Abstract
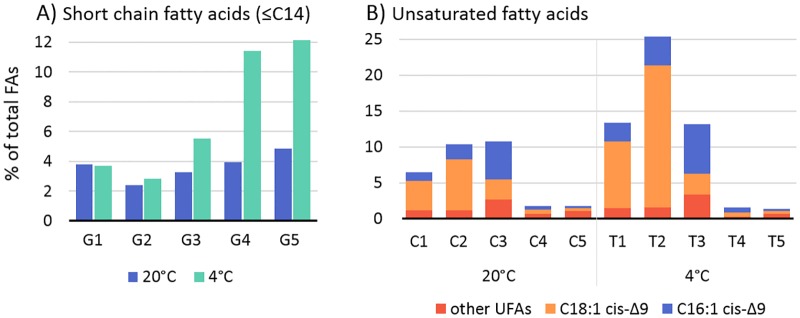
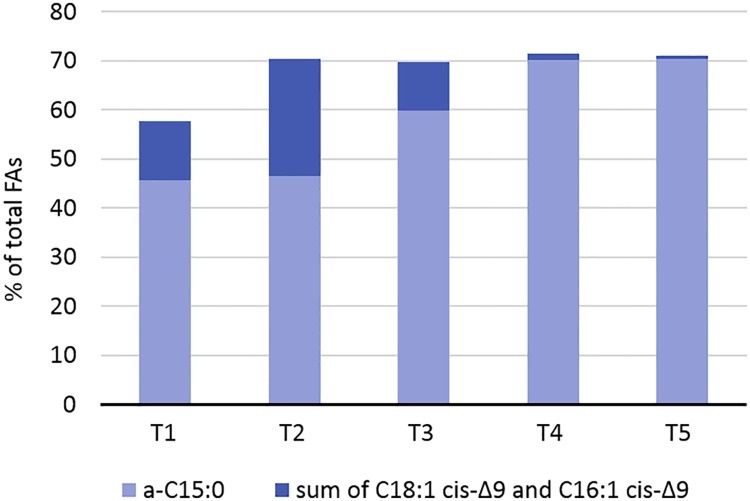
The human pathogen Listeria monocytogenes continues to pose a challenge in the food industry, where it is known to contaminate ready-to-eat foods and grow during refrigerated storage. Increased knowledge of the cold-stress response of this pathogen will enhance the ability to control it in the food-supply-chain. This study utilized strand-specific RNA sequencing and whole cell fatty acid (FA) profiling to characterize the bacterium’s cold stress response. RNA and FAs were extracted from a cold-tolerant strain at five time points between early lag phase and late stationary-phase, both at 4°C and 20°C. Overall, more genes (1.3×) were suppressed than induced at 4°C. Late stationary-phase cells exhibited the greatest number (n = 1,431) and magnitude (>1,000-fold) of differentially expressed genes (>2-fold, p<0.05) in response to cold. A core set of 22 genes was upregulated at all growth phases, including nine genes required for branched-chain fatty acid (BCFA) synthesis, the osmolyte transporter genes opuCBCD, and the internalin A and D genes. Genes suppressed at 4°C were largely associated with cobalamin (B12) biosynthesis or the production/export of cell wall components. Antisense transcription accounted for up to 1.6% of total mapped reads with higher levels (2.5×) observed at 4°C than 20°C. The greatest number of upregulated antisense transcripts at 4°C occurred in early lag phase, however, at both temperatures, antisense expression levels were highest in late stationary-phase cells. Cold-induced FA membrane changes included a 15% increase in the proportion of BCFAs and a 15% transient increase in unsaturated FAs between lag and exponential phase. These increases probably reduced the membrane phase transition temperature until optimal levels of BCFAs could be produced. Collectively, this research provides new information regarding cold-induced membrane composition changes in L. monocytogenes, the growth-phase dependency of its cold-stress regulon, and the active roles of antisense transcripts in regulating its cold stress response.
Introduction
The human pathogen Listeria monocytogenes continues to pose a challenge in the food industry, where it is known to contaminate ready-to-eat foods and to grow during refrigerated storage. Ingestion of L. monocytogenes by susceptible individuals can cause potentially fatal food-borne infections with mortality rates as high as 40% reported [1]. The ubiquitous nature of this pathogen makes it difficult to eliminate from our food systems however, post-processing levels of L. monocytogenes contamination are often low [2–4] and unlikely to cause disease [5, 6]. Furthermore, the risks associated with L. monocytogenes contamination can be addressed by heating foods to an appropriate temperature [7]. Therefore, the ability of L. monocytogenes to grow to unsafe levels in ready-to-eat foods during the shelf-life of products, pose the greatest concern to consumer health.
When subjected to low temperatures, all living cells will experience similar challenges that stem from a reduction in molecular dynamics, which leads to decreased rates of diffusion and structural changes to molecular structures [8]. Cold-stress adaptation mechanisms among microbes include membrane compositional changes, compatible solute uptake, and the synthesis of nucleic acid stabilization proteins, DNA-unwinding enzymes, and general stress response and cold shock proteins [9, 10]. Currently, most of our knowledge regarding bacterial cold stress response (CSR) mechanisms comes from the model organisms Escherichia coli and Bacillus subtilis. However, unlike L. monocytogenes, neither of these bacteria can multiply at temperatures close to 0°C. In the last decade, L. monocytogenes outbreaks associated with ready-to-eat foods in both North America and Europe have increased interest in the specific mechanisms employed by this psychrotrophic pathogen to adapt and grow at low temperatures.
The CSR of L. monocytogenes has been studied using microarrays [11–13], quantitative real-time PCR (qPCR) [14–17], mutagenesis [18–24] and other genetic and proteomic techniques [25–27]. Collectively, these methods have identified a large pool of genes with putative or known roles in cold tolerance. However, although a small portion of qPCR-based studies have looked at the differential expression (DE) of select genes during the early stages of the L. monocytogenes CSR (i.e., lag phase) [16, 17], global transcriptome studies have only been conducted on cold-adapted exponential- and stationary-phase cells to date, leaving much to be discovered regarding the initial CSR of L. monocytogenes. Furthermore, since CSRs become more pronounced in response to more dramatic changes in temperature, many studies have focused on abrupt temperature down shifts from 37°C to 15°C or lower [11, 14, 15, 17, 28]. Such conditions may not represent the food industry, where more commonly L. monocytogenes is typically transferred from an ambient-temperature environment to a food product or plant environment that is maintained at 4–10°C. Though exposure to ambient temperature (20–25°C) can also be considered a low temperature stress [15, 29], the maximum growth rate (μmax) of L. monocytogenes’ is approximately 10× faster at 20°C than at 4°C whereas there is much less of a difference between 15°C and 37°C (~4.5×) [30]. Moreover, at 37°C, the transcriptional landscape of L. monocytogenes undergoes a drastic change in preparation for intracellular survival [31].
Recent advances in molecular and sequencing technologies now allow us to detect several forms of non-coding RNA (ncRNA). Studies of ncRNA have increased our understanding of gene regulation and opened a new area in bacterial stress response research.
ncRNAs exist in several different forms and thus participate in a wide range of functions. Most ncRNAs can be divided into three main categories: 1) Cis-regulatory RNAs, 2) trans-encoded small RNAs (sRNAs), and 3) antisense RNAs (asRNAs) [32]. Cis-regulatory RNAs are located at the 5’-ends of mRNA and fold into alternative structures in response to physicochemical cues. These transcripts are often referred to as riboswitches or thermosensors. One of the best-known examples in L. monocytogenes is the riboswitch that blocks the translation of the major virulence regulator prfA at low temperatures (<30°C) [33]. Trans-encoded sRNAs, on the other hand, are not located adjacent to their target and share only limited complementarity allowing them to regulate multiple mRNAs. Lastly, asRNAs are transcribed from the DNA strand opposite of a gene and thus have perfect complementarity. asRNAs can be short (<100 nt) or long (>1000 nt) and in some cases correspond to the 5’- or 3’- extension of an mRNA transcribed from an adjacent gene [34]. The prevalence of genes with reported asRNA ranges from 20–75% depending on the organism [35, 36]. To date, more than 100 asRNAs have been described in L. monocytogenes [31, 36–38]. Compared with the numbers reported for other bacteria this number seems rather low. However, it will presumably rise with further study.
The objective of this study was to gain a comprehensive view of the L. monocytogenes CSR, with an emphasis on elucidating mRNA and asRNA expression patterns across multiple growth phases following cold stress. To gain a better understanding of the mechanisms employed by L. monocytogenes to grow during refrigerated storage, we aimed to characterize the CSR from cells during the initial lag phase following cold stress, throughout exponential growth and long-term stability at low temperatures, using conditions similar to those present in a food-contamination scenario. To accomplish these goals, strand-specific RNA sequencing and membrane lipid profiling were conducted on L. monocytogenes cell cultures at five distinct growth phases, following a downshift in temperature from 20°C to 4°C. As the phenotypic and genetic properties of L. monocytogenes isolates differ, we chose to elucidate the CSR mechanisms of a strain with relevance to the food industry and consumer health: a fast cold-growing, serotype 1/2a, food plant isolate containing full length versions of important virulence genes: inlA, inlB, inlC, prfA, plcA, hly, mpl, actA, plcB. Here, we report novel findings regarding cold-induced membrane compositional changes in L. monocytogenes, the growth phase-dependent cold-stress regulon, and the active roles of antisense transcripts in regulating the CSR of this pathogen.
Materials and methods
Culture conditions and time point selection
A previously evaluated L. monocytogenes environmental isolate (BioSample SAMN05256775) from a food-processing plant in British Columbia (Lm1, serotype 1/2a, sequence type 7) was selected for use in this study [39]. The strain displayed enhanced cold tolerance (μmax at 4°C) relative to a large collection of isolates [39]. Bacterial cultures were grown for 24 h in brain heart infusion broth (BHIB; Difco, Fisher Scientific, Canada) at 20°C, re-suspended in pre-tempered BHIB at a cell density of 107 CFU/ml and incubated at 4 or 20°C. RNA and lipids were extracted at five time points from cells grown at 4°C (treatment, T) and 20°C (control, C). Three biological replicates were conducted for each treatment and control sample. Each time point corresponded to a specific growth phase (G) (Fig 1). The time points were as follows: G1 –early lag phase, G2 –transition to exponential growth phase, G3 –mid-exponential growth phase, G4 –transition to stationary phase, and G5 –late-stationary phase. G1 corresponded to 20% of the lag phase duration at each temperature. The timing was determined by modelling previously obtained growth curve data using the Baranyi and Roberts model [40] in DMfit (v3.5) (http://browser.combase.cc/DMFit.aspx) [30]. G2 was marked by a doubling in cell numbers, confirmed using plate counts. G3 corresponded to mid-exponential growth (108 CFU/ml), and G4 corresponded to the transition to stationary phase; both time points were identified spectrophotometrically (A600nm) and confirmed with plate counts. Lastly, G5 was selected to correspond to G4 plus 8× the lag phase duration (h) at each temperature.
Fig 1. Cell growth phase sampling strategy at 4°C and 20°C.
Fatty acid analysis
At each of the five growth phases for cells grown at both 20°C and 4°C, cultures were pelleted (10–45 mg net weight), rinsed twice with one ml phosphate buffered saline (PBS, Fisher Scientific), and stored at -80°C. One sample per time point was collected. The frozen pellets were later sent to MIDI labs (Microbial ID, Inc., Newark DE, USA) where the cell lipids were extracted, followed by methylation of the fatty acids (FAs), and then loaded onto a gas chromatograph for analysis. Fatty acid methyl ester (FAME) profiles were then generated and analyzed using Sherlock® pattern recognition software.
RNA isolation and sequencing
At each sample point, growth in cultures was stopped by adding 10% phenol:chloroform (Fisher Scientific) in ethanol solution pre-chilled to -80°C in a 1:10 volume to the sample. The tubes were vortexed briefly and then centrifuged immediately for 10 min at 4,696×g and 0°C. The supernatants were removed and the resulting pellets were stored at -80°C for less than 2 weeks. Total RNA was isolated and purified using the PowerMicrobiome™ RNA Isolation kit (MO BIO Laboratories, CA, USA) per the manufacturer’s protocol. RNA integrity numbers (RINs) were determined using the 2100 Bioanalyzer (Agilent, CA, USA). Samples with a RIN between 9.7 and 10 and an RNA concentration >100 ng/μl were sent to Genome Québec (Montréal, QC, Canada) for rRNA-depleted Illumina TruSeq RNA library prep and TruSeq stranded total RNA 100 bp paired-end sequencing on the Illumina HiSeq 2000 platform.
RNA-seq data analysis
Sequencing quality was assessed using FastQC [41], and Illumina adapter sequences and low-quality base pairs were removed using default parameters in Trimmomatic v 0.36 [42]. After the removal of low-quality reads, 22–39 million reads remained for each dataset. Reads were mapped to the complete sequenced genome of L. monocytogenes EGD (NCBI RefSeq NC_022568.1) using Bowtie 2 v 2.2.6 [43] and allowing zero mismatches. L. monocytogenes EGD was selected as the reference genome as it allowed for the highest percent of successfully mapped reads compared to the more commonly employed reference strains EGDe and 10403S. The mapping efficiency ranged from 97.8–99.6% for individual reads and 92.1–95.6% for successfully paired reads. BAM alignment files were used as input for read counting using featureCounts v 1.5.0-p1 [44]. The default counting mode ‘union’ was used, and separate count files were generated for sense and antisense transcripts. Differential expression (DE) analyses were performed using DESeq2 [45] in R v 3.1.1 [46], and the DE was reported as log2 fold changes. p-values were adjusted by the DESeq2 default Benjamini-Hochberg (BH) adjustment method and genes with a >2-fold (>1 log2) change in expression and an adjusted p-value < 0.05 were considered as DE. A principle component analysis (PCA) was conducted using DESeq2 to determine the overall reproducibility of our RNA-seq biological replicates. To evaluate the overall transcription levels for individual genes at both 4°C and 20°C, raw read counts were normalized based on library size. No p-values were calculated for genes with low mean normalized counts, zero counts, or extreme outlier counts.
Clustering of gene expression profiles
Clustering of genes with similar expression patterns across the five growth phases, was performed using the Mfuzz package [47] in R. Default filtering and standardization methods (based on standard deviation) were applied to the log2 data from all five growth phases. Soft clustering was then performed using the fuzzy c-means algorithm and the following parameters: c = 20, m = 1.25. Genes were assigned to clusters of given expression patterns across the growth-phases based on having a membership value >0.5.
Functional categorization of differentially expressed genes
To investigate the roles of DE genes at each growth phase, we determined the overrepresentation of molecular and biological pathways (Fisher exact test, p<0.05) using SmartTables based on the BioCyc database (https://biocyc.org/) [48]. SmartTables were also used to map L. monocytogenes EGD genes to the equivalent genes in L. monocytogenes 10403S for which the BioCyc database contains transcription regulation and gene ontology information. Additional enrichment analyses were then performed (Fisher’s exact test, p<0.05) for gene ontology (GO) terms, and genes regulated by certain transcription regulators (i.e., σB, PrfA, RpoD, VirR, MogR, CodY, HrcA and CtsR).
Quantitative PCR validation of RNA-seq data
RNA-seq results were validated using qPCR amplification of three genes: 1) leuA which exhibited >4-fold higher expression at 4°C at all five growth phases, 2) cspB which exhibited >4-fold lower expression at 4°C at all growth phases, and 3) recJ which was chosen as a reference gene because it showed very little variation in expression across all growth phases at either temperature (S2 Table). Up to 1 μg of RNA from each of our 30 samples was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA) per the manufacturer’s protocol. Primers were designed using Primer3 Plus (Table 1) and our draft whole genome sequence for Lm1 (GenBank Accession number GCA_001709805.1). qPCR was conducted in a CFX96 Touch™ Real-Time PCR Detection System (BioRad) using SsoAdvanced ™ Universal SYBR Green ® Supermix (BioRad). The relative expression levels of leuA and cspB were calculated using the 2-ΔΔCT method [49], with recJ as the reference gene.
Table 1. Primers used for quantitative PCR validation of RNA-seq data.
| Gene abbr. | Primer sequence | Opt. Anneal. Temp | Source |
|---|---|---|---|
| cspB | Fw-CAAACAGGTACAGTTAAATGGTTTA | 55°C | [17] |
| Rv-ACGATTTCAAATTCAACGCTTTGA | |||
| leuA | Fw-TTGTCGGGTATGCCTGTTCC | 55°C | This study |
| Rv-GGGTTTTTCAGCACGCCATC | |||
| recJ | Fw-CTCGACCGGCAATTGTGTTG | 55°C | This study |
| Rv- GTCCACACTTCGACCAGACC |
Accession numbers
FastQ files of HiSeq runs were deposited into the NCBI Sequence Read Archive under BioProject PRJNA384077.
Results and discussion
mRNA transcriptome of L. monocytogenes cold-stress response
A total of 11–17 million paired-end mRNA reads per sample were successfully assigned to L. monocytogenes EGD open reading frames (ORFs). The number of reads mapped to each EGD ORF ranged from 0 to 630,000. No counts were observed for 27 ORFs, of which, 26 were confirmed to be absent in our strain, leaving one gene (LMON_0476) with no detectable expression at 20°C or 4°C. Overall, >99% of EGD ORFs were expressed by Lm1 at 20 and 4°C. This percentage is in line with the findings of Toledo-Arana et al. [31], who reported that Listeria spp. express more than 98% of their ORFs at 30°C and 37°C.
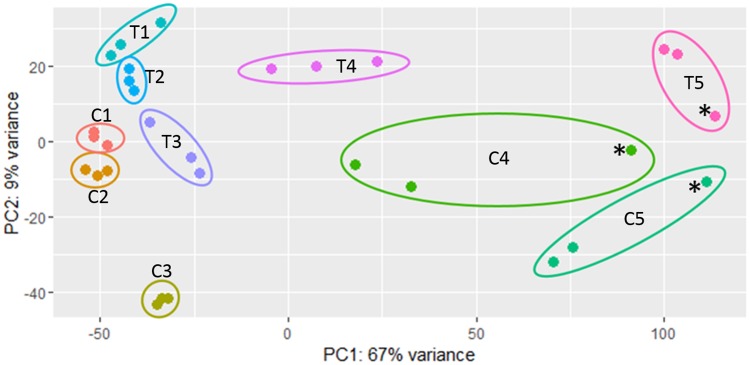
Three of the 30 sequenced samples were excluded from further analysis as they were deemed to be outliers as visualized in the PCA plot in Fig 2. The excluded samples included one biological replicate each from the T4, C4, and C5 treatments. Two of the three samples were from G4 which represents the transition from exponential growth to stationary phase. Given the short window of time available for extracting RNA from this very specific physiological phase, it is likely that these samples were processed either too early or too late relative to the other two biological replicates. The excluded C5 sample belonged to the same biological replicate as the excluded C4 sample. The PCA plot (Fig 2) also shows that the level of transcriptome variance is much greater (67%) among growth phases than between the two temperatures (9%), highlighting the importance of growth phase selection in determining the outcomes of an experiment.
Fig 2. Principle component analysis plot of RNA sequencing biological replicates.
C1 through C5 refer to the L. monocytogenes control cultures grown at 20°C, and T1 through T5 represent the treated cultures grown at 4°C (see Fig 1 for information about the sampling points). Samples marked with * were excluded from our analyses due to an abundance of outlier data points.
