Abstract
The classification of melanoma into four histological subtypes has been questioned regarding its clinical validity in providing relevant information for treatment for metastatic tumors. Specific genetic alterations are associated with particular clinical and histopathological features, suggesting that these could be helpful in refining existing melanoma classification schemes. We analyzed BRAF V600E mutated melanomas to explore the Reflectance confocal microscopy (RCM) utility as a screening aid in the evaluation of the most appropriate patients for genetic testing. Thus, 32 melanomas were assessed regarding their BRAF V600E mutational status. Experts blinded to dermoscopic images and V600E immunohistochemistry results evaluated RCM images regarding previously described melanoma features. BRAF positive melanomas were related to younger age (p = 0.035), invasive melanomas (p = 0.03) and to the presence of hiporreflective cells (p = 0.02), epidermal nests (p = 0.02), dermal-epidermal junction nests (p = 0.05), edged papillae (p = 0.05), and bright dots (p = 0.05), and to absence of junctional thickening due to isolated cells (p = 0.01) and meshwork (p = 0.02). This study can not characterize other mutations in the BRAF, because the immunohistochemistry is specific to the type V600E. The findings should encourage the genetic evaluation of BRAF mutation. This study highlights the potential of RCM as a supplementary tool in the screening of BRAF-mutated melanomas.
Introduction
The fact that melanomas can present with varied morphologies and biologic behaviors including differences in rate of growth, anatomical location and propensity to metastasize has been known for quite some time. Initial attempts at subtyping melanomas based on their clinical and histopathology characteristics lead to the classification of these tumors into superficial spreading, nodular, acro-lentiginous, and lentigo maligna, among others. Some have tried to simplify this classification based on the degree of UV exposure into melanomas on chronic sun damaged skin, melanomas on intermittent sun exposed skin and melanomas on UV protected sites [1]. However, evidence is now starting to emerge that the underpinnings responsible for the morphology and biology of melanoma may primarily be due to their molecular profile (gene expression profile) [2]. It has been shown that approximately 50% of melanomas have a BRAFV600 mutation [3]. Few recent studies have sought to correlate BRAF mutational status with features observed via non-invasive evaluation techniques, such as dermoscopy [4,5,6]. Researches have noted that BRAF mutated melanomas tend to be of the superficial spreading type and often reveal peppering on dermoscopy [5]. Ruini et al. used Reflectance Confocal Microscopy (RCM) to evaluate 8 melanomas and suggested that RCM may provide more specific information on the cytoarchitectural structure of BRAF mutated melanomas [7]. Our present study was undertaken to determine whether the RCM characteristics of the BRAF V600E mutated melanomas differ from the BRAF wild type melanomas.
Material and methods
This study was approved by the Institutional Review Board of A C Camargo Cancer Center, São Paulo, Brazil and it was registered under no. 1685/12.
Subjects
This retrospective cohort study included 32 consecutively diagnosed melanomas; all of which were imaged with RCM prior to biopsy. Only thin melanomas were included as RCM is not able to reach deeper skin layers. Each of the 32 melanomas came from a different patient and all cases were diagnosed between 2011 and 2013. Informed consent was not necessary because the data were analyzed anonymously.
Routine histopathology
The histopathology slides were stained with hematoxylin-eosin and the pathologist (C.P.) was required to comment on the following parameters according to the Pathology Department’s protocol: histological subtype, growth pattern, Breslow thickness, Clark's level, peritumoral inflammatory infiltrate, intratumoral inflammatory infiltrate, regression, lymphatic invasion, vascular invasion, perineural invasion, existence of associated nevus, microscopic satellites, and mitotic rate. Histopathology images were obtained of all cases using ScanScope Digital Slide Scanner (Aperio, Vista, CA, USA).
BRAF immunohistochemistry
The archived specimens were retrieved and processed by deparaffinization followed by antigen retrieval and dilution using the anti-VE1 antibody, which was performed on Ventana Benchmark XT immunostainer (Roche Diagnostics, Burges Hill—UK), according to specified standard protocol. OptiView DAB IHC Detection Kit (Roche, Burges Hill-UK) was used to detect V600E BRAF protein expression. The slides were stained with anti-V600E BRAF (VE1) Mouse Monoclonal Primary Antibody (Roche, Burges Hill-UK). Positive control with BRAF V600E mutated colorectal adenocarcinoma was used for procedure standardization. The omission of this antibody in tissue known to be positive for the BRAF V600E mutation served as the negative control. All immunohistochemistry stained slides were evaluated by the same pathologist (PCA). Immunostaining was interpreted as positive or negative according to criteria proposed by Capper et al. [8] and Long et al. [9], VE1 antibody staining was considered positive if tumor cells showed unambiguous cytoplasmic staining. The staining reaction was considered negative if there was no evidence of staining or if there was only weak, focal staining of isolated cells.
Molecular biology testing is still the gold standard method for BRAF V600E status assessment. However, a plenty of studies have shown that VE1 antibody staining is a less expensive approach to rapidly evaluate the BRAF V600E mutation in melanomas [10].
All study lesions proved to be relatively thin invasive melanomas or in situ melanomas. Hence the degree of BRAF staining in the BRAF mutated tumors was mostly weak to moderate and distributed focally or multifocally (Fig 1).
