the host species was chosen appropriately to allow for co-staining with other antibodies.
Oligonucleotides
Oligonucleotides used for METTL3 knockdown were sh#1: CTCAGTGGATCTGTTGTGATA; sh#2: CAGGAGATCCTAGAGCTATTA; sh#3: CGTCAGTATCTTGGGCAAGTT. Guide RNA sequences used for METTL3 knock-out were guide#1: AGAGTCCAGCTGCTTCTTGT or guide#2: GAAGCAGGACTCGGGGCACT. METTL3 guide#1 was used for all KO studies, unless indicated. Oligonucleotides used for METTL14 knock-down were sh#1: GCAAATACCTATCCCTAAATT or sh#2: GCAGCAAGTAAACAGTGTGAT. METTL14 sh#1 was used for all studies, unless indicated. Guide RNA sequence for FTO KO was GAAGCGCACCCCGACTGCCG. The following oligonucleotides were used for generating knock-downs-- PCNA: GCTGTTACCATAGAGATGAAT; YTHDF1: GGGGGTTGAGTGTTGCATCTT; YTHDF2: AAGGCTAAGCAGGTGTTGAAA; YTHDC1: CCAGAGAGTGAACAAGATAAA; HNRNPA2B1: TTTGAGGAACTACTACGAACA; XPA: GAAGAGGTCTCTTGAAGTTTG; ALKBH5: sh#1 GAAAGGCTGTTGGCATCAATA or sh#2 CCTCAGGAAGACAAGATTAGA.
For qPCR quantification, the following primer pairs were used: ALKBH5F (CGGCTGCAAGTTCCAGTTCAAGCCTATTC) and ALKBH5R (CGTATGCAGTGAGTGATTTCATCAGCAGC); HNRNPA2B1F (GCGACTGAGTCCGCGATGGAGAAAACTTTAG) and HNRNPA2B1R (GTCTGTAAGCTTTCCCCATTGTTCGTAG); YTHDC1F (CACCAGAGACCAGGGTATTTAAAGGATC) and YTHDC1R (CATTCCTTGCCAAGGTGGTGGTGGTCCCATG); YTHDF2F (GCCAGCTACAAGCACACCACTTCCATT) and YTHDF2R (GTAGAACTGCCTTTTATTTCCCACGACCTTGACG); PCNA-F (CTTCGACACCTACCGCTGCGACCGCAACC) and PCNA-R (CTTCATTTCATAGTCTGAAACTTTCTCCTGG).
meRIP-seq
Total RNA from U2OS cells was subjected to two rounds of polyA selection. Immunoprecipitation of m6A-containing RNA fragments was performed as previously described34, in 3 or 4 biological replicates for each cell line and condition. Reads were aligned to the reference genome (hg38) using Tophat (v2.0.1435 with parameter -g 1 --library-type=fr-firststrand). RefSeq Gene structure annotations were downloaded from UCSC Table Browser. The longest isoform was used for genes with multiple isoforms. Aligned reads were extended to 200 bp (average fragments size) and converted from genome-based coordinates to isoform-based coordinates, in order to eliminate the interference from intron in peak calling.
The peak calling method was modified from Dominissini et al.34 To call m6A peaks, the longest isoform of each human gene was scanned using a 100 bp sliding window with 10 bp step. To reduce bias from potential inaccurate gene structure annotation and the arbitrary usage of the longest isoform, windows with reads counts less than 1/20 of the top window in both m6A-IP and input sample were excluded. For each gene, the read count in each window was normalized by the median count of all windows of that gene. A Fisher exact test was used to identify the differential windows between IP and input samples. The window was called as positive if the fdr<0.01 and log2(Enrichment Score) >= 1. Overlapping positive windows were merged. The following four numbers were calculated to obtain the enrichment score of each peak (or window): reads count of the IP sample in the current peak/window (a), median reads count of the IP sample in all 100bp windows on the current mRNA (b), reads count of the input sample in the current peak/window (c) and median reads count of the input sample in all 100bp windows on the current mRNA (d). The enrichment score of each window was calculated as (a×d)/(b×c).