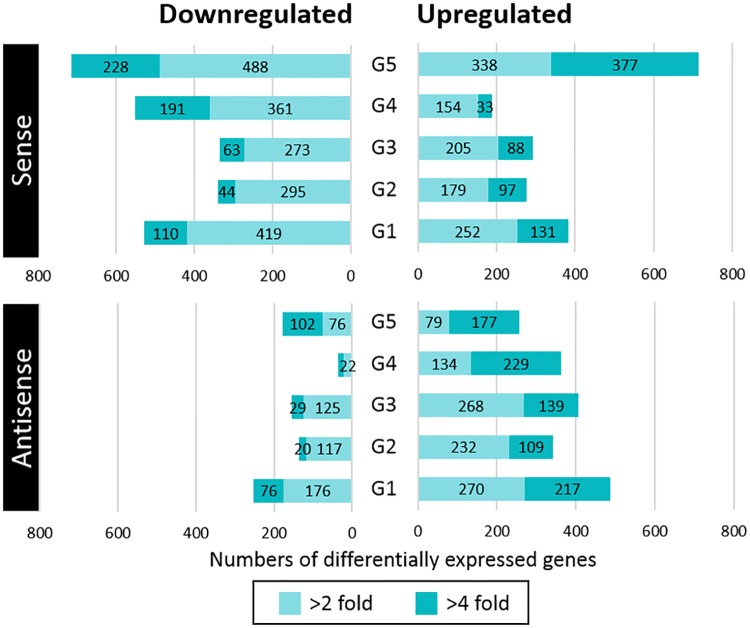
Overall, 1.3× more genes were suppressed than induced in L. monocytogenes under cold stress (Fig 3). This finding contrasts with the results reported in a microarray study conducted by Chan et al. [11], where 30–40% fewer genes were downregulated than upregulated in L. monocytogenes at 4°C compared to 37°C. However, in agreement with their work, we observed that cold-adapted stationary-phase cells exhibited the greatest number of DE genes (G5, Fig 3). From here on, the DE of genes at 4°C vs. 20°C will be discussed with respect to the growth phases (G1–G5) and C1–C5 and T1–T5 will be reserved for when referring to overall levels of transcription or changes in the membrane lipid profiles that occur at 20°C and 4°C, respectively.
Fig 3. Numbers of L. monocytogenes sense and antisense RNAs differentially expressed at five growth phases in response to cold stress (4°C relative to 20°C).
Differential expression (DE) analyses were performed using DESeq2 and DE was reported as log2 fold changes. Genes with a >2-fold change (> 1 log2) in expression and an adjusted p-value < 0.05 were considered DE. See Fig 1 for information about growth phases.
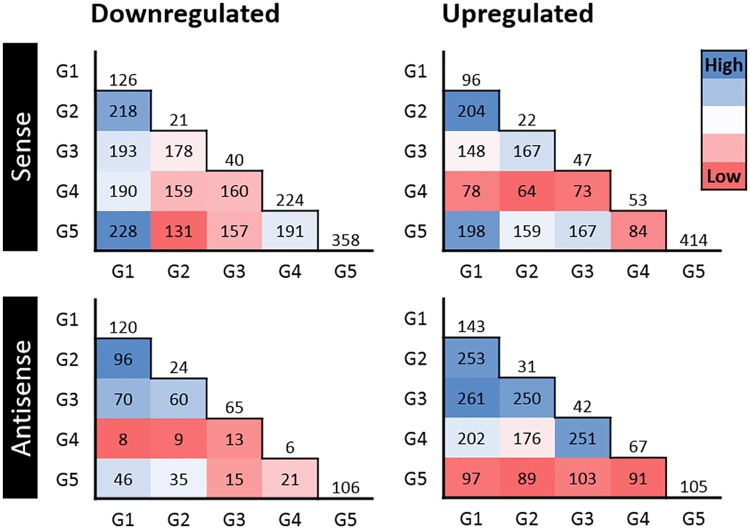
Fig 4 showed the pairs of growth phases that shared the highest and lowest numbers of the same up or downregulated genes at 4°C relative to 20°C. G4 had the lowest number of upregulated genes in common with the other growth phases, whereas G1 and G2 (G1:G2) and G1:G5 shared many of the same upregulated genes. With respect to downregulated genes, G4 and G5 had the smallest in common with G2 and G3 whereas G1:G2 and G1:G5 shared a high number of downregulated genes.
Fig 4. Heatmaps showing the number of L. monocytogenes sense and antisense RNA co-upregulated or co-downregulated (>2-fold) between pairs of growth phases at 4°C.
Numbers outside of the pyramids represent the number of genes uniquely upregulated or downregulated at each growth phase. See Fig 1 for information about growth phases.
To confirm our DE findings, we used RT-qPCR to quantify the expression levels of a gene that was upregulated for >4 fold in response to cold at all growth phases (cspB), and a gene that was downregulated for >4 fold in response to cold at all growth phases (leuA), according to the RNA-seq data. We observed a strong positive correlation (R2 = 0.96, y = 1.20x-0.41) between the mRNA levels detected by the two methods. Nevertheless, the fold changes calculated using RNA-seq data were consistently larger than those obtained using RT-qPCR (S1 Fig).
Core set of genes induced by cold stress
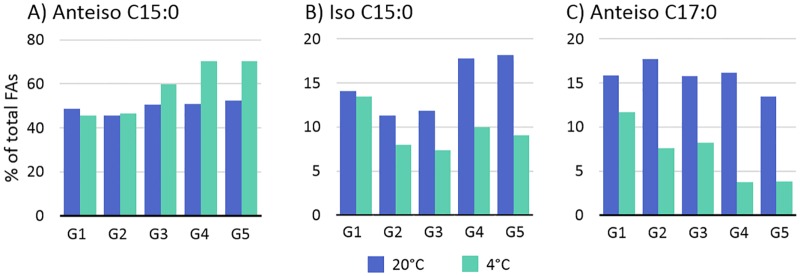
The DE analyses revealed a core set of 22 genes induced at 4°C relative to 20°C at all five growth phases (Table 2). These genes fall into the category of cold acclimation proteins (CAPs), which are defined as proteins encoded by genes that show continuous increased expression throughout prolonged cold-stress exposure [8, 50]. Among these CAPs were nine genes involved in the production of branched-chain amino acids (BCAAs) (ilvDBHC-leuABCD-ilvA). These BCAA synthesis genes were statistically overrepresented among the upregulated genes at all growth phases (Table 3). The BCAAs leucine, isoleucine, and valine, are critical for BCFA production, with isoleucine being the precursor for anteiso BCFAs. Listeria and similar bacteria, such as Bacillus species, alter their membrane lipid composition in response to temperature downshifts, to include higher percentages of branched-chain fatty acids (BCFAs) with anteiso configurations [24, 51–53]. Anteiso BCFAs have lower melting points than their iso-branched and straight-chain counterparts. They are thus more effective at increasing membrane fluidity, which is needed to maintain membrane-transport functions at low temperatures.
Table 2. Core set of genes upregulated (>2-fold) across multiple (≥4) growth phases in L. monocytogenes cells at 4°C vs. 20°C.
| EGD ORF | EGD description | Gene abbr. | Strand | EGD-e ORF | Cluster memb-ership | Log2 DE across five growth phases | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||||||
| LMON_0187 | Veg protein | + | lmo0189 | 16/14 | 2.96 | 2.45 | 2.28 | 2.38 | 1.54 | |
| LMON_0209 | LSU ribosomal protein L25p | ctc | + | lmo0211 | 10 | 2.16 | 3.43 | 2.72 | 0.87 | 1.58 |
| LMON_0260 | Internalin H | inlH | + | lmo0263 | 12 | 2.21 | 3.67 | 2.11 | -1.54 | 1.57 |
| LMON_0261 | Internalin D | inlD | + | 9 | 1.86 | 2.79 | 2.18 | 1.23 | 2.70 | |
| LMON_0350 | Transaldolase | lmo0343 | 5 | 1.88 | 2.03 | 1.56 | 0.27 | 4.16 | ||
| LMON_0351 | 3-oxoacyl-[acyl-carrier protein] reductase | lmo0344 | 5 | 2.35 | 1.71 | 1.30 | 0.62 | 4.34 | ||
| LMON_0352 | Ribose 5-phosphate isomerase B | lmo0345 | 5 | 2.75 | 2.30 | 1.25 | 0.26 | 4.61 | ||
| LMON_0353 | Triosephosphate isomerase | lmo0346 | 5 | 2.38 | 1.95 | 1.75 | 0.45 | 4.38 | ||
| LMON_0354 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, ADP-binding subunit DhaL | lmo0347 | 5 | 2.21 | 2.08 | 1.70 | 0.03 | 4.21 | ||
| LMON_0355 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, dihydroxyacetone binding subunit DhaK | lmo0348 | 5 | 2.34 | 1.85 | 1.60 | 0.26 | 4.50 | ||
| LMON_0356 | Hypothetical protein | lmo0349 | 5 | 1.94 | 2.17 | 1.35 | -0.16 | 5.37 | ||
| LMON_0357 | Hypothetical protein | lmo0350 | 5 | 3.01 | 1.62 | 1.45 | -0.25 | 5.55 | ||
| LMON_0358 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, subunit DhaM | lmo0351 | 5 | 2.49 | 1.79 | 1.57 | -0.37 | 5.50 | ||
| LMON_0360 | Acetyltransferase, GNAT family | + | lmo0353 | 12 | 1.85 | 1.50 | 1.23 | 0.67 | 1.72 | |
| LMON_0441 | Internalin A | inlA | + | lmo0433 | 5 | 2.01 | 1.82 | 1.92 | 1.62 | 2.30 |
| LMON_0486 | LSU ribosomal protein L32p | rpmF | + | lmo0486 | 19 | 1.43 | 1.46 | 1.76 | 1.75 | 1.22 |
| LMON_0515 | Universal stress protein | + | lmo0515 | 12 | 1.34 | 1.76 | 1.12 | -2.06 | 1.98 | |
| LMON_0561 | Phosphoribosyl-ATP pyrophosphatase | hisE | lmo0561 | 2 | 1.70 | 1.64 | 1.01 | 1.53 | 2.98 | |
| LMON_0611 | Internalin-like protein | lmo0610 | 12 | 2.50 | 2.85 | 2.06 | -0.73 | 2.19 | ||
| LMON_0625 | Acetyltransferase, GNAT family | lmo0624 | 5 | 2.07 | 2.19 | 1.67 | 1.33 | 4.46 | ||
| LMON_0626 | Hypothetical protein | lmo0625 | 5 | 2.18 | 2.19 | 1.59 | 1.23 | 4.43 | ||
| LMON_0785 | PTS system, mannose-specific IID component | lmo0781 | 12 | 1.81 | 2.76 | 1.32 | -0.54 | 1.97 | ||
| LMON_0786 | PTS system, mannose/fructose-specific IIC component | lmo0782 | 12 | 1.60 | 2.72 | 1.43 | -0.76 | 1.64 | ||
| LMON_0787 | PTS system, mannose-specific IIB component | lmo0783 | 12 | 1.86 | 2.80 | 1.43 | -1.11 | 1.86 | ||
| LMON_0788 | PTS system, mannose-specific IIB component / IIA component | lmo0784 | 12 | 2.05 | 3.10 | 1.49 | -1.23 | 1.70 | ||
| LMON_0920 | Succinate-semialdehyde dehydrogenase [NAD] | + | lmo0913 | 10 | 4.64 | 5.74 | 2.96 | 0.40 | 2.35 | |
| LMON_0944 | Hypothetical protein | 10 | 2.77 | 3.84 | 4.22 | 1.39 | 2.00 | |||
| LMON_1488 | Osmotically activated L-carnitine/choline ABC transporter, permease protein OpuCD | opuCD | lmo1425 | 10 | 2.34 | 5.37 | 3.52 | 1.79 | 1.29 | |
| LMON_1489 | Osmotically activated L-carnitine/choline ABC transporter, substrate-binding protein OpuCC | opuCC | lmo1426 | 10 | 2.45 | 5.23 | 3.78 | 1.82 | 1.24 | |
| LMON_1490 | Osmotically activated L-carnitine/choline ABC transporter, permease protein OpuCB | opuCB | lmo1427 | 10 | 2.51 | 5.28 | 3.78 | 1.58 | 1.10 | |
| LMON_1491 | Osmotically activated L-carnitine/choline ABC transporter, ATP-binding protein OpuCA | opuCA | lmo1428 | 10 | 2.63 | 5.54 | 3.94 | 1.30 | -0.01 | |
| LMON_1932 | Predicted membrane protein hemolysin III | + | lmo1864 | 14 | 2.21 | 1.58 | 1.78 | 1.84 | 1.06 | |
| LMON_2054 | Dihydroxy-acid dehydratase | ilvD | + | lmo1983 | 4* | 3.52 | 3.89 | 3.34 | 4.29 | 3.62 |
| LMON_2055 | Acetolactate synthase large subunit | ilvB | + | lmo1984 | 3/15 | 3.67 | 4.17 | 3.41 | 4.41 | 4.37 |
| LMON_2056 | Acetolactate synthase small subunit | ilvH | + | lmo1985 | 15/1 | 3.67 | 4.02 | 3.39 | 4.41 | 4.03 |
| LMON_2057 | Ketol-acid reductoisomerase | ilvC | + | lmo1986 | 4 | 2.98 | 4.08 | 3.00 | 3.96 | 3.28 |
| LMON_2058 | 2-isopropylmalate synthase | leuA | + | lmo1987 | 11 | 3.41 | 3.68 | 2.74 | 3.56 | 3.87 |
| LMON_2059 | 3-isopropylmalate dehydrogenase | leuB | + | lmo1988 | 3 | 3.18 | 3.60 | 2.86 | 3.84 | 4.72 |
| LMON_2060 | 3-isopropylmalate dehydratase large subunit | leuC | + | lmo1989 | 3 | 3.29 | 3.48 | 2.72 | 3.72 | 5.25 |
| LMON_2061 | 3-isopropylmalate dehydratase small subunit | leuD | + | lmo1990 | 3 | 3.16 | 3.64 | 2.71 | 3.96 | 5.70 |
| LMON_2062 | Threonine dehydratase | ilvA | + | lmo1991 | 3 | 3.16 | 3.65 | 2.50 | 3.86 | 5.58 |
| LMON_2234 | Hypothetical protein | lmo2158 | 12 | 3.80 | 5.25 | 3.18 | 0.77 | 3.03 | ||
| LMON_2407 | Internalin-like protein | + | lmo2396 | 3 | 1.13 | 1.52 | 0.92 | 1.30 | 2.67 | |
| LMON_2696 | Universal stress protein | + | lmo2673 | 12 | 3.09 | 3.64 | 1.90 | -0.77 | 4.24 | |
| LMON_2718 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, dihydroxyacetone binding subunit DhaK | + | lmo2695 | 12 | 2.73 | 3.82 | 2.25 | -0.43 | 2.36 | |
| LMON_2719 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, ADP-binding subunit DhaL | + | lmo2696 | 12 | 2.76 | 3.76 | 2.13 | 0.31 | 2.61 | |
| LMON_2720 | Phosphoenolpyruvate-dihydroxyacetone phosphotransferase, subunit DhaM; | + | lmo2697 | 12 | 2.46 | 3.79 | 2.13 | 0.58 | 2.82 | |
| LMON_2735 | Secreted protein | lmo2713 | 1 | 2.10 | 1.16 | 1.12 | 1.69 | 2.03 | ||
| LMON_2770 | general stress protein 26 | lmo2748 | 12 | 2.51 | 3.72 | 1.71 | -1.81 | 1.98 | ||
| LMON_2800 | PTS system, cellobiose-specific IIA component | lmo2780 | 12 | 1.79 | 1.57 | 1.45 | -0.55 | 2.57 | ||
| LMON_2801 | Beta-glucosidase | lmo2781 | 12 | 2.06 | 1.38 | 1.45 | -1.18 | 2.27 | ||
| LMON_2802 | PTS system, cellobiose-specific IIB component | lmo2782 | 12 | 2.11 | 1.49 | 1.81 | -1.48 | 2.44 | ||
| LMON_2803 | PTS system, cellobiose-specific IIC component | lmo2783 | 12 | 2.32 | 1.24 | 1.80 | -1.99 | 2.05 | ||
* indicates <0.50 cluster membership; G1–G5 refer to differential expression in L. monocytogenes cells grown at 4°C relative to 20°C and across five specific growth phases (see Fig 1); Blue shading indicates genes with significantly increased (> 1log2, p<0.05) gene expression and yellow shading indicates genes with significantly decreased (< -1log2, p<0.05) expression at 4°C relative to 20°C; Bolded values indicate significant (p<0.05) differential expression changes.
Table 3. Pathways and gene ontology processes significantly (p<0.05*) enriched among genes upregulated (>2-fold) at 4°C vs. 20°C.
| Biological process or pathway | Gene examples | p-value (# of contributing genes) | ||||
|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||
| Branched-chain amino acid synthesis | ilvABCDH, leuABCD, | 1.18E-6 (9) | 8.04E-8 (9) | 9.14E-8 (9) | 7.46E-11 (9) | 5.75E-4 (9) |
| Arginine biosynthesis | argBDGHJ, carbB | 3.42E-2 (4) | 4.77E-4 (6) | 4.25E-5 (7) | 4.37E-3(8) | |
| Glycerol degradation | LMON_0354–355, 0358, 2718–2720 | 2.87E-2 (6) | 1.41E-5 (8) | 9.82E-7 (9) | 4.06E-3 (9) | |
| Histidine biosynthesis | hisABDEGIZ | 5.58E-5 (9) | 7.20E-3 (4) | 5.99E-9 (7) | 1.16E-2 (6) | |
| Methionine biosynthesis | metX, LMON_0595, 0847 | 4.99E-3 (3) | 3.57E-2 (3) | |||
| Oxidation-reduction process (GO:0055114) | qoxABCD, gabD | 4.37E-2 (4) | 1.54E-6 (8) | 2.46E-4 (9) | ||
| Response to stress (GO:0006950) | recF, hrcA, grpE, dnaK, ltrC, uspA, csbD, ctsR, clpP, clpE | 1.96E-5 (13) | ||||
| Biological regulation (GO:0065007) | yycF, ctsR, hrcA, frvA, opuCD | 2.89E-3 (10) | ||||
| Pyrimidine ribonucleotide biosynthesis | pyrABC, carB | 1.24E-3 (4) | ||||
| Pentose phosphate pathway | LMON_2682–2685, 2697, 0349, 0350, 0352 | 1.23E-3 (13) | ||||
| Carboxylates degradation | fruA, bvrB, LMON_0095–0097, 2706–2708, 0785–0788 | 7.60E-6 (44) | ||||
| Tryptophan biosynthesis | trpBCDFG) | 2.96E-2 (5) | ||||
| Purine ribonucleotide biosynthesis | purMQDFN | 2.96E-2 (5) | ||||
| Active transporter activity (GO:0022804) | opuCD, mpoABCD, mptACD, PTS systems | 1.56E-8 (42) | ||||
| Structural constituent of ribosome (GO:0003735) | rplEFNOPVX, rpsEHNP, rpmC | 2.90E-2 (20) | ||||
* Statistical overrepresentation of gene sets were determined using Fisher’s exact test and the BioCyc database.
Other genes with increased expression in response to cold stress at all growth phases, included those encoding the L-carnitine transporter OpuC (opuCB-opuCC-opuCD), internalins A and D, a ribosomal protein (RpmF), a histidine biosynthesis protein (HisE), a GNAT family acetyltransferase (LMON_0625), and Veg protein (LMON_0187). The roles of carnitine and ribosomal proteins in bacterial adaptation to certain stresses have been previously reported [18, 54–56]. In B. subtilis Veg protein has been shown to activate extracellular matrix genes and contribute to biofilm formation [57]. In L. monocytogenes, increased expression of Veg has been observed following exposure to acid and cold stress [11, 58], but its function remains unknown.
