Abstract
Background
SDHD promoter mutations were reported in 4–10% of cutaneous melanomas. The advanced clinico-pathological and patient survival association with SDHD mutation and/or expression in cutaneous melanoma remains controversial.
Objectives
To evaluate the presence of SDHD promoter mutations and SDHD protein expression in a melanoma series and its possible association with prognosis and survival of the patients.
Methods
We assessed SDHD promoter status in cutaneous melanomas (CM), ocular melanomas (OM) and melanoma cell lines, and the expression of SDHD protein by immunohistochemistry in CM and OM, and by western blot in melanoma cell lines. We explored the putative association between SDHD protein expression and clinico-pathological and prognostic parameters of melanoma.
Results
We detected 2% of SDHD promoter mutations in CM, but none in OM and cell lines. SDHD protein expression was present in all CM, in OM and in all CM and OM derived cell lines analysed. A significant association between lower SDHD mean protein expression and presence of ulceration and higher pT stage was found.
Conclusions
SDHD promoter mutation seems to be a rare event in CM but SDHD lower expression might associate with worst prognostic features in CM.
Introduction
SDHD is one of the four subunits that compose the Succinate Dehydrogenase (SDH) complex [1]. SDH complex has a central role in mitochondrial metabolism, being a component of the tricarboxylic acid cycle (TCA) by catalysing the oxidation of succinate to fumarate, and of the electron transport chain by transferring electrons to ubiquinone [2].
The SDH genes act as tumour suppressor genes, showing loss of heterozygosity (LOH) in combination with germline inactivating mutations in several tumours [1]. SDHD alterations where reported in sporadic and familial paraganglioma, phaeochromocytoma and gastrointestinal stromal tumour (GIST) [3].
Following the identification of TERT promoter mutations in cancer [4–7], several authors have screened other promoter regions searching for mutations that can be relevant in cancer. Promoter mutations in SDHD where recently described by Weinhold et al in 10% of cutaneous melanoma, based on data mining, using the whole-genome sequences of human tumours collected from The Cancer Genome Atlas and other public sources. The mutations in SDHD promoter associated with reduced gene expression and decrease patient survival [8]. Scholz et al reported 4% of SDHD promoter mutations in a cohort of cutaneous melanomas, but not related with clinico-pathologic factors or patient survival, and no mutations were found in ocular, mucosal and occult melanomas [9].
Cutaneous melanomas (CM) are very aggressive and, although CM represent <5% of all skin cancers, they are responsible for most of skin cancer-related deaths [10]. Most CM are diagnosed in early stage, with a 5-year survival rate reaching 98% [10], but for patients with metastatic melanoma the median survival is 8–9 months [11]. The risk factors of melanoma development include environmental causes (e.g. sunlight/UV exposure) and genetic predisposition, namely fair complexion, red hair and multiple nevi [12]. CM is a very heterogeneous tumour and many cell signalling pathways are deregulated in melanomagenesis [13, 14]. Genetically, CM harbour a high frequency of activating mutations in oncogenes, such as BRAF, NRAS and c-KIT; loss of tumour suppressors, such as CDKN2A and PTEN [15], and also the recently discovered TERT promoter mutations [4, 5, 16].
Ocular melanomas (OM) are the most frequent primary eye tumour in adults, and account for approximately 5% of all melanomas [17]. The aetiology of uveal and conjunctival melanomas remains elusive and the role of sunlight/UV exposure remains controversial [18, 19]. Mutations in genes associated with CM are less frequently reported in OM [20–23]. BRAF and NRAS mutations were reported in 14–40% and 0–18%, respectively, of conjunctival melanomas [21, 23–26], whereas they seem to be absent in uveal melanomas [20, 22], in which GNAQ and GNA11 activating mutations [27–29] and loss of BAP1 [30] are prevalent. TERT promoter mutations were reported in conjunctival melanomas, ranging from 0 to 32% and, so far, only one case of uveal melanoma harbouring a TERT promoter mutation was reported [7, 31].
In this study, we assessed the presence of SDHD promoter mutations and SDHD protein expression in CM, OM and in melanoma cell lines, in which we already determined BRAF, NRAS, GNAQ and TERT promoter mutational status [16, 28, 32]. In addition, we evaluated the possible association between SDHD protein expression, prognosis and survival of patients with CM.
