Abstract
Millions of cis-regulatory elements are predicted in the human genome, but direct evidence for their biological function is still scarce. Here we report a high-throughput method, Cis-Regulatory Element Scan by Tiling-deletion and sequencing (CREST-seq), for unbiased discovery and functional assessment of cis regulatory sequences in the genome. We use it to interrogate the 2Mbp POU5F1 locus in the human embryonic stem cells and identify 45 cis-regulatory elements of POU5F1. A majority of these elements display active chromatin marks, DNase hypersensitivity and occupancy by multiple transcription factors, confirming the utility of chromatin signatures in cis elements mapping. Notably, 17 of them are previously annotated promoters of functionally unrelated genes, and like typical enhancers, they form extensive spatial contacts with the POU5F1 promoter. Taken together, these results support the utility of CREST-seq for large-scale cis regulatory element discovery and point to commonality of enhancer-like promoters in the human genome.
Introduction
Millions of candidate cis-regulatory elements have been annotated in the human genome based on histone modification, transcriptional factor binding, and DNase I hypersensitivity1–6. These putative regulatory sequences harbor a disproportionally large number of sequence variants associated with diverse human traits and diseases, supporting the hypothesis that non-coding sequence variants contribute to common traits and diseases by disrupting transcriptional regulation7–9. However, research on the role of these putative functional elements in human development and disease has been hindered by a dearth of direct evidence for their biological function in the native genomic context.
High-throughput CRISPR/Cas9-mediated mutagenesis using single guide RNAs (sgRNAs) has been used to functionally characterize cis-regulatory elements in mammalian cells 10–15. However, current approaches are limited because: (1) Not all sequences are suitable for CRISPR/Cas9-mediated genome editing due to the lack of protospacer adjacent motifs (PAMs) that are required for targeting and DNA cutting by CRISPR/Cas9 16–18; (2) CRISPR/Cas9 mediated genome editing with individual sgRNAs tends to cause point mutations or short insertions or deletions, necessitating the use of an unrealistically large number of sgRNAs to interrogate the human genome; (3) it has been challenging to distinguish the cis- and trans-regulatory elements. To overcome these limitations, we developed CREST-seq, short for Cis-Regulatory Elements Scan by Tiling-deletion and Sequencing, which enables efficient discovery and functional characterization of cis-regulatory elements by introducing massively parallel, kilobase-long deletions to the genome. Below, we provide evidence supporting the utility of CREST-seq for large-scale cis-regulatory element identification in the human embryonic stem cells (hESC). We report the discovery of 45 regulatory sequences of POU5F1 and a surprisingly large number of enhancer-like promoters.
Results
CREST-seq identified cis-regulatory elements of POU5F1
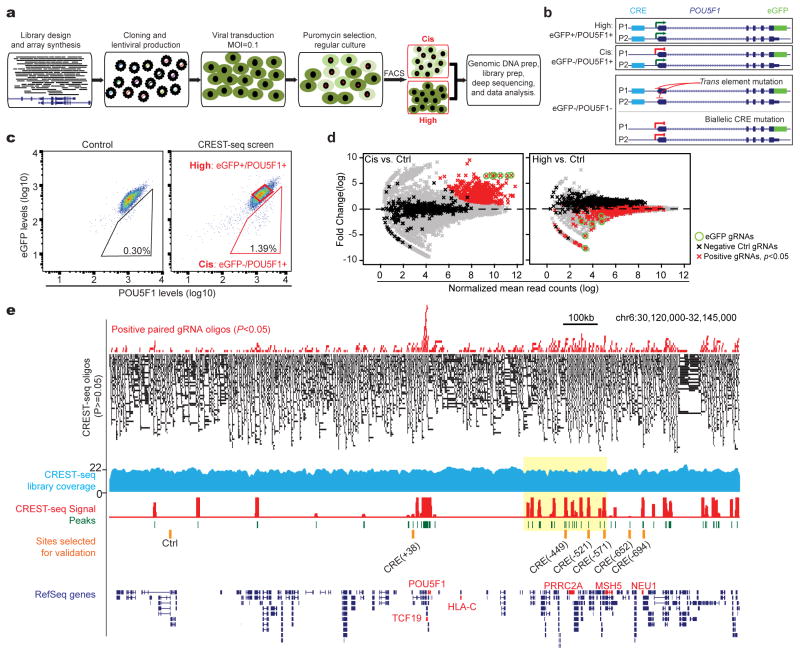
In a CREST-seq experiment, a large number of overlapping genomic deletions are first introduced to a genomic locus by CRISPR/Cas9-mediated genome editing using paired sgRNAs 16 (Fig. 1A). Cells with lowered expression of the gene of interest (Fig. 1B) are then isolated and the enriched sgRNA pairs determined by high-throughput sequencing. The enriched sgRNA-pair sequences are then used to infer the functional cis-regulatory sequences of the gene (Fig. 1A). To demonstrate the utility of CREST-seq, we applied it to the 2Mbp POU5F1 locus. As a model cell system we used a hESC line in which one POU5F1 allele was genetically tagged by eGFP, allowing transcription level of this allele to be monitored by eGFP expression 19 (Fig. 1B).
Fig 1. CREST-seq experimental design and application to the POU5F1 locus in hESC.
(A) Workflow of CREST-seq. A total of 11,570 oliogs containing dual sgRNA sequences were cloned into a lentiviral library that was in turn transduced into the H1 POU5F1-eGFP cells with MOI=0.1. After Puromycin selection, the cells were stained with antibodies specifically recognizing POU5F1 (PE) or eGFP (APC), respectively. The indicated “Cis” and “High” populations were sorted by FACS, and the integrated sgRNA pairs were amplified by PCR from genomic DNA followed by high-throughput sequencing.
(B) Schematic illustration of mono-allelic or bi-allelic deletions of cis-regulatory elements of POU5F1. The eGFP-tagging allele is designated as P1 and the wild-type allele as P2. Mono-allelic disruption of a POU5F1 CRE on the P1 allele would lead to reduced eGFP expression while POU5F1 protein levels remain relatively unchanged (eGFP-/POU5F1+). Bi-allelic disruption of a POU5F1 CRE would lead to reduction of both eGFP and POU5F1 protein level.
(C) FACS analysis of H1 POU5F1-eGFP cells transduced with control lentivirus expressing Cas9 but not sgRNA (left) or the CREST-seq lentiviral library (right) 14-day post transduction.
(D) The read counts of sgRNA from “Cis” (left) and “High” (right) are compared to those from a non-sorted control population (Ctrl). The fold changes represent the ratios between read counts in the “Cis” or “High” populations and the “Ctrl” population, with the significance of enrichment calculated by a negative binomial test. Green circles denote eGFP targeting sgRNA pairs; Red dots correspond to sgRNA pairs enriched in the “Cis” population with P-value < 0.05 and log(fold change) > 1. Black dots denote the negative control sgRNA pairs and grey dots for the rest of pairs.