In addition to the upregulation of inlA and inlD at all growth phases at 4°C, inlH and two genes encoding internalin-like proteins (LMON_2407, LMON_0611) were upregulated at four growth phases (Table 2). The most recognized roles of internalins A and B are in L. monocytogenes’ virulence. Nevertheless, the Listeria spp. family of internalin proteins all share a leucine-rich-repeat domain that allows them to bind structurally unrelated ligands, thereby implicating them in a wide range of functions [59]. Recent studies have shown that isolates containing a full-length version of inlA exhibit enhanced cold tolerance relative to those with a truncated version [39, 60]. Further research is needed to confirm the potential role(s) of internalins in the L. monocytogenes CSR.
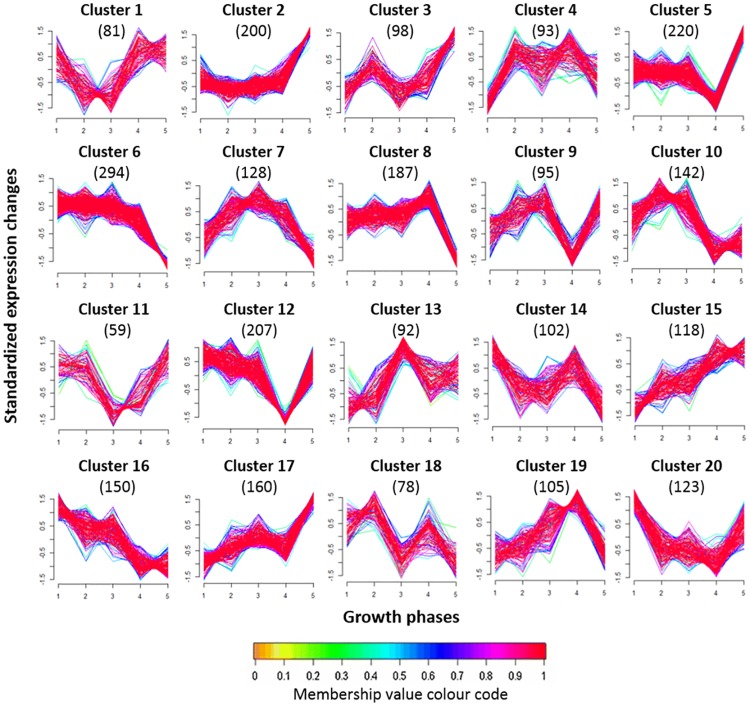
Many more genes (n = 104) were upregulated at four growth phases at 4°C relative to 20°C, with the majority being induced at G1, G2, G3, and G5 (G1–G3:G5, n = 70). These genes shared similar DE patterns across all growth phases (Fig 5. Cluster 5 and 12), where levels of DE dramatically decrease at G4. Present in this group were genes involved in arginine biosynthesis (dapE, argGHJ), general stress response (LMON_0515, LMON_2234, LMON_2696, LMON_2770), FA synthesis (LMON_0351), and phosphotransferase system (PTS) uptake of cellobiose (LMON_2800–2803) and mannose (LMON_0785–0788) (Table 2). Additionally, the genes encoding succinate-semialdehyde dehydrogenase (LMON_0920), 11 genes from the pentose phosphate pathway (LMON_0350–0360), and two sets of genes (LMON_0354–0356, LMON_2718–2720) encoding dihydroxyacetone kinase (DhaKLM) were upregulated in these growth phases (Table 2).
Fig 5. Differential gene expression patterns observed in L. monocytogenes cells grown across five growth phases at 4°C (see Fig 1).
Clusters were formed using fuzzy c-means soft clustering and standardized log2 fold change values for 4°C expression levels vs. 20°C expression levels at each growth phase. Numbers in parentheses denote the number of genes in each cluster core. Represented genes had cluster membership values >0.5. See Fig 1 for information about growth phases.
An additional 100 genes displayed induced transcription at three growth phases, with the largest number co-upregulated at G1–G2:G5 (n = 29), followed by G1–G3 (n = 24). The similarities between these sets of growth phases are visible in Fig 4. At G1–G2:G5, L. monocytogenes growth is inhibited, and such conditions would be expected to induce genes encode proteins associated with 1) secondary membrane-transport systems that are less energy demanding, 2) the utilization of alternative energy sources, and 3) cell detoxification. It appears that the demand for these proteins is further increased in cold stressed cells as we observed the upregulation of genes involved in utilizing ethanolamine as an alternative energy source (LMON_1167–1180), oligopeptides transport (oppC), transcription regulation (5 genes), general stress response (clpC), and ribosomal subunit assembly (LMON_2523). At G1–G3, several of the shared upregulated genes encoded reductases (LMON_1496, LMON_2403, LMON_0798, LMON_0643, LMON_2843) with known or putative roles in maintaining redox balance within the cell and managing oxidative stress.
Core set of genes suppressed under cold stress
A total of 42 genes were downregulated at 4°C in all growth phases, including genes involved in cold shock (cspB), virulence (actA, plcB, capA, mpl), nucleotide degradation (LMON_0129, LMON_1234, guaB), vitamin B9 and B12 synthesis (LMON_1163, LMON_2771–2772), glycerophospholipid export (LMON_0106–0107), and sugar uptake (LMON_0770–0772, LMON_1451–1452). Many of the genes suppressed at 4°C contributed to either cobalamin (B12) biosynthesis and subsequent utilization, or the production and export of cell wall components (Table 4).
Table 4. Pathways and gene ontology processes significantly (p<0.05*) enriched among genes downregulated (>2-fold) at 4°C vs. 20°C.
| Biological process or pathway | Gene examples | p-value (# of contributing genes) | ||||
|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||
| Cobalamin biosynthesis | cbiKA, cbiP | 5.82E-4 (5) | 5.14E-6 (6) | 4.01E-3 (6) | 4.33E-4 (5) | |
| Biosynthesis of cell wall macrostructures (GO:0070589) | dltABCD, pgdA, murABCE, pgi, tagD | 5.76E-4 (6) | 3.94E-4 (5) | 1.67E-3 (5) | 1.02E-4 (20) | |
| Pathogenesis | inlK, inlJ, actA, arcB, flaA, ctaP | 1.13E-2 (9) | 4.83E-3 (7) | 2.62E-2 (7) | ||
| Pyrimidine nucleotides/UMP biosynthesis | pyrCBFGHR, comEB, udk, carB | 2.60E-3 (8) | 1.64E-6 (9) | 3.03E-6 (11) | ||
| Localization (GO:0051179) | dtpT, phoU, pstB, ctaP | 1.03E-2 (13) | 2.52E-3 (10) | 4.59E-2 (12) | ||
| Transport (GO:006810) | ctpA, pstB, phoU | 1.03E-2 (13) | 2.52E-3 (10) | 4.59E-2 (12) | ||
| Carboxylate degradation | PdhABC | 1.11E-4 (21) | 3.92E-8 (43) | |||
| UDP biosynthesis | murABCE, racE, ddl | 3.32E-3 (6) | 2.74E-2 (6) | |||
| tRNA charging | metS, pheS, serS, tyrS, aspS, alaS, argS | 1.80E-2 (8) | ||||
| Myo-inositol degradation | LMON_2235, 2238–2239 | 1.54E-2 (4) | ||||
| Nucleotide degradation | pdp, dra, drm, pnp, guaB | 1.29E-2 (10) | ||||
| Biosynthesis | folD, cbiP, gatAC, comEB, hemABCD, cycEK, tagD, cdsA | 5.50E-5 (128) | ||||
| Cofactors and electron carrier biosynthesis | hemABCD, menEF, nadE, cbiP, ubiE | 2.20E-3 (40) | ||||
* Statistical overrepresentation of gene sets were determined using Fisher’s exact test and the BioCyc database.
Cobalamin derivatives function as essential cofactors for important enzymes that catalyze a variety of transmethylation and rearrangement reactions [61]. In bacteria, cobalamin is necessary to degrade ethanolamine, propanediol, glycerol, and pyruvate and to produce carbon and energy [61–63]. L. monocytogenes and other bacteria induce the transcription of both cobalamin biosynthesis and ethanolamine utilization genes under a variety of stress conditions [64–68]. However, in the present study, we observed the downregulation of cobalamin biosynthesis genes and others encoding cobalamin-dependent enzymes and pathways (eutB, LMON_1144–1154, LMON_1803, LMON_0942), whereas the ethanolamine utilization operon remained upregulated at several growth phases.
Genes involved in the biosynthesis and export of peptidoglycan, teichoic acids, cell wall proteins, and membrane lipids were also suppressed at 4°C (Table 4). Both peptidoglycan and teichoic acid biosynthesis stem from the precursor molecule UDP-N-acetyl-α-D-glucosamine (UDP-GlcNAc) and require the attachment of D-alanine residues, and for peptidoglycan also glutamate. Correspondingly, genes involved in UDP-GlcNAc biosynthesis and transport, alanine attachment (dltABCD), and glutamate transport (LMON_0849–0850) and attachment (racE) were downregulated. Other suppressed genes included several fatty-acid-coA ligases, lipid export proteins, and enoyl-acyl-carrier proteins and reductases, which collectively assist in the production and export of phospholipids, cell wall-associated lipoproteins, and lipoteichoic acids. These findings suggest a reduced rate of peptidoglycan/cell envelope turnover at 4°C relative to 20°C. This reduced turnover rate may reflect the reduced cellular growth rate at this temperature, or conservation of carbohydrates for alternative uses.
Other genes suppressed at 4°C had roles in localization and pathogenicity, heme biosynthesis, tRNA processing and charging, nucleotide degradation and salvage, and carbohydrate transport and catabolism (Table 4). Not surprising, many of these genes also have alternative roles in cell wall biogenesis. Notably, over 50 genes from PTS operons were downregulated at one or more growth phases, far exceeding the number of PTS genes upregulated in response to cold stress. PTSs utilize phosphate to facilitate the uptake of simple sugars and thus consume more energy than other membrane kinases with the same sugar specificity [69]. This suggests that L. monocytogenes may benefit from employing alternative sugar uptake systems when exposed to cold stress to conserve energy for more critical CSR mechanisms.
L. monocytogenes cold-stress response at individual growth phases
Early lag phase (G1)
Previous studies have described the lag phase following cold stress as the acclimatization phase. In this phase, bacteria suppress bulk protein synthesis, and dramatically increase production of transient cold induced stress response proteins (CIPs) [8]. Once the cells are cold adapted, CIP synthesis ends and bulk protein synthesis and cell growth resume. Depending on the degree of cold stress, the types and expression levels of CIPs will differ. Some of the functions associated with CIPs include DNA unwinding, nucleic acid stabilization, and compatible solute uptake [70, 71].
A total of 96 genes were uniquely induced at G1, accounting for 25% of the genes upregulated at this growth phase. Most of these genes had DE patterns that belonged to clusters 14, 16, or 20 (Fig 5), and a large number encoded transcription regulators (n = 19), including HrcA, CtsR, DegU, and YycF. Both HrcA and CtsR are negative regulators of several heat-shock and general stress-response genes, a number of which were also upregulated at G1 (grpE, dnaK, clpB, gmpA, mscAB, clpC, clpE, and clpP). Many of these upregulated genes, including groEL and groES, encode chaperone proteases capable of degrading the misfolded or damaged proteins that would probably occur following a rapid temperature downshift. By contrast, the transcription regulator DegU contributes to motility, growth at high temperatures, and efficient biofilm formation [72]. Co-transcribed with degU is yviA, which encodes a DegV family protein with putative functions in FA transport or metabolism [72, 73]. As we did not observe upregulation of genes known to be activated by DegU, the role of this operon in early cold adaptation may be primarily associated with the functions of YviA. In support of this hypothesis, YycF, a transcription regulator of FA biosynthesis and cell wall metabolism genes [74], was also upregulated at G1.
Additional genes upregulated at G1 encode proteins involved in DNA repair (recF, gyrAB, recX, LMON_0230, LMON_1225), ethanolamine utilization (eutABCM, LMON_1166–1180), carnitine uptake (opuCABCD), phosphate transport and metabolism (pstABCS, phoU, phoB), RNA stability (LMON_0869, LMON_1037), nitrogen utilization (LMON_1582–1583), and histidine and arginine biosynthesis. Two genes with >20-fold increased expression at G1 encode succinate-semialdehyde dehydrogenase [NAD] (LMON_0920) and pyruvate phosphate dikinase (LMON_1936) (Table 5). These proteins function in energy production, and glycerol metabolism, respectively.
Table 5. Top 10 most highly induced genes in L. monocytogenes cells at 4°C vs 20°C at five different growth phases.
| EGD ORF | EGD description | Gene abbr. | Strand | EGD-e ORF | Cluster memb-ership | Log2 DE across five growth phases | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||||||
| G1 –early lag phase | ||||||||||
| LMON_0762 | Hypothetical protein | + | lmo0758 | 20 | 3.53 | -0.35 | 0.48 | -1.47 | 1.75 | |
| LMON_0763 | glyoxalase family protein | + | lmo0759 | 20 | 3.55 | 0.04 | 1.28 | -0.61 | 1.78 | |
| LMON_0765 | Nitrilotriacetate monooxygenase component B | + | lmo0761 | 20 | 3.66 | 0.53 | 1.49 | 0.70 | 1.99 | |
| LMON_0920 | Succinate-semialdehyde dehydrogenase [NAD] | + | lmo0913 | 10 | 4.64 | 5.74 | 2.96 | 0.40 | 2.35 | |
| LMON_1761 | Hypothetical protein | + | lmo1694 | 10 | 3.78 | 5.06 | 2.55 | -0.95 | 0.67 | |
| LMON_1936 | Pyruvate,phosphate dikinase | + | lmo1867 | 20 | 4.31 | 2.17 | 1.90 | 0.47 | 2.16 | |
| LMON_2054 | Dihydroxy-acid dehydratase | ilvD | + | lmo1983 | 4* | 3.52 | 3.89 | 3.34 | 4.29 | 3.62 |
| LMON_2055 | Acetolactate synthase large subunit | ilvB | + | lmo1984 | 3/15 | 3.67 | 4.17 | 3.41 | 4.41 | 4.37 |
| LMON_2056 | Acetolactate synthase small subunit | ilvH | + | lmo1985 | 15/1 | 3.67 | 4.02 | 3.39 | 4.41 | 4.03 |
| LMON_2234 | Hypothetical protein | lmo2158 | 12 | 3.80 | 5.25 | 3.18 | 0.77 | 3.03 | ||
| G2 –transition to exponential growth phase | ||||||||||
| LMON_0020 | Hypothetical protein | lmo0019 | 10 | 2.62 | 5.25 | 3.85 | 0.65 | 1.64 | ||
| LMON_0920 | Succinate-semialdehyde dehydrogenase [NAD] | + | lmo0913 | 10 | 4.64 | 5.74 | 2.96 | 0.40 | 2.35 | |
| LMON_1003 | Hypothetical protein | lmo0994 | 12 | 3.09 | 5.04 | 2.48 | -0.56 | 2.59 | ||
| LMON_1488 | L-carnitine/choline ABC transporter, permease protein | opuCD | lmo1425 | 10 | 2.34 | 5.37 | 3.52 | 1.79 | 1.29 | |
| LMON_1489 | L-carnitine/choline ABC transporter, substrate-binding protein | opuCC | lmo1426 | 10 | 2.45 | 5.23 | 3.78 | 1.82 | 1.24 | |
| LMON_1490 | L-carnitine/choline ABC transporter, permease protein | opuCB | lmo1427 | 10 | 2.51 | 5.28 | 3.78 | 1.58 | 1.10 | |
| LMON_1491 | L-carnitine/choline ABC transporter, ATP-binding protein | opuCA | lmo1428 | 10 | 2.63 | 5.54 | 3.94 | 1.30 | -0.01 | |
| LMON_1761 | Hypothetical protein | + | lmo1694 | 10 | 3.78 | 5.06 | 2.55 | -0.95 | 0.67 | |
| LMON_2234 | Hypothetical protein | lmo2158 | 12 | 3.80 | 5.25 | 3.18 | 0.77 | 3.03 | ||
| LMON_2306 | Arsenate reductase | + | lmo2230 | 10 | 3.36 | 4.77 | 2.95 | -1.12 | 1.49 | |
| G3 –mid-exponential growth phase | ||||||||||
| LMON_0369 | Twin-arginine translocation protein | tatC | lmo0361 | 13 | -0.08 | 0.57 | 4.35 | 0.25 | 3.49 | |
| LMON_0370 | Twin-arginine translocation protein | tatA | lmo0362 | 13 | -0.06 | 0.94 | 5.30 | 0.65 | 3.26 | |
| LMON_0373 | Ferrous iron transport permease | efeU | + | lmo0365 | 13 | -0.66 | 0.87 | 4.80 | 2.03 | 3.72 |
| LMON_0374 | Ferrous iron transport periplasmic protein | efeO | + | lmo0366 | 13 | -1.18 | 0.87 | 4.60 | 1.12 | 3.74 |
| LMON_0375 | Ferrous iron transport peroxidase | efeB | + | lmo0367 | 13 | -1.08 | 1.04 | 4.55 | 1.33 | 3.74 |
| LMON_0541 | ABC transporter, substrate-binding protein | lmo0541 | 13 | -0.95 | -0.08 | 4.34 | 0.95 | 2.00 | ||
| LMON_0944 | Hypothetical protein | 10 | 2.77 | 3.84 | 4.22 | 1.39 | 2.00 | |||
| LMON_1016 | Hypothetical protein | + | lmo1007 | 13 | 0.17 | 1.14 | 4.89 | 1.41 | 3.19 | |
| LMON_2261 | Cell surface protein IsdA, transfers heme from hemoglobin to apo-IsdC | isdA | lmo2185 | 13 | -1.77 | -0.61 | 4.21 | 0.36 | 3.24 | |
| LMON_2262 | NPQTN cell wall anchored protein | isdC | lmo2186 | 13 | -1.50 | -0.56 | 4.38 | 0.95 | 4.24 | |
| G4 –transition to stationary phase | ||||||||||
| LMON_1312 | Membrane protein | lmo1252 | 19/15 | -0.07 | 0.15 | -0.01 | 4.16 | 1.28 | ||
| LMON_2054 | Dihydroxy-acid dehydratase | ilvD | + | lmo1983 | 4* | 3.52 | 3.89 | 3.34 | 4.29 | 3.62 |
| LMON_2055 | Acetolactate synthase large subunit | ilvB | + | lmo1984 | 3/15 | 3.67 | 4.17 | 3.41 | 4.41 | 4.37 |
| LMON_2056 | Acetolactate synthase small subunit | ilvH | + | lmo1985 | 15/1 | 3.67 | 4.02 | 3.39 | 4.41 | 4.03 |
| LMON_2057 | Ketol-acid reductoisomerase | ilvC | + | lmo1986 | 4 | 2.98 | 4.08 | 3.00 | 3.96 | 3.28 |
| LMON_2059 | 3-isopropylmalate dehydrogenase | leuB | + | lmo1988 | 3 | 3.18 | 3.60 | 2.86 | 3.84 | 4.72 |
| LMON_2061 | 3-isopropylmalate dehydratase small subunit | leuD | + | lmo1990 | 3 | 3.16 | 3.64 | 2.71 | 3.96 | 5.70 |
| LMON_2062 | Threonine dehydratase | ilvA | + | lmo1991 | 3 | 3.16 | 3.65 | 2.50 | 3.86 | 5.58 |
| LMON_2286 | Hypothetical protein | lmo2210 | 19 | 1.59 | 0.71 | 2.28 | 3.79 | 2.33 | ||
| LMON_2534 | Cell wall-binding protein | lmo2522 | 4 | -1.33 | 2.20 | 0.95 | 4.50 | 1.70 | ||
| G5 –late-stationary phase | ||||||||||
| LMON_0740 | D-allulose-6-phosphate 3-epimerase | + | lmo0735 | 2 | -0.81 | -1.52 | -2.11 | -1.19 | 10.05 | |
| LMON_0741 | D-allose-6-phosphate isomerase | + | lmo0736 | 2 | -0.52 | -1.36 | -1.52 | -0.35 | 10.41 | |
| LMON_0742 | Hypothetical protein | + | lmo0737 | 2 | 0.04 | -0.98 | -1.86 | 1.08 | 10.49 | |
| LMON_0743 | PTS system, D-allose-specific | + | lmo0738 | 2 | -0.61 | -0.29 | -0.59 | 1.52 | 9.56 | |
| LMON_0744 | 6-phospho-beta-glucosidase | + | lmo0739 | 2 | -0.85 | -0.15 | -0.41 | 0.86 | 8.78 | |
| LMON_2673 | PTS system, lactose/cellobiose specific IIB subunit | lmo2650 | 2 | -2.19 | -3.61 | -1.53 | -1.74 | 8.51 | ||
| LMON_2674 | PTS system, IIA component | lmo2651 | 2 | -2.41 | -4.12 | -1.54 | -1.80 | 9.19 | ||
| LMON_2706 | PTS system, cellobiose-specific IIB component | + | lmo2683 | 17 | -1.46 | -1.30 | 1.32 | -2.04 | 10.66 | |
| LMON_2707 | PTS system, cellobiose-specific IIC component | + | lmo2684 | 17 | -1.57 | -1.40 | 1.44 | -2.83 | 9.36 | |
| LMON_2708 | PTS system, beta-glucoside/cellobiose-specific IIA component | + | lmo2685 | 17 | -1.38 | -1.28 | 1.33 | -2.43 | 9.28 | |
* indicates <0.50 cluster membership; G1–G5 refer to differential expression in L. monocytogenes cells grown at 4°C relative to 20°C and across five specific growth phases (see Fig 1); Blue shading indicates genes with significantly increased gene expression (> 1 log2, p<0.05) and yellow shading indicates genes with significantly decreased expression (< -1 log2, p<0.05) at 4°C relative to 20°C; Bolded values indicate significant (p<0.05) differential expression changes. Genes shaded in yellow are highly expressed at more than one growth phase.