Materials and methods
Sample selection, clinical-pathological and prognostic parameters
Formalin-fixed, paraffin-embedded tissues from 107 CM and 35 OM (29 uveal and 6 conjunctival melanomas) were retrieved from the Department of Anatomic Pathology of the Hospital S. João, Porto, and of Hospital S. Marcos, Braga. Clinico-pathological (Tables 1 and 2). Follow-up data were obtained from the patients’ records, the Oncology Registries of Hospital S. João and of Hospital S. Marcos, and from RORENO (Oncology Registry of North Region). All cases were revised and staged according to the 7th edition of AJCC [11]. In CM, follow-up data included time of recurrences and metastases (disease-free survival; DFS) (n = 96) and death due to melanoma (overall survival; OS) (n = 105). The mean follow-up time of the patients for DFS was 52 months (SE±3.94, range 1–195) and for OS was 56 months (SE±3.82, range 1–207). This work was approved by the Local Ethical Committee (CES) and was in accordance with the National ethical rules and Helsinki declaration.
Table 1. Clinico-pathological features of cutaneous melanomas.
| Clinico-pathological features | |
|---|---|
| Number of cases (n) | 107 |
| Median age (range) | 61.7 (7–95) |
| Gender [n (%)] | |
| Female | 61 (57.0) |
| Male | 46 (43.0) |
| Sun exposure (body site) | |
| absent | 25 (23.6) |
| intermittent | 64 (60.4) |
| chronic | 17 (16.0) |
| Histological subtype [n (%)] | |
| LMM | 13 (12.1) |
| ALM | 22 (20.6) |
| NM | 18 (16.8) |
| SSM | 54 (50.5) |
| Pigmentation | |
| absent | 8 (7.9) |
| present | 93 (92.1) |
| Median thickness (range) [mm] | 3.9 (0–70) |
| Epidermal ulceration [n (%)] | |
| absent | 70 (65.4) |
| present | 37 (34.6) |
| Clark level (≤ 1mm) [n (%)] | |
| I | 18 (36.0) |
| II | 17 (34.0) |
| III | 14 (28.0) |
| IV | 1 (2.0) |
| V | 0 (0.0) |
| Mitotic rate [n (%)] | |
| < 1/mm2 | 36 (33.6) |
| ≥ 1/mm2 | 71 (66.4) |
| pT [n (%)] | |
| ≤ pT2 | 54 (50.5) |
| > pT2 Mutation status wt BRAFV600 NRASQ61 TERT promoter BRAFV600/ TERT promoter |
53 (49.5) 56 (52.3) 20 (18.7) 8 (7.5) 11 (10.3) 12 (11.2) |
Table 2. Clinico-pathological features of ocular melanomas.
| Clinico-pathological features | Uveal melanomas | Conjunctival melanomas |
|---|---|---|
| Number of cases [n (%)] | 29 (82.9.4) | 6 (17.1) |
| Median age (range) | 55 (14–90) | 63 (28–90) |
| Cytological type [n (%)] | ||
| epithelioid | 5 (17.2) | 4 (80.0) |
| spindle | 12 (41.4) | 1 (20.0) |
| mixed | 12 (41.4) | 0 |
| pT [n (%)] | ||
| ≤ pT2 | 21 (72.4) | 5 (83.3) |
| > pT2 | 8 (27.6) | 1 (16.7) |
| Mitotic rate [n (%)] | ||
| ≤ 1/mm2 > 1/mm2 |
21 (72.4) 8 (27.6) |
3 (60.0) 2 (40.0) |
| Median thickness [mm (range)] |
NA | 3 (0.3–7) |
| Median basal tumour diameter [mm (range)] |
10.9 (3–18) | NA |
| Tumour scleral involvement [n (%)] | ||
| present | 8 (27.6) | NA |
| absent | 21 (72.4) |
NA–Not Applicable
Cell lines and culture conditions
BLM, G361 and Mewo skin melanoma cell lines were kindly provided by Dr. Marc Mareel, from the Department of Radiotherapy and Nuclear Medicine, Ghent University Hospital, Belgium. A375 skin melanoma cell line was kindly provided by Dr. Madalena Pinto, from CEQUIMED, Faculty of Pharmacy, University of Porto, Portugal. 92.1 [33], OCM1 [34], OMM1 [35], OMM2.3 [36], Mel202 [37], Mel270 [36] and Mel285 [37] uveal melanoma cell lines were kindly provided by Dr. Martine Jager, from the Laboratory of Ophthalmology, Leiden University, Netherlands. All the cell lines were tested for mycoplasma.