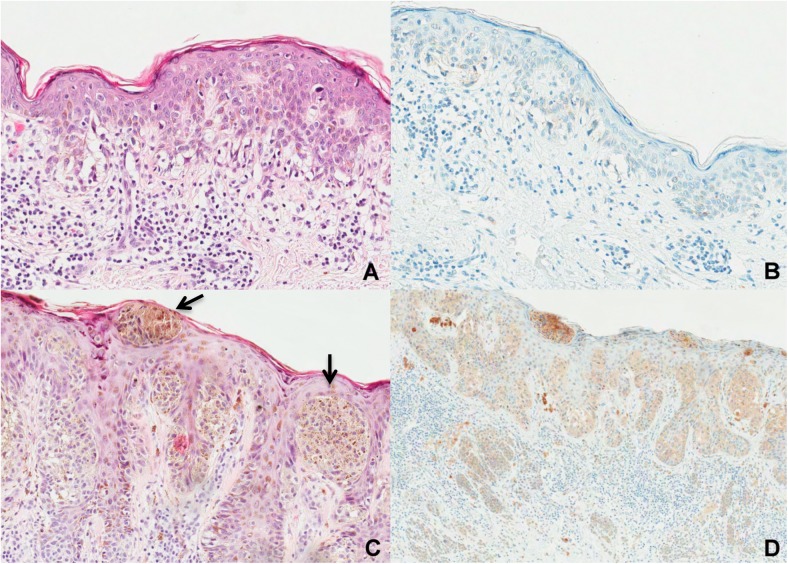
Fig 1. Tumor tissues with hematoxylin-eosin staining and with immunohistochemistry reaction for anti-VE1 antibody.
(A) Histopathological features of in situ superficial spreading melanoma, exhibiting mainly scattered melanocytes along the basal layer and only few pagetoid cells spreading in the epidermis (H&E staining, original magnification 400x) (B) BRAF V600E IHC negative case characterized by complete lack of tumor cell immunostaining (original magnification 400x)—A and B corresponding to the case in Fig 2 (2A). (C) Histopathological features of superficial spreading melanoma, 1 mm in Breslow thickness showing a predominantly nested pattern of large intraepidermal and junctional nests (arrows) (H&E staining, original magnification 400x). (D) BRAF V600E cytoplasmic immunostaining positive (original magnification 200x)—C and D corresponding to the case in Fig 2D.
Instruments
RCM images were acquired using the near-infrared reflectance confocal laser microscope (Vivascope 1500; Caliber I.D., Rochester, NY, USA). The RCM scanning technique and the acquisition of RCM images was the same as published by Pellacani et al. [11]. RCM “mosaic images”, ranging in size from 4x4 to 8x8 mm, were captured in all cases ("Vivablock"). A minimum of three mosaics was obtained for each lesion with at least one mosaic acquired at the superficial epidermal layer, one at dermal-epidermal junction (DEJ) and one at the papillary dermis. In addition, focal areas of interest were imaged via the “stack” method, which captures images of the area of interest by acquiring sequential images from the stratum corneum up to papillary dermis.
Parameter evaluation
RCM: Two dermatologists with experience in reading RCM images (TBJC and RGG) independently evaluated all of the RCM images blinded to both dermoscopic and histopathological features. The findings were categorized in a binary fashion for the following parameters [12,13,14].
Epidermal layer: presence or absence of atypical honeycomb pattern, atypical cobblestone pattern, pagetoid pattern and hyporreflective cells. If within any 1x1mm field of view there were more then 10 pagetoid cells per mm2 we considered this parameter to be present and if ≤ 10 pagetoid cells it was considered as absent. Hyporreflective cells were considered present if more than 5 cells were seen in any high power field (1x1).
Dermal-epidermal junction (DEJ): presence or absence of junctional nests, edged papillae, non-edged papillae, non-visible papillae, sheet-like structures, mitochondria-like structures, meshwork pattern and junctional thickening (enlargement of the inter-papillary spaces by bright sparse cells or in aggregates <5 cells).
Superficial dermis: presence or absence of bright dots, sparse plump cells, aggregated plump cells, reticulated collagen, and collagen in bundles.
Statistical analysis
The results were tabulated and the frequency distributions, standard deviations, and range of measurements calculated. The Mann -Whitney test was used to compare the mean age as a function of BRAF V600E expression. The Fisher's Exact Test was used to evaluate the association between the expression of BRAF V600E and the presence or absence of: growth phase (radial or vertical), hyporreflective cells, epidermal nests, dermal-epidermal junction (DEJ) nests, edged papillae, nonedged papillae, junctional thickening, meshwork pattern and bright dots.
Multiple logistic regression was employed to identify the independent RCM factors predictive of positive BRAF V600E mutation status. All RCM variables seen in BRAF V600E mutated and BRAF wild type melanomas were compared and any variable that was found to be different between these two groups, with p value of less than 0.20, in univariate analysis were selected for a stepwise forward selection model and only variables with a p value less than or equal to 0.05 remained in the final multiple logistic model. The predictive value of the final model was assessed by calculating the area under the ROC curve (AUC).
Microsoft Excel 2007 (www.microsoft.com) and the IBM SPSS version 22.0.0.0 were used for all analyses.
Results
A total of 32 melanomas from 16 women and 16 men with ages ranging between 28 and 85 years were evaluated. Four melanomas (12.5%) were located on the head and neck area, 13 (40.6%) on the torso, 12 (37.5%) on the arm and 3 on the leg (Table 1).