(E) Genome browser screenshot showing CREST-seq positive sgRNA pairs (P-value < 0.05, top) and CREST-seq negative sgRNA pairs (P-value>0.05, black bars); genomic coverage of the CREST-seq library (blue track); the computed CREST-seq signals (see Methods), and the genomic regions identified as cis-regulatory sequences of POU5F1 (peaks, green), along with the CRE sites selected for further in-depth validation (orange bars). Yellow box highlighted a region enriched for CREs with a close-up view in Figure 2B.
We designed a total of 11,570 sgRNA pairs (Table S1) to introduce the same number of genomic deletions (Fig. 1A and Fig. S1A) to the POU5F1 locus. The average size of each deletion is ~2kb, with an overlap of 1.9kb between two adjacent deletions (Fig. S1B) such that each nucleotide in this locus is covered by ~20 distinct genomic deletions on average. As negative controls, we included 424 sgRNA oligos lacking the PAM sequence necessary for effective dsDNA breaks. As positive controls, we included six sgRNA pairs that target the eGFP coding sequence (Table S1). We constructed a lentiviral library that express these sgRNA pairs (Fig. S2A–2E) and transduced it into the hESC line at low multiplicity of infection (MOI = 0.1), which ensures that the majority of cells receives one or no lentiviral particle (detailed in Supplementary protocol).
To isolate mutant cells with deletion in POU5F1’s cis-regulatory sequences, we used FACS to sort out cells showing lowered POU5F1 expression from the eGFP-tagged allele but relatively unchanged expression from the non-tagged allele (Fig. 1C). We refer to this eGFP-/POU5F1+ subpopulation as “Cis” population (Fig. 1B, middle and Fig. 1C). As a control, we also collected a sample of cells before FACS sorting (referred to as “Ctrl”). Finally, we collected the eGFP+/POU5F1+ population (referred to as “High”) (Fig. 1B, top; Fig. 1C). Genomic DNA was purified from each cell populations, and the sgRNA pairs present in each subpopulation were then determined by massively parallel sequencing (Table S2). The experiment was conducted in multiple replicates (Table S2 and Fig. S3A), with the abundance of sgRNA pairs highly reproducible between replicates (Pearson Correlation Coefficients R=0.90 for “Cis”, R=0.92 for “Ctrl” and R=0.97 for “High”, respectively) (Fig. S3B).
To identify cis-regulatory elements of POU5F1, we first compared the abundance of sgRNA pairs between the “Cis” population and the “Ctrl” population (Table S2) using a negative binomial test and computed the fold enrichment and P-value of each sgRNA pair (Table S3 and Fig. S3C). We found 495 sgRNA pairs to be significantly enriched (P < 0.05 and log(fold change) > 1) in the “Cis” samples (Fig. 1D, red dots; Fig. 1E red bars, and Table S3). As expected, all six sgRNA pairs targeting the eGFP sequence were highly enriched in the “Cis” population (Fig. 1D, green circles). By contrast, only 2 of the 424 negative control sgRNAs were enriched, corresponding to an empirical FDR smaller than 0.5%. Further supporting the effectiveness of our experimental design, the sgRNA pairs with significant enrichment in the “Cis” population were generally depleted in the “High” samples (Table S3 and Fig. 1D, right panel). Next, we sought to identify cis-regulatory sequences by taking full advantage of the tiling deletion design (Fig. 1E). We began by ranking all sgRNA pairs based on their enrichment levels in the “Cis” population relative to the “Ctrl” (Table S3). We then partitioned the 2MB POU5F1 locus into 50bp bins, and used Robust Rank Aggregation (RRA) 20 to calculate a score for each bin to indicate whether the ranks of deletions spanning that bin are skewed toward top of the sorted list (Detailed in methods; Table S4). Altogether, we identified 45 genomic regions with a significant score (Fig 1E and Table S5). Using the same criteria, no genomic region was identified as positive in the “High” cell population (Fig. S4A). We named each of the 45 CREST-positive elements (referred to hereafter as “CRE”) using its relative genomic distance (kb) to the transcription start site (TSS) of POU5F1, with a negative sign denoting upstream of POU5F1 and a positive for downstream (Table S5). The 45 CREs include 4 previously identified POU5F1-regulatory elements that act in cis: its promoter (Fig. S4B), an upstream enhancer21 (Fig. S4B) and two temporarily phenotypic (TEMP) enhancers13 (Fig. S4C, DHS_65 and DHS_108). The remaining 41 CREs are novel POU5F1-regulatory sequences found in this study.
CREs are enriched with active chromatin marks and dense TF clusters
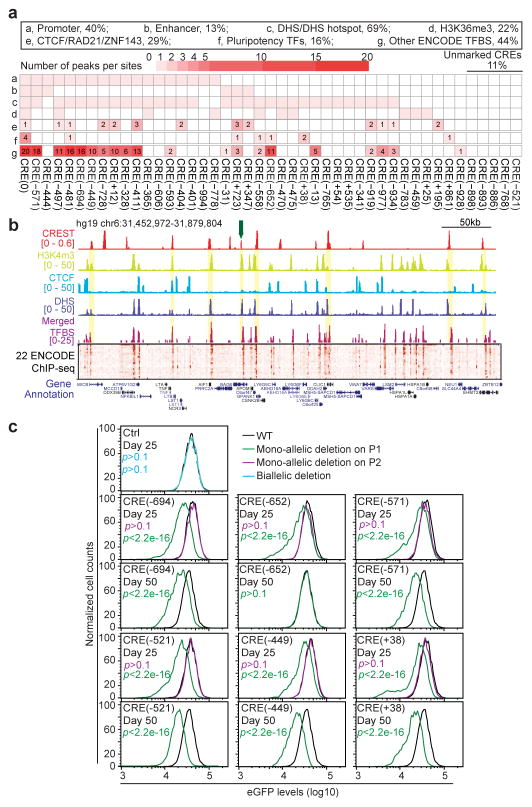
In order to determine chromatin features of the CREs, we examined the publically available chromatin accessibility data, transcription factor binding profiles and chromatin modification datasets from the H1 hESC cell line 3,5. We also generated ATAC-seq 22 and CTCF ChIP-seq with the cell line used in the present study and ensured that the data highly resembles the previous datasets from the same parental cell line5 (Fig. S5A and S5B). As expected, a majority of CREs were associated with biochemical features characteristic of cis-regulatory elements, including DNase Hypersensitivity (69%), transcription factor occupancy, active chromatin marks such as H3K27ac (22%), H3K4me3 (31%), and H3K4me1 (22%) 5. Notably, CREs are also enriched for binding sites of CTCF/RAD21 (29%), which have been linked to DNA looping and topologically associating domain (TAD) boundaries 23,24 (Fig. 2A, 2B, and Table S5). It has been reported that transcription factor binding in human cells tend to form dense clusters 25-27. Accordingly, we found that the CREST-positive regions overlap with dense clusters of TF binding sites (16% CREs are bound by essential pluripotency master regulators and 44% by other TFs; Fig. 2A–2C) and are bound by more transcription factors on average than DNase hypersensitive sites (DHS) (Fig. 2D, Wilcoxon tests P-value<6e-11). In general, CREST-positive regions are significantly associated with active histones modifications and transcription factor binding (Fig. 2E), and depleted for repressive chromatin marks H3K9me3 and H3K27me3 28 (Fig. 2E , and see Fig. S5C for other features), consistent with previous studies highlighting the role of clustered TF binding sites in gene regulation 25,29. Interestingly, five CREs lack any canonical chromatin signatures associated with active cis-regulatory sequences (Fig. 2A, Unmarked region, 11%), suggesting existing of elements without canonical epigenetic signatures, as recently reported12.