Metagene profiles were generated as described15. To search for sequence motifs, the findMotifs program from the HOMER package was then run using the settings -rna -len 5,6,7 –noweight.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The MeRIP-seq datasets generated in this study have been deposited in the Gene Expression Omnibus database under accession number GSE92867.
Extended Data
Extended Data Figure 1. 6A-modified RNA accumulates at damage sites in response to UV irradiation.
a, U2OS cells were subjected to the indicated doses of UVC irradiation, incubated at 37°C for 2 min, and costained for m6A and γH2A.X. Relative m6A intensity is indicated on the right. b, U2OS cells were subjected to 50 J UVC irradiation through a micropore filter, incubated at 37°C for 2 or 5 min, and costained for m6A and DNA (DAPI). c, A375 (melanoma) or HeLa cells were subjected or not (0′) to 25 J UVC irradiation, incubated at 37°C for the indicated times, and then costained for m6A and γH2A.X. d, U2OS cells were subjected or not (0′) to 20 Gray of γ-irradiation, incubated at 37°C for 4 min, then costained for m6A and γH2A.X. e, U2OS cells were either irradiated with 25 J UVC or treated with DMSO, mitomycin C, hydroxyurea, or arabinoside-C, then costained for m6A and γH2A.X. c–e, relative m6A intensity is indicated on the right. f, Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) cells were subjected to 25 J UVC irradiation (top panel) or microirradiation by UVA laser (bottom panel), incubated at 37°C for 2 min, and stained for m6A. Top and bottom panels show m6A signal in representative S/G2/M or G1 phase cells, respectively. Arrows in middle panels denote representative G1-phase cells negative for m6A signal. The percentage of G1 (red) or S/G2/M (green) cells positive for m6A signal is indicated on the right. g, U2OS cells were microirradiated by UVA laser, permeabilized and treated with or without RNase A, and costained for m6A and γH2A.X (left panel). The percentage of γH2A.X-positive cells displaying colocalizing m6A signal is indicated. Nucleic acids (DNA and RNA) from cells treated with or without RNase A were isolated and analyzed on an agarose gel (right panel). Ladder (1kb DNA ladder). h, Poly(A)+ RNA was extracted from the samples in (a), and subjected to dot-blot analysis with an antibody recognizing m6A. Methylene blue staining was used as a loading control.
Extended Data Figure 2. METTL3/14, but not WTAP, regulate m6A RNA at damage sites.
a, U2OS cells stably expressing Flag-tagged METTL3 were microirradiated by UVA laser, incubated at 37°C for 2 min, and costained for m6A and Flag. The percentage of m6A-positive cells with colocalizing Flag signal is indicated on the right. Lower panel shows a representative cell with METTL3 localized at a damage site (arrow). b, Western blot for METTL3 in WT, METTL3 KO (M3 KO), or METTL3 KO U2OS cells stably expressing Flag-tagged METTL3 (M3 KO/WT-R) or its catalytic mutant (M3 KO/Cat-R). Actin is shown as a loading control. c, Western blot for METTL3 in U2OS cells expressing control (Control KD) or 2 independent shRNAs targeting METTL3 (M3 KD1, M3 KD2). Actin is shown as a loading control. d, WT or METTL3 KO (M3 KO) U2OS cells were irradiated or not (0′) with 15 J UVC, incubated at 37°C for 2 or 8 min, then subjected to poly(A)+ RNA extraction. Isolated poly(A)+ RNA was analyzed by dot-blot with an antibody recognizing m6A. Methylene blue staining was used as a loading control. e, Cells shown in (c) were subjected or not (0′) to 15 J UVC irradiation, incubated at 37°C for 1, 2, 4 or 10 min, and then subjected to poly(A)+ RNA extraction and dot-blot analysis as described in (d). f, WT cells were microirradiated by UVA laser, incubated at 37°C for 2 min, and stained for γH2A.X and METTL14. Arrows indicate representative γH2A.X-positive cells with colocalizing METTL14. The percentage of γH2A.X-positive cells showing colocalization with METTL14 is indicated on the right. g, WT U2OS cells expressing control (Control KD) or an shRNA targeting