Table 1. Clinical variables and BRAF V600E mutation.
| BRAF V600E | |||||
|---|---|---|---|---|---|
| Variable | Category | negativea | positiveb | Total | P value |
| Gender | Male | 13 (56.5%) | 3 (33.3%) | 16 | 0.4 |
| Female | 10 (43.4%) | 6 (66.7%) | 16 | ||
| *Age | Mean (SD) | 58.6 (14.3) | 58.1 (14.6) | 0.02 | |
| *Age < 50years old | Mean (SD) | 56.7 (14.1) | 58.1 (14.6) | 0.03 | |
| Topography | Head and neck | 3 (75.0%) | 1 (25.0%) | 4 | 0.9 |
| Superior limb | 8 (66.7%) | 4 (33.3%) | 12 | ||
| Trunk | 10 (76.9%) | 3 (23.1%) | 13 | ||
| Inferior limb | 2 (66.7%) | 1(33.3%) | 3 | ||
*Mann Whitney Test
a. Negative: absence of staining
b. Positive: any positive staining (focal or multifocal).
Histopathology variables were assessed and are summarized in Table 2.
Table 2. Histological variables and BRAF V600E mutation.
| BRAF V600E | |||||
|---|---|---|---|---|---|
| Variable | Category | negativea | positiveb | Total | P value |
| Histological subtype | S. Spreading | 19 (67.9%) | 9 (32.1%) | 28 | 1.0 |
| L. Maligna | 2 (100%) | 0 | 2 | ||
| not informed | 2 (100%) | 0 | 2 | ||
| **Growth | In situ | 17 (94.4%) | 1 (5.6%) | 18 | 0.03 |
| Radial | 3 (42.9%) | 4 (57.1%) | 7 | ||
| Vertical | 3 (42.9%) | 4 (57.1%) | 7 | ||
| Ulceration | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Clark | II | 3 (42.9%) | 4 (57.1%) | 7 | 1.0 |
| III | 3 (42.9%) | 4 (57.1%) | 7 | ||
| Breslow | Mean (SD) | 0.67 (0.23) | 0.64 (0.25) | 0.6 | |
| Mitotic Index mm2 | Mean (SD) | 0.40 (0.92) | 0.37 (0.89) | 0.6 | |
| Peritumoral Invasion | Present | 6 (46.2%) | 7 (53.8%) | 13 | 1.0 |
| Absent | 0 | 1 (100%) | 1 | ||
| Intratumoral Invasion | Present | 2 (28.6%) | 5 (71.4%) | 7 | 0.6 |
| Absent | 4 (57.1%) | 3 (42.9%) | 7 | ||
| Regression | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Lymphatic Invasion | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Vascular Invasion | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Perineural Invasion | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Previous nevus | Present | 6 (66.7%) | 3 (33.3%) | 9 | 0.7 |
| Absent | 17 (73.9) | 6 (26.1%) | 23 | ||
| Microscopic Satelitosis | Absent | 6 (42.9%) | 8 (57.1%) | 14 | + |
| Margins | Free | 20 (69.0%) | 9 (31.0%) | 29 | 0.5 |
| Positive | 3 (100%) | 0 | 3 | ||
**Fisher’s Exact Test
a. Negative: absence of staining
b. Positive: any positive staining (focal or multifocal).
Superficial spreading melanoma was the most common histological subtype, comprising 87.5% (n = 28) of the cases and they were located on the torso (n = 11), limbs (n = 13) and head and neck (n = 4). Two cases were classified as lentigo maligna melanomas and they were located on the upper trunk and upper limb. In two cases the histological subtype could not be determined, since there was an overlapping of features (superficial spreading melanoma and lentigo maligna). Most melanomas were in situ (18/32; 56.3%) and were located on the head&neck (n = 1), torso (n = 9), upper limbs (n = 7) and lower limbs (n = 1). The invasive melanomas had a median Breslow thickness of 0.66mm and most had a Clark level between II and III. Peritumoral and intratumoral infiltrates were seen in 92.7% and 50% of melanomas, respectively. No ulceration, regression, satellitosis, perineural invasion, vascular invasion, nor lymphatic invasion was observed. Nine melanomas (9/32; 28.1%) were found to have an associated melanocytic nevus.
RCM analysis
RCM images revealed that all 32 melanomas had an atypical honeycomb pattern, non-edged papillae and reticulated collagen. Other frequent features observed were atypical cobblestone (93.8%), sparse plump cells (90.4%), bright dots (62.5%), pagetoid cells (62.5%), edged papillae (62.5%), dermal-epidermal junction nests (59.4%) and sheet-like structures (53.1%). Most epidermal nests observed on RCM were of the dense variety. The distribution of RCM parameters is described further in Table 3.
Table 3. Confocal microscopy characteristics and BRAF V600E mutation.