Fig 2. CREs tend to be associated with canonical active chromatin markers of cis-regulatory elements and dense TF clusters.
(A) A matrix showing the chromatin features and transcription factor binding at the 45 CREs. “Pluripotency TFs” denotes POU5F1, SOX2, NANOG, and PRDM14.
(B) A close-up view of genome browser snapshot of the yellow highlighted region in Fig. 1E with tracks corresponding to chromatin modifications, DHS, merged TFBS ChIP-seq peaks and a heatmap of normalized ChIP-seq signals for 22 transcription factors in hESCs. The height of merged TFBS bars indicates the number of bound TF. Yellow bars highlighted regions where CREs overlap with active chromatin marks and TFBS clusters. The green arrow points to the CREs in (C).
(C) A close-up view of a 5kb CRE occupied by a cluster of TFs.
(D) A box plot shows that transcription factor binding sites more frequently cluster at CREs than at typical cis-regulatory elements represented by DHS. (Wilcoxon test P-value < 6e-11)
(E) A bar chart shows the degree of enrichment of each chromatin feature in the CREs. To calculate the “Enrichment Test Score”, we first calculated the fraction of CREST-seq peaks that intersected with sites associated with each feature as a ratio between the observed over expected. An average ratio is calculated from 1,000 random permutations of the CREs. The enrichment test score is defined as the percentage that observed ratio is greater than expected. (*χ2 P-value < 0.01).
(F) Six CREs and one CREST-seq negative site (Ctrl) were selected (orange bars in Fig. 1E) for individual validation. Mutant clones were generated harboring bi-allelic deletion (Ctrl, blue curves), mono-allelic deletion on the P1 allele (green curves), or mono-allelic deletion on the P2 allele (magenta curves) at the indicated genomic loci. P1 is the eGFP-containing allele and P2 is the non-eGFP allele. FACS analysis was performed for all the mutant clones and wide-type cells (WT: black curves) at day 25 and day 50 after CRISPR/Cas9 transfection. The FACS data was quantified with FlowJo and P-value is calculated with two-sample t-test. “p” in Green and magenta letter “p” represent the P-values for mono-allelic mutants harboring P1-specific or P2-specific deletion, respectively.
To validate the function of the novel POU5F1 CREs, we selected 6 for in-depth analysis (Fig. 1E, orange bars). The regions were chosen based on three criteria: 1) they are located at a wide range of genomic distances, from 38kb to 694kb, from POU5F1 TSS; 2) they are surrounded by phased SNPs so that allelic analysis of gene expression could be performed; and 3) they represent a wide range of CREST-seq signals, ranking 9th, 13th, 23rd, 24th, and 37th out of 45 (Table S5). Additionally, while five CREs, CRE (-694), CRE (-652), CRE (-571), CRE (-449) and CRE (+38), are marked by canonical chromatin marks (Fig. 2A, and Fig. S6A), one CRE, CRE (-521), is unmarked (Fig. 2A and Fig. S6A). As a control, we tested a CREST-negative region (Fig. 1E and Fig. S6A). We used the CRISPR/Cas9 genome-editing to introduce mono-allelic deletions of lengths 2-4kb to remove these regions in the hESC line (Fig. S6A). As shown in Fig. 2F, all cell clones with mono-allelic deletion (green curves) on the P1 allele showed significant reduction in eGFP expression (Fig. S6B, t-test P-value <2.2e-16, error bars, s.d.). By contrast, clones bearing mono-allelic deletions of the P2 allele showed normal eGFP expression (Fig. 2F, magenta curves), indicating that these sequences act in cis to regulate POU5F1 expression. No change in eGFP expression was observed in clones containing bi-allelic deletions of the negative control region (Fig. 2F, “Ctrl site”, solid and dash blue curves). Notably, deletion of CRE (-521), which lacks any canonical marks of regulatory sequences (Fig. S6A), also led to a decrease in POU5F1 expression in cis. Interestingly, while deletion of five CREs resulted in durable reduction of POU5F1, deletion of the CRE (-652) element led to only temporary reduction of eGFP expression that was fully recovered by day 50 (Fig. 2F and Fig. S6B), suggesting that it belongs to the type of temporarily phenotypic enhancers (TEMP-enhancer) that we recently reported 13. Taken together, these results provided strong evidence that CREST-seq can be used to identify cis-regulatory sequences of a specific target gene in an unbiased and high-throughput manner.