WTAP (WTAP KD) were subjected to UVA laser microirradiation, incubated at 37°C for 2 min, then costained for γH2A.X and WTAP, using three independent WTAP antibodies as indicated. The percentage of γH2A.X-positive cells displaying colocalizing WTAP signal is indicated on the right. h, Western blot for METTL14 in U2OS cells expressing control shRNA (Control KD) or shRNA targeting METTL14 (M14 KD). Actin is shown as a loading control. i, Western blot of METTL3 and METTL14 in WT, 2 independent METTL3 KO (M3 KO), or 2 independent METTL3 KO/METTL14 KD (M3 KO + M14 KD) U2OS cell lines. Actin is shown as a loading control. M3 KO1 and M3 KO1 + M14 KD1 were used for all subsequent experiments. j, WT, METTL3 KD (M3 KD), METTL14 KD (M14 KD), WTAP KD, METTL3 KO (M3 KO), or METTL3 KO/METTL14 KD (M3 KO/M14 KD) U2OS cells were microirradiated by UVA laser, incubated at 37°C for 2 min, and costained for m6A and γH2A.X. The percentage of γH2A.X-positive cells displaying colocalizing m6A signal is indicated on the right. All images are representative of at least 50 cells in triplicate. Scale bar, 20 μm.
Extended Data Figure 3. FTO, but not ALKBH5, modulates m6A RNA levels and duration at damage sites.
a, WT or FTO KO U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for 4 min, then stained for FTO and γH2A.X as indicated. Middle panel shows a representative γH2A.X-positive cell with FTO colocalizing at the damage site (arrow). The percentage of cells showing colocalization of signals is indicated on the right. b, Western blot of FTO in WT or FTO KO U2OS cells. Actin is shown as a loading control. c, U2OS cells expressing control (Control KD) or one of two independent shRNAs targeting ALKBH5 (ALKBH5 KD1, KD2) were subject to qPCR analysis of ALKBH5 mRNA levels and normalized to GAPDH. d, WT or FTO KO U2OS cells were microirradiated by UVA laser, incubated at 37°C for 4 min, and costained for m6A and γH2A.X. Two different exposures for m6A are shown. The percentage of γH2A.X-positive cells showing colocalizing m6A signal is indicated on the right. e, WT, FTO KO, and two ALKBH5 KD U2OS cell lines were irradiated with 50 J UVC, incubated at 37°C for 2 min, and costained for m6A and γH2A.X. Relative m6A intensity is indicated on the right. All images are representative of at least 50 cells in triplicate. Scale bar, 20 μm.
Extended Data Figure 4. METTL3, FTO, and PARP1 regulate m6A RNA at damage sites.
a–b, U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for the indicated time, then stained for γH2A.X and METTL3 (a) or FTO (b) as indicated. Cells in blue boxes are shown at higher zoom level in the second column. Arrows denote γH2A.X-positive damage sites with colocalizing METTL3 (a) or FTO (b). The percentage of γH2A.X-positive cells displaying colocalizing METTL3 or FTO signal is indicated on the right. c, WT or H2A.X KO MEF cells were microirradiated by UVA laser, incubated at 37°C for 2 min, and co-stained for m6A and TFIIH or γH2A.X as indicated. Where possible, the percentage of γH2A.X-positive cells showing colocalizing m6A signal is indicated on the right. d, U2OS cells pre-treated with DMSO or PARP inhibitors BYK, PJ-34, or Olaparib were microirradiated by UVA laser, incubated at 37°C for 2 min, and then costained for m6A and γH2A.X. e, U2OS cells pre-treated with DMSO or PARP inhibitor (Olaparib, 10 μM) were microirradiated by UVA laser, incubated at 37°C for 2 min, and then stained for PARP1, Poly(ADP-ribos)ylation (PAR), m6A, and γH2A.X as indicated. The percentage of γH2A.X-positive cells showing colocalizing signal is indicated on the right. f, U2OS cells pre-treated with DMSO or PARP inhibitor (Olaparib, 10 μM) were microirradiated by UVA laser, incubated at 37°C for 2 min, and then costained for γH2A.X and METTL3. Arrows denote representative γH2A.X-positive cells displaying colocalizing METTL3 signal. The percentage of γH2A.X-positive cells showing colocalizing METTL3 is indicated on the right. All images are representative of at least 50 cells in triplicate. Scale bar, 20 μm.