| BRAF V600E | |||||
|---|---|---|---|---|---|
| Variable | Category | negativea | positiveb | Total | P value |
| Atypical Honeycomb | Present | 23 (71.9%) | 9 (28.1%) | 32 | + |
| Atypical Cobblestone | Present | 22 (73.3%) | 8 (26.7%) | 30 | 0.5 |
| Absent | 1 (50.0%) | 1 (50.0%) | 2 | ||
| Pagetoid cells | Present | 12 (60.0%) | 8 (40.0%) | 20 | 0.1 |
| Absent | 11 (91.7%) | 1 (8.3%) | 12 | ||
| **Hyporreflective cells | Present | 3 (37.5%) | 5 (62.5%) | 8 | 0.02 |
| Absent | 20 (83.3%) | 4 (16.7%) | 24 | ||
| **Epidermal nests | Present | 7 (50.0%) | 7(50.0%) | 14 | 0.02 |
| Absent | 16 (88.9%) | 2 (11.1%) | 18 | ||
| **DEJ nests | Present | 11 (57.9%) | 8 (42.1%) | 19 | 0.05 |
| Absent | 12 (92.3%) | 1 (7.7%) | 13 | ||
| **Edged papillae | Present | 17 (85.0%) | 3 (15.0%) | 20 | 0.05 |
| Absent | 6 (50.0%) | 6 (50.0%) | 12 | ||
| Nonedged papillae | Present | 23 (71.9%) | 9 (28.1%) | 32 | + |
| **Abscence of papillae | Present | 1 (25.0%) | 3 (75.0%) | 4 | 0.06 |
| Absent | 22 (78.6%) | 6 (21.4%) | 28 | ||
| Sheet-like Structure | Present | 12 (70.6%) | 5 (29.4%) | 17 | 1.0 |
| Absent | 11 (73.3%) | 4 (26.7%) | 15 | ||
| **Meshwork | Present | 16 (88.9%) | 2 (11.1%) | 18 | 0.02 |
| Absent | 7 (50.0%) | 7 (50.0%) | 14 | ||
| Mitochondria | Present | 4 (80.0%) | 1 (20.0%) | 5 | 1.0 |
| Absent | 19 (70.4%) | 8 (29.6%) | 27 | ||
| Absent | 20 (69.0%) | 9 (31.0%) | 29 | ||
| **Bright dots | Present | 17 (85.0%) | 3 (15.0%) | 20 | 0.05 |
| Absent | 6 (50.0%) | 6 (50.0%) | 12 | ||
| Sparse Plump cells | Present | 22 (75.9%) | 7 (24.1%) | 29 | 0.2 |
| Absent | 1 (33.3%) | 2 (66.7%) | 3 | ||
| Agregated Plump cells | Present | 8 (72.7%) | 3 (27.3%) | 11 | 1.0 |
| Absent | 15 (71.4%) | 6 (28.6%) | 21 | ||
| Reticulated Collagen | Present | 23 (71.9%) | 9 (28.1%) | 32 | + |
| Collagen Bundles | Present | 5 (83.3%) | 1 (16.7%) | 6 | 0.6 |
| Absent | 18 (69.2%) | 8 (30.8%) | 26 | ||
**Fisher’s Exact Test
a. Negative: absence of staining
b. Positive: any positive staining (focal or multifocal).
BRAF analysis
Nine of 32 melanomas stained positive for the BRAF V600E mutation (28.1%; Figs 1 and 2).
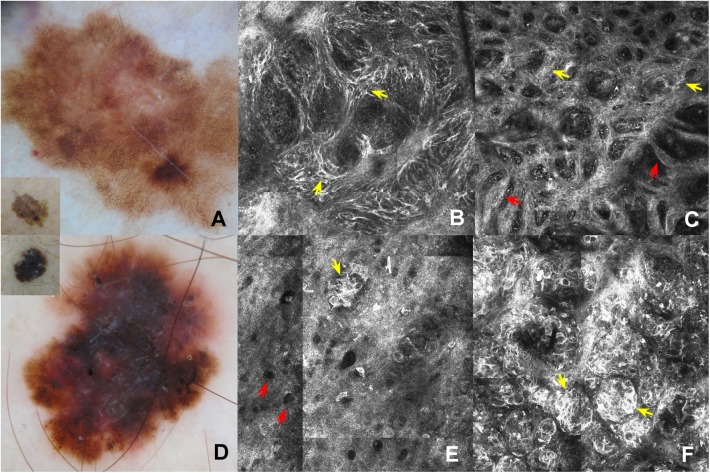
Fig 2. Comparison between findings of dermatoscopy and RCM.
(A) Dermoscopy of BRAF V600E negative in situ superficial spreading melanoma: broadened pigmented network (the inset corresponds to clinical image). (B) RCM mosaic image (1.5x1.5 mm) in DEJ shows junctional thickening due to isolated atypical cells (yellow arrows). (C) RCM mosaic image (1.5x1.5 mm) in DEJ showing non-edged papillae separated by loosely thick interpapillary spaces (yellow arrows) and meshwork pattern (red arrows). (D) Dermoscopy of BRAF V600E mutated superficial spreading melanoma, 1 mm in Breslow thickness): multicomponent pattern (the inset corresponds to clinical image). (E) RCM mosaic image (0.75x0.75 mm) at the level of the epidermis shows hyporeflective pagetoid cells (red arrows) and epidermal nests (yellow arrow). (F) RCM mosaic image (0.75x0.75 mm) at the level of the DEJ shows dermal-epidermal nests (yellow arrows).
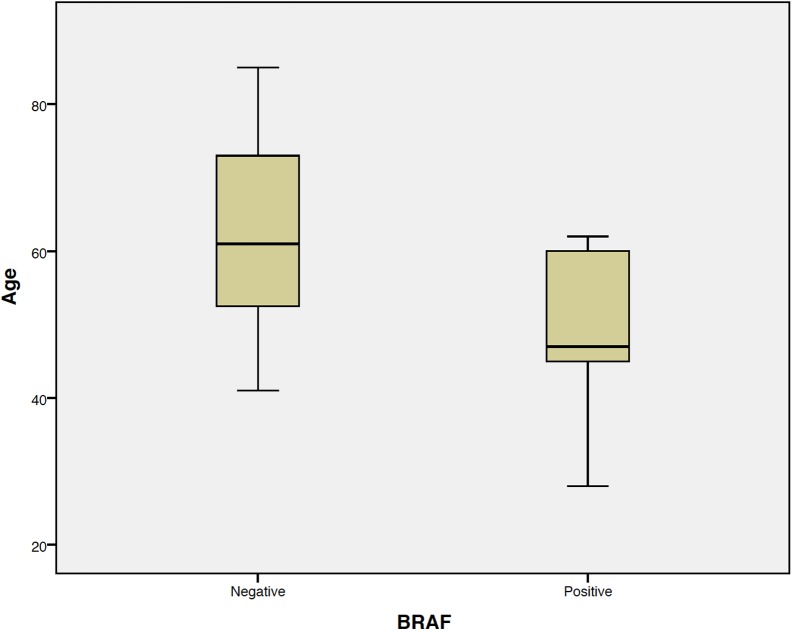
Patients with BRAF positive melanomas were significantly younger (dichotomized into those ≤50 vs >50 years of age) as compared to patients with BRAF wild type melanoma (p = 0.035) (Fig 3).
