Abstract
Ragweed pollen is primarily responsible for the hay fever allergies of sufferers throughout the world. A proteome study of three ragweed plants (Ambrosia artemisiifolia, Ambrosia trifida, and Ambrosia psilostachya) was undertaken to document and compare their protein profiles. Proteins extracted from the pollen of the three plants were subjected to one dimensional electrophoresis followed by tandem liquid chromatography-mass spectroscopy. Peptide sequence mapping permitted discovery of proteins not previously reported for all three plants and 45% of the identified proteins were shared by all three of them. Application of stringent criteria revealed not only a majority of known allergens for short ragweed but also allergens not previously reported for the other two plants. Additionally, potentially allergy inducing enolases are reported for the three plants. These results suggest that all three ragweed plants could contribute to the allergy malady.
Keywords: Proteomics, Ragweed, Pollen, LC-MS/MS, 1-D SDSPAGE
1. INTRODUCTION
According to the CDC, ragweed pollen is largely responsible for the allergic hay fever reaction suffered by approximately 16 percent of the United States population each year. Despite the prevalence of this disease, only 11 proteins of the total of 99 listed by UniProt as arising from Ambrosia artemisiifolia, short ragweed, have well annotated sequence entries. The sequences for most of those 99 proteins were translated from nucleotide sources. The nomenclature committee of the World Health Organization/International Union of Immunological Societies (WHO/IUIS) lists 11 known antigens of short ragweed; six of these sequences were reviewed by UniProt. Although three prominent ragweed plants, Ambrosia artemisiifolia (common/short), Ambrosia trifida (giant), and Ambrosia psilostachya (western), are found in the United States, the majority of the documented proteins are associated with A. artemisiifolia. For A. trifida, the UniProt list of 26 proteins includes 3 reviewed, one of which is the antigen Amb t 5. For A. psilostachya, having the fewest sequence entries, only 2 of 17 are reviewed.
Early work focused on the immunogenic properties of extracts from short ragweed [1]; however, there was some coverage of all three species [2]. Significant work expansion began in the 1960s with the isolation of 2 antigens of 32 kDa and 10 kDa by King and Norman [3]. Multiple antigens were discovered in a glycosylated protein extract of short ragweed [4]. Major allergen E, Amb a 1 in current nomenclature, with four isoforms, was also isolated [5]. Employing IEF, King [6] demonstrated multiple proteins in the short ragweed extract and isolated two acidic proteins, antigen E (Amb a 1) and antigen K (Amb a 2). Later, the size of Amb a 1 at 37.8 kDa and its alpha and beta forms were determined by MALDI/ TOF analyses [7]. Rafner [8] produced protein translates from three short ragweed cDNA clones (Amb a 1.1, 1.2, and 1.3) with 85% nucleotide sequence identity; polymorphism was confirmed for Amb a 1, and a fourth (Amb a 1.4) was identified by Griffith, et al. [9]. They also reported finding Amb a 2 RNA in flowers but not in pollen [9]. The cDNA based amino acid sequence of Amb a 2 was found to share 65% identity with the Amb a 1 group [10]. Subsequently, as reported on the WHO/IUIS page, Amb a 2 was renamed as a subcategory of Amb a 1.
Extensively investigated, the basic short ragweed pollen glycoprotein Ra3 (Amb a 3) reported as being15 kDa size presented differences in amino acid composition and immunological specificity when compared to antigen E [11]. Sequencing of a single isoform of Ra3 yielded 101 amino acids and confirmed glycosylation [12]. In a follow up study, two forms of Ra3 of 12.3 kDa were reported biochemically and immunologically similar [13].
Purified Ra5 (Amb a 5) from short ragweed, comprised of 45 amino acids [14], evinced amino acid sequence polymorphism [15]. Purified antigenic Amb t 5, a 4.4 kDa polypeptide, showed limited cross reactivity with Amb a 5 [16]. Likewise, Goodfriend et al. [17] demonstrated that Ra5G (Amb t 5) was biochemically similar but immunologically dissimilar to Ra5S (Amb a 5). Lowenstein and Marsh [18] detected 52 short ragweed antigens by immuno-electrophoresis and reported isoelectric points for some. In a refined procedure, specific antigens E (Amb a 1), K (Amb a 2), Ra5, and Ra6 (Amb a 6), but not Ra3, were identified amongst the 52 antigens [19], and nine of them were found to be significant allergens [20]. Purified Ra6 was shown to be composed of four 8 kDa (SDSPAGE) isoforms with equivalent immunological properties [21].
Metzler [22, 23] reported composite 3-dimensional structures for Amb t 5 and Amb a 5 with some variation in the beta sheet cores but similar C-terminal helices. Ghosh et al. [24] confirmed the entire protein sequence of Amb t 5 from its cDNA sequence.
Heightened interest in assigning the relative antigenicity and allergenicity among Amb 5 isoforms revealed immunological dissimilarities [16, 23, 25, 26] and similarities [16, 26]; Amb p 5 being the most similarity to Amb a 5 [27].
From short ragweed, two new endopeptidases, a chymotrypsin-like [28] and a trypsin-like [29], were purified and characterized. cDNA expressed sequences yielded pan allergens including two profilin isoforms (Amb a 8), two 2 EF-hand calcium binding proteins, (Amb a 9), and one 3 EF-hand calcium binding protein (Amb a 10) [30]. Newly isolated Amb a 4 allergen has a defensin domain and is a homolog of Mugwort Art v 1 [31]. The newest antigen Amb a 11, a 37 kDa cysteine protease, was discovered and characterized by a combination of electrophoretic, immunological, and mass spectroscopy methods [32].
Identification of proteins extracted from various tissue sources by mass spectroscopy has become a prominent proteomics tool increasingly applied to plant tissue, including the pollen of allergy inducers. Initial studies employed peptide mass fingerprinting (PMF) [33–37] that evolved to the more powerful peptide sequence mapping (PSM) [36–41], wherein the spectra of experimentally generated peptides are matched to those produced virtually from all of the database proteins. Bordas-Le Floch et al. [41] created a transcriptome-derived proteome database to determine identities of short ragweed proteins obtained in a shotgun PSM approach. One dimensional SDS PAGE coupled to liquid chromatography and tandem mass spectroscopy (LC-MS/MS) enhanced the powerful PSM method [42–45].
Justification for this project included the paucity of protein-based sequencing and limited-sequence entries for the giant and western plants. In addressing these aims, we undertook proteomic profiling of all three species. Based on our findings, we have increased the number of proteins attributed to each of them. Such knowledge enables profile comparison, reveals similarity in the allergens produced by the three plants, and perhaps suggests a basis for still unaccounted allergic reactions.
For this investigation, all profiles were generated by combining one dimensional SDS gel electrophoresis with liquid chromatography and tandem mass spectroscopy. The study yielded 257 proteins, including 28 orthologs that met stringent criteria. Of the 257 proteins identified, 117 were found in all three species, 21 were in common and giant, 29 in common and western, and 22 in giant and western. There were 68 proteins associated with only one species. Western was by far the most populated with 50; 12 were attributed to common and 6 to giant. To avoid duplicate protein names appearing in the data reported in the results section, it was decided to eliminate the orthologs. Ortholog elimination yielded 229 identified proteins.