Extended Data Figure 5. Sequencing analysis of m6A-methylated RNAs responding to UV.
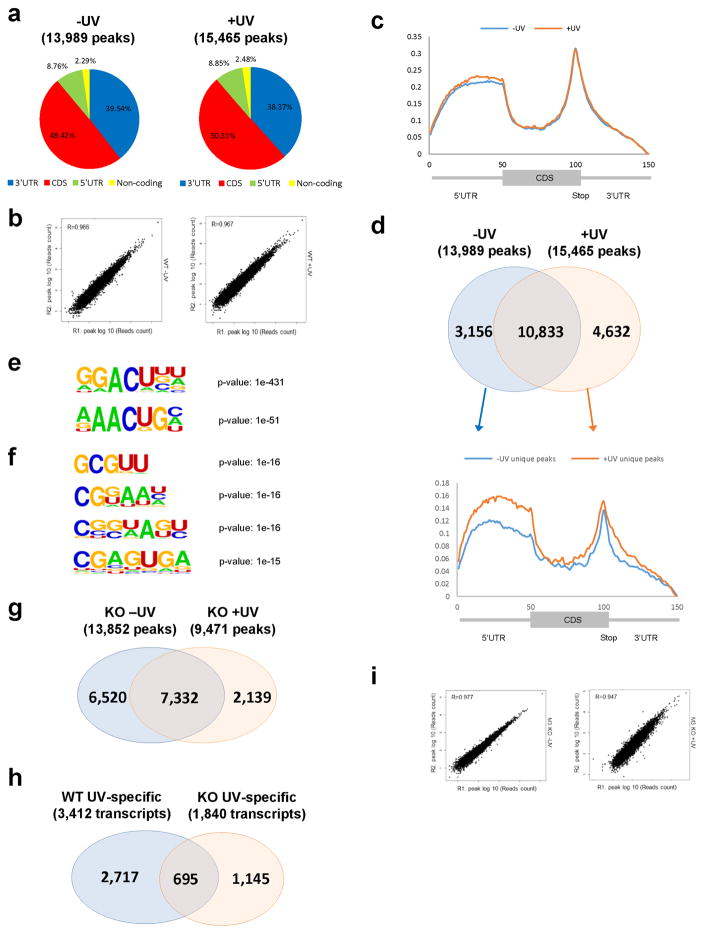
a, Pie-chart representation of the distribution of m6A peaks in different transcript segments in U2OS cells before (left panel) or 2 minutes after 50 J UVC irradiation (right panel). CDS, coding sequence region. b, Correlation of m6A peaks between two independent samples from U2OS cells before (top panel) or 2 minutes after (bottom panel) 50 J UVC irradiation. A total of 4 replicates was performed for unirradiated cells, and 3 for irradiated cells. c, Metagene profiles of m6A distribution across the transcriptome of U2OS cells before (−UV) or 2 minutes after 50 J UVC irradiation (+UV). d, Overlap of m6A peaks identified in mRNAs isolated from U2OS cells before (13,989 peaks) or 2 minutes after 50 J UVC irradiation (15,465 peaks) (top panel), and Metagene profiles of m6A peaks uniquely present in unirradiated (3,156 peaks) or irradiated cells (4,632 peaks) (bottom panel). e, Consensus motifs enriched in m6A peaks from transcripts identified in UV-irradiated cells (15,465 peaks). f, Consensus motifs enriched in m6A peaks from transcripts uniquely methylated in irradiated cells (4,632 peaks). g, Overlap of m6A peaks identified in mRNAs isolated from METTL3 KO cells before (13,852 peaks) or 2 minutes after 50J UVC irradiation (9,471 peaks). h, Overlap of transcripts that are uniquely methylated after 50 J UV irradiation in WT and METTL3 KO cells (3,412 and 1,840 transcripts respectively). i, Correlation of m6A peaks between two independent samples from METTL3 KO cells before (left panel) or 2 minutes after 50J UVC irradiation (right panel). A total of 3 replicates was performed for each condition.