Fig 3. Graphic age x BRAF V600E.
Melanomas that were BRAF mutated were more likely to be invasive (p = 0.03). No difference was observed in BRAF mutated vs BRAF wild type melanomas vis-a-vis patient gender, primary site of tumor, association with melanocytic nevus, nor any other histopathological parameter (Table 2).
Differences in RCM parameters between BRAF mutated and wild type melanoma was observed. BRAF positive melanomas were more likely to reveal hyporreflective cells (p = 0.02; Fig 2E), epidermal nests (p = 0.02), dermal-epidermal junction nests (p = 0.05), edged papillae (p = 0.05), and bright dots (p = 0.05; Table 3). In addition, BRAF mutated melanomas were less likely to reveal junctional thickening (p = 0.01) and a meshwork pattern (p = 0.02; Table 4).
Table 4. Clinical and histological characteristics of BRAF V600E mutated melanomas.
| Case | Gender | Age | Local | Histological Type |
Growth | Clark | Breslow | Mitotic index mm2 | Peritumoral invasion* | Intratumoral Invasion* |
Pre-existing. Nevus* | HRC (1)* | EN (2)* | DEJN (3)* | EP (4)* | AP (5)* | N (6)* | MS (7)* | BD (8)* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 45 | Upper limbs | SS | In situ | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | |||||
| 2 | Male | 60 | Upper limbs | SS | Radial | II | 0.26 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 3 | Female | 47 | Trunk | SS | Radial | III | 0.79 | 1.25 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 4 | Female | 42 | Head and neck | SS | Vertical | II | 0.52 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| 5 | Male | 61 | Lower limbs | SS | Vertical | III | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 6 | Female | 28 | Trunk | SS | Vertical | III | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| 7 | Female | 62 | Trunk | SS | Vertical | II | 0.5 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| 8 | Male | 47 | Upper limbs | SS | Radial | III | 0.71 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| 9 | Female | 45 | Upper limbs | SS | Radial | II | 0.7 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
(1)Hyporreflective cells
(2)Epidermal nests
(3)DEJ nests
(4)Edged papillae
(5)Abscence papillae
(6)Junctional thickening cell nests
(7)Meshwork
(8)Bright dots.
*0 means absence of parameter and 1 means presence of parameter.
SS: Superficial Spreading
Multiple logistic regression (Table 5) identified the following independent RCM features predictive of BRAF V600E positivity: absence of meshwork pattern (OR = 14.3; 95%CI: 1.3–156.8) and absence of junctional thickening (OR = 16.7; 95% CI: 1.6–175.0).
Table 5. Multivariate analysis for the identification of independent predictive factors for positive BRAF V600E.
| Variable | n | OR | 95%CI |
|---|---|---|---|
| Meshwork | |||
| Absent | 14 | 14.3 | 1.3–156.8 |
| Present | 18 | 1.0 | Reference |
| Junctional thickening cell agregate | |||
| Absent | 10 | 16.7 | 1.6–175.0 |
| Present | 22 | 1.0 | Reference |
OR: Odds ratio
95%CI: 95% Confidence interval
Number of events: 9 (BRAF V600E +)
The distribution of cases as a function of BRAF status and the number of cumulative predictive factors for BRAF V600E positivity are depicted in Table 6.
Table 6. Distribution of cases according to BRAF V600E status and number of cumulative predictive factors.
| Number of cumulative predictive factors (*) |
BRAF V600E | |||
|---|---|---|---|---|
| Negative | Positive | |||
| N | (%) | N | (%) | |
| None | 13 | (100.0) | 0 | (0.0) |
| One factor | 9 | (64.3) | 5 | (35.7) |
| Two factors | 1 | (20.0) | 4 | (80.0) |
Fisher’s exact test: P = 0.002; Area under the curve (AUC): 0.86 (95%CI: 0.73–0.99).
(*) Predictive factors: absence of meshwork and absence of junctional thickening cell agregate.