Twenty-nine genes were solely induced at G1 and G2, highlighting their importance in the earlier CSR stages of L. monocytogenes. Included among these genes were those encoding the low temperature requirement protein LtrC, cold-shock protein CspL, regulatory protein RecX, and several proteins associated with phosphate transport (PstCABB-PhoU-PstS).
Forty-seven genes were upregulated at both G1 and G5 (late-stationary phase), many of which followed the DE pattern shown in cluster 16 (Fig 5). As discussed, these growth phases both represent stages in cold stress survival, where the population size is static and their induced genes are probably specific to surviving adverse conditions. Among these genes were those encoding heat-shock proteins (clpP, clpE, groEL, LMON_2710–2711), DNA gyrase (gyrA), an SOS-response protein (lexA), an RNA helicase (secA), and seven transcription regulators. Interestingly, although these genes were upregulated in response to cold stress in this study, several of them were also reported by van der Veen et al. [75] as induced in L. monocytogenes following 3 min of heat stress (48°C). As both stresses are caused by a rapid change temperature which causes molecular structures to take on different conformations; it makes sense they would share similarly upregulated genes with functions in DNA and RNA repair (gyrA, lexA, secA), and protein degradation (clpP, clpE) or stabilization (groEL).
Transition to exponential growth phase (G2)
At T2, cells have had an adequate amount of time to repair cold-induced damage to molecular structures, and to reprogram their cellular machinery to support growth at low temperatures. As T2 shares characteristics with both the lag and exponential phases, only 22 genes were uniquely upregulated at G2, and only 21 genes were uniquely downregulated. The exclusively induced genes encoded the iron binding protein, Fri; a cold-shock protein, CspD; and a DNA repair and SOS response protein, RecA. Most of these genes exhibited slightly less than a 2-fold increase in expression at G1, maximum increased expression at G2, and no change in expression or decreased expression from G3–G5, as is characteristic of cluster 10 (Fig 5). The upregulation of fri, cspD, and recA have been previously reported in L. monocytogenes in response to cold stress [16, 26, 27, 50].
Among the genes upregulated at G2, there was a significant (p<0.05) overrepresentation of those involved in glycerol metabolism and the synthesis of arginine, histidine, and BCAAs (Table 3). Among the most induced genes (>36 fold) were those encoding the OpuC carnitine uptake system (opuCABCD), hypothetical proteins, and succinate-semialdehyde dehydrogenase (LMON_0920) which was also strongly induced at G1 (Table 5).
Mid-exponential growth phase (G3)
A total of 47 genes were uniquely induced during exponential growth at 4°C, with most genes exhibiting the DE pattern shown in cluster 13 (Fig 5). Aside from a few genes with roles in iron transport (fhuC, fhuB, LMON_2440–2441) and oxidoreductase activity (LMON_0560, LMON_2031, pdhD), most are not known to share any common functions. Among G3-upregulated genes, there was an overrepresentation (p<0.05) of genes involved in glycerol metabolism, aerobic respiration, iron transport, and the biosynthesis of arginine, histidine, and BCAAs (Table 3). Other upregulated genes included those involved in the pentose phosphate pathway (LMON_0349–0352, LMON_0647), osmolyte uptake (opuCABCD), and 12 genes fell within PTS operons that facilitate the uptake of mannose, cellobiose, and beta-glucoside. Genes highly induced at G3 (>16-fold) were associated with iron (efeUOB) and heme transport (isdCAEF), as well as protein export (tatAC) (Table 5). As expected, many of the proteins previously induced during cold acclimatization (G1–G2) were downregulated during exponential growth.
Transition to stationary phase (G4)
Upon entry into stationary phase, cold-adapted cells downregulated almost 3× more genes than they upregulated (Fig 3). Unique to G4 was the upregulation of 53 genes with roles in glycine betaine transport (gbuAB), tRNA processing (LMON_T39, T60, T66), nucleotide biosynthesis (pyrP, pyrB, pyrAa, pyrR, LMON_1838), and maintaining a rod cell shape (mreBD), among others. Most of these genes displayed DE patterns that belonged to clusters 8 or 19 (Fig 5), in which DE peaks at G4 and falls dramatically at G5. The five most strongly induced genes (>16-fold) at this growth phase encoded three isoleucine biosynthesis proteins (ilvDBH) and two cell wall-associated proteins (LMON_2534, LMON_1312) (Table 5).
G4 and G5 also shared a set of exclusively upregulated genes (n = 23) with functions in nucleotide biosynthesis (purAHNFE, pyrDII, carB, pyrC), tRNA processing (LMON_T51), and cell-wall recycling (LMON_0029, LMON_2776), among others. All genes exhibited patterns of DE that belonged to cluster 1 or 15 (Fig 5). Nucleotide biosynthesis genes are likely activated at these growth phases to accommodate the increased demand for ribonucleic acids imposed by the large abundance of strongly upregulated genes at G5.
Late-stationary phase (G5)
Cold-adapted stationary-phase cells exhibited both the largest number of DE expressed genes and the largest magnitude of expression changes (Fig 3). Similar results were noted by Chan et al. [11] in their L. monocytogenes microarray study, which compared the cold-stress regulons of exponential and stationary-phase cells. More than 50% of G5-upregulated genes exhibited >4-fold increased expression and 17 genes, predominantly associated with PTS mediated sugar uptake systems, were upregulated between 100- and >1,000-fold. Many of the genes strongly induced by cold stress at this growth phase, also exhibited some of the highest expression levels in stationary-phase cells at 20°C, highlighting their overall importance during this growth phase and their enhanced importance in cold-adapted stationary-phase cells. Additionally, 50–60% of G5 genes were exclusively DE at this growth phase. The exclusively induced genes had roles in tryptophan (trpBCDFG, aroE) and ATP synthesis (atpACDFGH, adk, LMON_0091), creatine and rhamnose degradation, and ribosome and phage-related processes. Other genes encoded motility-associated proteins (motB, cheV, flaA), an osmosensitive K+ channel histidine kinase (kdpCDE), an Na(+) H(+) antiporter (LMON_2393–2396), the transcription repressor CodY, and low temperature requirement protein B (ltrB). Many genes associated with transcription and translation were also strongly induced at G5 (polC, dnaC, dnaI, topB, rbfA, mfd), including 28 transcription regulators, and 14 genes involved in tRNA processing (S1 Table).
Biological processes enriched (p<0.05) among the genes induced at G5 included arginine, histidine, ribonucleotide, and BCAA biosynthesis genes, as well as genes associated with carbohydrate uptake, the pentose phosphate pathway, glycerol and peptidoglycan degradation, and aerobic respiration (Table 3). The DE patterns shown in clusters 1, 2, 3, 5, 9, 11, 12, 15, 17, and 20 all describe genes with increased expression in cold-adapted stationary-phase cells, while clusters 4, 6–8, 10, 14, 16, 18, and 19 describe genes with limited or no roles in stationary-phase viability at low temperatures (Fig 5).
L. monocytogenes cold stress regulon: A comparison with previous studies
Numerous previous studies have elucidated a large pool of genes with putative or known roles in the L. monocytogenes CSR [11–13, 16, 20, 27]. In this section, we will determine whether these genes are expressed in a cold-tolerant strain, whether a significant induction occurs when the downshift is from 20°C to 4°C (rather than from 37°C, as in most prior studies), and whether any growth-phase dependencies exist.
Osmolyte and oligopeptide uptake
In both L. monocytogenes and B. subtilis, low temperature and high osmolarity stress induce the intracellular accumulation of solutes and short peptides. These solutes and peptides function as osmoprotectants that facilitate cell growth under stressful conditions [18, 26, 54, 76–79]. In L. monocytogenes, carnitine and glycine betaine are the predominant solutes that accumulate [78–80], and their transport systems are encoded by the opuCABCD and gbuABC operons, respectively. Chan et al. [11] reported induction of opuCABCD and gbuC in cold-adapted L. monocytogenes exponential- but not stationary-phase cells, while Durack et al. [12] did not find that either system was induced in late exponential early stationary-phase cells. In the present study, opuCBCD was upregulated in all growth phases, while opuCA was upregulated at G1–G4 (Table 6). Regardless, all genes in the OpuC operon belonged to the DE pattern shown in cluster 10 (Fig 5), exhibiting the highest level of induction at G2 (up to 48-fold) and the lowest level at G5, which agrees with previous work [11, 12]. Similar to the findings of Chan et al. [11] gbuA and gbuB were only upregulated at G4 (Table 6).
Table 6. Genes commonly associated with the L. monocytogenes CSR.
| EGD ORF | EGD description | Gene abbr. | Strand | EGD-e ORF | Cluster memb-ership | Log2 DE across five growth phases | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||||||
| Osmolyte and oligopeptide uptake | ||||||||||
| LMON_1024 | Glycine betaine ABC transport system, ATP-binding protein | gbuA | + | lmo1014 | 8 | 0.15 | 0.56 | 0.82 | 1.68 | -0.69 |
| LMON_1025 | Glycine betaine ABC transport system, permease protein | gbuB | + | lmo1015 | 19 | 0.04 | 0.48 | 0.75 | 1.14 | 0.08 |
| LMON_1026 | Glycine betaine ABC transport system, glycine betaine-binding protein | gbuC | + | lmo1016 | 15 | 0.10 | 0.54 | 0.64 | 0.87 | 0.71 |
| LMON_1488 | Osmotically activated L-carnitine/choline ABC transporter, permease protein OpuCD | opuCD | − | lmo1425 | 10 | 2.34 | 5.37 | 3.52 | 1.79 | 1.29 |
| LMON_1489 | Osmotically activated L-carnitine/choline ABC transporter, substrate-binding protein OpuCC | opuCC | − | lmo1426 | 10 | 2.45 | 5.23 | 3.78 | 1.82 | 1.24 |
| LMON_1490 | Osmotically activated L-carnitine/choline ABC transporter, permease protein OpuCB | opuCB | − | lmo1427 | 10 | 2.51 | 5.28 | 3.78 | 1.58 | 1.10 |
| LMON_1491 | Osmotically activated L-carnitine/choline ABC transporter, ATP-binding protein OpuCA | opuCA | − | lmo1428 | 10 | 2.63 | 5.54 | 3.94 | 1.30 | -0.01 |
| LMON_2268 | Oligopeptide transport ATP-binding protein OppF | oppF | − | lmo2192 | 2 | -0.56 | -0.61 | -0.43 | -0.39 | 1.39 |
| LMON_2269 | Oligopeptide transport ATP-binding protein OppD | oppD | − | lmo2193 | 2 | -0.59 | -0.74 | -0.38 | -0.30 | 0.91 |
| LMON_2270 | Oligopeptide transport system permease protein OppC | oppC | − | lmo2194 | 15* | -0.66 | -0.91 | -0.35 | -0.15 | -0.29 |
| LMON_2271 | Oligopeptide transport system permease protein OppB | oppB | − | lmo2195 | 19 | -0.61 | -0.85 | -0.35 | 0.09 | -0.79 |
| LMON_2272 | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA | oppA | − | lmo2196 | 9 | -0.67 | 0.00 | -0.46 | -1.44 | -0.17 |
| RNA and DNA repair | ||||||||||
| LMON_0004 | Hypothetical protein | + | lmo0004 | 16 | 1.25 | 0.69 | 0.54 | -0.29 | -0.69 | |
| LMON_0005 | DNA recombination and repair protein RecF | recF | + | lmo0005 | 16 | 1.40 | 0.74 | 0.59 | 0.06 | -0.15 |
| LMON_0006 | DNA gyrase subunit B | gyrB | + | lmo0006 | 20 | 1.24 | 0.53 | 0.49 | -0.15 | 0.45 |
| LMON_0007 | DNA gyrase subunit A | gyrA | + | lmo0007 | 20 | 1.17 | 0.61 | 0.40 | -0.10 | 1.15 |
| LMON_0846 | Endoribonuclease L-PSP | + | lmo0844 | 7/6/10 | 0.53 | 1.18 | 1.11 | 0.24 | -0.41 | |
| LMON_0848 | Excinuclease ABC subunit C | + | lmo0846 | 15 | -0.44 | -0.04 | 0.15 | 1.76 | 1.12 | |
| LMON_0869 | Cold-shock DEAD-box protein A | + | lmo0866 | 14 | 1.97 | 0.85 | 0.38 | 1.87 | 0.45 | |
| LMON_1037 | Ribonuclease J1 (endonuclease and 5' exonuclease) | − | lmo1027 | 16 | 1.71 | 0.33 | -0.03 | -0.10 | -1.17 | |
| LMON_1225 | DNA polymerase X family | + | lmo1231 | 2 | 1.12 | 0.22 | 0.42 | 0.76 | 2.10 | |
| LMON_1305 | ATP-dependent RNA helicase YxiN | + | lmo1246 | 6 | 0.99 | 0.75 | 0.56 | 0.07 | -1.04 | |
| LMON_1317 | Hypothetical protein | + | lmo1256 | 12 | 2.04 | 1.68 | 1.00 | -1.20 | 2.14 | |
| LMON_1336 | DNA topoisomerase I | topA | + | lmo1275 | 4 | 0.02 | 0.74 | 0.30 | 1.00 | 0.31 |
| LMON_1348 | Topoisomerase IV subunit A | parC | + | lmo1287 | 2 | -0.02 | -0.06 | 0.19 | 0.10 | 1.39 |
| LMON_1461 | RecA protein | recA | + | lmo1398 | 12 | 0.73 | 1.27 | 0.34 | -0.27 | 0.46 |
| LMON_1513 | ATP-dependent RNA helicase YqfR | − | lmo1450 | 6 | 1.33 | 1.12 | 1.18 | 1.01 | -1.04 | |
| LMON_1787 | ATP-dependent RNA helicase YfmL | − | lmo1722 | 6 | 1.69 | 1.45 | 1.28 | 1.21 | 0.47 | |
| LMON_2342 | ATP-dependent nuclease, subunit A | − | lmo2267 | 17 | -0.99 | -0.77 | -0.35 | -0.62 | 1.77 | |
| LMON_2343 | ATP-dependent nuclease, subunit B | addB | − | lmo2268 | 17 | -0.78 | -0.45 | -0.11 | -0.60 | 1.42 |
| LMON_2778 | DNA topoisomerase III | topB | − | lmo2756 | 17 | -0.25 | -0.03 | 0.19 | -0.20 | 1.40 |
| LMON_2812 | DNA-binding protein | lmo2792 | 5 | 0.62 | 1.34 | 0.75 | -0.80 | 2.49 | ||
| Regulatory elements | ||||||||||
| LMON_0198 | Virulence regulatory factor PrfA | prfA | − | lmo0200 | 17 | -2.04 | -0.44 | 1.01 | 0.10 | 5.32 |
| LMON_0229 | Transcriptional regulator CtsR | ctsR | + | lmo0229 | 16 | 1.90 | 0.42 | 0.47 | -0.55 | 0.16 |
| LMON_0244 | RNA polymerase sigma factor SigH | sigH | + | lmo0243 | 7 | -1.29 | -0.60 | -0.49 | -0.59 | -2.35 |
| LMON_0431 | RNA polymerase sigma factor SigC | sigC | − | lmo0423 | 13 | -2.03 | -1.48 | -0.59 | -0.62 | -0.97 |
| LMON_0679 | Motility gene repressor MogR | mogR | − | lmo0674 | 16 | 0.62 | 0.02 | 0.03 | -1.20 | -0.72 |
| LMON_0900 | RNA polymerase sigma factor SigB | sigB | + | lmo0895 | 10 | 0.00 | 0.58 | 0.19 | -0.78 | -0.62 |
| LMON_1166 | Ethanolamine sensory transduction histidine kinase | + | lmo1173 | 5/2 | 1.03 | 0.71 | -0.03 | 0.06 | 2.43 | |
| LMON_1341 | GTP-sensing transcriptional pleiotropic repressor codY | codY | + | lmo1280 | 3 | -0.42 | 0.61 | 0.02 | 0.09 | 1.27 |
| LMON_1517 | RNA polymerase sigma factor RpoD | rpoD | − | lmo1454 | 5 | 0.08 | -0.24 | -0.60 | -0.88 | 0.54 |
| LMON_1539 | Heat-inducible transcription repressor HrcA | hrcA | − | lmo1475 | 16 | 1.42 | -0.02 | 0.10 | -0.28 | -0.43 |
| LMON_1807 | Two-component sensor histidine kinase BceS | cesK | − | lmo1741 | 2 | -0.17 | -0.17 | 0.02 | 0.03 | 2.05 |
| LMON_1811 | Two-component response regulator YvcP (VirR in L. monocytogenes 10403S) | virR | − | lmo1745 | 17 | -0.48 | -0.61 | -0.12 | -0.30 | 0.02 |
| LMON_2432 | Sensor histidine kinase | virS | − | lmo2421 | 14 | 1.27 | -0.03 | 0.28 | 0.89 | 0.17 |
| LMON_2472 | RNA polymerase sigma factor SigL | sigL | − | lmo2461 | 5 | -0.20 | -0.05 | -0.10 | -0.42 | 0.85 |
| LMON_2527 | Transcriptional regulator DegU | degU | − | lmo2515 | 16 | 1.61 | 0.62 | 0.54 | 0.04 | -0.94 |
| LMON_2702 | Osmosensitive K+ channel histidine kinase KdpD | kdpD | − | lmo2679 | 2 | 0.99 | 0.46 | 0.60 | 0.44 | 4.22 |
| Ribosome functions | ||||||||||
| LMON_0209 | LSU ribosomal protein L25p | ctc | + | lmo0211 | 10 | 2.16 | 3.43 | 2.72 | 0.87 | 1.58 |
| LMON_0486 | LSU ribosomal protein L32p | rpmF | + | lmo0486 | 19 | 1.43 | 1.46 | 1.76 | 1.75 | 1.22 |
| LMON_1388 | Translation initiation factor 2 | infB | + | lmo1325 | 2 | 0.55 | 0.09 | -0.01 | 0.39 | 1.16 |
| LMON_1390 | Ribosome-binding factor A | rbfA | + | lmo1327 | 2 | 0.59 | 0.16 | -0.03 | 0.25 | 1.75 |
| LMON_2523 | Ribosomal subunit interface protein | − | lmo2511 | 5 | 1.08 | 1.32 | 0.65 | -0.90 | 3.14 | |
| Cold-stress proteins | ||||||||||
| LMON_1427 | Cold-shock protein | cspL | + | lmo1364 | 16 | 1.58 | 1.34 | 0.45 | 0.12 | 0.27 |
| LMON_1947 | Cold shock protein CspD | cspD | + | lmo1879 | 12 | 0.96 | 1.00 | -0.09 | -3.93 | -1.96 |
| LMON_2087 | Cold shock protein CspB | cspB | − | lmo2016 | 16 | -3.67 | -4.42 | -5.85 | -7.83 | -2.97 |
| Additional proteins | ||||||||||
| LMON_0213 | Low temperature requirement B protein | ltrB | + | lmo0215 | 2 | 0.03 | -0.26 | -0.25 | 0.00 | 1.78 |
| LMON_0398 | Low temperature requirement protein A | ltrA | − | lmo0389 | 12* | 0.43 | -0.23 | 0.61 | -0.63 | 0.08 |
| LMON_0691 | Flagellar motor rotation protein MotB | motB | + | lmo0686 | 17 | -0.81 | -0.37 | 0.07 | -0.26 | 1.44 |
| LMON_0693 | Glycosyl transferase | gmaR | + | lmo0688 | 17 | -0.84 | -0.42 | -0.06 | -0.44 | 1.29 |
| LMON_0694 | Chemotaxis protein CheV | cheV | + | lmo0689 | 17 | -0.77 | -0.04 | -0.19 | -0.65 | 2.37 |
| LMON_0695 | Flagellin protein FlaA | flaA | + | lmo0690 | 5 | 0.58 | -1.03 | -0.70 | -2.00 | 1.16 |
| LMON_0950 | Non-specific DNA-binding protein Dps / Iron-binding ferritin-like antioxidant protein / Ferroxidase | fri | + | lmo0943 | 10 | 0.39 | 1.50 | 0.03 | -0.51 | -0.22 |
| LMON_2409 | Low temperature requirement C protein | ltrC | + | lmo2398 | 12 | 1.14 | 1.21 | 0.75 | -1.61 | 0.16 |
* indicates <0.50 cluster membership; G1–G5 refer to differential expression in L. monocytogenes cells grown at 4°C relative to 20°C and across five specific growth phases (see Fig 1); Blue shading indicates genes with significantly increased (> 1log2, p<0.05) gene expression and yellow shading indicates genes with significantly decreased (< -1log2, p<0.05) expression at 4°C relative to 20°C; Bolded values indicate significant (p<0.05) differential expression changes.