BLM, and Mewo cell lines were maintained in DMEM medium (Gibco/BRL–Invitrogen), G361 cell line was maintained in McCoy’s medium (Gibco/BRL–Invitrogen), and 92.1, OCM1, OMM1, OMM2.3, Mel202, Mel270 and Mel285 cell lines were maintained in RPMI medium (Gibco/BRL–Invitrogen). All media were supplemented with 10% of fetal bovine serum, 100U/mL Penicillin and 100ug/mL Streptomycin. Cell lines were maintained in a humidified atmosphere (5% CO2) at 37°C.
DNA extraction
Extraction of DNA from tumours smaller than 5mm was performed after microdissection with PALM MicroLaser Systems (PALM, Germany) and using the Quiamp DNA micro kit (Quiagen, Hilden). In tumours larger than 5mm, DNA extraction was done by manual dissection of 10μm whole sections of paraffin-embedded tissue using the Invisorb spin tissue mini kit (Invitek, Berlin). DNA extraction from the cell lines was also performed with the Invisorb spin tissue mini kit (Invitek, Berlin).
Mutation analysis
Fragments encompassing SDHD promoter region were amplified by polymerase chain reaction (PCR) of the tumour samples with the sets of primers: Fwd: 5’-CTCCGCCATTGTTCGCCTCA-3’, Rev: 5’-TTCCTGAGGGCTCAAGGTCAT-3’. Genomic DNA (25–100 ng) was amplified by PCR using the following cycling conditions: 30s at 94°C, 90s at 59°C and 60s at 72°C for 35 cycles. Products were enzymatically purified and directly sequenced in an ABI Prism 3130 xl Automatic sequencer (Perkin-Elmer, Foster City, CA) using the BigDye Terminator Sequencing kit (Perkin-Elmer). Cases with mutations were confirmed by an independent amplification.
BRAF, NRAS, GNAQ and TERT mutational analysis in the series was previously reported [16, 28, 32].
Immunohistochemical analysis
Paraffin sections were deparaffinised and rehydrated, followed by a microwave antigen retrieval procedure with 10 mM sodium citrate buffer pH 6.0. The sections were incubated overnight at 4°C in a humidified chamber with the primary antibody SDHD (polyclonal, rabbit, 1:100), from Santa Cruz Biotechnology. The antibody was validated by the manufacturer to ensure the antibody specificity to the target protein and IHC procedures were already published [38]. The detectionwas obtained with the alkaline phosphatase method (APAAP), with the EXPOSE Mouse and Rabbit Specific AP (red) Detection IHC Kit (ab94734; Abcam; Cambridge, UK), and the colour was developed with fast red chromogen, or with a streptavidin–biotin immunoperoxidase detection system with the Ultravision Quanto Detection System HRP (Thermo Scientific, Fremont, USA), and the immunohistochemical staining was developed with AEC substrate HIGHDEF® Red IHC (Enzo Life Sciences, Inc., New York, USA). The slides were counterstained with haematoxylin, and then mounted using a water-miscible mounting medium. A pancreatic endocrine tumour case, previously tested, was used as negative (omission of primary antibody) and positive control. pERKs and TERT expression in the series has been previously reported [16, 32].
Immunohistochemical evaluation
Two observers (J.M.L. and H.P.) evaluated tumour cell immunoreactivity without knowledge of any clinical and mutational data from the cases. An IHC score was settled for SDHD, and results from the multiplication of the intensity of staining (negative = 0, weak = 1, moderate = 2 and strong = 3) and the proportion of cells showing an unequivocal positive reaction (0–5% = 0, 6–25% = 1, 26–50% = 2, 51–75% = 3, 76–100% = 4).
Western blot analysis and antibodies
Cells were lysed for 15 min at 4°C using RIPA buffer (1% NP-40 in 150 mM NaCl, 50 mM Tris [pH 7.5], 2 mM EDTA) containing phosphatase and protease inhibitors. Proteins were quantified using a modified Bradford assay (Biorad). Protein samples (50 μg) were separated in 10% SDS/PAGE gels and electroblotted to Hybond ECL membrane (Amersham Biosciences). SDHD (polyclonal, rabbit, 1:200, Santa Cruz Biotechnology) was used. Secondary antibody was conjugated with peroxidase (Santa Cruz Biotechnology) and visualized by the ECL detection solution. Membrane was re-stained with a goat polyclonal anti-actin (Santa Cruz Biotechnology) for loading protein control. XTC1, a hurthle cell thyroid cell line, was used as positive control.