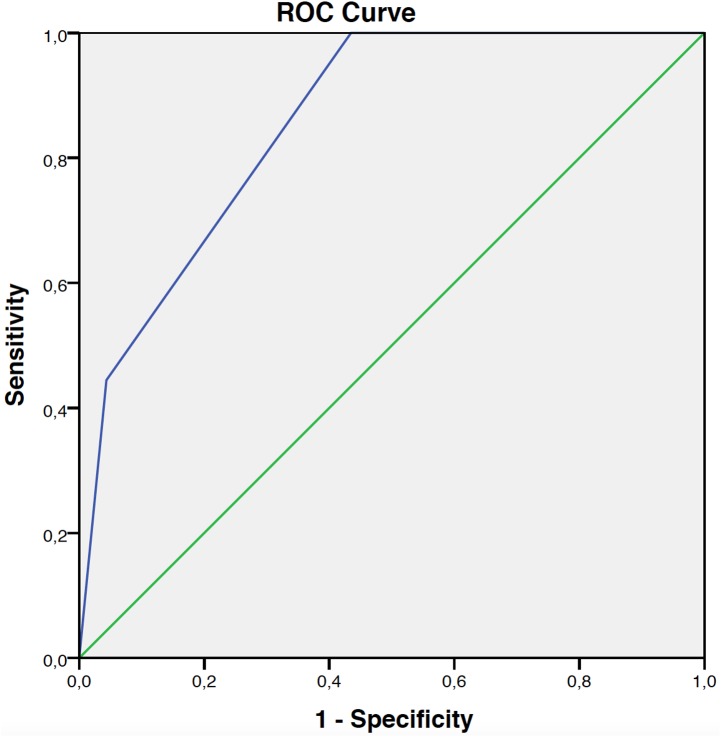
There was a statistically significant difference in BRAF positivity as a function of the number of cumulative factors (p = 0.002) present, ranging from zero up to 80%. This model had an AUC equal to 0.86 (95% CI: 0.73–0.99), as depicted in Fig 4.
Fig 4. ROC curve of positivity for BRAF V600E.
Discussion
The discovery that melanomas could be sub-classified based on their mutation profile provided the basis for the molecular classification of cutaneous melanoma [15]. While many mutations have been identified in melanoma, the most common is the BRAF V600E mutation, which is seen in approximately 50% of cutaneous melanomas [9,16,17] Studies have shown that BRAF mutated melanomas tend to develop in younger individuals as compared to BRAF wild type melanomas [18,2]. Indeed, 66.7% (6 of 9) of the patients with BRAF-mutated melanomas in our cohort were younger than 50 years of age. In contrast, 86.9% of the patients with BRAF wild type melanomas were older than 50; similar observations were also made by Menzies et al. [18]. Approximately one third of the BRAF mutated melanomas in our cohort were found to have an associated nevus in the histologic examination. Similar observations were made by Shitara et al. [19], who noted that 38.3% (23/60) of their V600E-mutated melanomas had an associated nevus that was also V600E-mutated.
RCM enables clinicians to visualize subsurface skin structures on the cellular level in vivo. The features seen on RCM correlate well with features seen on routine histological examination [20,21,22]. One previous study evaluated six BRAF V600E mutated primary melanomas and 2 metastasis [7] with RCM and found them to reveal pleomorphic pagetoid cells, disarrangement of the dermal-epidermal junction, discohesive junctional nests and bright particles at the DEJ. In our cohort, the presence of bright dots, which correlates with inflammation (leukocyte infiltration), was the only parameter found in common with Ruini et al.’s study being significantly associated with BRAF V600E status.
Since the presence of both bright dendritic and roundish cells in the spinous layer correlate with the presence of pagetoid cells on histopathology [23], we elected not to differentiate between these two subtypes of pagetoid cells seen on RCM. While Viros et al. observed on histopathology that increased upward scatter of intraepidermal melanocytes was associated with BRAF mutations [24], we did not find the same on RCM imaging. However, we did find that the presence of ‘hyporeflective pagetoid cells’ (HPCs), described as round dark structures (similar to ‘holes’) within the epidermis [25], was significantly correlated with BRAF V600E mutation. Of interest, these hyporeflective cells have been described in hypomelanotic melanomas, Paget's disease [26], and rarely in pigmented melanomas [24].
One of the novel findings of our study was that not only the presence of dermal-epidermal nest but also the presence of intraepidermal nests are associated with BRAF V600E mutations. This finding is consistent with Viros et al.’s histopathology study showing that intraepidermal melanocytes arranged in nests were positively associated with BRAF-mutation status [24]. Since BRAF V600E mutations are commonly seen in dermal nevi, it is tempting to speculate whether BRAF mutations can directly lead to nest formation [27,28,6]. It is interesting to note that while many melanomas reveal sparse nests, the nests in our BRAF mutated melanomas were dense; similar to nests seen in intradermal nevi, most of which also happen to harbor BRAF V600E mutations [29], No cerebriform nests were observed, most probably because the melanomas in our cohort were thin tumors.
Recently an attempt has been made to group melanomas based on their RCM features. Four types of melanomas have been identified: dendritic cell melanomas, round cell melanomas, dermal nest melanomas, and combined type melanomas. Melanomas with a meshwork pattern have been associated with both dendritic and round cell melanomas [30]. While dendritic cell melanomas were associated with the presence of thin meshwork pattern and dendritic cells, round cell melanomas were associated with the presence of large round cells distributed in a pagetoid fashion and with nest formation. This latter type melanoma was found to be associated with BRAF V600E mutations. Patients with this type of melanoma tend to be younger, have multiple nevi and rarely develop melanoma on chronic sun exposed skin. In contrast, dendritic cell melanomas tend to reveal junctional thickening resulting from isolated atypical cells without nest formation. These melanomas tend to be BRAF wild type [30]. Indeed, after multivariate analysis, the most important predictive factors for BRAF V600E positivity were the absence of meshwork and/or absence of junctional thickening. In other words, if both parameters are absent there is a high likelihood that the melanoma will harbor the BRAF V600E mutation.
The findings of the present study reveals that BRAF V600E mutated melanomas tend to have epidermal and DEJ nests, are of the superficial spreading type, occur at a younger age, and are located on intermittently sun exposed areas. In contrast, BRAF wild type melanomas develop at an older age, reveal a thin meshwork pattern and junctional thickening. The aforementioned supports the existence of different pathways to melanomas, leading to different subsets of melanomas, each depicting particular clinical, dermoscopic and RCM features (Table 7).