2. METHODS
2.1 Materials
The source of defatted pollen for all three Ambrosia species came from Greer Laboratories, Lenoir, NC. The following were purchased from Invitrogen (Life Technologies), Carlsbad, CA: NuPAGE Novex 4–12% Bis-Tris Protein Gels, 1.5 mm, 10-well; NuPAGE® MES SDS Running Buffer; NuPAGE® LDS Sample Buffer; and the BenchMark Protein Ladder 10–220 kDa. Iodoacetamide, Protease Inhibitor Cocktail, Trizma base , and Coomassie Brilliant Blue were obtained from Sigma-Aldrich, Saint Louis, MO. Dithiothreitol came from Bio-Rad, Hercules, CA, and sequencing grade Trypsin (V511) from Promega, Madison, WI. All general chemicals were acquired from Thermo Fisher Scientific, Waltham, MA.
2.2 Pollen Extraction
A Life Technologies procedure was modified to create an extraction process more proteomics suitable [46]. The pollen (50 mg) was suspended in 909 μL of Invitrogen LDS buffer (2% LDS and 2.5 mM Tris, pH 8.5), 3 μL of 1 M Tris Base, 10 μL of Sigma Protease Inhibitor Cocktail, 10 μL of 2 M dithiothretiol (DTT), and 18 μL of ultra-pure water. Placed on ice, it was sonicated at 20% power 5 times for 30 seconds followed by 15 minutes of shaking, while maintaining pH 8.5 by addition of 1 M Tris, if needed. Disulfide bonds were reduced by adding 10 μL of 2 M DTT to the lysate; sulfhydryl groups were alkylated by adding 121 μL of 0.4 M iodoacetamide to the lysate, and the samples were placed on the shaker for 30 minutes. The extracts were centrifuged at 4 °C and 16,000 x g for 30 minutes, with the resultant supernatant transferred to Amicon Ultra 0.5 mL centrifugal filters for concentrating while lowering the LDS concentration level to about 0.5% with water. The protein extracts were stored at −80 °C.
2.3 Electrophoresis
Samples of the protein extracts were subjected to gradient one dimensional electrophoresis on Invitrogen NuPAGE 4–12% Bis-Tris Gels, with 10 1.5 mm wells in an Invitrogen MES (2-(N-morpholino)ethane sufonic acid) SDS buffer (2.5 mM MES, 2.5 mM Tris base, 0.005% SDS, 0.05 mM EDTA, pH 7.3) for 40 minutes at 200 V using an Invitrogen XCell SureLock ® Mini-Cell. After fixation in 50% ethanol and 10% acetic acid, the gels were stained with Coomassie Colloidal Blue stain and destained with water.
2.4 Mass Spectroscopy
The protein bands were excised from the gel in roughly 2 x 5 mm slices and were cut into 3 or 4 pieces. A minimum of 20 slices was produced from each gel well. The gel slice pieces were washed twice with 200 μL of washing buffer (0.2 M ammonium bicarbonate and 50% acetonitrile) for 45 minutes at 37 °C in a water bath with gentle agitation. The washing buffer was discarded and the slices were dried with the Eppendorf Vacufuge for 5 minutes at 45 °C. The gel pieces were swelled with 19 μL of digestion buffer A (0.2 M ammonium bicarbonate and 5mM CaCl2), 1 μL of 0.4 μg/μL trypsin, and sufficient digestion buffer B (0.2 M ammonium bicarbonate and 10% acetonitrile) to bring the total liquid volume to 70 μL. In order to ensure proper mixing, the solution was centrifuged at 12,000 x g for 3 minutes prior to incubating the samples overnight in a water bath at 37 °C with gentle agitation. Twice the samples were acidified to 1% acetic acid, centrifuged at 12,000 x g for 3 minutes, the supernatant was removed, and the volume of combined supernatants was reduced to about 30 μL with the Vacufuge. Any remaining gel pieces were removed by microfiltration using Millipore Ultrafree-MC-HV centrifugal filters.
The digested samples were subjected to liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Analytical Proteomics Laboratory of the University of Kansas. Used was a NanoAcquity chromatographic system (Waters Corp., Milford, MA) with Binary Solvent Manager with mobile phase A and B of 0.1 % Formic acid (Water) 0.1 % Formic acid (ACN), respectively, to produce a gradient with solvents of highest purity (Optima LC-MS grade, Fisher Scientific) and delivering analytes through a reverse-phase column (ThermoScientific Acclaim PepMap300) to an electrospray source of an LTQ-FTICR mass spectrometer (ThermoFinnigan, Bremen, Germany). A linear gradient was developed in 50 minutes with a flow rate of 10 μL/min. The mass spectrometer was operated in a data-dependent acquisition mode.
XCalibur software was used to acquire the data. Mascot software was used for database search and protein mapping. It was setup to search SwissProt Viridiplantae database. Mass tolerance was 20 ppm for precursor ions and 0.60 Da for fragment ions. Carbamidomethyl (+57) was set as a fixed modification of cysteine residues, oxidation of methionine residues and acetylation of protein N-terminus were specified as variable modifications, and trypsin was the digestion enzyme, with a maximum of 2 missed cleavages. The results were exported into Scaffold software for statistical analysis and data sharing.
The limited number of proteins identified in the SwissProt database for these three ragweed species informed our decision to use the green plant database, Viridiplantae, which provided 35480 entries, including those from ragweed. It is not uncommon for researchers performing tandem mass spectroscopy proteomic analysis on plant sources to use the Viridiplantae database, when none exists for their particular species [34, 40, 47]. Sequence comparison is consistent with the long history of evolutionary homology discovered by protein sequence similarity and is the basis for BLASTP.
2.5 Data Validation
The criterion for naming found proteins involved Scaffold (Scaffold_4.3.4, Proteome Software Inc., Portland, OR) use to validate all MS/MS based peptide and protein identification results generated from Mascot, Sequest, and X! Tandem search engines [48, 49]. Scaffold employs the Peptide Prophet algorithm [50] to generate probability scores from the three search engines to enhance the accuracy of identifying peptides. Scaffold employs the resulting peptide probabilities to correctly match peptide sequence and mass to proteins with the Protein Prophet statistical algorithm [51].
2.6 Data Analysis
In Scaffold, the peptide threshold identification filter was established at 50.0% probability, the protein threshold identification filter was established at 95.0% probability, and the minimum number of peptides filter was set at 2. Using the Scaffold outcome, an additional criterion for including proteins in this report was 2 exclusively unique peptides (matched to no other protein in the database). Moreover, the protein had to be found in at least two separately extracted samples for a species. Additional criteria for reporting proteins were the total number of exclusively unique peptides and percentage of sequence coverage. When multiple entries were found for the same protein in a species, we selected those with the greatest number of exclusively unique peptides unless the coverage was greater with fewer. The same criteria were used when orthologs were excluded from the final data.
A minimum of 20 gel slices generated from each of 12 separately and independently extracted samples, four from each of the three plants, were subjected to LC-MS/MS analysis. To compare the protein profiles of the three plants, the Scaffold data was exported into Excel files and then converted into CSV format. The individual proteins from all 12 experiments were captured by name and accession number from the CSV file and aligned by protein name using a JAVA file designated as ProcessCSVCount, which also counted the incidences for a protein found in each of the ragweed plants. These counts were used to assign proteins among seven categories of All, Common, Giant, Western, Common & Giant, Common & Western, and Giant & Western. Visual Basic (VB) programs were written to accomplish the latter assignment. Additionally, other VB programs were created to manage the data, including eliminating entries that had fewer than 2 exclusively unique peptides and generating the non-exclusive assignment of proteins among the three plants
3. RESULTS and DISSCUSSION
The above criteria allowed protein assignment among the three kinds of ragweed pollen. A total of 257 unique proteins (redundancies removed) were found in the SwissProt database. When orthologs were eliminated, the number of found proteins reduced to the 229 that are displayed in Supplementary Table 1. UniProt lists a total of 142 proteins (99 short, 26 giant, and 17 western) found among the three plants. When the redundant UniProt entries and associated viruses and other pathogens were removed, the number of proteins for the plants became 57 (34 short, 18 giant, and 5 western). This investigation not only expands the protein profile of the ragweed species but also has the distinction of employing proteomics, an alternative to the often used nucleotide tools.