Oligopeptide uptake in L. monocytogenes is mediated by the membrane permease OppA (oppABCDF) which transports peptides containing up to eight residues [23]. This transporter appears to contribute to L. monocytogenes cold tolerance as an oppA null mutant displayed reduced growth at 5°C in BHI medium [23]. Previously, Durack et al. [12] observed upregulation of oppBCF in late exponential early stationary-phase cells at 4°C while Chan et al. [11] found that only oppA levels were elevated in stationary-phase cells. Contrary to these reports, we did not observe any notable DE of the opp operon, except for a 2.6-fold induction of oppF at G5 (Table 6). While the operon may be necessary for low temperature growth, it does not appear to have increased transcription at 4°C relative to 20°C in our cold tolerant strain.
RNA and DNA repair
At low temperatures, both DNA replication and transcription are hindered by cold-induced changes in nucleic acid structures. L. monocytogenes appears to respond to these challenges by upregulating genes encoding topoisomerases and DNA gyrases, which help maintain the superhelical tension of DNA; RNA helicases, which unwind secondary RNA structures; and exo- and endonucleases, which function in nucleic acid repair [11, 12, 15]. Markkula et al. [15] reported the upregulation of four DEAD-box RNA helicase genes (lmo0866, lmo1246, lmo1450, lmo1722) in L. monocytogenes for up to 7 h following a downshift from 37°C to 5°C. They also found that, compared with the wildtype strain, null mutants of lmo0866, lmo1450, and lmo1722 displayed restricted growth at 3°C. Similarly, we observed upregulation of lmo1450 (LMON_1513) and lmo1722 (LMON_1787) at G1–G4, and lmo0866 (LMON_0869) at G1:G4 (Table 6). Also in agreement with Markkula et al. [15], lmo1246 (LMON_1305) appeared to be the least important of the four genes, with <2-fold induced expression at G1–G3.
Several DNA gyrase and topoisomerase genes were also upregulated in our study. Notably, topA was upregulated at G4, and topB and parC were upregulated at G5 (Table 6). In L. monocytogenes, the DNA gyrase genes gyrB and gyrA are in a four-gene operon, along with a gene encoding a hypothetical protein, and recF, a DNA recombination and repair protein. All four genes were upregulated at G1, and gyrA was also induced at G5. Upregulation of another DNA recombination and repair protein, recA, was also observed at G2.
Among the nucleic acid repair-proteins induced in this study were ribonuclease J1 (LMON_1037) at G1, an endoribonuclease (LMON_0846) at G2–G3, two nuclease subunits (LMON_2342–2343) at G5, and an excinuclease (LMON_0848) at G4–G5 (Table 6). Additionally, an X family DNA polymerase (LMON_1225) was induced at G1:G5. These polymerases synthesize unusual DNA structures that may serve as indicators of the induction of DNA repair [81–83]. A DNA-binding protein (LMON_2812) with a putative role in oxidative-damage protection [84] was also upregulated at G2 and G5. Lastly, a hypothetical protein (LMON_1317) containing a nudix hydrolase domain was upregulated at G1–G3:G5 (Table 6). Some members of this protein family are known to degrade oxidatively damaged nucleoside di- and triphosphates, while other members control the levels of metabolic intermediates and signaling compounds [85].
Overall, genes associated with RNA and DNA repair were predominantly cold-induced in lag- and stationary-phase cells (G1 and G5). This is expected given that cold-induced damage to nucleic acids is highest directly following cold stress, and nutrient-depleted stationary-phase cultures experience higher mutation rates and increased levels of oxidative damage [86–89].
Regulatory elements
Many mutant characterization and transcriptome studies have evaluated the roles of alternative sigma factors (σB, σC, σH, σL), two-component regulatory systems (TCRSs), and negative regulators in the L. monocytogenes CSR. Of the four alternative sigma factors, σB has been the most extensively studied. It positively regulates over 100 genes when the organism enters stationary phase or is subjected to environmental stresses including low pH, high salt, or carbon starvation [90–94]. Although induced expression of sigB has been observed in L. monocytogenes up to 12 h following cold stress [11, 17, 19], the σB regulon is predominantly downregulated during cold stress [11, 12], demonstrating that increased sigB expression does not necessarily correlate with increased σB activity. Furthermore, compared with wildtype strains, sigB null mutants show little to no difference in cold tolerance [16, 19, 20, 95], indicating that sigB is not critical for cold-stress survival. In the present study, the DE pattern of sigB followed that of cluster 10 (Fig 5), with a maximum 1.5-fold increase in expression at G2 (Table 6). Similar expression patterns were observed for three additional sigB operon genes (rsbWVX) while the first four genes (rsbRSTU) of the eight-gene operon were not induced under cold stress. Despite low levels of sigB induction, we found that σB-dependent genes were overrepresented (p<0.05) among the cold-induced genes at all growth phases (Table 7). Examples of such genes include fri, bsh, dapE, uspA, gabD, opuCABCD, mpoBACD, phoU, csbD, ltrC, and hrcA. Although some of these genes are solely activated by σB, others are co-regulated by other unknown or known transcription factors such as σL, σH, and PrfA [16, 20, 96–98]. As previously mentioned, σB is also activated upon entry into stationary phase [99–101]. Consistent with this fact, we observed that sigB transcript levels during growth at 20°C increased from 4.5k mapped reads at C1 to 15k at C5 (S2 Table).
Table 7. Transcription regulators significantly (p<0.05*) overrepresented among genes differentially expressed at 4°C vs. 20°C.
| Regulators | Regulon gene examples | p-value (# of contributing genes) | ||||
|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | ||
| Regulons upregulated | ||||||
| σB | inlA, bsh, mpoABCD, gabDE, opuCABCD, ltrC, csbD, phoU, hrcA | 3.70E-10 (63) | 3.95E-23 (69) | 1.26E-2 (8) | 9.58E-4 (51) | |
| CodY | argBDFJ | 1.96E-2 (4) | 4.70E-2 (4) | |||
| CtsR | clpBCEP, mcsAB, ctsR, gpmA | 1.74E-4 (8) | ||||
| HrcA | hrcA, grpE, dnaK | 4.14E-2 (3) | ||||
| Regulons downregulated | ||||||
| RpoD | dltABCD, plcAB, hly, mpl, actA, pmk, pgdA, ctaP, arcB | 1.91E-2 (24) | 4.57E-2 (16) | 1.05E-2 (14) | 1.59E-2 (39) | |
| VirR | dltABCD | 2.15E-2 (4) | 3.50E-4 (5) | 1.66E-3 (4) | ||
| PrfA | prfA, mpl, actA, plcB, inlC, hly | 1.16E-2 (5) | 4.44E-4 (5) | 3.14E-2 (6) | ||
| σB | mogR, mpoAB, phoU, ltrC, arsC, uspA | 2.14E-3 (52) | ||||
| MogR | fliNPQRK, cheRY, flhB, flgDE | 1.35E-2 (11) | ||||
* Statistical overrepresentation of gene sets were determined using Fisher’s exact test and the BioCyc database.
The alternative sigma factor σL has been shown to contribute to the ability of L. monocytogenes to tolerate cold [20, 102], osmotic, and acid stress [102, 103]. This sigma factor positively regulates >400 genes, including those involved in cell envelope synthesis, motility, and PTS sugar uptake and catabolism [28, 104]. Although sigL induction has been reported in exponential and late exponential early stationary-phase cells of cold-adapted L. monocytogenes [12, 27, 102], deleting sigL does not impact the ability of L. monocytogenes ability to grow at cold temperatures [20, 28]. In the present study, the DE pattern of sigL followed that of cluster 5 (Fig 5), with baseline expression at G1–G4 and <2-fold increased expression at G5 (Table 6). Sixteen genes previously identified as being positively regulated by σL in L. monocytogenes at 3°C [28] also exhibited similar DE patterns.
The remaining alternative sigma factors, σH and σC, contribute to the survival of L. monocytogenes under alkaline and heat stress, respectively [105–107]. However, these sigma factors have limited roles in cold tolerance [20]. Consistent with previous findings, we did not observe the upregulation of sigH or sigC in our study.
RpoD is the principal RNA polymerase sigma factor in L. monocytogenes and regulates housekeeping genes associated with ribosome structure, protein synthesis, and rRNA and tRNA [108]. Like the DE pattern for sigL, the DE pattern of rpoD followed that of cluster 5 (Fig 5), with a maximum 1.5-fold increase in expression at G5 (Table 6). Accordingly, genes positively regulated by RpoD were overrepresented (p<0.05) among the downregulated genes at all growth phases except G4. Many genes in the RpoD regulon are co-activated by PrfA, and correspondingly, PrfA-regulated genes were also overrepresented among the downregulated genes at G1–G3:G5 (Table 7). Genes co-regulated by RpoD and PrfA include prfA, plcAB, hly, mpl, inlC, and actA, all of which are associated with virulence [109]. Other studies have also reported lower levels of PrfA-regulated virulence genes in cold-adapted exponential and stationary-phase cells [11, 12, 110]. In the present study, the DE pattern of prfA fit that of cluster 17 (Fig 5), with 2-fold and 40-fold increased expression at G3 and G5, respectively (Table 6). Interestingly, although the transcription of prfA increased dramatically at G5, the expression levels of many PrfA-dependent virulence genes remained strongly downregulated. Researchers have proposed that PrfA activity, but not prfA transcription, is inhibited by unphosphorylated forms of PTS permeases that occur during active sugar transport; these PTS permeases may bind directly to PrfA [111–113]. Our results support this hypothesis, as many PTS operons and other carbohydrate uptake and catabolism genes were upregulated at G5 and belonged to the same cluster profile as prfA (Fig 5, cluster 17). Whether PrfA is subsequently involved in regulating these genes is still unclear.
CodY is a pleiotropic transcriptional regulator that actively represses the transcription of genes involved in amino acid metabolism, nitrogen assimilation, mobility and chemotaxis, and sugar uptake, among others [114]. In agreement with previous work [11], we observed the highest induction (2.4-fold) of codY in at G5 following the DE pattern shown in cluster 3 (Fig 5).
DegU is another well-known response regulator that regulates the expression of motility-, virulence-, and biofilm-related genes in L. monocytogenes [29, 72, 115]. Our results showed that degU induction was greatest (3-fold) at G1 (Table 6), with a DE pattern consistent with that of cluster 16 (Fig 5). This probably explains why increased expression of degU was not observed in previous cold stress transcriptome studies of L. monocytogenes, which focused on exponential- and stationary-phase cells [11, 12]. Despite the induction of degU at G1, the flagella operon (LMON_0680–0694) that the DegU protein regulates remained suppressed, likely due to co-induction of the gene encoding the transcriptional repressor MogR, which shared the same DE pattern as degU. In B. subtilis, DegU is part of the two-component system DegS/U, which regulates the expression of genes encoding various extracellular enzymes [116]. This suggests that DegU may also contribute to the regulation of extracellular enzymes in L. monocytogenes that facilitate cold growth.
The negative regulators HrcA and CtsR repress expression of several genes for heat-shock proteins and cellular protein quality control (e.g. dnaK, grpE, groES, groEL, clpB, clpC, clpE, clpP) in L. monocytogenes and other bacteria [117–119]. Increased expression of many of these genes has also been reported in response to salt, cold, and ethanol stress [27, 120, 121]. In the present study, we found that hrcA and ctsR, like degU and mogR, were both maximally upregulated at G1, as were many of the genes they regulate. This was somewhat surprising given the inverse relationship expected between the DE patterns of transcription repressors and their gene targets. However, CtsR and HrcA regulons are known to have a considerable amount of overlap with PrfA and σB regulons, demonstrating the complexity and fine-tuning abilities of bacterial regulatory networks, which allow bacteria to survive a wide-range of conditions [98, 122, 123].
Two-component signaling systems (TCSs) are also important regulators of bacterial stress responses and typically consist of a transmembrane sensor histidine kinase (HK), and a cognate cytoplasmic response regulator [124–127]. The sequenced genome of L. monocytogenes EGD-e contains 16 known TCSs [128]. Using mutant characterization, Pöntinen et al. [129] showed that only the HKs LisK and YycG of the LisKR and YycGF TCSs, respectively, are important for L. monocytogenes growth under cold stress. However, increased expression of lisK was not observed in either the Pöntinen et al. [129] study or the present study, highlighting that the importance of a gene does not necessarily correlate with its level of induction. HKs with increased expression in our study included cesK and LMON_1166 at G1, and kdpD, LMON_1166, and virS at G5 (Table 6). Of these, kdpD, which encodes the HK for an osmosensitive K+ channel, exhibited the largest increase in expression (18.6-fold). KdpD, together with its response regulator KdpE, controls the expression of a high-affinity K+ translocating ATPase [130, 131]. Among the many K+ transport systems in bacteria, Kdp has the highest affinity for K+ and it is only expressed when other systems are unable to meet the cell’s K+ needs. Our results therefore suggest that activation of Kdp is an important mechanism used by L. monocytogenes to survive long-term exposure to cold stress.
Ribosome functions
When bacteria are subjected to a temperature downshift, their ribosome structures become compromised, resulting in translation inhibition and a prolonged lag phase. In fact, inhibition of ribosomal functions induces CSR proteins [132]. In E. coli, three ribosome-associated proteins (IF2, CsdA, RbfA) are required for protein synthesis and subsequent cell growth at low temperatures [133–135]. In contrast, 22 other ribosomal proteins are not believed to be essential for the growth of E. coli or B. subtilis at low temperatures [136, 137]. In L. monocytogenes, the role of ribosomes and their associated proteins in cold stress remains largely unknown. In a study by Durack et al. [12], ribosome protein genes were strongly activated in osmo- and cold-adapted L. monocytogenes cells. In our study, of the 58 ribosomal proteins identified in L. monocytogenes EGD, 22 were upregulated exclusively at G5 in the cold tolerant- strain we studied. Among these genes were those encoding L. monocytogenes homologs of IF2 (infB) and RbfA, previously mentioned for their roles in stabilizing cold-sensitive ribosomes in E. coli. An additional two ribosomal protein genes, ctc and rpmF, were significantly upregulated at all growth phases, and a ribosomal subunit interface protein (LMON_2523) was induced at G1–G2:G5 (Table 6). In B. subtilis, ctc is induced in response to osmotic, heat, and oxidative stress [138, 139], and ithas similarly been linked to osmo- and cold-tolerance in L. monocytogenes [12, 56].
In bacteria, rRNA and ribosomal protein synthesis are tightly controlled, to meet the translational needs of the cell. The increased transcription of ribosome proteins in cold-adapted stationary-phase cells may reflect an increased demand for protein synthesis, as suggested by the large number of strongly upregulated genes at G5. Alternatively, ribosomal proteins have been shown to participate in extra-ribosomal functions, as independent polypeptides with roles in transcription and DNA repair [140–142]. Thus, they might contribute to the L. monocytogenes CSR in yet undetermined ways.
Cold-stress proteins
CSPs are a conserved family of small (~70 aa) proteins containing a nucleic acid-binding domain. Found in many prokaryotic and eukaryotic organisms, CSPs can act as transcriptional activators, antiterminators, or as RNA chaperones that enhance translation at low temperatures by blocking the development of secondary mRNA structures [10, 70, 71]. Three CSPs have been identified in L. monocytogenes and are listed here in the order of functional importance: CspL>CspD>CspB [143]. Furthermore, L. monocytogenes CSPs appear to be only induced during the early stages following cold stress [11, 12, 143]. In the present study, cspB was downregulated at all growth phases, reaching a maximum 228-fold decrease at G5, while cspD and cspL were upregulated at G1–G2 and then exhibited no change or decreased expression at the later growth phases (Table 6).
Additional proteins with putative roles in the L. monocytogenes cold-stress response
A few other genes have also been associated with the L. monocytogenes CSR. Zheng and Kathariou [144, 145] identified three low temperature requirement proteins (LtrA, LtrB, and LtrC) necessary for growth at cold temperatures. However, varying results have been reported regarding the expression of these genes at low temperatures. Pieta et al. [14] observed higher levels of ltrC transcripts at 7°C than at 37°C in exponential-phase cells, whereas other researchers saw either no difference in expression or decreased expression in exponential- and stationary-phase cells of cold-adapted L. monocytogenes [11, 12, 27]. Increased expression of ltrC has also been reported in L. monocytogenes EGD-e, directly following an upshift in temperature from 37°C to 48°C [146]. Importantly, we observed increased expression of ltrC only in the early stages following cold stress (G1 and G2), which may partly explain why no differences were seen in studies that analyzed exponential- and stationary-phase cells. As for other low temperature requirement protein genes, ltrB was upregulated at G5 while no notable changes were seen for ltrA.