Extended Data Figure 6. m6A RNA is required for efficient DNA repair and cell survival after UV exposure.
a, U2OS cells pre-treated with DMSO or PARP inhibitor (Olaparib, 10 μM) were subjected or not to 15 J UVC irradiation and incubated at 37°C for 1, 2, 4 or 6 hours. Genomic DNA was purified from unirradiated or irradiated cells, and then subjected to dot blot analysis with an antibody recognizing CPDs. Methylene blue staining was used as a loading control. b, WT A375 cells, A375 cells expressing METTL3 shRNA (METTL3 KD), or METTL3 KD cells stably expressing shRNA-resistant METTL3 (M3 KD/WT-R) were subjected or not to 25 J UVC irradiation and incubated at 37°C for 12 or 24 hours. Nascent RNA was labeled in unirradiated or irradiated cells at each time-point by incubating for an additional 3 hours with 1 mM 5-ethynyl uridine (EU) (green). Images are representative of at least 50 cells in triplicate. Bar, 20 μm. c, WT, METTL3 KO (M3 KO), or METTL3 KO U2OS cells stably expressing Flag-tagged METTL3 (M3 KO/WT-R) or its catalytic mutant (M3 KO/Cat-R) were subjected to different dosages of UVC irradiation (0, 5, 10 and 15 J) and used to perform a colony formation assay. Colonies were stained by crystal violet solution 10–14 days after seeding, and the quantification is shown in Fig. 3d. d, U2OS cells infected with control (Control KD), two independent METTL3 shRNAs (M3 KD1 and M3 KD2), or M3 KD1 cells stably expressing shRNA-resistant WT METTL3 (KD1/WT-R) or its catalytic mutant (KD1/Cat-R), were subjected to different dosages of UVC irradiation (0, 5, 10 and 15 J) and used to perform a colony formation assay. Colonies were stained by crystal violet solution 10–14 days after seeding (top panel), and the number of colonies was determined (lower panel). Y-axis represents colony survival normalized to unirradiated control. Results are shown as mean ± SEM from at least three independent experiments. (**) P ≤ 0.001.
Extended Data Figure 7. m6A RNA does not interact with canonical NER or DSB repair pathways.
a, U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for 2 min and costained for γH2A.X and m6A, XPA, TFIIH, DDB2, XPC, CSA, or CPD as indicated. b–c, WT or METTL3 KO (M3 KO) U2OS cells were subjected to UVA laser, incubated at 37°C for 2 min, then costained for γH2A.X and XPA or TFIIH (b), and 53BP1 or BRCA1 (c) as indicated. a–c, The percentage of γH2A.X-positive cells displaying the indicated signal colocalizing with γH2A.X is indicated on the right.
Extended Data Figure 8. m6A RNA recruits Pol κ to damage sites.