Table 1 displays a selection of the 229 proteins found through this LCMS/MS investigation. The presented 59 proteins are among those not previously reported to be found in the pollen of any of the three ragweed species. Having an average sequence coverage of 28%, these selected proteins also represent a wide range of protein function, and 66% of them were found for all three species. The information presented in the table by column has headings of protein name, the name of the database plant which yielded the protein sequence, the accession number, molecular mass of the protein, the number of exclusively unique peptides, the percent probability of protein identification (PID), total spectra, percentage coverage of the database sequence by the peptides, whether the protein was identified for the common/short, giant, and western plants, and the putative function in the cell. A broad array of protein types was identified, such as, metabolic enzymes for carbohydrate, amino acids, lipids, citric acid cycle, proteins, and nucleic acids. The list includes, anti-oxidant, regulatory, signaling, structural and motion, electron transport and ATP synthases, nucleosomes, proteasomes, folding, stress response, chaperones, translation, transcription, and transport (intra and intercellular) functions. The entire list of 229 proteins can be found in the supplemental material as Table S1.
Table 1.
Proteins Newly Assigned to the Ragweed Plant and Their Properties^
| PROTEIN NAME | DBSP | Num | Mr | EUP | PID% | TS | COV% | C | G | W | PUTATIVE FUNCTION |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14-3-3 protein 6 | S. lycopersicum | P93211 | 28938 | 2 | 100 | 24 | 41 | 1 | 1 | 1 | Metabolic Regulation |
| 17.3 kDa class I heat shock protein | G. max | P02519 | 17347 | 5 | 100 | 5 | 31 | 1 | Protein Folding | ||
| 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 1 | A. thaliana | O04499 | 60580 | 4 | 100 | 9 | 12 | 1 | 1 | 1 | Glucose Metabolism |
| 26S protease regulatory subunit S10B homolog B | A. thaliana | Q9MAK9 | 44758 | 4 | 100 | 5 | 16 | 1 | 1 | Protein Catabolism | |
| 40S ribosomal protein S13 | G. max | P62302 | 17141 | 4 | 100 | 7 | 30 | 1 | 1 | Translation | |
| 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | C. roseus | Q42699 | 84860 | 2 | 100 | 14 | 14 | 1 | 1 | Amino Acid Metabolism | |
| 6-phosphogluconate dehydrogenase, decarboxylating 1 | S. oleracea | Q94KU1 | 53247 | 3 | 100 | 9 | 16 | 1 | 1 | 1 | Gluconate Metabolism |
| 60S ribosomal protein L11 | O. sativa. ind | A2YDY2 | 20836 | 5 | 100 | 6 | 28 | 1 | 1 | 1 | Translation |
| ADP,ATP carrier protein 1, mitochondrial | G. hirsutum | O22342 | 42095 | 2 | 99 | 8 | 11 | 1 | 1 | 1 | Membrane Transport |
| ADP-ribosylation factor 1 | S. bakko | O48649 | 20609 | 8 | 100 | 14 | 58 | 1 | 1 | 1 | Protein Transport |
| ATP synthase subunit alpha, mitochondrial | H. annuus | P18260 | 55488 | 5 | 100 | 33 | 52 | 1 | 1 | 1 | ATP Synthesis |
| ATPase 8, plasma membrane-type | A. thaliana | Q9M2A0 | 104136 | 5 | 100 | 35 | 18 | 1 | 1 | 1 | ATP Synthesis |
| Aconitate hydratase 1 | A. thaliana | Q42560 | 98153 | 4 | 100 | 18 | 20 | 1 | 1 | 1 | Citrate Metabolism |
| Adenosylhomocysteinase | T. aestivum | P32112 | 53438 | 2 | 100 | 10 | 18 | 1 | 1 | One Carbon Metabolism | |
| Adenylate kinase A | O. sativa.jap | Q08479 | 26407 | 2 | 100 | 2 | 10 | 1 | Nucleotide Metabolism | ||
| Alpha-1,4-glucan-protein synthase [UDP-forming] | Z. mays | P80607 | 41206 | 3 | 100 | 17 | 35 | 1 | 1 | 1 | Cellulose Biosynthesis |
| Aspartate aminotransferase, cytoplasmic | D. carota | P28734 | 44175 | 5 | 100 | 5 | 12 | 1 | 1 | Amino Acid Metabolism | |
| Calmodulin | T. aestivum | P04464 | 16847 | 6 | 100 | 20 | 74 | 1 | 1 | 1 | Enzyme Mediator |
| Calreticulin-2 | A. thaliana | Q38858 | 48159 | 3 | 100 | 8 | 17 | 1 | Protein Folding | ||
| Cell division cycle protein 48 homolog | G. max | P54774 | 89772 | 6 | 100 | 25 | 30 | 1 | 1 | 1 | Cell Cycle Metabolism |
| Citrate synthase, mitochondrial | F. ananassa | P83372 | 52284 | 3 | 100 | 8 | 17 | 1 | 1 | 1 | Citrate Metabolism |
| Clathrin heavy chain 2 | A. thaliana | Q0WLB5 | 193276 | 6 | 100 | 31 | 14 | 1 | 1 | 1 | Endocytosis |
| Cytochrome c oxidase subunit 2 | A. thaliana | P93285 | 29675 | 4 | 100 | 5 | 21 | 1 | 1 | 1 | Redox-Electron Transport |
| Dihydrolipoyl dehydrogenase (Fragment) | S. lycopersicum | P80503 | 3912 | 2 | 100 | 2 | 78 | 1 | 1 | Redox Alpha Keto Acids | |
| Elongation factor 1-alpha | O. sativa.jap | O64937 | 49294 | 3 | 100 | 25 | 40 | 1 | 1 | 1 | Protein Biosynthesis |
| Eukaryotic initiation factor 4A-1 | O. sativa.jap | P35683 | 47088 | 2 | 100 | 13 | 36 | 1 | 1 | Translation | |
| Fructokinase-2 | O. sativa. ind | A2YQL4 | 35518 | 2 | 100 | 5 | 12 | 1 | 1 | 1 | Starch Biosynthesis |
| GTP-binding protein SAR1A | A. thaliana | O04834 | 22032 | 5 | 100 | 12 | 54 | 1 | 1 | 1 | Vesicle-mediated Transport |
| Glutamate decarboxylase | P. hybrida | Q07346 | 56726 | 4 | 100 | 13 | 20 | 1 | 1 | 1 | Amino Acid Metabolism |
| Glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic | Z. mays | Q09054 | 36542 | 2 | 100 | 10 | 32 | 1 | 1 | 1 | Glycolysis |
| Guanosine nucleotide diphosphate dissociation inhibitor | A. thaliana | Q9LXC0 | 49551 | 6 | 100 | 19 | 21 | 1 | 1 | 1 | Regulate Transport |
| Heat shock protein 81-1 | O. sativa. ind | A2YWQ1 | 80198 | 4 | 100 | 35 | 36 | 1 | Stress Response | ||
| Histone H4 | G. max | P0CG89 | 11410 | 9 | 100 | 13 | 60 | 1 | 1 | 1 | Nucleosome Assembly |