Statistical analysis
Statistical analysis was performed using STAT VIEW-J 5.0 (SAS Institute, Inc., Cary, NC). The relationship between the average expression level (score) of the immunohistochemistry markers and clinical-pathological parameters was evaluated by ANOVA. When appropriate, multiple comparison corrections were performed using the post hoc Bonferroni or Tamhane tests. The correlation between the immunoreactivity score of the different markers was assessed using the Fisher’s exact test. The Kaplan-Meier method and log-rank test were used to evaluate the melanoma survival data. Univariate analyses were performed to determine the prognostic value of covariates regarding OS and DFS using the Cox regression model. OS and DFS were calculated from the time of diagnosis until death due to disease or metastasis, respectively, or censored at the time of the latest follow-up or death unrelated to the disease. A p value <0.05 was considered statistically significant.
Results
SDHD promoter mutations analysis in CM, OM and melanoma cell lines
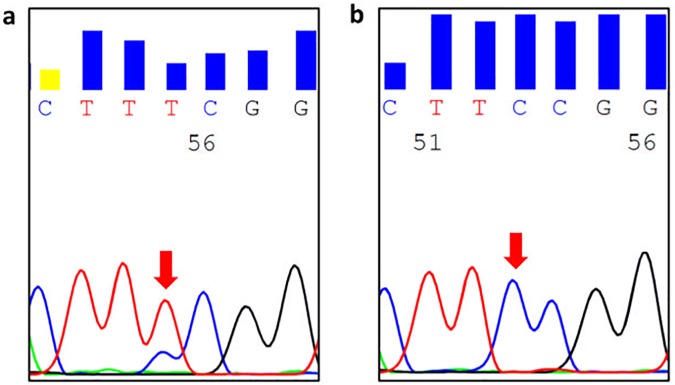
86 CM were analysed for SDHD promoter mutations. We found a chr. 11:111,957,523 (TTCC>TTTC) SDHD alteration [one of the mutations reported by Weinhold et al [8]] in two CM (2%) (Fig 1). One mutated case was a superficial spreading melanoma diagnosed in 1999, that did not display BRAF, NRAS and TERT promoter mutations, and the patient was alive at the last follow-up (180 months). The other mutated case was an acral melanoma diagnosed in 2009, displayed a -124:G>A TERT promoter mutation, but not a BRAF mutation; the patient died 24 months after diagnosis, in line with the poor prognosis of CM harbouring TERT promoter mutations. [4, 5, 16, 39]. None of the OM cases (26),cutaneous (n = 4) and ocular (n = 7) melanoma cell lines studied harboured any alteration in the promoter of the SDHD gene. Due to the lower number of mutated cases, we could not assess any association between the presence of the mutation and the clinico-pathological and prognostic parameters of CM.
Fig 1.
Representative electropherograms of SDHD promoter sequencing from a case with the chr. 11:111,957,523 (TTCC>TTTC) SDHD alteration (a) and a case with wild-type sequence (b).
Expression of SDHD protein in CM, OM and melanoma cell lines
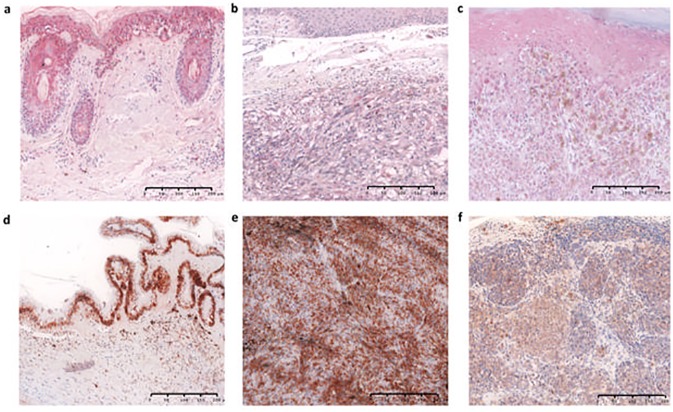
SDHD protein expression, evaluated in 107 CM, was cytoplasmic and present in all cases, including those (two cases) with SDHD promoter mutations (Fig 2). SDHD was expressed not only in melanocytes/melanoma cells, but also in keratinocytes and in the cells of sebaceous glands and hair follicles. Low staining score (score ≤2) was observed in 41% and moderate/high staining score (score >2) was observed in 59% of melanomas.