Promoters acting as distal enhancers
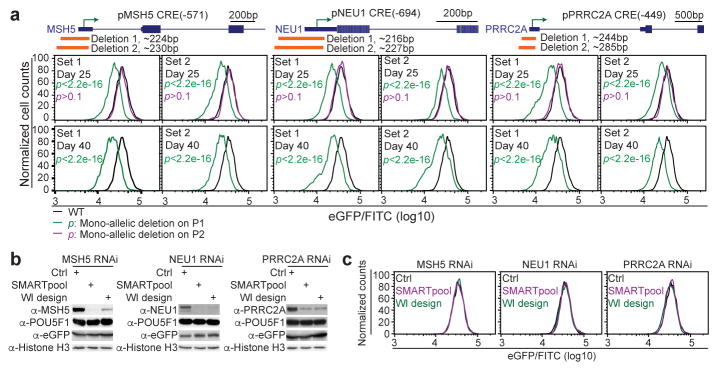
Results from the above CREST-seq experiments showed that 18 gene promoters, including the POU5F1 promoter, are necessary for optimal POU5F1 expression in hESC (Table S6). This is surprising because promoters are traditionally thought to mediate transcription of its immediate downstream sequences. Although recent reports indicated that some lncRNA and mRNA promoters may act as enhancers of their adjacent genes 12,30,31, definitive evidence illustrating a causative role of promoters acting as distal enhancers is still lacking. Identification of CRE(-449), CRE(-571) and CRE(-694) as cis-regulatory elements of POU5F1 suggests that promoters of PRRC2A, MSH5 and NEU1 genes may act as distal enhancers of POU5F1 in the hESC (Fig. S6A). To rule out the possibility that promoter-proximal elements in these genes were responsible for POU5F1 regulation, we deleted 216-285bp core promoter sequences containing the TSS of each gene and carried out allelic expression analysis in the resulting cell clones (Fig. 3A, Fig. S7). To avoid potential off-target effects, we used two sets of sgRNA pairs (Deletion 1 and Deletion 2, Fig. 3A, Fig. S7) for the genome editing, and recovered a total of 37 independent clones carrying mono-allelic deletions for in-depth analysis (Fig. S8 and Table S6). We found that all mutants with the P1 mono-allelic deletion displayed long-lasting reduction in eGFP expression (green curves in Fig. 3A, Fig. S8A and Fig. S8B; quantified in Fig. S8C and Table S6, error bars, s.d.), while in mutant clones with the P2 mono-allelic deletion eGFP levels were indistinguishable from WT (magenta curves in Fig. 3A, Fig. S8A and Fig. S8B; see Fig. S8C and Table S6 for quantification, error bars, s.d.). The reduced eGFP expression could not be due to loss of the PRRC2A, MSH5 or NEU1 gene products, because knockdown of each gene using two sets of siRNA (Fig. 3B, 3C) and shRNAs (Fig. S9A–9C) did not affect the POU5F1 mRNA or protein levels (Fig. 3B, 3C, and Fig. S9D). Thus, the core promoter sequences of PRRC2A, MSH5 and NEU1, but not their gene products, are required for optimal POU5F1 expression.
Fig 3. The core promoter regions of MSH5, NEU1, and PRRC2A are required for optimal POU5F1 expression in hESC.
(A) The core promoter regions of MSH5, NEU1, and PRRC2A were deleted by two sets of distinct sgRNAs (orange bars, Deletion 1 and 2). Mutant cell clones harboring mono-allelic deletions on the P1 allele (green curves), or P2 allele (magenta curves) were identified after genotyping and sequencing of the phased SNPs. FACS analysis was performed for all the mutant clones and wild-type cells (WT: black curves) at day 25 and day 40 after transfection. The FACS data is quantified with FlowJo. P-value is computed using two-sample t-test.
(B, C) The H1 POU5F1-eGFP cells were transfected with either control scrambled siRNA or siRNAs targeting the gene as indicated. Each gene is targeted by two sets of siRNAs (SMARTpool and WI design) with different sequences. The cells were analyzed 48 hours after transfection.
(B) Whole cell extract was collected and subjected to western blot analysis with indicated antibodies.
(C) An aliquot of cells were dissociated into single cells for FACS analysis. Black, magenta, and green curves represent the data from cells treated with Scrambled siRNA (Ctrl), SMARTpool siRNA and WI (http://sirna.wi.mit.edu/) designed siRNA, respectively.
To further show whether these gene promoters could function as enhancers in a traditional reporter assay, we constructed reporter plasmids that contain the 360-bp POU5F1 core promoter sequence driving a luciferase reporter gene, with the core promoter fragments of PRRC2A, MSH5 or NEU1 inserted downstream of the reporter 13,32. We transfected these plasmids into the H1 hESC cells, and assayed the luciferase activities 3 days after transfection. All elements exhibited significant enhancer activities compared to the control vector (Fig. S9E).
To rule out the possibility that CRISPR/Cas9-mediated genome editing impacts POU5F1 expression through locus-wide, non-specific mechanisms, we performed FACS analysis of the CRE deletion mutant clones to monitor levels of both POU5F1-eGFP and HLA-C, located 100kb upstream of POU5F1 TSS. We found that deletion of a CRE resulted in down-regulation of POU5F1-eGFP expression without affecting levels of HLA-C (Fig. S10A and S10B). To further exclude the possibility that CRISPR/Cas9 leads to double-strand-DNA-break (DSB)- induced transcriptional silencing in the cells, we examined phosphorylated H2AX (γH2AX, a DNA damage marker) in the mutant clones 33–35. We found that none of the mutant clones stained positive for γH2AX at the time of the experiments (25 days after transfection) (Fig. S10A) when down-regulation of POU5F1 was detected. Therefore, identification of multiple promoters serving as distal enhancers of POU5F1 by CREST-seq was unlikely due to artifacts of the experimental system.
The enhancer-like promoters are spatially close to POU5F1 TSS
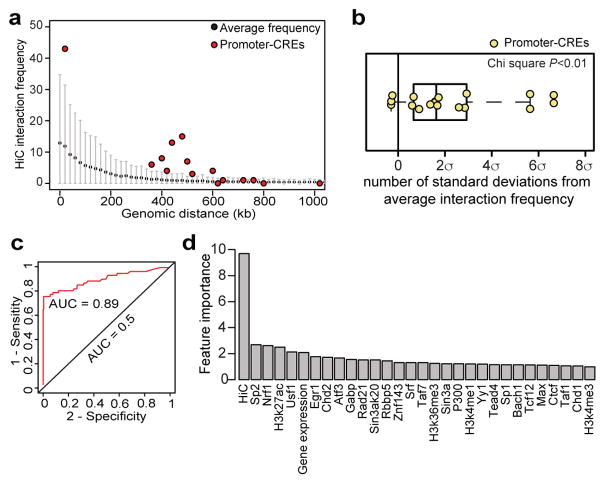
To understand potential mechanisms that allow the 17 CREST-positive promoters, among promoters of ~120 genes in this 2MB locus, to specifically regulate POU5F1, we examined the 3D chromatin organization of the locus, reasoning that long-range chromatin interactions may allow these enhancer-like promoters to act as distal cis-regulatory sequences. Indeed, analysis of H1 hESC Hi-C data36 indicate that 14 of the 17 POU5F1-regulating promoters display significantly higher levels of chromatin interactions with the POU5F1 TSS than expected by chance (Fig. 4A and 4B, Wilcoxon tests P-value < 0.01). The enhancer-like promoters are also characterized by other chromatin features that distinguish them from other promoters in the region, such as high levels of POL2 binding, H3K4me3, and H3K27ac (Fig. S11A and S11B, permutation P-value < 0.01). In addition, mRNA transcription from these promoters is significantly higher than other genes in the same region (Fig. S11C, Wilcoxon test, P-value < 0.01).
Fig 4. Analysis of chromatin interactions between the enhancer-like promoters and POU5F1 promoter in hESC.
(A) A dotplot shows the distribution of pairwise Hi-C contact frequencies within the 2Mbp locus, and between the POU5F1 TSS and the 17 POU5F1-regulating promoters (red dots, promoter-CREs). The black dots and the gray bar represent the average and standard deviation of Hi-C read counts at a given genomic distance, respectively.