| Isocitrate dehydrogenase [NADP], chloroplastic (Fragment) | M. sativa | Q40345 | 48385 | 4 | 100 | 9 | 21 | 1 | 1 | 1 | Glyoxylate Cycle |
| Luminal-binding protein | S. lycopersicum | P49118 | 73237 | 2 | 100 | 41 | 41 | 1 | 1 | 1 | ER Chaperone |
| Malate dehydrogenase 1, mitochondrial | A. thaliana | Q9ZP06 | 35805 | 5 | 100 | 7 | 21 | 1 | 1 | TCA Cycle | |
| Mediator of RNA polymerase II transcription subunit 37f | A. thaliana | Q39043 | 73563 | 2 | 100 | 48 | 38 | 1 | 1 | 1 | Transcription Regulation |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial | A. thaliana | Q9FGI6 | 81526 | 6 | 100 | 6 | 12 | 1 | 1 | 1 | Electron Transport |
| Nucleoside diphosphate kinase B | F. bidentis | P47920 | 16200 | 2 | 100 | 10 | 36 | 1 | 1 | 1 | Nucleotide Biosynthesis |
| Peptidyl-prolyl cis-trans isomerase | C. roseus | Q39613 | 18285 | 4 | 100 | 8 | 44 | 1 | 1 | 1 | Protein Folding |
| Phosphoenolpyruvate carboxylase 1 | F. trinervia | Q9FV66 | 110418 | 2 | 100 | 13 | 15 | 1 | 1 | Carbon Fixation | |
| Phosphoglucomutase, cytoplasmic 2 | Z. mays | P93805 | 63042 | 4 | 100 | 10 | 21 | 1 | 1 | 1 | Glucose Metabolism |
| Phosphoglycerate kinase, cytosolic | N. tabacum | Q42962 | 42365 | 4 | 100 | 17 | 36 | 1 | 1 | 1 | Glycolysis |
| Phospholipase D alpha 1 | C. cardunculus | Q70EW5 | 91786 | 5 | 100 | 15 | 16 | 1 | 1 | 1 | Lipid Catabolism |
| Probable mediator of RNA polymerase II transcription subunit 37c | A. thaliana | P22954 | 71388 | 3 | 100 | 27 | 31 | 1 | 1 | 1 | Transcription Regulation |
| Proteasome subunit alpha type-5-A | A. thaliana | O81149 | 25947 | 4 | 100 | 4 | 23 | 1 | 1 | Protein Catabolism | |
| Putative vesicle-associated membrane protein 726 | A. thaliana | Q9MAS5 | 24874 | 4 | 100 | 6 | 17 | 1 | 1 | Exocytosis | |
| Pyruvate decarboxylase 2 | N. tabacum | P51846 | 67057 | 2 | 100 | 7 | 10 | 1 | 1 | 1 | Aerobic Fermentation |
| Ras-related protein RABB1c | A. thaliana | P92963 | 23165 | 7 | 100 | 7 | 42 | 1 | 1 | 1 | Protein Transport |
| Ribulose bisphosphate carboxylase small chain, chloroplastic | H. annuus | P08705 | 20212 | 4 | 100 | 7 | 30 | 1 | Reductive Pentose Phosphate Pathway | ||
| Soluble inorganic pyrophosphatase | O. sativa. ind | A2X8Q3 | 24160 | 3 | 100 | 8 | 16 | 1 | 1 | Phosphate Metabolism | |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial | A. thaliana | O82663 | 69658 | 3 | 100 | 9 | 15 | 1 | TCA Cycle | ||
| Superoxide dismutase [Cu-Zn] | S. canadensis | O04996 | 15436 | 3 | 100 | 4 | 22 | 1 | 1 | 1 | Antioxidant Defense |
| Triosephosphate isomerase, cytosolic (Fragment) | L. sativa | P48493 | 20540 | 5 | 100 | 9 | 50 | 1 | 1 | 1 | Glucose Metabolism |
| Tubulin alpha chain | P. dulcis | P33629 | 49653 | 11 | 100 | 13 | 26 | 1 | 1 | 1 | Microtubule Based Process |
| UDP-glucose 6-dehydrogenase 4 | A. thaliana | Q9FM01 | 53098 | 2 | 100 | 6 | 15 | 1 | 1 | Carbohydrate Metabolism | |
| UTP--glucose-1-phosphate uridylyltransferase | S. tuberosum | P19595 | 51876 | 2 | 100 | 10 | 18 | 1 | 1 | 1 | UDP-Glucose Metabolism |
| V-type proton ATPase catalytic subunit A | G. hirsutum | P31405 | 68524 | 2 | 100 | 33 | 48 | 1 | 1 | ATP Metabolism | |
| Vesicle-fusing ATPase | A. thaliana | Q9M0Y8 | 81490 | 10 | 100 | 13 | 16 | 1 | 1 | 1 | Golgi Vesicle Docking |
Column abbreviations: DBSP, Num, Mr, EUP, PID%, TS, COV%, C, G, W are: database species, accession number, molecular mass, number of exclusively unique peptides, percent probable protein identification, total spectrum count, percentage coverage, and common/short, giant, and western ragweed plants; numbers in columns C, G, W indicate protein presence.
The PID values presented in Table 1 are calculated protein identification probabilities, rounded to the ones-digit. The calculations done in Scaffold used Bayesian statistics to combine the scores of three search engines (Mascot, Sequest, X! Tandem). The outcome operated on by the Scaffold version of the Protein Prophet yields the PID score.
Certificates of analysis from Greer Labs for each of the three species show less than 1% contamination from other sources. The three plants were harvested from different locales and different environments. There are preferences in natural environment; giant tends to grow in moist areas, common in multiple types of drier soils, and western in dry uplands [52]. The pollen from all sources was processed and stored in the same manner. Results of studies done on a specific plant grown at the same location but subjected to different environmental treatments have found differential proteome effects [47, 53]. To our knowledge, the three species investigated in this research grew under native conditions of the site and were not subjected to environment experiments. Still, it is not impossible the proteome reported here bears witness to some effects by growing at different sites. Whether the differences found are more a reflection of the species native genome expression or the locale environment is not determinable by this work. Protein expression response to an identical stress could be similar or individualistic and perhaps a reflection of the native habitat.
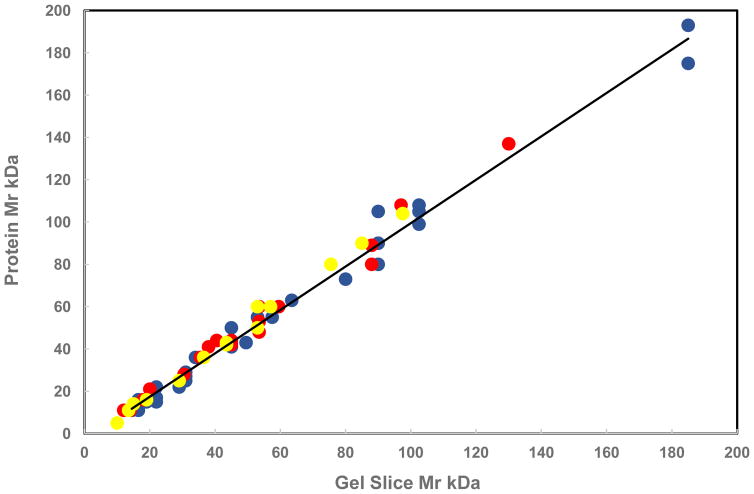
As shown in Figure 1, the molecular masses of database proteins assigned to the ragweed species match very well those of the gel slices from which the data arose. The scatter about calculated lines is quite small with an average R2 value of 0.9885, especially considering that the plotted data came from three different gels and species. Such excellent agreement further supports the findings of the presented proteome results.