Fig 2.
Representative microphotographs of SDHD protein expression in adjacent skin (a), a cutaneous case with the chr. 11:111,957,523 (TTCC>TTTC) SDHD alteration (b), a cutaneous case with wild-type SDHD promoter sequence (c), adjacent ocular structures (d), a uveal case with wild-type SDHD promoter sequence (e) and a conjunctival case with wild-type SDHD promoter sequence (f). APAAP×200 (a-c), HRP×200 (d-f).
SDHD protein expression was also evaluated in 33 OM. Cytoplasmic immunoreactivity was observed in 67% of conjunctival melanomas and 30% of uveal melanoma (Fig 2). SDHD was expressed also in adjacent non-tumour eye structures, mostly iris, retina and ciliary pigment epitheliums. Low staining score (score ≤2) was observed in all the positive conjunctival melanomas and 19% of uveal melanoma, and moderate/high staining score (score >2) was observed in 11% of uveal melanoma.
No association was found between the expression of SDHD and CM histological subtypes or type of sunlight/UV exposure. Concerning the prognostic factors of CM, a significant association was found between lower mean SDHD protein expression and the presence of ulceration (p<0.01) and higher pT stage (p<0.01) (Table 3). No significant association was found between SDHD protein expression and other clinico-pathological parameters (age, sex, mitotic index and tumour thickness), although the same tendency, lower mean SDHD protein expression, was observed with higher thickness and mitotic rate. To evaluate whether SDHD protein expression correlates with disease-free (DFS) and overall (OS) survival in CM, Kaplan Meier curves and univariate Cox regression were performed. No significant association between SDHD protein expression and DFS and OS of the patients was found, although it was observed in the Kaplan Meier curves that patients with low expression of SDHD displayed non-significant reduced DFS and OS compared with patients with high expression of SDHD (S1 Fig). Regarding OM, the low number of conjunctival and uveal cases did not allow the statistical analysis.
Table 3. Summary of the statistical associations between SDHD expression and the clinico-pathological parameters of cutaneous melanomas.
| Clinico-pathological features | SDHD mean expression level (±SD) | p-value |
|---|---|---|
| Pigmentation | ||
| absent | 4.37 (3.07) | ns |
| present | 3.74 (2.79) | |
| Thickness | ||
| ≤ 1/mm | 4.72 (3.56) | ns |
| > 1/mm | 3.91 (2.78) | |
| Epidermal ulceration | ||
| absent | 4.80 (3.34) | <0.01 |
| present | 3.11 (2.17) | |
| Clark level (≤ 1mm) [n (%)] | ||
| I/II | 4.05 (3.14) | ns |
| III/IV | 4.21 (3.33) | |
| Mitotic rate [n (%)] | ||
| < 1/mm2 | 4.26 (3.76) | ns |
| ≥ 1/mm2 | 4.18 (2.75) | |
| pT | ||
| ≤ pT2 | 5.02 (3.63) | <0.01 |
| > pT2 | 3.40 (2.21) |
We also evaluated if the expression of SDHD protein was related to the activation of the MAPK pathway, and the presence of TERT promoter mutation and TERT protein expression. No association was found between the expression of SDHD and BRAF/NRAS mutations, pERK expression (the readout of MAPK pathway activation) or with TERT promoter mutations and TERT protein expression. Due to the low number of mutated cases, we cannot infer if there is a reduction in the protein expression associated with the presence of SDHD promoter mutation, but moderate/high staining scores (6 and 9) were found in the two cases with SDHD promoter mutations. All the melanoma cell lines studied expressed SDHD protein (S2 Fig).
Discussion
In this work we established, for the first time, that SDHD protein expression associates with prognostic features of CM.
SDH enzyme has a central role in mitochondrial metabolism [2]. SDHD alterations result in the disruption of the SDH complex and loss of SDH enzymatic activity [40, 41]. In paragangliomas, SDHB alterations were linked to malignancy and SDHD alterations are more frequent in head-and-neck localized tumours [42, 43]. Recently, SDHD promoter mutations were reported in melanomas, associated with reduced gene expression and reduced patient survival [8, 9].