(B) A boxplot shows the number of standard deviation of the Hi-C read counts between POU5F1 TSS and the promoter-CREs (yellow dots) compared to the expected (0, black line) (χ2 P-value < 0.01).
(C) ROC curve shows that POU5F1-regulating promoters can be separated from the other promoters in the 2Mbp region with a high accuracy (AUC=0.89) using a random forest model built from binding sites of 52 TFs, seven histone modifications profiles, gene expression profile and maps of long-range chromatin interactions (Table S7, see Supplementary Methods for more details).
(D) A bar chart shows the relative importance of each feature to the Random Forest classifier in predicting enhancer-like promoters.
To further characterize the features of enhancer-like promoters, we developed a random forest based classifier capable of predicting which promoters are cis-regulatory sequences of POU5F1. As input, we used datasets of transcription factor binding sites (TFBS) (Table S7), histone modification5 profiles, gene expression profiles, and the long-range chromatin contacts centered at POU5F136. The performance of the classifier was evaluated using leave-one-out cross validation. Strikingly, our model can distinguish POU5F1-regulating promoters from control promoters in the 2Mbp screen region with high accuracy (Fig. 4C, AUC = 0.89, error rate = 6.3% and PPV=97.2%). We next determined feature importance by estimating the average decrease in node impurity after permuting each predictor variable, finding that the chromatin interaction frequency is the single most important predictor (Fig. 4D and Fig. S12, “Hi-C” for normalized HiC interacting frequency). This result provides strong evidence that the enhancer-like promoters specifically affect POU5F1 expression through chromatin interactions. This observation promoted us to use spatial proximity alone to make a single-variable random forest model, which also achieves high accurate prediction (AUC=0.93, error rate=9.0%) but lower PPV (74.5%), suggesting the physical proximity is an important predictor for predicting regulatory relationship, but other factors are also crucial.
Discussion
In summary, we have developed a high-throughput method for functional screening of cis-regulatory elements in their native genomic context. We demonstrated the utility of this method by applying it to the 2Mbp POU5F1 gene locus in human ES cells, and validated the results by extensive experiments using allelic gene expression analysis.
Our finding that nearly 40% of the cis-regulatory sequences of POU5F1 correspond to promoters of other genes reveals the commonality and widespread use of promoters as distal enhancers. Previous studies have suggested that promoters and enhancers share common properties in terms of transcription factor binding and ability to produce RNA transcripts37. Recently, it was shown that the promoters of lncRNAs and mRNAs could act as enhancers of adjacent genes 12,31,38. The current study adds to the accumulating literature that distal promoters can regulate the expression of a gene other than the immediate downstream gene. Our results further showed that one potential mechanism for promoters to act as enhancers is via long-range chromatin interactions. This is consistent with previous studies showing extensive promoter-promoter interactions in mammalian cells 30,36,39–46, and reports that many promoters indeed show enhancer activity in heterologous ectopic luciferase reporter assay 30,47.
CREST-seq is a highly scalable tool for unbiased discovery of cis-regulatory sequences in the human genome. Compared to the previous CRISPR/Cas9 screens, which typically require more than 100 gRNAs-expressing oligos to “saturate” a targeted region, CREST-seq achieved 20x coverage for the entire 2Mbp POU5F1 locus with less than six sgRNAs per kilobase (Table 1). CREST-seq also outperforms the dCas9-KRAB based CRISPRi screen15 in which the size of H3K9me3 peaks generated by dCas9-KRAB is less than 850bp 48. Although the size of positive hits identified by CREST-seq are usually larger than the size of element/motif identified by single sgRNA approach, by generating overlapping deletions in a massively parallel fashion, CREST-seq allows functional interrogation of a large fraction of the genome with high sensitivity and specificity. More importantly, CREST-seq can distinguish cis- and trans-regulatory sequences by monitoring the allelic expression of a reporter gene, without the knowledge of haplotypes of the genome. Finally, it is feasible to design nested tiling deletions across a whole chromosome or even the genome. Combination of CREST-seq and single sgRNA screen approaches would allow us to achieve both high coverage and high resolution, thereby enabling truly comprehensive discovery of transcriptional regulatory sequences in the human genome.
Table 1. Comparison of CREST-seq to published CRISPR/Cas9 screens of non-codin gregulatory sequences.
CREST-seq is compared to the published screening of non-coding regulatory elements. The following aspects are compared: the size of screen region; the total number of oligos required to construct the library; the average number of oligos per kilobase in each screen; and the estimated coverage of the target region. To estimate the coverage of target region, we assume that the PAM motifs are equally distributed across the genome and each gRNA creates a mean indel size 9.5bp ± 13.7bp. To compute the coverage of CRISPRi screen using dCas9-KRAB, we assume that the average size of H3K9me3 peaks introduced by dCas9-KRAB is about 850bp.
| Reference | Target region | Total oligo # | Oligo density (per KB) | Coverage | Distinguish Trans or Cis? |
|---|---|---|---|---|---|
| Canver et al, 2015 10 | 4.2kb, 3 DHS and 1 exon | 582 | 137 | ~1x | No |
| Korkmaz et al, 2016 11 | 685 p53 ChIP-seq peaks | 1116 | NA | 1.3-1.6 oligos per ChIP-seq peak | No |
| 73 eRNA expressing ERa ChIP-seq peaks | 97 | NA | |||
| 2kb, deCDKN1A locus | 197 | 98.5 | <93.6% | ||
| Rajagopal et al, 2016 12 | 40kb Tdgf1 locus | 3908 | 98 | <93.1% | No |
| Rpp25, Nanog, and Zfp42 loci | 3908 | NA | NA | ||
| Diao et al, 2016, 13 | 37.6kb, 174 putative enhancers in 1Mbp POU5F1 locus | 1964 | 52 | <49.4% | No |
| Sanjana et al, 2016 14 | 200kb NF1 locus | 6682 | 33.4 | <31.7% | No |
| 200kb NF2 locus | 6934 | 34.6 | <32.9% | ||
| 200kb CUL3 locus | 4699 | 23.5 | <22.3% | ||
| Fulco et al, 2016 15 | 1.29Mbp, GATA1 and MYC loci | 98,000 | 76 | ~64x | No |
| CREST-seq | 2Mbp, POU5F1 locus | 11,600 | 5.7 | 20x | Yes |
Online Methods
A step-by-step protocol of CREST-seq is available in the Protocol Exchange and as a Supplementary Protocol.
Cell culture
The POU5F1-eGFP H1 hESC line was purchased from WiCell (Log number: DL-02) and described previously19. The cells were cultured on Matrigel-coated (Corning, Cat #354277) plates and maintained in TeSR-E8 media (STEMCELL Technologies, Cat#05940), and passaged by Accutase (STEMCELL Technologies, Cat#A1517001) with 10uM ROCK inhibitor Y-27632 (STEMCELL Technologies, Cat# 72302) supplement. The cells have been tested by WiCell Research Institute and UCSD human Stem Cell Core facility to confirm no mycoplasma contamination.