Fig. 1.
Assigned Protein and Gel Slice Molecular Mass from
 western,
western,
 giant,
giant,
 common/short
common/short
Considering the results reported in Table S1, assessment of exclusive protein distribution among the species reveals both similarities and distinctions. Of the 229 unique proteins, 104 are assigned to all three plants. Common and western are the most similar, sharing 26 proteins; also shared are 19 and 21 proteins for common and giant and giant and western, respectively. Fifty-nine proteins are assigned exclusively to a single species: 10 to common, 4 to giant, and 45 to western, the only perennial of the three. A non-exclusive assignment yields a more comprehensive protein profile for each plant, with western being the most complex with 196, followed by common with 159, and giant with 148 proteins.
It is well known that the pollen of A. artemisiifolia contains several allergens of which UniProt lists 11 reviewed proteins. Among the reviewed are five pectate lyases better known as Amb a 1.1, 1.2, 1.3, 1.4, and Amb a 2, now Amb a 1.5; all are isoallergens. The remaining six reviewed proteins include Amb a 3, a plastocyanin; Amb a 5, an allergen; Amb a 6, a non-specific lipid transfer protein; and Amb a 8, having three forms of profilin. The UniProt list of non-reviewed proteins includes three calcium-binding proteins that appear to be the equivalent of the allergens, polcalcins Amb a 9, composed of 2 isoforms, and Amb a 10. A cysteine protease, probably Amb a 11, and an Art v 1 homolog, also known as Amb a 4 are among the non-reviewed allergens. In contrast, the known allergens are limited for the other two ragweed plants; UniProt shows Amb t 5 for A. trifida and Amb p 5a and 5b for A. psilostachya
The Allergen Nomenclature site (Allergen.org) lists the following ragweed pollen allergens: 11 for short (Amb a 1- Amb a 11), 1 for giant (Amb t 5), and 2 for western (Amb p 5a and 5b isoforms). Allergen.org manages the allergens somewhat differently from UniProt, subsuming all isoforms of Amb a 1 (the pectate lyases) under the major numeral. The site also contains allergen Amb a 7 a plastocyanin, not found in UniProt.
For the three plants, our results of found allergens, shown in a Table 2, are somewhat broader. All five of the pectate lyases, Amb a 3, Amb a 5, Amb a 6, and profilin (Hel a 2) and profilin-3 (Amb a 8), were found for short ragweed. If a less restrictive criterion were applied, we also found profilin-2 once in all three species. For giant ragweed Amb t 5 was found, but also were some of the pectate lyases (Amb a 1.2, 1.4, and Amb a 2), profilin and profilin-3. Pectate lyases Amb a 1.2, 1.3, and Amb a 2 were found for western ragweed as well as Amb a 6 and profilin-3. These findings imply that although short ragweed may be the primary source of allergens, giant and western might also contribute to the symptoms of hay fever.
Table 2.
Pollen Allergen Species Distribution
| Amb^ | IUIS% | Protein Name | Common* | Giant* | Western* |
|---|---|---|---|---|---|
| 1.1 | 1.0101 | Pectate Lyase 5 | 2 | ||
| 1.2 | 1.0201 | Pectate Lyase 1 | 2 | 2 | 2 |
| 1.3 | 1.0301 | Pectate Lyase 2 | 2 | 2 | |
| 1.4 | 1.0401 | Pectate Lyase 3 | 2 | 2 | 1 |
| 2 | 1.0501 | Pectate Lyase 4 | 2 | 2 | 2 |
| 3 | 3.0101 | Plastocyanin | 3 | ||
| 5 | 5.0101 | Pollen Allergen Amb a 5 | 3 | ||
| 5 t | 5.0101 | Pollen Allergen Amb t 5 | 2 | ||
| 6 | 6.0101 | NS Lipid Transfer Protein | 4 | 4 | |
| Hel a 2 | 2.0101 | Profilin# | 2 | 2 | |
| 8 | & | Profilin-3 | 4 | 4 | 4 |
| Hev b 9 | 9.0101 | Enolase-1$ | 1 | 4 |
Amb a is the database species unless otherwise specified; 5 t is giant; NS is non-specific;
International Union of Immunological Societies;
from Helianthus annuus;
from Hevea brasiliensis;
the numbers correspond to occurrences of the proteins for each of the ragweed plants; & not defined in IUIS
We also found a probable calcium-binding protein (CBP) with a 3 EF-hand, which may be related to the polcalcin allergens of short ragweed. The evidence for the presence of the CBP is very compelling, having been found for all three ragweed plants in all twelve experiments. An NCBI Blastp search with this 3 EF-hand protein of 148 residues as the query sequence revealed a sequence identity of 83% with an Artemisia vulgaris polcalcin, a protein with similar length of 149 residues. This striking similarity of the 3 EF-hand protein with the Artemisia vulgaris polcalcin suggests the discovered probable calcium- binding protein might be a member of the polcalcin family. The CBP (148 residues) might be viewed Amb a 10-like (160 residues) as the two proteins show 24% identity in a CLUSTAL Omega alignment done at UniProt.org. Interestingly, an additional alignment query revealed the polcalcin-like Amb a 10 allergen with a 4 EF-hand and 160 residues shared 23% identity with the polcalcin allergenic protein Amb a 9, a 2 EF-hand protein with 83 residues. Consideration of these sequence similarities might suggest a possible allergenic character for the probable CBP, even though its sequence shows just 15% identity to the much shorter Amb a 9 protein. Whether CBP has allergenic properties will require immunological evidence. Both Amb a 9 and Amb a 10 are listed on Allergen.org as separate short ragweed allergens.
Our results show the well-known latex Hevea brasiliensis allergen enolase-1 was assigned to giant ragweed multiple times but only once in the short plant. We also found in all three ragweed plants, the Zea mays enolase-1 having 89% identity to that of H. b. Additionally, the enolase of Ricinus communis was found multiple times in the short and western plants but only once in giant; it displayed a 95% identity to the H. b. enolase-1. This high sequence identity is compelling evidence to consider these two enolases as possible allergens, worthy of further investigation. Our results are consistent with those of Bordas-Le Floch et al. [41] who demonstrated an enolase allergen in short ragweed. Babbitt et al. [54] defined an enolase fingerprint motif, which can be found in H. b. [55]. In a UniProt alignment of all three enolases, we found the fingerprint motif runs from S380 to D388 for H. b. and R. c. and includes active site residue H381; the motif is shifted one residue toward the C-terminal residue for Z. m., having a sequence one residue longer. The significant identity R. c. and Z. m. demonstrate with the H. b. enolase-1 could imply an enolase contribution to allergic reactions inducible by all three ragweed plants.