The fri (flp) gene, which encodes ferritin, is also commonly discussed in cold stress studies of L. monocytogenes and is hypothesized to play a role in iron storage [96, 146, 147]. Previous studies have reported that fri is induced upon entry into stationary phase in L. monocytogenes subjected to low temperatures [27, 50, 96]. Additionally, fri null mutants display reduced growth at 4°C in BHIB and increased sensitivity to oxidative stress [146, 148]. In our study, fri was exclusively induced at G2 (Table 6). However, in agreement with the findings of Polidoro et al. [96], the number of fri transcripts at 20°C was nine times higher in stationary-phase cells than in lag-phase cells.
Liu et al. [22] identified a membrane-associated phosphohydrolase (pgpH) as the interrupted gene in an L. monocytogenes cold-sensitive transposon mutant. Compared with the parent strain, this mutant also showed increased intracellular levels of the phosphorylated guanosine nucleotide (p)ppGpp, suggesting that PgpH may be critical for (p)ppGpp degradation at low temperatures. Cellular (p)ppGpp inhibits RNA synthesis when a shortage of amino acids is present. Such conditions can occur during cold stress, because of structural damage to membrane transporters and intracellular enzymes. Arguedas-Villa et al. [17] reported increased pgpH expression in cold-tolerant but not cold-sensitive strains of L. monocytogenes at 4°C compared to 37°C. Though we also evaluated the gene expression of a cold-tolerant strain, we observed no significant induction of pgpH (LMON_1529) at 4°C relative to 20°C.
The induction of flagella biosynthesis and motility-related genes has frequently been reported in L. monocytogenes following a temperature downshift [11, 14, 27]. However, the reliability of these observations is debatable as 37°C was uniformly used as the control temperature and L. monocytogenes is not typically motile above 30°C [149, 150]. Recently, Cordero et al. [13] reported that fast cold-growing L. monocytogenes strains are less motile than slow cold-growing strains. They hypothesized that low motility might allow cold-tolerant strains to proliferate more rapidly at low temperature. In the present study, our cold-tolerant strain exhibited decreased expression of most flagella operons at all growth phases. However, five motility-specific genes (LMON_0691–0695) were upregulated at G5, including motB, gmaR, cheV, and flaA (Table 6). The role of motility genes in prolonged cold-stress survival remain speculative, but one hypothesis is that it is beneficial for cells to be able to move to environments more suitable in terms of nutrition or other requirements.
Cold-induced membrane lipid composition changes
To gain a better understanding of both the timing associated with cold-induced membrane FA changes in L. monocytogenes and the types of changes that occur, we analyzed FAs extracted from cells at the same time points used in our RNA-seq experiment.
Increase in anteiso C15:0
The L. monocytogenes lipid membrane consists predominantly of BCFAs (~90% of total FAs). The four most abundant BCFAs, listed in decreasing order, are anteiso-C15:0 (a-C15:0), a-C17:0, iso-C15:0 and i-C17:0 [24, 51, 151, 152]. When L. monocytogenes experiences a decrease in temperature, it increases the relative proportion of a-C15:0 at the expense of a-C17:0 [24, 52]. Depending on the length of the BCFA and the bacterial strain studied, a switch from iso to anteiso FAs can also occur (ie. i-C17:0 to a-C17:0).
To date, the membrane FA profile of L. monocytogenes under cold stresses has primarily been investigated using late exponential to early stationary-phase cells [24, 51, 151, 152]. In most cases, researchers have observed a ~20% increase in the relative proportion of a-C15:0 among cold-grown cells (5–10°C), with maximum levels ranging from 66–80%. Correspondingly, a-C17:0 levels decrease by 20–25% with minimum levels ranging from 3–14%.
In the present study, a-C15:0 levels increased from 46% at T1 to a maximum of 70% at T4 (Fig 6A), whereas levels remained constant around 50% in 20°C-grown cells. Thus, in this study, the first one to look at cold-induced membrane FA changes at multiple growth phases, we show that L. monocytogenes continues to make alterations until it transitions into stationary phase (T4), at which point minimal further adjustments are made. Correspondingly, a-C17:0 levels decreased from 12% at T1 to 4% at T4, whereas at 20°C they ranged from 13–18% (Fig 6C). Levels of i-C16:0 and iC15:0 also decreased by 2.3% and 9%, respectively at 4°C (Fig 6B). Additional changes in the BCFA composition of cold-stressed cells included a 2.5–2.9% increase in the proportion of i-C14:0 and the appearance of i-C13:0 and a-C13:0.
Fig 6. Relative proportions of specific branched-chain fatty acids (FAs) out of total FAs found in L. monocytogenes cells harvested across five growth phases at 4°C and 20°C.
A) anteiso C15:0, B) iso C15:0, and C) anteiso C17:0. Lipids were extracted from cell concentrates and resulting fatty acid methyl esters (FAME) were analyzed by gas chromatography. See Fig 1 for information about growth phases.
As previously discussed, anteiso BCFAs are synthesized from the BCAAs isoleucine, and iso BCFAs are synthesized from leucine and valine [52]. These BCAAs are produced by proteins encoded by a nine-gene operon (LMON_2054–2062). At 4°C, we found that all genes in this operon were induced >4-fold at all growth phases. Interestingly, genes from the BKD operon (LMON_1432–1437) which encodes enzymes that convert BCAAs into BCFAs, showed either no change in expression or decreased expression at 4°C. Other researchers have also noted a lack of bkd induction in L. monocytogenes under cold-stress conditions [24, 51]. A BKD-independent pathway may exist, in which BCFA synthesis uses branched-chain α-keto acids instead of branched-chain acyl-CoA esters as primers [52]. Alternatively, Nickel and colleagues [153] showed that in B. subtilis the promoter regions of five operons, including BKD, are not cold-inducible but exhibited increased mRNA stability at low temperatures.
In addition to the BKD operon, many other FA synthesis genes were also downregulated at 4°C (e.g. fabD, fabG, fabZ, acpP, plsX, plsY). However, a fabG isozyme (LMON_0351) encoding a 3-oxoacyl-[acyl-carrier protein] reductase remained upregulated at G1–G3:G5. Other upregulated genes with known or putative functions in FA biosynthesis included two short-chain dehydrogenases (LMON_0674 and LMON_1898), an oxidoreductase (ylbE), and a two-component response regulator (LMON_0289) with homology to a membrane FA regulator (YccF) in Streptococcus pneumoniae [154, 155].
Shortening of fatty acid chain lengths
Following a temperature downshift, L. monocytogenes incorporates FAs with shorter chain lengths to further decrease the phase-transition temperature of its membrane [156]. Overall, we observed an 8% increase in ≤C14 FAs at 4°C relative to 20°C, which became noticeable at T3 (Fig 7A). Moreover, by T5, 92% of Lm1’s membrane FAs contained fewer than 15 carbons; in 20°C grown cells, this percentage was 75%. Our results suggest that L. monocytogenes begins degrading unfavorable membrane phospholipids during lag phase but cannot synthesize its preferred FAs until growth resumes. This poses the question of how the cell retains adequate membrane fluidity during the early stages of cold stress. The answer to this question appears to involve the desaturation of existing FAs.
Fig 7. Relative proportion of short chain and unsaturated fatty acids (UFAs) out of total FAs found in L. monocytogenes cells harvested across five growth phases at 4 and 20°C.
A) Proportion of short chain fatty acids containing ≤14 carbons, B) Proportion of C16:1 cis-Δ9 (palmitoleic acid), C18:1 cis-Δ9 (oleic acid), and all remaining UFAs. Lipids were extracted from cell concentrates and resulting fatty acid methyl esters (FAME) were analyzed by gas chromatography. See Fig 1 for information about growth phases.
Increase in unsaturated fatty acids
Several bacterial species including E. coli, some species belonging to the phylum cyanobacteria, and some species belonging to the genus Bacillus, increase their proportion of membrane unsaturated FAs (UFAs) directly following a temperature downshift [157–159]. This process occurs rapidly due to the presence of membrane-associated desaturases that introduce double bonds into preexisting saturated FAs (SFAs). For example, in E. coli, cis-vaccenate is produced 30 seconds following a shift from 42°C to 24°C [160].
To date, the roles of UFAs in the L. monocytogenes CSR have been largely overshadowed by interests in BCFA modulation. Although some papers have reported the absence of UFAs in the L. monocytogenes membrane [24, 161–163], others have detected their presence in small amounts under various conditions [51, 151, 152, 162, 164–168]. In a study of L. innocua, levels of C18:1 FAs increased to 6.2% following a downshift from 35°C to 10°C [169]. Similarly, another study found that the Bacillus megaterium membrane contained a maximum of 22% C16:1 cis-Δ9 (palmitoleic acid) at 10°C, whereas the maximum was 2% at 35°C [53]. In the present study, the proportions of C16:1 cis-Δ9 and C18:1 cis-Δ9 (oleic acid) peaked at T3 (7%) and T2 (20%), respectively (Fig 7B). By contrast, no difference was detected between the proportions of oleic and palmitoleic acid in T4 and C4 cells, or T5 and C5 cells. Very small percentages (<1.2%) of other UFAs were also detected in our samples, but no notable differences were observed between the temperatures treatments (S3 Table). The fact that membrane UFA proportions only increased in cold-stressed lag- and exponential-phase cells likely explains why studies that have investigated late exponential- and stationary-phase cells have failed to detect any changes.
Unlike the rapid conversion of SFAs to UFAs, switching from iso to anteiso BCFAs and from longer to shorter acyl chain FAs requires de novo synthesis of whole lipid molecules by cytoplasmic enzymes that are usually linked to growth [170]. Sato and Murata [171] showed that following a 10–15°C downshift in temperature, cyanobacterial cells could only resume growth and FA biosynthesis once a certain degree of membrane unsaturation was reached. Based on our findings, we suggest that in response to cold stress, L. monocytogenes converts C18:0 and C16:0 to oleic and palmitoleic acid, respectively, to rapidly lower its membrane phase-transition temperature. Fig 8 shows how UFA levels appear to compensate for a-C15:0 until its optimal levels are reached at T4. The centre placement of the cis-double bond in oleic and palmitoleic acid makes these FAs highly efficient at increasing membrane fluidity. For example, the average melting points of oleic and palmitoleic acid are 13°C and 1.22°C, respectively, compared to 24°C and 52°C for a-C15:0 and i-C15:0, respectively [172]. While oleic and palmitoleic acids are effective at rapidly increasing the membrane fluidity of L. monocytogenes, this membrane profile is probably less stable than one that includes a high proportion of a-C15:0. In support of this idea, Juneja and Davidson [173] showed that when provided with an external source of oleic acid, L. monocytogenes was able to increase the relative proportion of this acid in its membrane from 0.72 to 28%. However, these cells were highly susceptible to salt and several antimicrobials.
Fig 8. Proportions of anteiso C15:0 and the combined sum of the unsaturated fatty acids 16:1 cis-Δ9 and 18:1 cis-Δ9 out of total FAs found in L. monocytogenes cells harvested across five growth phases at 4°C.
Lipids were extracted from cell concentrates and resulting fatty acid methyl esters (FAME) were analyzed by gas chromatography. See Fig 1 for information about growth phases.
In addition to increasing in response to cold, UFA levels also increased up to 11% in lag and exponential-phase cells grown at 20°C (Fig 7B), suggesting a role for UFAs in regular cell growth. In Saccharomyces cerevisiae, oleic acid and ergosterol supplementation mitigate oxidative stress [174]. Similarly, bacteria transferred from a stationary-phase environment into a fresh oxygenated medium also experience oxidative stress [175] and may benefit from increased levels of membrane UFAs.
In Bacillus, phospholipid desaturases, encoded by des, are induced upon cold stress and controlled by a two-component regulator that senses changes in membrane fluidity [176, 177]. These desaturases introduce double bonds at the Δ5 or Δ10 positions of acyl chains attached to existing phospholipids. Pseudomonas has a similar desaturase system that can introduce double bonds at the Δ9 position [178]. Currently, no lipid desaturases have been identified in Listeria. However, Vadyvaloo et al. [166] showed that when L. monocytogenes is treated with a desaturase inhibitor, it exhibits decreased levels of membrane UFAs, demonstrating that such a system does exist. The synthesis of oleic (18:1 cis-Δ9) and palmitoletic (16:1 cis-Δ9) acids by L. monocytogenes suggests that it contains a desaturate system like that present in Pseudomonas. In the present study, we demonstrate the presence of additional UFAs, which contain double bonds in several other locations (S3 Table) highlighting the possibility that more than one desaturate system exists in the bacterium.
Comparison of antisense transcription at 20°C and 4°C
A total of 92–261k paired-end (PE) reads were successfully mapped to the antisense strand of L. monocytogenes EGD ORFs, accounting for 0.61–1.63% of the total number of mapped reads. Overall, 70% of genes had >10 mapped PE asRNA reads, however, only 56 genes had >1,000 asRNA mapped PE reads with a maximum of 40,000 PE reads observed for a LMON_R5 (vs. a maximum of 591,000 observed for a single mRNA transcript). These results are in line with previous findings from other bacteria, from which asRNAs were detected for 20–75% of genes [35, 179–181].
Overall, more asRNA transcripts were upregulated than downregulated under cold stress (Fig 3). The largest number of upregulated asRNA transcripts was observed at G1 (Fig 3), which makes sense given that asRNAs act as gene regulators. At T1, cells are actively restructuring their transcriptional activity and cellular processes to prepare for growth in their new environment [175]. Around 10% of the asRNAs currently described for L. monocytogenes are thought to be involved in regulating transcription regulators [34]. This idea is supported by the overall low levels of antisense expression observed in this study.
While many asRNA transcripts were upregulated at G1, total antisense transcription was generally highest in late stationary-phase cells at both temperatures (Table 8), highlighting the importance of antisense regulation at this growth phase. Compared with regulatory proteins, asRNA transcripts offer the advantage of being able to provide a rapid connection between quorum sensing and direct destabilization of target mRNAs; moreover, as they can act post-transcriptionally, they can more tightly control the repression of proteins under environmental stress conditions [34, 182, 183]. Finally, unnecessary regulatory RNAs can be cleared quickly and via a process that requires less energy consumption than the removal of regulatory proteins. This may in part explain the abundance of asRNAs during late stationary-phase, in which energy sources are limited.
Table 8. Top 20 most highly expressed antisense transcripts in L. monocytogenes at 20°C and 4°C.
| Overlapped EGD ORF | Description of EGD ORF | ORF strand | ORF length (bp) | Type of antisense transcript | % of ORF covered | Normalized paired-end read counts* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | T1 | T2 | T3 | T4 | T5 | ||||||
| LMON_0226 | Hypothetical protein | - | 161 | 5’ UTRof LMON_0227 | 100 | 1693 | 1739 | 1200 | 730 | 618 | 738 | 1205 | 960 | 673 | 666 |
| LMON_0347 | Hypothetical protein | + | 113 | 3’ UTR of LMON_0348 | 62 | 75 | 49 | 54 | 118 | 593 | 120 | 89 | 109 | 370 | 3838 |
| LMON_0384a | Transcriptional regulator, DeoR family | - | 920 | 3' UTR of LMON_0383 | 100 | 421 | 291 | 415 | 952 | 6745 | 816 | 449 | 494 | 1467 | 21581 |
| LMON_0652abc | Magnesium and cobalt transport protein CorA | + | 950 | 5' UTR of LMON_0651 | 38 | 33 | 17 | 42 | 874 | 2380 | 289 | 242 | 344 | 1411 | 12635 |
| LMON_0681d | Flagellar biosynthesis protein FliP | + | 767 | IT | 100 | 12 | 18 | 107 | 1715 | 5087 | 69 | 169 | 256 | 603 | 1644 |
| LMON_0682d | Flagellar biosynthesis protein FliQ | + | 272 | IT | 81 | 2 | 8 | 69 | 1642 | 2650 | 40 | 121 | 218 | 442 | 422 |
| LMON_0738a | Transcriptional regulator, XRE family | + | 509 | 3' UTR of LMON_0739 | 100 | 205 | 336 | 321 | 406 | 289 | 1044 | 574 | 612 | 590 | 17856 |
| LMON_1143 | Propanediol utilization transcriptional activator | - | 884 | IT | 100 | 239 | 120 | 818 | 1024 | 11556 | 222 | 106 | 176 | 241 | 364 |
| LMON_1780a | Cell-shape determining protein MreBH | + | 992 | IT with LMON_T39 | 100 | 975 | 938 | 895 | 1403 | 2973 | 2575 | 1257 | 1311 | 1453 | 19114 |
| LMON_2216 | Hypothetical protein | - | 665 | Unclear | 100 | 5 | 3 | 3 | 55 | 460 | 16 | 6 | 3 | 39 | 3567 |
| LMON_2351 | Hypothetical protein | - | 443 | 3' UTR of pepC | 100 | 115 | 108 | 183 | 307 | 1147 | 87 | 127 | 259 | 517 | 7613 |
| LMON_2352 | Pseudouridine 5'-phosphate glycosidase | - | 911 | 3' UTR of pepC | 100 | 86 | 61 | 66 | 68 | 624 | 107 | 107 | 156 | 293 | 7949 |
| LMON_2353 | Pseudouridine kinase | - | 1118 | 3' UTR of pepC | 100 | 61 | 48 | 43 | 37 | 442 | 102 | 81 | 98 | 179 | 6452 |
| LMON_2550 | Hypothetical protein | + | 206 | 5' UTR of atpI | 100 | 1932 | 3007 | 8743 | 17166 | 11120 | 631 | 2379 | 6384 | 15637 | 13462 |
| LMON_2699ab | ImpB/MucB/SamB family protein | + | 1256 | 3' UTR of LMON_2700 | 100 | 919 | 978 | 1240 | 1568 | 1422 | 873 | 841 | 968 | 1175 | 6461 |
| LMON_R1 | 5S rRNA | + | 1520 | IT | 100 | 1515 | 2188 | 4954 | 8066 | 9881 | 5305 | 5839 | 4863 | 6117 | 16314 |
| LMON_R2 | 23S rRNA | + | 2931 | IT | 100 | 3196 | 4648 | 10041 | 15825 | 19762 | 9955 | 11248 | 11037 | 14694 | 39471 |
| LMON_R4 | 16S rRNA | + | 1520 | IT | 100 | 1561 | 2162 | 5015 | 8089 | 9955 | 5323 | 5919 | 4922 | 6091 | 16331 |
| LMON_R5 | 23S rRNA | + | 2931 | IT | 100 | 3188 | 4578 | 10055 | 15765 | 19809 | 9819 | 11148 | 11018 | 14626 | 39964 |
| LMON_T39 | tRNA | + | 71 | IT | 100 | 73 | 73 | 71 | 170 | 255 | 193 | 105 | 113 | 146 | 1458 |
* The normalized read counts presented represent the average of the 2–3 biological replicates. ORFs with the highest abundance of antisense transcription were determined by dividing the number of PE reads per ORF by the length of each ORF. ORFs highlighted in yellow have been previously shown or are assumed in the present study to be long antisense transcripts covering multiple ORFs. The following superscripts denote the study in which a transcript was identified:
a Wehner et al. [38];
b Wurtzel et al. [183];
c Mraheil et al. [36];
d Toledo-Arana et al. [31]. C1–C5 and T1–T5 represent the five growth phases at 20 and 4°C, respectively. Abbreviations: UTR = untranslated region; IT = individually transcribed.
Most genes with no or <10 asRNA transcripts encoded tRNAs (S2 Table). By contrast, rRNA had the highest levels of antisense transcription (Table 8). Similarly, Wehner et al. [38] have reported high levels of antisense transcription for rRNA in L. monocytogenes under intracellular growth conditions. Given the critical importance of ribosomes in cell functioning, it makes sense that bacteria would take advantage of the tight means of regulation offered by antisense transcripts. Other highly expressed antisense transcripts targeted genes encoding the cell-shape determining protein, MreBH; flagellar biosynthesis protein, FliP; magnesium and cobalt transport protein, CorA; and two transcription regulators (LMON_0384, LMON_0738), among others (Table 8). Again, most of these genes exhibited maximum levels of asRNA transcription in stationary-phase cells with higher levels evident at 4°C than 20°C.