Design of sgRNA pairs for CREST-Seq
CREST-seq library design is available online (http://crest-seq.ucsd.edu/web/) and includes the following steps: 1) all 20-bp potential sgRNA sequences followed by PAM motif ‘NGG’ within the 2-MB screened region were first identified; 2) Bowtie 54 was used to map these 20-bp sgRNA sequences to the reference genome (hg19) with following parameter ‘-t -a -f -m 1000 --tryhard -v 3’ which outputs alignments up to 1000 candidates with less than 4 mismatches; 3) In order to prevent off-target binding, a sgRNA sequence was filtered out if it a) perfectly maps to another region on the genome; or b) has suboptimal alignment with 1 or 2 mismatched bases outside the sgRNA “seed” region, i.e. the 10bp sequence adjacent to PAM motif 55; or d) has suboptimal alignment with 3 mismatches but all three mismatched bases are 17-bp further to the PAM sequence; 4) the identified sgRNA sites were paired in order to generate 2kb-deletions evenly across the 2 Mbp-region. Based on the distribution of the filtered sgRNA, a chain of unique single guide RNAs were selected as follows: First, the initial sgRNA was picked, and the next sgRNA was chosen based on a pre-determined distance cutoff (D, for example 100bp) and an odd number of step size (S, for example 15) such that the distance between the target sequences of the two sgRNAs is no less than D; the procedure was repeated until no more unique sgRNA was found. Next, the first sgRNA pair was designed using the 1st sgRNA and the 16th (1+S) sgRNA, then the second pair using 3rd and 18th (3+S), the procedure was repeated to the end of the chain. The distance cutoff D and step S were both adjustable to allow for different deletion sizes and genomic coverage. For example, using D=100, and S=15, the deletion size would be a minimum of 1,500 bp, an average of 2,000 bp in the current design. The average coverage was (1+S)/2, 8 times with S=15, since there were 8 sgRNAs (relatively 1st, 3rd, … 15th) crossover to 8 guide RNAs on other side (relatively 16th, 18th, … 30th) for any region in the middle. Three different sets of deletion/steps were used: 100/15, 200/13, 500/13. An unique guide RNA was not used if it has been used in previous selection. After a pair of dual CRISPR guide RNAs, namely {a, b}, were selected, we used the following template to link two guide RNAs: TGTGGAAAGGACGAAACACC{a}GTTTAGAGACG{rnd}CGTCTCACCTT{b}GTTTTAG AGCTAGAAATAGCAAGTT, note that if a guide RNA start with A, C, or T, a G was added in front. The ${rnd} was selected from all combinations of 9-bp nucleotide sequence excluding either number of GC less than 4 or more than 6, or include any subsequence within: {"AAAA", "CCCC", "TTTT", "GGGG", "GAGACG", or "CGTCTC"}.
Oligo synthesis and library cloning
The CREST-seq oligo library with sequences shown in Fig. S2a was amplified with the following primers:
Forward primer: CTTGTGGAAAGGACGAAAC
Reverse primer: TTTTAACTTGCTATTTCTAGCTCTAAAAC
The PCR product was size selected and gel-purified with NucleoSpin Gel and PCR Clean-Up Kit (Clontech, Cat# 740609), and then inserted into BsmbI digested lentiCRISPRv2 plasmid by Gilbson Assembly (Addgene plasmid #52961). The end product was electro-transformed into 5-alpha Electrocompetent E. coli (NEB, Cat#C2989K) and grown on Agar plates. About 20 million independent bacterial colonies were collected and the plasmids were extracted with QIAGEN Plasmid Giga Kit (Cat#12191). The resulting plasmid DNA was linearized by BsmbI digestion, gel purified and ligated with a DNA fragment (see complete IDT gBlocks sequence in Table S8) containing tracRNA(E/F) and the mouse U6 promoter (mU6). The ligates was electro-transformed into 5-alpha Electrocompetent E. coli and plated on Agar plates. About 20 million bacterial colonies were collected and purified with EndoFree Plasmid Giga Kit (QIAGEN, Cat#12391)
Lentiviral library production
The CREST-seq lentiviral library was prepared as previously described 56 with minor modifications. Briefly, 5ug of lentiCRISPR plasmid library was co-transfected with 4 ug PsPAX2 and 1 ug pMD2.G (Addgene #12260 and #12259) into a 10-cm dish of HEK293T cells in DMEM (Life Technologies) containing 10% FBS (Life Technologies) by PolyJet transfection reagents (Signagen, Cat# SL100688). Growth medium was replaced 6 hours after transfection. The supernatant of cell culture media was harvested at 24 hours and 48 hours after transfection, and filtered by Millex-HV 0.45 μm PVDF filters (Millipore, Cat# SLHV033RS). The viruses were further concentrated with 100, 000 NMWL Amicon Ultra-15 Centrifugal Filter Units (Amicon, Cat#UFC910008).
For viral titration, 0.5 million hESC POU5F1-eGFP cells were seeded per well on 6-well plate. 12 hours later, different amount (1ul, 2ul, 4ul, 8ul) of concentrated viral-containing media were added to the cell culture media to infect the hESC following the same protocol described in the lentiviral screening section. The same amount of non-infected cells was seeded and not treated with puromycin as the control. 24 hours post-infection, the viral infected cells were treated with 500ng/ml Puromycin (Life Technologies, Cat#A1113802) for another 72 hours. We counted the number of Puromycin resistant cells and the control cells to calculate the ration of infected cells, and then viral titer. In the screening, about 10 million POU5F1-eGFP hESCs were used in each independent screening replicate and infected with viral particles at low MOI (0.1) to make sure each infected cell gets one viral particle.
Lentiviral transduction and FACS
Briefly, the screening was performed following previous protocol described earlier 13 with minor modifications. In each independent screen, about 10 million cells per 12-well plates were spin infected with CREST-seq lentiviral library at MOI=0.1. 24 hours post infection, the cells were dissociated with Accutase, and plated into l5cm culture dish coated with Matrigel (4 million cells per dish). The cells were treated with E8 media containing 250ng/ml Puromycin for 7 days, followed by another 7-day culture without Puromycin treatment. For CREST-seq screen FACS sort, the cells were dissociated and co-immunostained with PE-POU5F1 antibody and APC-eGFP antibody. The eGFP-/POU5F1+, eGFP+/POU5F1+, and non-sorted control cells were collected by FACS sort for further analysis.