Table 7. Clinical, dermoscopic, confocal microscopy and histopathological features of positive and negative BRAF V600E melanomas.
| BRAF V600E | Clinic | Dermoscopy | Confocal microscopy | Histopathology |
|---|---|---|---|---|
| POSITIVE | Age < 50 (p = 0,022); | Globular pattern with irregular globules; Blotches | Hyporeflective cells in epidermis (p = 0,21); Epidermal nests (p = 0,019); Junctional nests (p = 0,035); | Radial growth (p = 0,003); Vertical growth (p = 0,003) |
| NEGATIVE | Age > 50 (p = 0,022); | Atypical network; Sparse irregular globules; Radial projections; Irregular hypopigmentation; Veil; Peppering | Edged papillae (p = 0,038); Junctional thickening cell aggregates (p = 0,012); Meshwork (p = 0,019); | In situ (p = 0,003) |
Melanomas occurring at a younger age, on intermittently sun-exposed areas, with tendency to nest formation are clearly different from melanomas in older patients, on chronically sun exposed areas, with histopathological tendency of more lentiginous proliferation.
The present study highlights the potential of RCM to be used as a supplementary tool in the screening for BRAF-mutated melanomas. The findings of the presence of intraepidermal nests, DEJ nest, bright dots, in the absence of junctional thickening and absence of meshwork is highly suggestive that the melanoma in question will harbor a BRAF V600E mutation.
Data Availability
All relevant data have been uploaded to Dryad at: http://doi.org/10.5061/dryad.s4f13.
Funding Statement
The research was funded by Grants from FAPESP (São Paulo Research Foundation/ Fundação de Amparo à Pesquisa do Estado de São Paulo) process number 2012 /13090-5, URL: http://www.fapesp.br/, to JPD.
References
- 1.Whiteman DC, Watt P, Purdie DM, Hughes MC, Hayward NK, Green AC. Melanocytic nevi, solar keratoses, and divergent pathways to cutaneous melanoma. J Natl Cancer Inst. 2003;95:806–12. . [DOI] [PubMed] [Google Scholar]
- 2.Whiteman DC, Pavan WJ, Bastian BC. The melanomas: a synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 2011;24:879–97. doi: 10.1111/j.1755-148X.2011.00880.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- 4.Ponti G, Pellacani G, Tomasi A, Loschi P, Luppi G, Gelsomino F, et al. Molecular targeted approaches for advanced BRAF V600, N-RAS, c-KIT, and GNAQ melanomas. Dis Markers. 2014;671283. doi: 10.1155/2014/671283. Erratum in: Dis Markers. 2014;2014:246751 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pozzobon FC, Puig-Butillé JA, González-Alvarez T, Carrera C, Aguilera P, Alos L, et al. Dermoscopic criteria associated with BRAF and NRAS mutation status in primary cutaneous melanoma. Br J Dermatol. 2014;171:754–9. doi: 10.1111/bjd.13069 . [DOI] [PubMed] [Google Scholar]
- 6.Zalaudek I, Guelly C, Pellacani G, Hofmann-Wellenhof R, Trajanoski S, Kittler H, et al. The dermoscopical and histopathological patterns of nevi correlate with the frequency of BRAF mutations. J Invest Dermatol. 2011;131:542–5. doi: 10.1038/jid.2010.332 . [DOI] [PubMed] [Google Scholar]
- 7.Ruini C, Manfredini M, Pellacani G, Mandel VD, Tomasi A, Ponti G. Confocal microscopy characterization of BRAFV600E mutated melanomas. Melanoma Res. 2015;25:367–71. doi: 10.1097/CMR.0000000000000147 . [DOI] [PubMed] [Google Scholar]
- 8.Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–9. doi: 10.1007/s00401-011-0841-z . [DOI] [PubMed] [Google Scholar]
- 9.Long GV, Wilmott JS, Capper D, Preusser M, Zhang YE, Thompson JF, et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol. 2013;37:61–5. doi: 10.1097/PAS.0b013e31826485c0 . [DOI] [PubMed] [Google Scholar]
- 10.Long E, Ilie M, Lassalle S, Butori C, Poissonnet G, Washetine K, et al. Why and how immunohistochemistry should now be used to screen for the BRAFV600E status in metastatic melanoma? The experience of a single institution (LCEP, Nice, France). J Eur Acad Dermatol Venereol. 2015;29:2436–43. doi: 10.1111/jdv.13332 . [DOI] [PubMed] [Google Scholar]
- 11.Pellacani G, Guitera P, Longo C, Avramidis M, Seidenari S, Menzies S. The impact of in vivo reflectance confocal microscopy for the diagnostic accuracy of melanoma and equivocal melanocytic lesions. J Invest Dermatol. 2007;127:2759–65. doi: 10.1038/sj.jid.5700993 [DOI] [PubMed] [Google Scholar]
- 12.Braga JC, Macedo MP, Pinto C, Duprat J, Begnami MD, Pellacani G, et al. Learning reflectance confocal microscopy of melanocytic skin lesions through histopathologic transversal sections. PLoS One. 2013;8:81205 doi: 10.1371/journal.pone.0081205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellacani G, Farnetani F, Gonzalez S, Longo C, Cesinaro AM, Casari A, et al. In vivo confocal microscopy for detection and grading of dysplastic nevi: a pilot study. J Am Acad Dermatol. 2012;66:109–21. doi: 10.1016/j.jaad.2011.05.017 . [DOI] [PubMed] [Google Scholar]
- 14.Rezze GG, Casagrande J. Atlas de microscopia confocal na dermatologia, 1st ed. São Paulo, SP: Lemar, 2015.