Some L. monocytogenes genes exhibited high levels of antisense transcription with no or very low levels of mRNA transcription. Not surprisingly, this occurred when an antisense transcript was an extension of a 3’ or 5’ untranslated region (UTR) of an adjacent gene (Fig 9A, 9B and 9F), suggesting that the asRNA probably prevented RNA polymerase from binding to the promoter region of the gene on the opposite strand. On the contrary, antisense transcripts that appeared to be individually transcribed, frequently had high levels of mRNA expression (Fig 9E, 9G and 9M). These forms of asRNA have been shown to alter transcript stability by forming a sense/antisense RNA duplex leading to RNase-mediated degradation [183–186] or by stabilizing mRNA transcripts by inducing cleavage of unstable polycistronic transcripts [187–189]. Future research aimed at validating the mechanisms and functions of specific L. monocytogenes antisense transcripts will further enhance our understanding of gene regulation in this pathogen and possibly lead to the development of novel intervention strategies that can be used in the food industry.
Fig 9. Coverage maps of select highly expressed antisense transcripts in L. monocytogenes cells grown at 20 or 4°C (refer to Table 6).
The coverage maps shown are from stationary phase cultures grown at 4°C. Panels A-F shows individual paired-end, reads while panel G shows overall coverage trends for rRNA (5, 16, and 23S). Reads with the same start and end alignment positions are overlaid and depicted in green. Areas shaded in blue and orange denote sense and antisense transcripts of target genes, respectively.
Conclusions
Here we present results from the first time-course study to investigate the transcriptional response and associated cold-induced membrane FA changes of L. monocytogenes, from early growth phases through late stationary-phase. To the best of our knowledge this is also the first study to look at asRNA expression in L. monocytogenes during cold stress and across multiple growth phases. Our results revealed that the L. monocytogenes transcriptomic response to cold stress is most active during late stationary-phase survival. Similarly, we show that the L. monocytogenes cold-stress regulon differs greatly across growth phases, highlighting the importance of carefully selecting appropriate time points when designing and conducting transcriptome studies.
Overall, more genes were suppressed than induced in L. monocytogenes under cold stress conditions. A core set of 22 genes was upregulated at all growth phases, including nine genes required for BCFA synthesis, the osmolyte transporter genes opuCBCD, and genes encoding the internalins A and D. This reflects the cell’s need to synthesize BCFAs to maintain membrane fluidity at low temperatures, and to support proper protein-folding through the uptake of structural-stabilizing osmolytes. Genes suppressed at 4°C were largely associated with cobalamin (B12) biosynthesis or the production/export of cell wall components. While is unclear how L. monocytogenes may benefit from downregulating cobalamin biosynthesis genes during cold stress, a reduced rate of peptidoglycan/cell envelope turnover at 4°C relative to 20°C may reflect the reduced cellular growth rate at this temperature, or allow carbohydrates to be conserved for alternative uses.
Notable cold-induced membrane FA changes included a 15% increase in the proportion of BCFAs and a 15% transient increase in UFAs between the lag and exponential phases. Such information may be useful for improving intervention strategies that target the food industry, as increased membrane UFA levels are known to increase the susceptibility of L. monocytogenes to salt and several antimicrobials.
Overall, around 70% of L. monocytogenes genes exhibited antisense transcription but only a small portion of antisense transcripts exhibited expression levels comparable to that of mRNA. On average, antisense transcription was higher at 4°C than at 20°C, highlighting the importance of antisense regulation in the L. monocytogenes CSR. The largest number of upregulated antisense transcripts was observed during early lag phase; however, at both temperatures antisense transcription was generally highest in late stationary-phase cells. Stationary-phase cells likely benefit from the tight control of protein expression that is offered by antisense RNA, thereby reducing the overall energy requirement of the cell while faced with nutrient-limiting conditions.
Collectively, our results reveal novel gene expression patterns and membrane FA alterations that occur in L. monocytogenes following cold stress, and highlight the abundance of antisense transcription in this microorganism at both 20°C and 4°C. We believe that this research will serve as a platform for future cold-stress studies by facilitating the selection of candidate genes and antisense transcripts for further functional validation.
Supporting information
Genes belonging to a single cluster exhibited ≥0.50 membership. For ORFs with 0.2–0.5 membership to multiple clusters, the clusters are listed in decreasing order of membership. A * denotes ORFs with <0.5 membership to one cluster while a “0” denotes ORFs with <0.2 membership to one or more clusters. NC = average normalized count for control treatment.
(XLSX)
Paired-end read counts were averaged across biological replicates. “as” denotes antisense RNA transcripts.
(XLSX)
Values are presented as percentages relative to the total amount of extracted fatty acids.
(XLSX)
Points represent the average differential expression values for the genes cspB (▲) and leuA (●) at all five growth phases evaluated in this study. The y-axis represents the differential expression levels obtained using RNA sequencing while the x-axis represents the levels obtained using qPCR.
(TIF)
Acknowledgments
We would like to thank Darren Wong for providing technical assistance in analyzing our RNA sequencing data and Maksym Opushnyev for providing system administration support.
Data Availability
FastQ files of HiSeq runs were deposited into the NCBI Sequence Read Archive under the accession number BioProject ID PRJNA384077.
Funding Statement
This work was funded by an investment agreement between Alberta Innovates – Bio Solutions and the University of British Columbia (FSC-12-030, http://bio.albertainnovates.ca). P. Hingston was funded by an Alexander Graham Bell Canada Graduate Scholarships-Doctoral Program Scholarship (http://www.nserc-crsng.gc.ca/Students-Etudiants/PG-CS/BellandPostgrad-BelletSuperieures_eng.asp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ramaswamy V, Cresence VM, Rejitha JS, Lekshmi MU, Dharsana KS, Prasad SP, et al. Listeria-review of epidemiology and pathogenesis. J Microbiol Immunol. 2007;40(1):4. [PubMed] [Google Scholar]
- 2.Fenlon DR, Wilson J, Donachie W. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J Appl Bacteriol. 1996;81(6):641–50. [DOI] [PubMed] [Google Scholar]
- 3.Kozak J, Balmer T, Byrne R, Fisher K. Prevalence of Listeria monocytogenes in foods: incidence in dairy products. Food Control. 1996;7(4):215–21. [Google Scholar]
- 4.Cabedo L, Picart i Barrot L, Teixidó i Canelles A. Prevalence of Listeria monocytogenes and Salmonella in ready-to-eat food in Catalonia, Spain. J Food Prot. 2008;71(4):855–9. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan RL, Damert WG, Whiting RC, van Schothorst M. Use of epidemiologic and food survey data to estimate a purposefully conservative dose-response relationship for Listeria monocytogenes levels and incidence of listeriosis. J Food Prot. 1997;60(8):918–22. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Ross WH, Scott VN, Gombas DE. Listeria monocytogenes: low levels equal low risk. J Food Prot. 2003;66(4):570–7. [DOI] [PubMed] [Google Scholar]
- 7.Doyle ME, Mazzotta AS, Wang T, Wiseman DW, Scott VN. Heat resistance of Listeria monocytogenes. J Food Prot. 2001;64(3):410–29. [DOI] [PubMed] [Google Scholar]
- 8.Thieringer HA, Jones PG, Inouye M. Cold shock and adaptation. Bioessays. 1998;20(1):49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N [DOI] [PubMed] [Google Scholar]
- 9.Weber MHW, Marahiel MA. Bacterial cold shock responses. Sci Prog. 2003;86(1–2):9–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phadtare S. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol. 2004;6(2):125–36. [PubMed] [Google Scholar]
- 11.Chan YC, Raengpradub S, Boor KJ, Wiedmann M. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl Environ Microbiol. 2007;73(20):6484–98. AEM.00897-07 [pii]. doi: 10.1128/AEM.00897-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durack J, Ross T, Bowman JP. Characterisation of the transcriptomes of genetically diverse Listeria monocytogenes exposed to hyperosmotic and low temperature conditions reveal global stress-adaptation mechanisms. PloS ONE. 2013;8(9):e73603 doi: 10.1371/journal.pone.0073603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordero N, Maza F, Navea-Perez H, Aravena A, Marquez-Fontt B, Navarrete P, et al. Different transcriptional responses from slow and fast growth rate strains of Listeria monocytogenes adapted to low temperature. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieta L, Garcia FB, Riboldi GP, de Oliveira LA, Frazzon APG, Frazzon J. Transcriptional analysis of genes related to biofilm formation, stress-response, and virulence in Listeria monocytogenes strains grown at different temperatures. Ann Microbiol. 2014;64(4):1707–14. [Google Scholar]
- 15.Markkula A, Mattila M, Lindström M, Korkeala H. Genes encoding putative DEAD‐box RNA helicases in Listeria monocytogenes EGD‐e are needed for growth and motility at 3°C. Environ Microbiol. 2012;14(8):2223–32. doi: 10.1111/j.1462-2920.2012.02761.x [DOI] [PubMed] [Google Scholar]
- 16.Chan YC, Boor KJ, Wiedmann M. SigmaB-dependent and sigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol. 2007;73(19):6019–29. AEM.00714-07 [pii]. doi: 10.1128/AEM.00714-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arguedas-Villa C, Stephan R, Tasara T. Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol. 2010;27(5):653–60. doi: 10.1016/j.fm.2010.02.009 [DOI] [PubMed] [Google Scholar]
- 18.Angelidis AS, Smith LT, Hoffman LM, Smith GM. Identification of opuC as a chill-activated and osmotically activated carnitine transporter in Listeria monocytogenes. Appl and Environ Microbiol. 2002;68(6):2644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker LA, Evans SN, Hutkins RW, Benson AK. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J Bacteriol. 2000;182(24):7083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan YC, Hu Y, Chaturongakul S, Files KD, Bowen BM, Boor KJ, et al. Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J Food Prot. 2008;71(2):420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chassaing D, Auvray F. The lmo1078 gene encoding a putative UDP-glucose pyrophosphorylase is involved in growth of Listeria monocytogenes at low temperature. FEMS Microbiol Lett. 2007;275(1):31–7. FML840 [pii]. doi: 10.1111/j.1574-6968.2007.00840.x [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Bayles DO, Mason TM, Wilkinson BJ. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl Environ Microbiol. 2006;72(6):3955–9. 72/6/3955 [pii]. doi: 10.1128/AEM.02607-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun. 2000;68(12):7069–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu K, Bayles DO, Xiong A, Jayaswal RK, Wilkinson BJ. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain α-keto acid dehydrogenase. Microbiol. 2005;151(2):615–23. [DOI] [PubMed] [Google Scholar]
- 25.Cacace G, Mazzeo MF, Sorrentino A, Spada V, Malorni A, Siciliano RA. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J Proteomics. 2010;73(10):2021–30. doi: 10.1016/j.jprot.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 26.Bayles DO, Wilkinson BJ. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett Appl Microbiol. 2000;30(1):23–7. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Graham JE, Bigelow L, Morse PD, Wilkinson BJ. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl Environ Microbiol. 2002;68(4):1697–705. doi: 10.1128/AEM.68.4.1697-1705.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattila M, Somervuo P, Rattei T, Korkeala H, Stephan R, Tasara T. Phenotypic and transcriptomic analyses of Sigma L-dependent characteristics in Listeria monocytogenes EGD-e. Food Microbiol. 2012;32(1):152–64. doi: 10.1016/j.fm.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 29.Knudsen GM, Olsen JE, Dons L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett. 2004;240(2):171–9. S0378-1097(04)00715-3 [pii]. doi: 10.1016/j.femsle.2004.09.039 [DOI] [PubMed] [Google Scholar]
- 30.Baranyi J, Tamplin ML. ComBase: a common database on microbial responses to food environments. J Food Prot. 2004;67(9):1967–71. [DOI] [PubMed] [Google Scholar]
- 31.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459(7249):950–6. doi: 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 32.Mellin JR, Cossart P. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 2012;9(4):372–8. doi: 10.4161/rna.19235 [DOI] [PubMed] [Google Scholar]
- 33.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139(4):770–9. doi: 10.1016/j.cell.2009.08.046 [DOI] [PubMed] [Google Scholar]
- 34.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet. 2010;44:167–88. doi: 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiß S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–5. doi: 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- 36.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011;39(10):4235–48. doi: 10.1093/nar/gkr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behrens S, Widder S, Mannala GK, Qing X, Madhugiri R, Kefer N, et al. Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. PLoS ONE. 2014;9(2):e83979 doi: 10.1371/journal.pone.0083979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wehner S, Mannala GK, Qing X, Madhugiri R, Chakraborty T, Mraheil MA, et al. Detection of very long antisense transcripts by whole transcriptome RNA-seq analysis of Listeria monocytogenes by semiconductor sequencing technology. PloS ONE. 2014;9(10):e108639 doi: 10.1371/journal.pone.0108639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hingston P, Chen J, Dhillon BK, Laing C, Bertelli C, Gannon V, et al. Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front Microbiol. 2017;8:369 doi: 10.3389/fmicb.2017.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baranyi J, Roberts TA. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol. 1994;23(3–4):277–94. [DOI] [PubMed] [Google Scholar]
- 41.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. http://www.Bioinformaticsbabraham.ac.uk/projects/fastqc.
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Team RC. R: A language and environment for statistical computing. R Foundation. 2015. www.R-project.org. 2016.
- 47.Kumar L, E Futschik M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44(D1):D471–D80. doi: 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 50.Hebraud M, Guzzo J. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol Lett. 2000;190(1):29–34. S0378-1097(00)00310-4 [pii]. [DOI] [PubMed] [Google Scholar]
- 51.Annous BA, Becker LA, Bayles DO, Labeda DP, Wilkinson BJ. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol. 1997;63(10):3887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55(2):288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suutari M, Laakso S. Unsaturated and branched chain-fatty acids in temperature adaptation of Bacillus subtilis and Bacillus megaterium. BBA-Lipid Lipid Met. 1992;1126(2):119–24. [DOI] [PubMed] [Google Scholar]
- 54.Wemekamp-Kamphuis HH, Sleator RD, Wouters JA, Hill C, Abee T. Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol. 2004;70(5):2912–8. doi: 10.1128/AEM.70.5.2912-2918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmalisch M, Langbein I, Stulke J. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J Mol Microbiol Biotechnol. 2002;4(5):495–501. [PubMed] [Google Scholar]
- 56.Gardan R, Duche O, Leroy-Setrin S, Labadie J, European Listeria Genome C. Role of ctc from Listeria monocytogenes in osmotolerance. Appl Environ Microbiol. 2003;69(1):154–61. doi: 10.1128/AEM.69.1.154-161.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei Y, Oshima T, Ogasawara N, Ishikawa S. Functional analysis of the protein Veg, which stimulates biofilm formation in Bacillus subtilis. J Bacteriol. 2013;195(8):1697–705. doi: 10.1128/JB.02201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balachandran P, Cummings C, Petrauskene O, Pawar H, Tebbs R, Furtado M. Inducible nucleic acid targets for detection of pathogens, methods and compositions thereof. United States patent US20120009575. 2012.