Sequencing library construction
Genomic DNA was extracted from the eGFP-/POU5F1+, eGFP+/POU5F1+ or the non-sorted control cells populations. The sgRNAs inserts were then amplified from genomic DNA PCR using the following primers:
Forward: AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG
Reverse: GGACTGTGGGCGATGTGCGCTCTG
The PCR products were gel purified and subjected to the 2nd PCR reaction to add Illumina TruSeq adaptor sequence with the following primers:
Forward: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC TctTGTGGAAAGGACGAAAC
Reverse (N indicate the index sequence): CAAGCAGAAGACGGCATACGAGANNNNNNGTGACTGGAGTTCAGACGTGTGCTCTT CCGATCTTTTTAACTTGCTATTTCTAGCTCTAAAAC
Sequencing and processing of CREST-seq libraries
CREST-seq libraries were sequenced using HiSeq 4000 in pair-ended mode with 100bp read length. A sgRNA pair {a, b} was considered valid if it matched the initial sgRNA design and met the following criteria: (1) a subsequence of the read1 matched GGACGAAACACCG, followed by 19 or 20 nucleotides (namely, {a'}), and GTTTAAGAGCTATGCTG, (2) a subsequence of read2 matched AAAC, followed by 19 or 20 nucleotides (namely, {b'}), and followed by CAA; (3) {a} exactly matched {a'} if length of {a'} was 20, or {a} exactly matched G+{a'} if length of {a'} was 19; (4) {b} exactly matched reverse complementary of {b'} if length of {b'} was 20, or {b} exactly matched G+reverse complementary {b'} if length of {b'} was 19. Those sgRNA pairs with total read count less than 30 among all samples were filtered out. In the end, we kept 10,159 sgRNA pairs for further analysis (Table S4).
Peak calling in CREST-seq data
For each sgRNA pair, the MAGeCK algorithm 20 was used to estimate the statistical significance (using Negative Binomial test) of enrichment in the cell population relative to the control population. Next, sgRNAs pairs were ranked by log (NBP – value) x sign(log(exp/control)) in an increasing order. Third, we partitioned the 2-MB screened region into a set of non-overlapping 50-bp bins B = (b1, … ,bn) , and a bin was considered positive if many of the sgRNA pairs spanning it rank near the top of the sorted list. A Robust Rank Aggregation (RRA) algorithm 57 was then used to identify the positive bins. Specifically, let Rt = (ri1, … ,rik , be the vector of ranks of sgRNA pairs that span bin bi , we normalized Ri into percentiles Ui = (ui1, … ,uik) where uij = rij/M (M is the total number of sgRNA pairs). The goal was to identify the bins whose normalized rank vector Ui is strongly skewed toward zero. Under null hypothesis where the normalized ranks follow a uniform distribution between 0 and 1, the j-th smallest value among (ui1, … ,uik) is an order statistics ρ(uij) which can be calculated by a beta distribution Beta (j,k + 1−j). . We defined the final score for the rank vector Ui as the minimum of -score:
ρ(Ui) score was converted to P-value by permutation test as proposed by Li et al 20 and finally P-value was finally adjusted to FDR. A bin was considered as significant if its FDR was smaller than a given threshold.
Calculation of Enrichment Test Score
We downloaded DNase Hypersensitive Sites (DHSs) and peaks of ChIP-seq datasets from H1 hESC from ENCODE data portal5. Enhancers were predicted using RFECS 58, and promoter coordinates were based on RefSeq gene annotation. The observed overlap ratio oi of feature i was computed as the fraction of CREST-seq peaks that overlapped with this feature. We then randomly shuffled CREST-seq peaks in this region using ‘shuffleBed’ 59, and the expected overlap rate e1 was counted as the fraction of shuffled peaks that overlapped with feature i. Fold enrichment was computed as oi/ei . We repeated this process 1000 times for each feature and defined the enrichment test score as the fraction of tests where the fold enrichment was greater than 1. The significance of enrichment was derived using the χ2 test.
Analysis of chromatin signatures of POU5F1-regulating promoters
We randomly shuffled CREST-seq peaks in the 2Mbp POU5F1 region using ‘shuffleBed’ 59 and only kept those permutations with 18 peaks overlapping promoter regions. The expected overlap rate for each shuffle was counted as the fraction of permutations that contain active promoter signature (Pol2/H3k4m3/H3k27ac). We repeated this process 1000 times and calculated permutation P-value as the percentage of tests in which the overlap rate is above 0.78.
Classification of POU5F1-regulating promoters by Random Forest
We downloaded RefSeq annotated promoters (2,000bp upstream TSS) from UCSC genome browser within the screened region. Promoters were divided into positive and control groups based on their overlap with CREs. RNA-seq data was downloaded from previously work and gene expression was estimated using software Cufflinks for each transcript. Random forest implemented by R package “randomForest” was applied to classify positive promoters from the negative ones with default parameter setting without further model selection. Prediction performance was evaluated by leave-one-out cross validation. Feature importance was estimated by the average decrease of node purity by permuting each variable.
CRISPR/Cas9-mediated deletion
CRISPR/Cas9 constructs targeting genomic loci indicated on Fig. S6A was made following the protocol described earlier13. The oligos used for cloning are listed in Table S8. The designed sgRNAs sequence was cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid #42230) vector. After validating the sgRNA sequences by Sanger sequencing, a pair of plasmids targeting 5’- and 3’-boundary of the same element, were mixed at 1:1 ratio and co-transfected with plasmid expressing mCherry into POU5F1-eGFP cells by hESCs Nuclearfector Kits 2 (Lonzo, Cat#VPH-5022) according to the manufacture’s instruction. To knockout POU5F1-regulatory core promoters, we used in vitro synthesized CRISPR crRNA and CRISPR tracrRNA (IDT) with the sequence specified in Table S8. The Cas9 recombinant protein was purchased from NEB (Cat M0386M) and the Cas9/crRNA/tracRNA was assembled in vitro by following a protocol60. The RNP complex was electro-transfeced into POU5F1-eGFP hESC reporter line with Neon Transfection System 10μl kit (ThermoFisher Scientific, Cat#: MPK1096) with the default electrotranfection protocol #9. 72 hours post-transfection, the mCherry positive cells were collected by FACS. The mCherry positive single cells were plated into Matrigel-coated plate at low density (about 1000 cells per 10 cm coated petri-dish), and cultured in E8 media supplemented with 10uM ROCK inhibitor. After 10 to 14 days, the surviving sorted single cells formed colonies. Individual colonies were picked and expanded, followed by genotyping and in-depth analysis.
Genotyping of mutant clones
The cells from mutant clones were collected and treated with QuickExtract™ DNA Extraction Solution (Epicentre, Cat# QE0905T), followed by genotyping PCR using primers listed in Table S8. Then Topo cloning (Life Technologies, Cat#K2800-20) and Sanger sequencing were conducted to verify the sequences.