At variance with Weinhold et al [8], who reported frequent noncoding alterations in SDHD promoter in melanomas, we only found two cases (2%) with SDHD alteration in our cutaneous melanoma series, similar to the 4% reported by Scholz et al [9]. Several factors may explain these discrepant results when compared with Weinhold et al work which was exclusively based on data mining, without sequencing validation, namely the use of different methodologies with different sensitivities, and differences in the cutaneous melanoma subtypes and melanomas staging.
Although C>T alterations are generally considered a marker of UV-exposure, we found a 523C>T SDHD mutation in an acral melanoma case that occur in skin without sun exposure, at variance with Scholz et al [9] that did not find SDHD mutations in acral melanomas. We did not find also any mutations in uveal melanomas, that are considered not associated with exposure to sunlight/UV radiation [19]. No alterations were found by us in cutaneous and ocular melanoma cell lines; these results reinforce the possibility that these alterations are rare in melanomas. The rarity of SDHD mutations detected in our series does not allow us to validate the association between the presence of SDHD promoter mutation and a reduction in the expression of SDHD gene or the association between this mutation and prognostic parameters of CM, as reported by Weinhold et al [8].
SDHD protein was expressed in all melanoma cell lines analysed and in all CM cases, including the two cases with SDHD mutation. We found a significant association between low mean SDHD protein expression and the presence of ulceration and high pT stage. Our results indicate an association between the reduction of SDHD protein expression and worst prognosis, but without a significant relation with survival of patients with CM. Although the molecular and cellular mechanisms linking SDH inactivation and tumorigenesis it is not completely understood, SDHD mutation/inactivation results in a loss of electron transport chain complex II activity and in the activation of the hypoxia-angiogenic pathway, namely an increase of EPAS-1, HIF-1 and VEGF expression, which may be the involved mechanism in tumorigenesis [40, 44]. The presence of pigmentation is also linked to increase HIF-1 expression and shorter DFS and OS [45, 46], however in our series, no relation between SDHD expression and pigmentation status was found.
Diverse etiopathogenic mechanisms may operate in the development of cutaneous, conjunctiva and uveal melanomas and it seems that conjunctiva melanomas share more comparable pathogenesis with cutaneous melanomas than with uveal melanomas [47, 48]. We still dispute the biological meaning of absence of SDHD expression in OM, as no SDHD promoter mutations were found in the OM cases analysed.
In conclusion, our results indicate that SDHD promoter mutation is a rare event in CM and is absent in OM. Yet, SDHD expression might have prognostic relevance in CM. Larger studies are necessary to validate if low expression of SDHD (related or not with the presence of the promoter mutation) might associate with worst prognostic features of CM. Importantly, the metabolic reshape created by SDHD alteration may open a possible therapeutic window that can benefit CM patients, through drugs that can revert this metabolism shift.
Supporting information
Kaplan Meier curves demonstrating the correlation between SDHD protein expression and disease-free (a) and overall (b) survival of cutaneous melanoma patients. Patients with low expression of SDHD protein (solid line) display reduced disease-free and overall survival compared with patients high expression of SDHD protein (dashed line).
(TIF)
All the cell lines analysed express SDHD protein.