- 59.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11(6):725–32. [DOI] [PubMed] [Google Scholar]
- 60.Kovacevic J, Arguedas-Villa C, Wozniak A, Tasara T, Allen KJ. Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl Environ Microbiol. 2013;79(6):1915–22. doi: 10.1128/AEM.03341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matthews RG. Cobalamin-dependent methyltransferases. Acc Chem Res. 2001;34(8):681–9. [DOI] [PubMed] [Google Scholar]
- 62.Jeter RM. Cobalamin-dependent 1, 2-propanediol utilization by Salmonella typhimurium. Microbiol. 1990;136(5):887–96. [DOI] [PubMed] [Google Scholar]
- 63.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8(4):290–5. doi: 10.1038/nrmicro2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeter RM, Olivera BM, Roth JR. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol. 1984;159(1):206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowman JP, Bittencourt CR, Ross T. Differential gene expression of Listeria monocytogenes during high hydrostatic pressure processing. Microbiol. 2008;154(2):462–75. [DOI] [PubMed] [Google Scholar]
- 66.Joseph B, Goebel W. Life of Listeria monocytogenes in the host cells' cytosol. Microb Infect. 2007;9(10):1188–95. [DOI] [PubMed] [Google Scholar]
- 67.Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188(2):556–68. doi: 10.1128/JB.188.2.556-568.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, et al. Glycerol-metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meadow ND, Fox DK, Roseman S. The bacterial phosphoenol-pyruvate: glycose phosphotransferase system. Ann Rev Biochem. 1990;59(1):497–542. [DOI] [PubMed] [Google Scholar]
- 70.Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol. 2006;188(1):240–8. 188/1/240 [pii]. doi: 10.1128/JB.188.1.240-248.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272(1):196–202. [DOI] [PubMed] [Google Scholar]
- 72.Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, Msadek T. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiol. 2008;154(8):2251–64. [DOI] [PubMed] [Google Scholar]
- 73.Schulze‐Gahmen U, Pelaschier J, Yokota H, Kim R, Kim SH. Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima, reveals its function as a fatty acid—binding protein. Proteins. 2003;50(4):526–30. doi: 10.1002/prot.10305 [DOI] [PubMed] [Google Scholar]
- 74.Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, et al. Genes controlled by the essential YycG/YycF two‐component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol. 2003;49(6):1639–55. [DOI] [PubMed] [Google Scholar]
- 75.van der Veen S, Hain T, Wouters JA, Hossain H, de Vos WM, Abee T, et al. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiol. 2007;153(10):3593–607. [DOI] [PubMed] [Google Scholar]
- 76.Angelidis AS, Smith GM. Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium. Appl Environ Microbiol. 2003;69(12):7492–8. doi: 10.1128/AEM.69.12.7492-7498.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko R, Smith LT. Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl Environ Microbiol. 1999;65(9):4040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ko R, Smith LT, Smith GM. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol. 1994;176(2):426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beumer RR, Te Giffel MC, Cox LJ, Rombouts FM, Abee T. Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl Environ Microbiol. 1994;60(4):1359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerhardt PN, Tombras Smith L, Smith GM. Osmotic and chill activation of glycine betaine porter II in Listeria monocytogenes membrane vesicles. J Bacteriol. 2000;182(9):2544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramadan K, Shevelev I, Hübscher U. The DNA-polymerase-X family: controllers of DNA quality? Nat Rev Mol Cell Bio. 2004;5(12):1038–43. [DOI] [PubMed] [Google Scholar]
- 82.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22(14):5194–202. doi: 10.1128/MCB.22.14.5194-5202.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Showalter AK, Byeon I-JL, Su M-I, Tsai M-D. Solution structure of a viral DNA polymerase X and evidence for a mutagenic function. Nat Struct Mol Biol. 2001;8(11):942–6. [DOI] [PubMed] [Google Scholar]
- 84.Nair S, Finkel SE. Dps protects cells against multiple stresses during stationary phase. J Bacteriol. 2004;186(13):4192–8. doi: 10.1128/JB.186.13.4192-4198.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63(2):123–43. doi: 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llorens JMN, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34(4):476–95. doi: 10.1111/j.1574-6976.2010.00213.x [DOI] [PubMed] [Google Scholar]
- 87.Fredriksson Å, Nyström T. Conditional and replicative senescence in Escherichia coli. Curr Opin Microbiol. 2006;9(6):612–8. doi: 10.1016/j.mib.2006.10.010 [DOI] [PubMed] [Google Scholar]
- 88.Ballesteros M, Fredriksson Å, Henriksson J, Nyström T. Bacterial senescence: protein oxidation in non‐proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20(18):5280–9. doi: 10.1093/emboj/20.18.5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Díaz-Acosta A, Sandoval ML, Delgado-Olivares L, Membrillo-Hernández J. Effect of anaerobic and stationary phase growth conditions on the heat shock and oxidative stress responses in Escherichia coli K-12. Arch Microbiol. 2006;185(6):429–38. doi: 10.1007/s00203-006-0113-9 [DOI] [PubMed] [Google Scholar]
- 90.Becker LA, Çetin MS, Hutkins RW, Benson AK. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and tts role in osmotolerance. J Bacteriol. 1998;180(17):4547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fraser KR, Sue D, Wiedmann M, Boor K, O'Byrne CP. Role of sigmaB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is sigma B dependent. Appl Environ Microbiol. 2003;69(4):2015–22. doi: 10.1128/AEM.69.4.2015-2022.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol. 2003;185(19):5722–34. doi: 10.1128/JB.185.19.5722-5734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferreira A, O'Byrne CP, Boor KJ. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol. 2001;67(10):4454–7. doi: 10.1128/AEM.67.10.4454-4457.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180(14):3650–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moorhead SM, Dykes GA. Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int J Food Microbiol. 2004;91(1):63–72. doi: 10.1016/S0168-1605(03)00332-5 [DOI] [PubMed] [Google Scholar]
- 96.Polidoro M, De Biase D, Montagnini B, Guarrera L, Cavallo S, Valenti P, et al. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene. 2002;296(1):121–8. [DOI] [PubMed] [Google Scholar]
- 97.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, et al. Listeria monocytogenes bile salt hydrolase is a PrfA‐regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45(4):1095–106. [DOI] [PubMed] [Google Scholar]
- 98.Chaturongakul S, Raengpradub S, Palmer ME, Bergholz TM, Orsi RH, Hu Y, et al. Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes. Appl Environ Microbiol. 2011;77(1):187–200. doi: 10.1128/AEM.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boylan SA, Redfield AR, Price CW. Transcription factor sigma B of Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993;175(13):3957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kullik I, Giachino P. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167(2–3):151–9. [DOI] [PubMed] [Google Scholar]
- 101.DeMaio J, Zhang Y, Ko C, Young DB, Bishai WR. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. P Natl Acad Sci USA. 1996;93(7):2790–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raimann E, Schmid B, Stephan R, Tasara T. The alternative sigma factor σL of L. monocytogenes promotes growth under diverse environmental stresses. Foodborne Pathog Dis. 2009;6(5):583–91. doi: 10.1089/fpd.2008.0248 [DOI] [PubMed] [Google Scholar]
- 103.Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett. 2006;263(1):54–60. FML405 [pii]. doi: 10.1111/j.1574-6968.2006.00405.x [DOI] [PubMed] [Google Scholar]
- 104.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hebraud M, et al. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiol. 2004;150(5):1581–90. [DOI] [PubMed] [Google Scholar]
- 105.Rea RB, Gahan CG, Hill C. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72(2):717–27. doi: 10.1128/IAI.72.2.717-727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Phan‐Thanh L, Mahouin F. A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis. 1999;20(11):2214–24. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2214::AID-ELPS2214>3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 107.Zhang C, Nietfeldt J, Zhang M, Benson AK. Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode sigmaC and LstR, a lineage II-specific heat shock system. J Bacteriol. 2005;187(21):7243–53. 187/21/7243 [pii]. doi: 10.1128/JB.187.21.7243-7253.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Metzger R, Brown DP, Grealish P, Staver MJ, Versalovic J, Lupski JR, et al. Characterization of the macromolecular synthesis (MMS) operon from Listeria monocytogenes. Gene. 1994;151(1):161–6. [DOI] [PubMed] [Google Scholar]
- 109.Farber J, Peterkin P. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55(3):476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ivy RA, Wiedmann M, Boor KJ. Listeria monocytogenes grown at 7 degrees C shows reduced acid survival and an altered transcriptional response to acid shock compared to L. monocytogenes grown at 37 degrees C. Appl Environ Microbiol. 2012;78(11):3824–36. doi: 10.1128/AEM.00051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aké F, Joyet P, Deutscher J, Milohanic E. Mutational analysis of glucose transport regulation and glucose‐mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol. 2011;81(1):274–93. doi: 10.1111/j.1365-2958.2011.07692.x [DOI] [PubMed] [Google Scholar]
- 112.Stoll R, Mertins S, Joseph B, Müller-Altrock S, Goebel W. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiol. 2008;154(12):3856–76. [DOI] [PubMed] [Google Scholar]
- 113.Mertins S, Joseph B, Goetz M, Ecke R, Seidel G, Sprehe M, et al. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J Bacteriol. 2007;189(2):473–90. JB.00972-06 [pii]. doi: 10.1128/JB.00972-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, et al. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63(5):1453–67. doi: 10.1111/j.1365-2958.2007.05597.x [DOI] [PubMed] [Google Scholar]
- 115.Williams T, Joseph B, Beier D, Goebel W, Kuhn M. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol Lett. 2005;252(2):287–98. S0378-1097(05)00626-9 [pii]. doi: 10.1016/j.femsle.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 116.Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, et al. A proteomic view on genome-based signal peptide predictions. Genome Res. 2001;11(9):1484–502. doi: 10.1101/gr.182801 [DOI] [PubMed] [Google Scholar]
- 117.Hanawa T, Kai M, Kamiya S, Yamamoto T. Cloning, sequencing, and transcriptional analysis of the dnaK heat shock operon of Listeria monocytogenes. Cell Stress Chaperon. 2000;5(1):21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karatzas KAG, Wouters JA, Gahan CGM, Hill C, Abee T, Bennik MHJ. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol Microbiol. 2003;49(5):1227–38. [DOI] [PubMed] [Google Scholar]
- 119.Nair S, Derré I, Msadek T, Gaillot O, Berche P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol Microbiol. 2000;35(4):800–11. [DOI] [PubMed] [Google Scholar]
- 120.Kilstrup M, Jacobsen S, Hammer K, Vogensen FK. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63(5):1826–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salotra P, Singh DK, Seal KP, Krishna N, Jaffe H, Bhatnagar R. Expression of DnaK and GroEL homologs in Leuconostoc esenteroides in response to heat shock, cold shock or chemical stress. FEMS Microbiol Lett. 1995;131(1):57–62. 037810979500235W [pii]. [DOI] [PubMed] [Google Scholar]
- 122.Hu Y, Oliver HF, Raengpradub S, Palmer ME, Orsi RH, Wiedmann M, et al. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and sigmaB in Listeria monocytogenes. Appl Environ Microbiol. 2007;73(24):7981–91. AEM.01281-07 [pii]. doi: 10.1128/AEM.01281-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu Y, Raengpradub S, Schwab U, Loss C, Orsi RH, Wiedmann M, et al. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and Sigma B in Listeria monocytogenes. Appl Environ Microbiol. 2007;73(24):7967–80. AEM.01085-07 [pii]. doi: 10.1128/AEM.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117(3):723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni M-E, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Ann Rev of Microbiol. 2010;64:539–59. [DOI] [PubMed] [Google Scholar]
- 126.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Ann Rev Biochem. 2000;69(1):183–215. [DOI] [PubMed] [Google Scholar]
- 127.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–76. [DOI] [PubMed] [Google Scholar]
- 128.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, et al. Comparative genomics of Listeria species. Science (New York, NY). 2001;294(5543):849–52. doi: 10.1126/science.1063447 [DOI] [PubMed] [Google Scholar]
- 129.Pontinen A, Markkula A, Lindstrom M, Korkeala H. Two-component-system histidine kinases involved in growth of Listeria monocytogenes EGD-e at low temperatures. Appl Environ Microbiol. 2015;81(12):3994–4004. doi: 10.1128/AEM.00626-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Csonka LN, Epstein W. Osmoregulation Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC: 1996:1210–23. [Google Scholar]
- 131.Jung K, Tjaden B, Altendorf K. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J Biol Chem. 1997;272(16):10847–52. [DOI] [PubMed] [Google Scholar]
- 132.Jones PG, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold‐shock protein whose absence triggers the cold‐shock response. Mol Microbiol. 1996;21(6):1207–18. [DOI] [PubMed] [Google Scholar]
- 133.Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochem. 1990;29(25):5881–9. [DOI] [PubMed] [Google Scholar]
- 134.Dammel CS, Noller HF. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Gene Dev. 1995;9(5):626–37. [DOI] [PubMed] [Google Scholar]
- 135.Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173(11):3291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol. 2011;413(4):751–61. doi: 10.1016/j.jmb.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, et al. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol. 2012;194(22):6282–91. doi: 10.1128/JB.01544-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hecker M, Völker U. Non‐specific, general and multiple stress resistance of growth‐restricted Bacillus subtilis cells by the expression of the σB regulon. Mol Microbiol. 1998;29(5):1129–36. [DOI] [PubMed] [Google Scholar]
- 139.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, et al. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiol. 1994;140(4):741–52. [DOI] [PubMed] [Google Scholar]
- 140.Zimmermann RA. The double life of ribosomal proteins. Cell. 2003;115(2):130–2. [DOI] [PubMed] [Google Scholar]
- 141.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115(2):187–98. [DOI] [PubMed] [Google Scholar]
- 142.Lindström MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Bioph Res Co. 2009;379(2):167–70. [DOI] [PubMed] [Google Scholar]
- 143.Schmid B, Klumpp J, Raimann E, Loessner MJ, Stephan R, Tasara T. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl Environ Microbiol. 2009;75(6):1621–7. doi: 10.1128/AEM.02154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zheng W, Kathariou S. Transposon-induced mutants of Listeria monocytogenes incapable of growth at low temperature (4 degrees C). FEMS Microbiol Lett. 1994;121(3):287–91. 0378-1097(94)90305-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 145.Zheng W, Kathariou S. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4 degrees C). Appl Environ Microbiol. 1995;61(12):4310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Olsen KN, Larsen MH, Gahan CGM, Kallipolitis B, Wolf XA, Rea R, et al. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiol. 2005;151(3):925–33. [DOI] [PubMed] [Google Scholar]
- 147.Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Mol Biol. 2000;7(1):38–43. [DOI] [PubMed] [Google Scholar]
- 148.Dussurget O, Dumas E, Archambaud C, Chafsey I, Chambon C, Hebraud M, et al. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol Lett. 2005;250(2):253–61. S0378-1097(05)00476-3 [pii]. doi: 10.1016/j.femsle.2005.07.015 [DOI] [PubMed] [Google Scholar]
- 149.Peel M, Donachie W, Shaw A. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. Microbiol. 1988;134(8):2171–8. [DOI] [PubMed] [Google Scholar]
- 150.Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, Freitag NE, et al. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 2004;6(3):235–42. [DOI] [PubMed] [Google Scholar]
- 151.Juneja VK, Foglia TA, Marmer BS. Heat resistance and fatty acid composition of Listeria monocytogenes: effect of pH, acidulant, and growth temperature. J Food Prot. 1998;61(6):683–7. [DOI] [PubMed] [Google Scholar]
- 152.Verheul A, Russell NJ, Van'T Hof R, Rombouts FM, Abee T. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl Environ Microbiol. 1997;63(9):3451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nickel M, Homuth G, Böhnisch C, Mäder U, Schweder T. Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability. Mol Genet Genomics. 2004;272(1):98–107. doi: 10.1007/s00438-004-1038-0 [DOI] [PubMed] [Google Scholar]
- 154.Ng WL, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53(4):1161–75. doi: 10.1111/j.1365-2958.2004.04196.x [DOI] [PubMed] [Google Scholar]
- 155.Mohedano ML, Overweg K, de la Fuente A, Reuter M, Altabe S, Mulholland F, et al. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J Bacteriol. 2005;187(7):2357–67. 187/7/2357 [pii]. doi: 10.1128/JB.187.7.2357-2367.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhu K, Ding X, Julotok M, Wilkinson BJ. Exogenous isoleucine and fatty acid shortening ensure the high content of anteiso-C15: 0 fatty acid required for low-temperature growth of Listeria monocytogenes. Appl Environ Microbiol. 2005;71(12):8002–7. doi: 10.1128/AEM.71.12.8002-8007.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aguilar PS, Cronan JE, De Mendoza D. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J Bacteriol. 1998;180(8):2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weber MHW, Klein W, Müller L, Niess UM, Marahiel MA. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol Microbiol. 2001;39(5):1321–9. [DOI] [PubMed] [Google Scholar]
- 159.Sakamoto T, Murata N. Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr Opin Microbiol. 2002;5(2):206–10. [DOI] [PubMed] [Google Scholar]
- 160.Garwin JL, Cronan JE, Thermal modulation of fatty acid synthesis in Escherichia coli does not involve de novo enzyme synthesis. J Bacteriol. 1980;141(3):1457–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miladi H, Bakhrouf A, Ammar E. Cellular lipid fatty acid profiles of reference and food isolates Listeria monocytogenes as a response to refrigeration and freezing stress. J Food Biochem. 2013;37(2):136–43. [Google Scholar]
- 162.Mastronicolis SK, Berberi A, Diakogiannis I, Petrova E, Kiaki I, Baltzi T, et al. Alteration of the phospho-or neutral lipid content and fatty acid composition in Listeria monocytogenes due to acid adaptation mechanisms for hydrochloric, acetic and lactic acids at pH 5.5 or benzoic acid at neutral pH. Anton Leeuw. 2010;98(3):307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mastronicolis SK, Arvanitis N, Karaliota A, Litos C, Stavroulakis G, Moustaka H, et al. Cold dependence of fatty acid profile of different lipid structures of Listeria monocytogenes. Food Microbiol. 2005;22(2):213–9. [Google Scholar]
- 164.Gianotti A, Serrazanetti D, Kamdem SS, Guerzoni ME. Involvement of cell fatty acid composition and lipid metabolism in adhesion mechanism of Listeria monocytogenes. Int J Food Microbiol. 2008;123(1):9–17. [DOI] [PubMed] [Google Scholar]
- 165.Bisbiroulas P, Psylou M, Iliopoulou I, Diakogiannis I, Berberi A, Mastronicolis SK. Adaptational changes in cellular phospholipids and fatty acid composition of the food pathogen Listeria monocytogenes as a stress response to disinfectant sanitizer benzalkonium chloride. Lett Appl Microbiol. 2011;52(3):275–80. doi: 10.1111/j.1472-765X.2010.02995.x [DOI] [PubMed] [Google Scholar]
- 166.Vadyvaloo V, Hastings JW, van der Merwe MJ, Rautenbach M. Membranes of class IIa bacteriocin-resistant Listeria monocytogenes cells contain increased levels of desaturated and short-acyl-chain phosphatidylglycerols. Appl Environ Microbiol. 2002;68(11):5223–30. doi: 10.1128/AEM.68.11.5223-5230.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Raines LJ, Moss CW, Farshtchi D, Pittman B. Fatty acids of Listeria monocytogenes. J Bacteriol. 1968;96(6):2175–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Juneja VK, Davidson PM. Influence of temperature on the fatty acid profile of Listeria monocytogenes. J Rapid Met Aut Mic. 1993;2(1):73–81. [Google Scholar]
- 169.Moorman MA, Thelemann CA, Zhou S, Pestka JJ, Linz JE, Ryser ET. Altered hydrophobicity and membrane composition in stress-adapted Listeria innocua. J Food Prot. 2008;71(1):182–5. [DOI] [PubMed] [Google Scholar]
- 170.Gounot AM, Russell NJ. Physiology of cold-adapted microorganisms Cold-adapted organisms: Springer; 1999. p. 33–55. [Google Scholar]
- 171.Sato N, Murata N. Temperature shift-induced responses in lipids in the blue-green alga, Anabaena variabilis: the central role of diacylmonogalactosylglycerol in thermo-adaptation. BBA-Lipid Lipid Met. 1980;619(2):353–66. [DOI] [PubMed] [Google Scholar]
- 172.Knothe G, Dunn RO. A comprehensive evaluation of the melting points of fatty acids and esters determined by differential scanning calorimetry. J Am Oil Chem Soc. 2009;86(9):843–56. [Google Scholar]
- 173.Juneja VK, Davidson PM. Influence of altered fatty acid composition on resistance of Listeria monocytogenes to antimicrobials. J Food Prot. 1993;56(4):302–5. [DOI] [PubMed] [Google Scholar]
- 174.Landolfo S, Zara G, Zara S, Budroni M, Ciani M, Mannazzu I. Oleic acid and ergosterol supplementation mitigates oxidative stress in wine strains of Saccharomyces cerevisiae. Int J Food Microbiol. 2010;141(3):229–35. doi: 10.1016/j.ijfoodmicro.2010.05.020 [DOI] [PubMed] [Google Scholar]
- 175.Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron AD, et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J Bacteriol. 2012;194(3):686–701. doi: 10.1128/JB.06112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Diomande SE, Doublet B, Vasai F, Guinebretiere MH, Broussolle V, Brillard J. Expression of the genes encoding the CasK/R two-component system and the DesA desaturase during Bacillus cereus cold adaptation. FEMS Microbiol Lett. 2016;363(16): Epub 2016 Jul 18. doi: 10.1093/femsle/fnw174 [DOI] [PubMed] [Google Scholar]
- 177.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001;20(7):1681–91. doi: 10.1093/emboj/20.7.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu K, Choi KH, Schweizer HP, Rock CO, Zhang YM. Two aerobic pathways for the formation of unsaturated fatty acids in Pseudomonas aeruginosa. Mol Microbiol. 2006;60(2):260–73. doi: 10.1111/j.1365-2958.2006.05088.x [DOI] [PubMed] [Google Scholar]
- 179.Voigt K, Sharma CM, Mitschke J, Lambrecht SJ, Voß B, Hess WR, et al. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014;8(10):2056–68. doi: 10.1038/ismej.2014.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev: MMBR. 2011;75(2):286–300. doi: 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Passalacqua KD, Varadarajan A, Weist C, Ondov BD, Byrd B, Read TD, et al. Strand-specific RNA-seq reveals ordered patterns of sense and antisense transcription in Bacillus anthracis. PloS ONE. 2012;7(8):e43350 doi: 10.1371/journal.pone.0043350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Izar B, Mraheil MA, Hain T. Identification and role of regulatory non-coding RNAs in Listeria monocytogenes. Int J Mol Sci. 2011;12(8):5070–9. doi: 10.3390/ijms12085070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583 doi: 10.1038/msb.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci. 2006;103(18):7054–8. doi: 10.1073/pnas.0600927103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76(4):1020–33. doi: 10.1111/j.1365-2958.2010.07161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lasa I, Toledo-Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. RNA Biol. 2012;9(8):1039–44. doi: 10.4161/rna.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Opdyke JA, Kang J-G, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186(20):6698–705. doi: 10.1128/JB.186.20.6698-6705.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol. 2011;406(1):29–43. doi: 10.1016/j.jmb.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tramonti A, De Canio M, De Biase D. GadX/GadW‐dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY—gadW divergent promoters and identification of four novel 42 bp GadX/GadW‐specific binding sites. Mol Microbiol. 2008;70(4):965–82. doi: 10.1111/j.1365-2958.2008.06458.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes belonging to a single cluster exhibited ≥0.50 membership. For ORFs with 0.2–0.5 membership to multiple clusters, the clusters are listed in decreasing order of membership. A * denotes ORFs with <0.5 membership to one cluster while a “0” denotes ORFs with <0.2 membership to one or more clusters. NC = average normalized count for control treatment.
(XLSX)
Paired-end read counts were averaged across biological replicates. “as” denotes antisense RNA transcripts.
(XLSX)
Values are presented as percentages relative to the total amount of extracted fatty acids.
(XLSX)
Points represent the average differential expression values for the genes cspB (▲) and leuA (●) at all five growth phases evaluated in this study. The y-axis represents the differential expression levels obtained using RNA sequencing while the x-axis represents the levels obtained using qPCR.
(TIF)
Data Availability Statement
FastQ files of HiSeq runs were deposited into the NCBI Sequence Read Archive under the accession number BioProject ID PRJNA384077.