FACS analysis
To directly monitor the eGFP expression levels, the wild type or mutant POU5F1-eGFP cells were dissociated with Accutase and subjected to FACS analysis with BD FACSAria II. To examine the levels of HLA-C protein, the cells were stained with PE-conjugated antibody specifically recognizing HLA-C (Millpore, Cat#MABF233). To carry out immunostaining of eGFP, POU5F1, or γH2AX, the cells were fixed with 2% PFA for 30 minutes, followed by overnight permeabilization in Methanol at −20°C. The treated cells were stained with the antibodies. PerCP-cy5.5-conjugated mouse anti-H2AX(pS139) was purchased BD Biosciences (Cat#564718); PE-conjugated anti-human OCT4(OCT3) antibody was from STEMCELL Technologies (Cat# 60093PE.1) and APC-conjugated anti-GFPuv/eGFP antibody is available from R&D Systems (Cat# IC4240A)
Luciferase reporter assays
Luciferase assays were conducted as previously described 61. Briefly, to test the enhancer activity of CREs with native POU5F1 promoter, the 360bp POU5F1 minimal promoter 32 (hg18 Chr 6: 31,246,377-31,246,736) was synthesized as gblock by IDT, and cloned into pGL3-promoter vector to replace the original SV-40 promoter. The core promoter regions of pPRRC2A, pMSH5, pNEU1 and pTFC19 were PCR amplified from H1 hESC genomic DNA, and cloned into a modified pGL3-POU5F1 vector (Promega), in which the SV40 promoter has been replaced by a 360bp minimal POU5F1 promoter by In-fusion cloning. The primer sequences are listed in Table S8. After validation by Sanger sequencing, the constructs were co-transfected with pRL-SV40 Renilla reporter vector in H1 hESCs with Fugene HD (Roche) at a 4:1 reagent to DNA ratio. The transfected cells were cultured for an additional 2 days prior to harvest for reporter assay. The Dual-Luciferase Reporter Assay kit (Promega Cat#:E1960) was used according to manufacturer’s protocol. The adjusted firefly luciferase activity of each sample was normalized to the average of activities of 3 negative control regions.
RNA interference
The siRNAs were purchased from Dharmacon in the format of ON-TARGETplusSMARTpool-Human targeting MSH5, NEU1 and PRRC2A, respectively. We also designed siRNAs by using WI siRNA selection program and the sequence of siRNA are listed in Table S8. The siRNAs were transfected into hESC with Human Stem Cell Nucleofector Kit 2 (LONZA) per manufacturer’s instruction.
Western blotting
Western blotting was performed by following the protocol described previously 62. Briefly, whole cell extracts (WCE) were collected and quantified with Pierce™ BCA Protein Assay Kit (Cat#23225). 30μg WCE of each sample was subjected to Western blot analysis with antibodies specifically recognizing NEU1(Thermo Scientific, Cat#PA5-42552), PRRC2A (Abcam, Cat#ab188301), MSH5 (Abcam, Cat#ab130484), Histone-H3(Abcam, Cat#ab1791), POU5F1 (Abcam, Cat#ab19875), and eGFP (Abcam, Cat#ab190584).
ATAC-seq experiment and analysis
ATAC-seq was performed by following the protocol described earlier 22. Briefly, each library starts with 100k cells which were permeabilized with NPB (0.2% NP-40, 5%BSA, 1Mm DTT in PBS with one complete proteinase inhibitor) at 4 degree for 10min, followed by spin down at 500g for 5min. The resulting nuclei were resuspended in 20ul 1xDMF (33mM Tris-acetate (pH=7.8), 166mM K-Acetate, 10mM Mg-Acetate, 16 % DMF). The chromatin tagmentation was done by adding 0.5ul Tn5 into 10ul solution for 30min at 37 degrees.
We processed our ATAC-seq data in the following steps: 1) ATAC-seq sequencing reads were mapped to hg19 reference genome using Bowtie(61) in pair-end mode; 2) poorly mapped, improperly paired and mitochondrial reads were filtered; 3) PCR duplications were further removed using Picards MarkDuplicates (http://broadinstitute.github.io/picard.); 4) Mapping positions of reads were adjusted accounting for Tn5 insertion; 6) Reads were next shifted for 75bp followed by peak calling using MACS263 with following parameters “-q 0.01 --nomodel --shift 175 –B --SPMR --keep-dup all --call-summits”; 7) ATAC-seq signal was normalized into RPKM using deeptools64 for visualization.
PCA analysis
We first extracted all 478 H1 DHS sites within the screened regions and counted the average RPKM for each site using 122 public DHS datasets (Table S8) and our own ATAC-seq dataset. Pair-wise Pearson correlation between the datasets were calculated and used as input for PCA analysis. We found the first two principle components accounted for 80% of the variance and therefore used for 2D visualization as shown in Figure S5B.
Code availability
The computer code used in this study is available https://github.com/r3fang/crest
Supplementary Material
Acknowledgments
We thank D. Gorkin and J. Yan for feedbacks on previous versions of the manuscript. We thank Z. Ye and S. Kuan for technical assistance. This work is supported by National Institutes of Health (NIH) (U54 HG006997, U01 DK105541, R01HG008135, 1UM1HG009402 and 2P50 GM085764) and by the Ludwig Institute for Cancer Research (to B.R.). Y.D. is supported by the Human Frontier Science Program (HFSP) Long Term Postdoctoral Fellowship.
Footnotes
Accession Codes and Data Availability.
Sequencing data have been deposited to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE81026. Additional materials, data, code, and associated protocols are available upon request.
Author contributions:
Y.D. and B.R. conceived the idea for CREST-seq; R.F., Y. D., and B. L., conducted integrative data analysis with help from Y.Q., H.H., and I.J.; B.L. and Y. D., designed paired sgRNA libraries; Y.D., Z.M., J.Y., K.C.L, T.L., H.H., R.J.M. and Y.S. performed the experiment; Z.M., K.C.L. and K.L.G. packaged the lentiviral library; Y.D., R.F., B.L. and B.R. wrote the paper.
Competing Financial Interests Statement
B.R. is a co-founder of Arima Genomics, Inc.
References
- 1.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 12.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diao Y, et al. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9–mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco CP, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016 doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 20.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware CB, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 26.MacArthur S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yip KY, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandra T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016 doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chia NY, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 33.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 35.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 36.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paralkar VR, et al. Unlinking an lncRNA from Its Associated cis Element. Mol Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeMare LE, et al. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji X, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Z, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 48.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu S, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 51.Feuerborn A, Cook PR. Why the activity of a gene depends on its neighbors. Trends Genet. 2015;31:483–490. doi: 10.1016/j.tig.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng Z, et al. Berbamine inhibits the growth of liver cancer cells and cancer-initiating cells by targeting Ca(2)(+)/calmodulin-dependent protein kinase II. Mol Cancer Ther. 2013;12:2067–2077. doi: 10.1158/1535-7163.MCT-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajagopal N, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley ML, Strezoska Z, He K, Vermeulen A, Smith A. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J Biotechnol. 2016;233:74–83. doi: 10.1016/j.jbiotec.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 62.Diao Y, et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.