(TIF)
Acknowledgments
We are grateful to all of the patients who participated in this study as well as the physicians that provided clinical, pathological and follow-up information.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by the Portuguese Foundation for Science and Technology through Post-Doc grant to HP (Ref.: SFRH/BPD/85249/2012). IPATIMUP integrates the i3S Research Unit, which is partially supported by FCT. This work was financed by FEDER - Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 - Operacional Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through FCT - Fundação para a Ciência e a Tecnologia/ Ministério da Ciência, Tecnologia e Inovação in the framework of the project "Institute for Research and Innovation in Health Sciences" (POCI-01-0145-FEDER-007274). Further funding was obtained from the project "Advancing cancer research: from basic knowledgment to application";NORTE-01-0145-FEDER-000029; “Projetos Estruturados de I&D&I”, funded by Norte 2020 – Programa Operacional Regional do Norte. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 2011;1807(11):1432–43. doi: 10.1016/j.bbabio.2011.07.003 [DOI] [PubMed] [Google Scholar]
- 2.Huang S, Millar AH. Succinate dehydrogenase: the complex roles of a simple enzyme. Current opinion in plant biology. 2013;16(3):344–9. doi: 10.1016/j.pbi.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 3.Barletta JA, Hornick JL. Succinate dehydrogenase-deficient tumors: diagnostic advances and clinical implications. Advances in anatomic pathology. 2012;19(4):193–203. doi: 10.1097/PAP.0b013e31825c6bc6 [DOI] [PubMed] [Google Scholar]
- 4.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61. doi: 10.1126/science.1230062 [DOI] [PubMed] [Google Scholar]
- 5.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9. doi: 10.1126/science.1229259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr., et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6. doi: 10.1073/pnas.1303607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinagre J, Almeida A, Populo H, Batista R, Lyra J, Pinto V, et al. Frequency of TERT promoter mutations in human cancers. Nature communications. 2013;4:2185 doi: 10.1038/ncomms3185 [DOI] [PubMed] [Google Scholar]
- 8.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46(11):1160–5. doi: 10.1038/ng.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz SL, Horn S, Murali R, Moller I, Sucker A, Sondermann W, et al. Analysis of SDHD promoter mutations in various types of melanoma. Oncotarget. 2015;6(28):25868–82. doi: 10.18632/oncotarget.4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- 11.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–206. doi: 10.1200/JCO.2009.23.4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chudnovsky Y, Khavari PA, Adams AE. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115(4):813–24. doi: 10.1172/JCI24808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Bergami P, Fitchman B, Ronai Z. Understanding signaling cascades in melanoma. Photochemistry and photobiology. 2008;84(2):289–306. doi: 10.1111/j.1751-1097.2007.00254.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolotto C. Melanoma: from melanocyte to genetic alterations and clinical options. Scientifica. 2013;2013:635203 doi: 10.1155/2013/635203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Populo H, Soares P, Lopes JM. Insights into melanoma: targeting the mTOR pathway for therapeutics. Expert Opin Ther Targets. 2012;16(7):689–705. doi: 10.1517/14728222.2012.691472 [DOI] [PubMed] [Google Scholar]
- 16.Populo H, Boaventura P, Vinagre J, Batista R, Mendes A, Caldas R, et al. TERT promoter mutations in skin cancer: the effects of sun exposure and X-irradiation. J Invest Dermatol. 2014;134(8):2251–7. doi: 10.1038/jid.2014.163 [DOI] [PubMed] [Google Scholar]
- 17.Chang AE, Karnell LH, Menck HR. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83(8):1664–78. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher RP, Lee TK. Adverse effects of ultraviolet radiation: a brief review. Prog Biophys Mol Biol. 2006;92(1):119–31. doi: 10.1016/j.pbiomolbio.2006.02.011 [DOI] [PubMed] [Google Scholar]
- 19.Singh AD, Rennie IG, Seregard S, Giblin M, McKenzie J. Sunlight exposure and pathogenesis of uveal melanoma. Surv Ophthalmol. 2004;49(4):419–28. doi: 10.1016/j.survophthal.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 20.Cruz F 3rd, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63(18):5761–6. [PubMed] [Google Scholar]
- 21.Gear H, Williams H, Kemp EG, Roberts F. BRAF mutations in conjunctival melanoma. Invest Ophthalmol Vis Sci. 2004;45(8):2484–8. doi: 10.1167/iovs.04-0093 [DOI] [PubMed] [Google Scholar]