Table 2 contains a list of some Ambrosia allergens found in Allergen.org and revealed through our LCMS/MS study of the pollen from all three ragweed species. Most are widely considered known allergens of short/common ragweed; however, as observed in this table some also appear in the data for the other two species. Enolase-1, a known allergen in Hevea brasiliensis, shown as Hev b 9, is included in Table 2 based on the data providing evidence for its presence in these ragweed plants as well. Profilin, Amb a 8, is represented in the table by Helianthus annuus, in the same family as Ambrosia, and appears as allergen Hel a 2. The table also includes Profilin-3, known allergen of A. artemisiifolia, and is assigned to all three species. Interestingly, Profilin-3 occurs in the UniProt and NCBI protein databases, but is not found at Allergen.org. The numbers given in the last three columns of Table 2 are the number of occurrences of the protein for each species.
The Bordas-Le Floch paper reporting shotgun LC MS/MS of short ragweed pollen found 573 proteins using the criterion of 1 unique peptide. Their list, somewhat different from ours, seems to include identical UniProt homolog entries as well as homologs of the same name but different UniProt identification entries. Our list eliminates all duplicates by name whether found in the ragweed plants or homologs in other species, as well as name identical UniProt entries. When the authors used the more stringent criterion of the total number of sequenced peptides being equal to or greater than 5 their list reduced to 328 proteins [41].
Employing the 1 exclusively unique peptide criterion on our short ragweed data we report 681 homologs, which represent 378 distinct proteins. It is difficult to compare these two sets of data as we seem to be using somewhat different criteria. Although it is possible these two ways of reporting data are the same, the Bordas-Le Floch paper appears to be using total unique sequenced peptides, whereas our results contain exclusively unique peptides. The outcome of applying exclusivity and elimination of homologs is our reported 229 distinct proteins.
Although The Bordas-Le Floch paper and ours report many of the same proteins, missing from both lists are both RNA and DNA polymerases; however, we do report finding transcription factors. Both papers, theirs and ours, report initiation and translation proteins. They report the inability to find Amb a 3 by either transcriptome or proteome analysis, but, instead, an Amb a 3-like protein was identified. Our proteomic analysis revealed the presence of Amb a 3 in short pollen, but not giant or western.
We used Gene Ontology (GO) Annotations from NCBI to display broad categories of biological processes associated with greater than 98% of the distinct 229 proteins. Slightly under 50% of GO designations fell into a broad metabolism category including biosynthesis of cell wall, proteins, and nucleotides among others but not including regulation. Regulation (metabolic, cell cycle, enzyme chromatin silencing, transport, transcription, and so forth), transport (protein, ion, membrane, electron, cytosis, and so forth) protein folding (heat shock, chaperone, isomerase, and others) and translation (ribosomes and initiation factors) constituted another 50% with additional contributions from microtubule processes and nucleosome assembly. There were only three entries -Amb a 5, Amb t 5, and a protein fragment- for which no GO assignment could be found.
The proteome for all three ragweed species shows important similarities and distinctions. Similarities are demonstrated by 45% of the found proteins shared by all. Notable are the differences whether one considers what is the unique for each or the number of proteins in the non-exclusive distribution category among the plants. The finding of a protein only in one of the three species could have multiple explanations. It is possible the other two plants may not express the protein; alternately, there could be differences in expression level; also harvesting of pollen granules might be time and environment dependent; although the pollen extraction protocol was common and samples of all species were simultaneously extracted on four different occasions, variations are not impossible. The somewhat higher similarity shown between short and western ragweed may reflect their likeness of appearance. It is difficult to distinguish them on the basis of foliage, unless one is expert in the field; however, it is possible to do so by the root system. Being an annual short has a tap root not shared by the perennial west.
We found seven categories of allergens distributed among the three plants. The categories include all five pectate lyases, Amb a 3, Amb a 5, Amb t 5, Amb a 6, profilin and profilin-3 (Amb a 8). Although we were unable to find Amb p 5 at the two peptide criterion, we did find it in two experiments at the one peptide level. Evidence for the enolase-1 of H.b. or its equivalent was found for giant and common ragweed. Sequence similarity evidence for the presence of possible new allergens, enolases and polcalcin-like, was found for all three species. The identity information for the enolases is quite strong; that for the CBP polcalcin-like protein is less so.
4. CONCLUSION
Overall, in comparison to UniProt, this work has added to the protein knowledge for all three plants: short 159, giant 148, and western 196. The contribution to the protein knowledge base of the giant and western plants is most notable. Not only did we report a new antigen enolase-1 for some ragweed plants but also we found not previously reported antigens for giant and western; both were found to have some pectate lyases, Amb a 2, and profilin; Amb a 6 was found in western. Although short ragweed may still be the most important source of allergies, there is now evidence for a greater role of giant and western plants in the hay fever malady.
Supplementary Material
Highlights.
Total of 229 proteins representing multiple cellular protein functions identified
Found a minimum of 59 proteins not previously reported for any ragweed plant
Known allergens of short ragweed identified for giant and western species
Newly found enolase-1 allergen for some ragweed species
Potential allergens proposed based on sequence similarity
Acknowledgments
We thank Dr. Bruce Mechtly of the Washburn University Department of Computer Information Sciences for the JAVA Program, ProcessCSVCount, for aligning proteins by name for all 12 experiments. We appreciate the work of Dr. Nadya Galeva of the University of Kansas, Analytical Proteomics Laboratory, for the LC MS/MS, PSM, and Validation Analysis.