- 15.Flaherty KT, Fisher DE. New strategies in metastatic melanoma: oncogene-defined taxonomy leads to therapeutic advances. Clin Cancer Res. 2011;17:4922–8. doi: 10.1158/1078-0432.CCR-10-2612 . [DOI] [PubMed] [Google Scholar]
- 16.Singh M, Lin J, Hocker TL, Tsao H. Genetics of melanoma tumorigenesis. Br J Dermatol. 2008;158:15–21. doi: 10.1111/j.1365-2133.2007.08316.x [DOI] [PubMed] [Google Scholar]
- 17.Tschandl P, Berghoff AS, Preusser M, Burgstaller-Muehlbacher S, Pehamberger H, Okamoto I, et al. NRAS and BRAF mutations in melanoma-associated nevi and uninvolved nevi. PLoS One. 2013;8:69639 doi: 10.1371/journal.pone.0069639 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menzies AM, Haydu LE, Visintin L, Carlino MS, Howle JR, Thompson JF, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012;18:3242–9. doi: 10.1158/1078-0432.CCR-12-0052 . [DOI] [PubMed] [Google Scholar]
- 19.Shitara D, Tell-Martí G, Badenas C, Enokihara MM, Alós L, Larque AB, et al. Mutational status of naevus-associated melanomas. Br J Dermatol. 2015;173:671–80. doi: 10.1111/bjd.13829 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langley RG, Walsh N, Sutherland AE, Propperova I, Delaney L, Morris SF, et al. The diagnostic accuracy of in vivo confocal scanning laser microscopy compared to dermoscopy of benign and malignant melanocytic lesions: a prospective study. Dermatology. 2007;215:365–72. doi: 10.1159/000109087 [DOI] [PubMed] [Google Scholar]
- 21.Pellacani G, Cesinaro AM, Seidenari S. In vivo confocal reflectance microscopy for the characterization of melanocytic nests and correlation with dermoscopy and histology. Br J Dermatol. 2005;152:384–6. doi: 10.1111/j.1365-2133.2005.06348.x [DOI] [PubMed] [Google Scholar]
- 22.Zalaudek I, Leinweber B, Hofmann-Wellenhof R, Scope A, Marghoob AA, Ferrara G, et al. The epidermal and dermal origin of melanocytic tumors: theoretical considerations based on epidemiologic, clinical, and histopathologic findings. Am J Dermatopathol. 2008;30:403–6. doi: 10.1097/DAD.0b013e3181734e9a . [DOI] [PubMed] [Google Scholar]
- 23.Longo C, Rito C, Beretti F, Cesinaro AM, Piñeiro-Maceira J, Seidenari S, et al. De novo melanoma and melanoma arising from pre-existing nevus: in vivo morphologic differences as evaluated by confocal microscopy. J Am Acad Dermatol. 2011;65:604–14. doi: 10.1016/j.jaad.2010.10.035 . [DOI] [PubMed] [Google Scholar]
- 24.Viros A, Fridlyand J, Bauer J, Lasithiotakis K, Garbe C, Pinkel D, et al. Improving melanoma classification by integrating genetic and morphologic features. PLoS Med. 2008;5:120 doi: 10.1371/journal.pmed.0050120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losi A, Longo C, Cesinaro AM, Benati E, Witkowski A, Guitera P, et al. Hyporeflective pagetoid cells: a new clue for amelanotic melanoma diagnosis by reflectance confocal microscopy. Br J Dermatol. 2014;171:48–54. doi: 10.1111/bjd.12781 . [DOI] [PubMed] [Google Scholar]
- 26.Pan ZY, Liang J, Zhang QA, Lin JR. Zheng ZZ. In vivo reflectance confocal microscopy of extramammary Paget disease: diagnostic evaluation and surgical management. J Am Acad Dermatol. 2012;66:47–53. doi: 10.1016/j.jaad.2010.09.722 . [DOI] [PubMed] [Google Scholar]
- 27.Gill M, Celebi JT. B-RAF and melanocytic neoplasia. J Am Acad Dermatol. 2005;53:108–14. doi: 10.1016/j.jaad.2005.04.013 [DOI] [PubMed] [Google Scholar]
- 28.Marchetti MA, Kiuru MH, Busam KJ, Marghoob AA, Scope A, Dusza SW, et al. Melanocytic naevi with globular and reticular dermoscopic patterns display distinct BRAF V600E expression profiles and histopathological patterns. Br J Dermatol. 2014;171:1060–5. doi: 10.1111/bjd.13260 . [DOI] [PubMed] [Google Scholar]
- 29.Benati E, Argenziano G, Kyrgidis A, Moscarella E, Ciardo S, Bassoli S, et al. Melanoma and naevi with a globular pattern: confocal microscopy as an aid for diagnostic differentiation. Br J Dermatol. 2015;173:1232–8. doi: 10.1111/bjd.14049 . [DOI] [PubMed] [Google Scholar]
- 30.Pellacani G, De Pace B, Reggiani C, Cesinaro AM, Argenziano G, Zalaudek I, et al. Distinct melanoma types based on reflectance confocal microscopy. Exp Dermatol. 2014;23:414–8. doi: 10.1111/exd.12417 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data have been uploaded to Dryad at: http://doi.org/10.5061/dryad.s4f13.