a, WT or METTL3 KO/METTL14 KD (M3 KO/M14 KD) U2OS cells were microirradiated by UVA laser, incubated at 37°C for 30 seconds to 10 min, and then stained for the indicated polymerases and γH2A.X. Staining for each polymerase was conducted at the peak of its localization, which occurred at 4 min post-UV irradiation for Pol κ, and 8 min post-UV irradiation for Pol δ, Pol ε, and Pol η. Pol ι was not detected during the first 10 min following irradiation, using either of the two antibodies listed in Methods section (ABclonal A1942 shown). Arrows denote representative γH2A.X-positive damage sites with colocalizing polymerase. The percentage of γH2A.X-positive cells displaying the indicated signal colocalizing with γH2A.X is indicated on the right. b, Table summarizing the observed localization patterns of the tested polymerases (from (a) and Fig. 4a). c, U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for indicated time, then costained for Pol κ and γH2A.X as indicated. Arrows denote representative γH2A.X-positive damage sites with colocalizing Pol κ. The percentage of γH2A.X-positive cells displaying colocalizing Pol κ is indicated on the right. The 10 min timepoint displayed greater cell-to-cell heterogeneity in the intensity of Pol κ staining, as compared to the other timepoints. d, Western blot of Pol κ in WT or METTL3 KO (M3 KO) U2OS cells. Actin is shown as a loading control. e, U2OS cells pre-treated with DMSO or PARP inhibitor (Olaparib, 10 μM) were microirradiated by UVA laser, incubated at 37°C for 2 min, and then costained for γH2A.X and Pol κ, as indicated. Arrows denote representative γH2A.X-positive damage sites with colocalizing Pol κ, and the percent of cells showing colocalization is indicated on the right. All images are representative of at least 50 cells in triplicate. Scale bar, 20 μm. f, Western blot of Pol κ, Flag, and METTL3 in WT, METTL3 KO, and METTL3 KO U2OS cells stably expressing Flag-tagged Pol κ (M3 KO/Flag- Pol κ). g, METTL3 KO (M3 KO) or METTL3 KO U2OS cells stably expressing Flag-tagged Pol κ (M3 KO/Flag- Pol κ) were irradiated or not (0 h) with UVC, and incubated at 37 °C, at which point genomic DNA was extracted. DNA was subjected to dot-blot analysis with an antibody recognizing CPDs. Methylene blue staining was used as a loading control.
Extended Data Figure 9. m6A readers and known Pol κ interactors are not responsible for early Pol κ recruitment to UV damage sites.
a, WT U2OS cells, stably expressing Flag-tagged YTHDC1, YTHDF1, or YTHDF2, were subjected to UVA laser microirradiation, incubated at 37°C for 2 min and costained for Flag and m6A or γH2A.X as indicated. b, WT, HNRNPA2B1 KD, YTHDC1 KD, YTHDF1 KD, or YTHDF2 KD U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for 2 min and costained for Pol κ and γH2A.X. c, XPA KD or PCNA KD U2OS cells were subjected to UVA laser microirradiation, incubated at 37°C for 2 min and costained for Pol κ and γH2A.X. a–c, data from one representative experiment shown. d, U2OS cells were subjected to UVA laser microirradiation and incubated at 37°C for 2 or 10 min, then costained for γH2A.X and m6A, Pol κ, or RAD18 as indicated. Arrows denote representative γH2A.X-positive damage sites with colocalizing Pol κ. n=3, 50 cells per replicate. a–d, The percentage of cells displaying the indicated signal colocalizing with γH2A.X (or with m6A, panel a, left) is indicated on the right. Scale bar, 20 μm.
Extended Data Figure 10. Knockdown efficiencies of candidates tested for their effects on Pol κ recruitment.
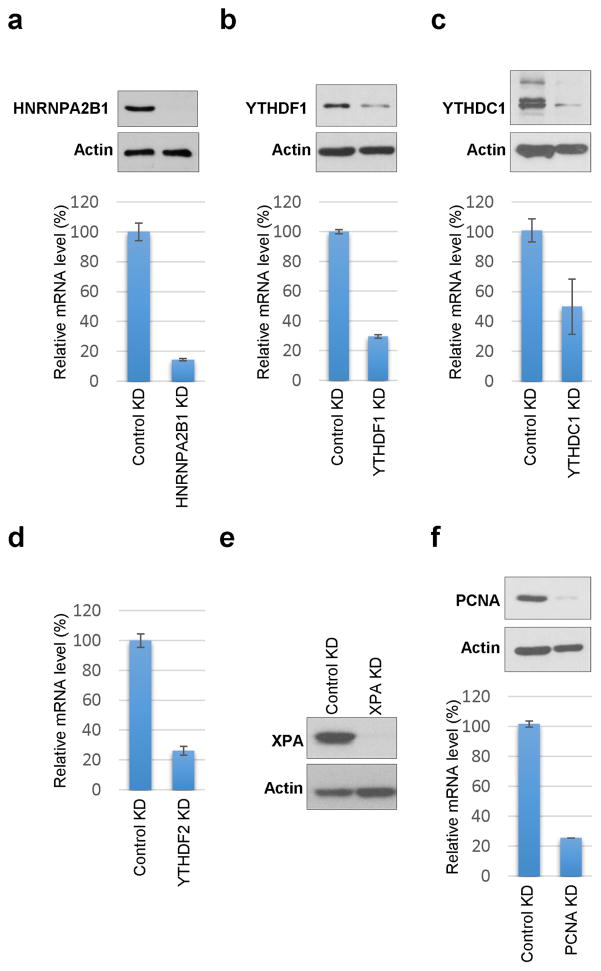
a–f, U2OS cells were infected with control (Control KD) or shRNA targeting (a) HNRNPA2B1 (HNRNPA2B1 KD), (b) YTHDF1 (YTHDF1 KD), (c) YTHDC1 (YTHDC1 KD), (d) YTHDF2 (YTHDF2 KD), (e) XPA (XPA KD), or (f) PCNA (PCNA KD). mRNA levels were measured by qPCR and normalized to that of GAPDH. Results are shown as mean ± SEM from at least 3 independent experiments. Protein expression levels were examined in control and KD cells by qPCR and western blot; actin is shown as a western blot loading control.