This project was supported by grants from the National Center for Research Resources (P20 RR016475) and the National Institute of General Medical Sciences (P20 GM103418) from the National Institutes of Health. The contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heyl FW. The Protein Extract of Ragweed Pollen. J Am Chem Soc. 1919;41:670–682. [Google Scholar]
- 2.Johnson CA, Rappaport BZ. The Proteins of Ragweed Pollens. J Infect Dis. 1932;50:290–309. [Google Scholar]
- 3.King TP, Norman PS. Isolation Studies of Allergens from Ragweed Pollen. Biochemistry. 1962;1(4):709–720. doi: 10.1021/bi00910a027. [DOI] [PubMed] [Google Scholar]
- 4.Robbins KC, WU H, Baram P, Mosko MM. Isolation of Non-Dialyzable Allergens and Antigens from Low Ragweed Pollen (Ambrosian elatoir)) J Immunol. 1963;91:354–361. [PubMed] [Google Scholar]
- 5.King TP, Norman PS, Connell JT. Isolation and Characterization of Allergens from Ragweed Pollen. II. Biochemistry. 1964;3(3):458–468. doi: 10.1021/bi00891a026. [DOI] [PubMed] [Google Scholar]
- 6.King TP. Separation of Proteins by Ammonium Sulfate Gradient Solubilization. Biochemistry. 1972;1(3):367–371. doi: 10.1021/bi00753a010. [DOI] [PubMed] [Google Scholar]
- 7.Wopfner N, Jahn-Schmid B, Schmid G, Christ T, Hubinger G, Briza P, Radauer C, Bohle B, Vogel L, Ebner C, Asero R, Ferreira F, Schwarzenbacher R. The Alpha and Beta Subchain of Amb A 1, The Major Ragweed-Pollen Allergen Show Divergent Reactivity at the IgE and T-Cell Level. Mol Immunol. 2009;46(10):2090–2097. doi: 10.1016/j.molimm.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Rafnar T, Griffith IJ, Kuo MC, Bond JF, Rogers BL, Klapper DG. Cloning of Amb a I (Antigen E), the Major Allergen Family of Short Ragweed Pollen. J Biol Chem. 1991;266(2):1229–1236. [PubMed] [Google Scholar]
- 9.Griffith IJ, Pollock J, Klapper DG, Rogers BL, Nault AK. Sequence Polymorphism of Amb a I and Amb a II, the Major Allergens in Ambrosia artemisiifolia (Short Ragweed) Int Arch Allergy Appl Immunol. 1991;96(4):296–304. doi: 10.1159/000235512. [DOI] [PubMed] [Google Scholar]
- 10.Rogers BL, Morgenstern JP, Griffith IJ, Yu XB, Counsell CM, Brauer AW, King TP, Garman RD, Kuo MC. Complete Sequence of the Allergen Amb Alpha II. Recombinant Expression and Reactivity with T Cells from Ragweed Allergic Patients. J Immunol. 1991;147(8):2547–2552. [PubMed] [Google Scholar]
- 11.Underdown BJ, Goodfriend L. Isolation and Characterization of an Allergen from Short Ragweed Pollen Biochemistry. 1969;8(3):980–989. doi: 10.1021/bi00831a031. [DOI] [PubMed] [Google Scholar]
- 12.Klapper DG, Goodfriend L, Capra JD. Amino Acid Sequence of Ragweed Allergen Ra3. Biochemistry. 1980;19(25):5729–5734. doi: 10.1021/bi00566a010. [DOI] [PubMed] [Google Scholar]
- 13.Goodfriend L, Roebber M, Lundkvist U, Choudhury AM. Two Variants of Ragweed Allergen Ra3. J Allergy Clin Immunol. 1981;67(4):299–304. doi: 10.1016/0091-6749(81)90025-7. [DOI] [PubMed] [Google Scholar]
- 14.Lapkoff CB, Goodfriend L. Isolation of a Low Molecular Weight Ragweed Pollen Allergen: Ra5. Int Arch Allergy Appl Immunol. 1974;46:215–229. doi: 10.1159/000231125. [DOI] [PubMed] [Google Scholar]
- 15.Mole LE, Goodfriend L, Lapkoff CB, Kehoe M, Capra JD. The Amino Acid Sequence of Ragweed Pollen Allergen Ra5. Biochemistry. 1975;14(6):1216–1220. doi: 10.1021/bi00677a019. [DOI] [PubMed] [Google Scholar]
- 16.Roebber M, Klapper DG, Goodfriend L, Bias WB, HSU SH, Marsh DG. Immunochemical and Genetic Studies of Amb.T. V (Ra5G), an Ra5 Homologue from Giant Ragweed Pollen. J Immunol. 1985;134:3062–3069. [PubMed] [Google Scholar]
- 17.Goodfriend L, Choudhury AM, Klapper DG, Coulter KM, Dorval G, Del Carpio J, Osterland CK. Ra5G, A Homologue of Ra5 in Giant Ragweed Pollen: Isolation, HLA-DR-Associated Activity and Amino Acid Sequence. Mol Immunol. 1985;22(8):899–906. doi: 10.1016/0161-5890(85)90076-8. [DOI] [PubMed] [Google Scholar]
- 18.Lowenstein H, Marsh DG. Antigens of Ambrosia elatior (Short Ragweed) pollen. I. Crossed Immunoelectrophoretic Analyses. J Immunol. 1981;126(3):943–948. [PubMed] [Google Scholar]
- 19.Lowenstein H, King TP, Goodfriend L, Hussain R, Roebber M, Marsh DG. Antigens of Ambrosia Elatior (Short Ragweed) Pollen. II. Immunochemical Identification of Known Antigens by Quantitative Immunoelectrophoresis. J Immunol. 1981;127(2):637–642. [PubMed] [Google Scholar]
- 20.Lowenstein L, Marsh DG. Antigens of Ambrosia elatior (Short Ragweed) Pollen. III. Crossed Radioimmunoelectrophoresis of Ragweed-Allergic Patients' Sera with Special Attention to Quantification Of IgE Responses. J Immunol. 1983;130(2):727–731. [PubMed] [Google Scholar]
- 21.Roebber M, Hussain R, Klapper DG, Marsh DG. Isolation and Properties of A New Short Ragweed Pollen Allergen, Ra6. J Immunol. 1983;31(2):706–711. [PubMed] [Google Scholar]
- 22.Metzler WJ, Valentine K, Roebber M, Friedrichs MS, Marsh DG, Mueller L. Determination of the Three-Dimensional Solution Structure of Ragweed Allergen Amb t V by Nuclear Magnetic Resonance Spectroscopy. Biochemistry. 1992;31:5117–5127. doi: 10.1021/bi00137a005. [DOI] [PubMed] [Google Scholar]
- 23.Metzler WJ, Valentine K, Roebber M, Marsh DG, Mueller L. Proton Resonance Assignments and Three-Dimensional Solution Structure of the Ragweed Allergen Am6 a V by Nuclear Magnetic Resonance Spectroscopy. Biochemistry. 1992;31:8697–8705. doi: 10.1021/bi00152a003. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh B, Perry MP, Marsh DG. Cloning the cDNA Encoding the AmbtV Allergen From Giant Ragweed (Ambrosia trifida) Pollen. Gene. 1991;101(2):231–238. doi: 10.1016/0378-1119(91)90416-9. [DOI] [PubMed] [Google Scholar]
- 25.Huang SK, Marsh DG. Human T-cell responses to ragweed allergens: Amb V homologues. Immunology. 1991;73:363–365. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Greenstein JL, Rogers BL, Kuo MC. T Cell Epitope Mapping of Ragweed Pollen Allergen Ambrosia artemisiifolia (Amb a 5) and Ambrosia trifida (Amb t 5) and the Role of Free Sulfhydryl Groups in T Cell Recognition. J Immunol. 1995;155(10):5064–5073. [PubMed] [Google Scholar]
- 27.Ghosh B, Rafnar T, Perry MP, Bassolino-Klimas D, Metzler WJ, Klapper DG, Marsh DG. Immunologic and Molecular Characterization of Amb p V Allergens from Ambrosia psilostachya (Western Ragweed) Pollen. J Immunol. 1994;152(6):2882–2889. [PubMed] [Google Scholar]
- 28.Bagarozzi BA, Jr, Pike R, Potempa J, Travis J. Purification and Characterization of a Novel Endopeptidase in Ragweed (Ambrosia artemisiifolia) Pollen. J Biol Chem. 1996;271(42):26227–26232. doi: 10.1074/jbc.271.42.26227. [DOI] [PubMed] [Google Scholar]
- 29.Bagarozzi BA, Jr, Potempa J, Travis J. Purification and Characterization of an Arginine- Specific Peptidase from Ragweed (Ambrosia artemisiifolia) Pollen. Am J Respir Cell Mol Biol. 1998;18(3):363–369. doi: 10.1165/ajrcmb.18.3.2825. [DOI] [PubMed] [Google Scholar]
- 30.Wopfner N, Grube P, Wallner M, Briza P, Ebner C, Mari A, Richter K, Vogel L, Ferreira F. Molecular and Immunological Characterization of Novel Weed Pollen Pan-Allergens. Allergy. 2008;63(7):872–881. doi: 10.1111/j.1398-9995.2008.01635.x. [DOI] [PubMed] [Google Scholar]
- 31.Léonard R, Wopfner N, Pabst MA, Stadlmann J, Petersen BO, Duus JO, Himly M, Radauer C, Gadermaier G, Razzazi-Fazeli E, Ferreira F, Altmann F. A New Allergen from Ragweed (Ambrosia artemisiifolia) with Homology to Art v 1 from Mugwort. J Biol Chem. 2010;285(35):27192–27200. doi: 10.1074/jbc.M110.127118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouley J, Groeme R, Le Mignon M, Jain K, Chabre H, Bordas-Le Floch V, Couret MN, Bussières L, Lautrette A, Naveau M, Baron-Bodo V, Lombardi V, Mascarell L, Batard T, Nony E, Moingeon P. Identification of the Cysteine Protease Amb a 11 as a Novel Major Allergen from Short Ragweed. J Allergy Clin, Immun. 2015;136(4):1055–1064. doi: 10.1016/j.jaci.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Noir S, Brautigam A, Colby T, Schmidt J, Panstruga R. A Reference Map of the Arabidopsis thaliana Mature Pollen Proteome. Biochem Biophys Res Commun. 2005;337:1257–1266. doi: 10.1016/j.bbrc.2005.09.185. [DOI] [PubMed] [Google Scholar]
- 34.Sheoran IS, Ross AR, Olson DJ, Sawhney VK. Proteomic Analysis of Tomato (Lycopersicon esculentum) Pollen. J Exp Bot. 2007;58(13):3525–3535. doi: 10.1093/jxb/erm199. [DOI] [PubMed] [Google Scholar]
- 35.Zou J, Song L, Zhang W, Wang Y, Ruan S, Wu WH. Comparative Proteomic Analysis of Arabidopsis Mature Pollen and Germinated Pollen. J Integr Plant Biol. 2009;51(5):438–455. doi: 10.1111/j.1744-7909.2009.00823.x. [DOI] [PubMed] [Google Scholar]
- 36.Sheoran IS, Ross AR, Olson DJ, Sawhney VK. Differential Expression of Proteins in the Wild Type and 7B-1 Male-Sterile Mutant Anthers of Tomato (Solanum lycopersicum): A Proteomic Analysis. J Proteomics. 2009;71(6):624–636. doi: 10.1016/j.jprot.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Goliáš J, Humlová Z, Halad P, Hábová V, Janatková I, Tučkova L. Identification of Rice Proteins Recognized by the IgE Antibodies of Patients with Food Allergies. J Agric Food Chem. 2013;61:8851–8860. doi: 10.1021/jf402759f. [DOI] [PubMed] [Google Scholar]
- 38.Ge P, Ma C, Wang S, Gao L, Li X, Guo G, Ma W, Yan Y. Comparative Proteomic Analysis of Grain Development in two Spring Wheat Varieties under Drought Stress. Anal Bioanal Chem. 2012;402:1297–1313. doi: 10.1007/s00216-011-5532-z. [DOI] [PubMed] [Google Scholar]
- 39.Sarhadi E, Bazargani MM, Sajise AG, Abdolahi S, Vispo NA, Arceta M, Nejad GM, Singh RK, Salekdeh GH. Proteomic Analysis of Rice Anthers under Salt Stress. Plant Physiol Biochem. 2012;58:280–287. doi: 10.1016/j.plaphy.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Abou Chakra OR, Sutra JP, Thomas DE, Vinh J, Lacroix G, Poncet P, Sénéchal H. Proteomic Analysis of Major and Minor Allergens from Isolated Pollen Cytoplasmic Granules. J Proteome Res. 2012;11:1208–1216. doi: 10.1021/pr200923f. [DOI] [PubMed] [Google Scholar]
- 41.Bordas-Le Floch V, Le Mignon M, Bouley J, Groeme R, Jain K, Baron-Bodo V, Nony E, Mascarell L, Moingeon P. Identification of Novel Short Ragweed Pollen Allergens Using Combined Transcriptomic and Immunoproteomic Approaches. PLOSONE. 2012 doi: 10.1371/journal.pone.0136258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cargile BJ, Bundy JL, Grunden AM, Stephenson JL., Jr Synthesis/Degradation Ratio Mass Spectrometry for Measuring Relative Dynamic Protein Turnover. Anal Chem. 2004;76(1):86–97. doi: 10.1021/ac034841a. [DOI] [PubMed] [Google Scholar]
- 43.Thiede B, Treumann A, Kretschmer A, Söhlke J, Rudel T. Shotgun Proteome Analysis of Protein Cleavage in Apoptotic Cells. Proteomics. 2005;5(8):2123–30. doi: 10.1002/pmic.200401110. [DOI] [PubMed] [Google Scholar]
- 44.Piersma SR, Fiedler U, Span S, Lingnau A, Pham TV, Hoffmann S, Kubbutat MHG, Jimenez CR. Workflow Comparison for Label-Free, Quantitative Secretome Proteomics for Cancer Biomarker Discovery: Method Evaluation, Differential Analysis, and Verification in Serum. J Proteome Res. 2010;9(4):1913–1922. doi: 10.1021/pr901072h. [DOI] [PubMed] [Google Scholar]
- 45.Lee MY, Huang CH, Kuo CJ, Lin CL, Lai WT, Chiou SH. Clinical Proteomics Identifies Urinary CD14 as a Potential Biomarker for Diagnosis of Stable Coronary Artery Disease. PLOSONE. 2015 doi: 10.1371/journal.pone.0117169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Life Technologies. Protocol A: Prepare Cell/Tissue Lysate and Protocol B: Prepare Plant Lysate. Zoom 2D Protein Solubilizer Manuel. 2012:2–3. [Google Scholar]
- 47.Sarhadi E, Bazargani MM, Sajise AG, Abdolahi S, Vispo AZ, Arceta M, Nejad GM, Singh RK, Salekdeh GH. Proteomic Analysis of Rice Anthers under Salt Stress. Plant Physiol Biochem. 2012;58:280–287. doi: 10.1016/j.plaphy.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Searle BC, Turner M, Nesvizhskii AI. Improving Sensitivity by Probabilistically Combining Results from Multiple MS/MS Search Methodologies. J Proteome Res. 2008;7:245–253. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 49.Searle BC. Scaffold: A Bioinformatic Tool for Validating MS/MS-Based Proteomic Studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 50.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 51.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 52.Barkley TM. Field Guide to the Common Weeds of Kansas. University Press of Kansas; Lawrence: 1983. [Google Scholar]
- 53.Mirzaei M, Soltani N, Sarhadi E, Pascovici D, Keighley T, Salekdeh GH, Haynes PA, Atwell BJ. Shotgun Proteomic Analysis of Long-distance Drought Signaling in Rice Roots. J Proteome Res. 2012;11:348–358. doi: 10.1021/pr2008779. [DOI] [PubMed] [Google Scholar]
- 54.Babbitt PC, Hasson MS, Wedekind JE, Palmer DR, Barrett WC, Reed GH, Rayment I, Ringe D, Kenyon GL, Gerlt JA. The Enolase Superfamily: A General Strategy for Enzyme-Catalyzed Abstraction of the Alpha-Protons of Carboxylic Acids. Biochemistry. 1996;35:16489–16501. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 55.Wagner S, Breiteneder H, Simon-Nobbe B, Susani M, Krebitz M, Niggemann B, Brehler R, Scheiner O, Hoffmann-Sommergruber K. Hev b 9, an Enolase and a New Cross-Reactive Allergen from Hevea Latex and Molds. Purification, Characterization, Cloning and Expression. Eur J Biochem. 2000;267:7006–7014. doi: 10.1046/j.1432-1327.2000.01801.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.