Supplementary Material
Acknowledgments
We thank members of the Shi lab for helpful discussions. We are especially indebted to Dr. Lei Li (MD Anderson, USA) for insightful advice, discussion, and sharing reagents; to Rita Meganck and Dr. Aziz Sancar (University of North Carolina Chapel Hill, USA) for providing reagents and helpful discussions; to Dr. François Huetz (Institut Pasteur, France) for the cell cycle analysis and Dr. Steve Elledge (Harvard Medical School, USA) for helpful advice and the use of laser micro-irradiation microscope at the early stage of this study; and to Lee Kraus for reagents and advice. We thank Abclonal and Santa Cruz Biotechnology for providing antibodies, and Dr. Nima Mosammaparast and Dr. Barry Sleckman for the H2A.X KO MEF cells. B.L. was supported by a fellowship from Association pour la Recherche sur le Cancer (ARC) (France) and NIH 4 T32 HD 7466-20. This work was supported by a grant from the NIH to Y.S. (R01 CA118487) and funds from Boston Children’s Hospital. Y.S. is an American Cancer Society Research Professor.
Footnotes
Author Contributions
YS conceived the project and provided advice for experimental designs and data interpretation. Y.X., B.L. and C.H.H. designed, performed and analyzed experiments as described below with input from Y.S., A.J. and all authors. Y.X. and C.H.H. performed the microirradiation and global UV irradiation IF experiments with help from W.S., C.X., H.C., J.O. and S.W. who cultured and prepared the cells. J.O. carried out microfilter irradiation experiment under the supervision of L.Z. Y.X., B.L., and C.H.H. established KD, KO, and rescue cell lines with help from C.X., W.S. and D.L. RNA preparation and MeRIP-seq experiments were performed by B.L. and S.N., and the data analyzed by Z.L. L.Z. provided discussions and input throughout the project, while C.H. provided advice on MeRIP-seq data analysis carried out by B.L. and S.N. Y.S. and A.J. wrote the manuscript with input from C.H., L.Z. and P.H.H.
Competing financial interests
Y.S. is a cofounder of Constellation Pharmaceuticals and a member of its scientific advisory board, and a consultant for Active Motif, Inc.
References
- 1.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 4.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 6.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 10.Munns TW, Liszewski MK, Sims HF. Characterization of antibodies specific for N6-methyladenosine and for 7-methylguanosine. Biochemistry. 1977;16:2163–2168. doi: 10.1021/bi00629a019. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m6A Promotes Cap-Independent Translation. Cell. 2015 doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 16.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarcon CR, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell DL. The relative cytotoxicity of (6–4) photoproducts and cyclobutane dimers in mammalian cells. Photochem Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 27.Jambhekar A, et al. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Bi X, et al. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada T, et al. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J Biol Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 32.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 35.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The MeRIP-seq datasets generated in this study have been deposited in the Gene Expression Omnibus database under accession number GSE92867.