- 22.Rimoldi D, Salvi S, Lienard D, Lejeune FJ, Speiser D, Zografos L, et al. Lack of BRAF mutations in uveal melanoma. Cancer Res. 2003;63(18):5712–5. [PubMed] [Google Scholar]
- 23.Spendlove HE, Damato BE, Humphreys J, Barker KT, Hiscott PS, Houlston RS. BRAF mutations are detectable in conjunctival but not uveal melanomas. Melanoma Res. 2004;14(6):449–52. [DOI] [PubMed] [Google Scholar]
- 24.Goldenberg-Cohen N, Cohen Y, Rosenbaum E, Herscovici Z, Chowers I, Weinberger D, et al. T1799A BRAF mutations in conjunctival melanocytic lesions. Invest Ophthalmol Vis Sci. 2005;46(9):3027–30. doi: 10.1167/iovs.04-1449 [DOI] [PubMed] [Google Scholar]
- 25.El-Shabrawi Y, Radner H, Muellner K, Langmann G, Hoefler G. The role of UV-radiation in the development of conjunctival malignant melanoma. Acta Ophthalmol Scand. 1999;77(1):31–2. [DOI] [PubMed] [Google Scholar]
- 26.Griewank KG, Westekemper H, Murali R, Mach M, Schilling B, Wiesner T, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19(12):3143–52. doi: 10.1158/1078-0432.CCR-13-0163 [DOI] [PubMed] [Google Scholar]
- 27.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Populo H, Vinagre J, Lopes JM, Soares P. Analysis of GNAQ mutations, proliferation and MAPK pathway activation in uveal melanomas. Br J Ophthalmol. 2011;95(5):715–9. doi: 10.1136/bjo.2009.174417 [DOI] [PubMed] [Google Scholar]
- 29.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–9. doi: 10.1056/NEJMoa1000584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–3. doi: 10.1126/science.1194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dono M, Angelini G, Cecconi M, Amaro A, Esposito AI, Mirisola V, et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br J Cancer. 2014;110(4):1058–65. doi: 10.1038/bjc.2013.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Populo H, Soares P, Faustino A, Rocha AS, Silva P, Azevedo F, et al. mTOR pathway activation in cutaneous melanoma is associated with poorer prognosis characteristics. Pigment Cell Melanoma Res. 2011;24(1):254–7. doi: 10.1111/j.1755-148X.2010.00796.x [DOI] [PubMed] [Google Scholar]
- 33.De Waard-Siebinga I, Blom DJ, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, et al. Establishment and characterization of an uveal-melanoma cell line. Int J Cancer. 1995;62(2):155–61. [DOI] [PubMed] [Google Scholar]
- 34.Kan-Mitchell J, Mitchell MS, Rao N, Liggett PE. Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci. 1989;30(5):829–34. [PubMed] [Google Scholar]
- 35.Luyten GP, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, Van Drunen E, et al. Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer. 1996;66(3):380–7. doi: 10.1002/(SICI)1097-0215(19960503)66:3<380::AID-IJC19>3.0.CO;2-F [DOI] [PubMed] [Google Scholar]
- 36.Chen PW, Murray TG, Uno T, Salgaller ML, Reddy R, Ksander BR. Expression of MAGE genes in ocular melanoma during progression from primary to metastatic disease. Clin Exp Metastasis. 1997;15(5):509–18. [DOI] [PubMed] [Google Scholar]
- 37.Ksander BR, Rubsamen PE, Olsen KR, Cousins SW, Streilein JW. Studies of tumor-infiltrating lymphocytes from a human choroidal melanoma. Invest Ophthalmol Vis Sci. 1991;32(13):3198–208. [PubMed] [Google Scholar]
- 38.Xekouki P, Pacak K, Almeida M, Wassif CA, Rustin P, Nesterova M, et al. Succinate dehydrogenase (SDH) D subunit (SDHD) inactivation in a growth-hormone-producing pituitary tumor: a new association for SDH? J Clin Endocrinol Metab. 2012;97(3):E357–66. doi: 10.1210/jc.2011-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gimenez-Roqueplo AP, Favier J, Rustin P, Mourad JJ, Plouin PF, Corvol P, et al. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am J Hum Genet. 2001;69(6):1186–97. doi: 10.1086/324413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douwes Dekker PB, Hogendoorn PC, Kuipers-Dijkshoorn N, Prins FA, van Duinen SG, Taschner PE, et al. SDHD mutations in head and neck paragangliomas result in destabilization of complex II in the mitochondrial respiratory chain with loss of enzymatic activity and abnormal mitochondrial morphology. J Pathol. 2003;201(3):480–6. doi: 10.1002/path.1461 [DOI] [PubMed] [Google Scholar]
- 42.Pawlu C, Bausch B, Neumann HP. Mutations of the SDHB and SDHD genes. Fam Cancer. 2005;4(1):49–54. doi: 10.1007/s10689-004-4227-4 [DOI] [PubMed] [Google Scholar]
- 43.Mediouni A, Ammari S, Wassef M, Gimenez-Roqueplo AP, Laredo JD, Duet M, et al. Malignant head/neck paragangliomas. Comparative study. European annals of otorhinolaryngology, head and neck diseases. 2014;131(3):159–66. doi: 10.1016/j.anorl.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 44.Rustin P, Munnich A, Rotig A. Succinate dehydrogenase and human diseases: new insights into a well-known enzyme. European journal of human genetics: EJHG. 2002;10(5):289–91. doi: 10.1038/sj.ejhg.5200793 [DOI] [PubMed] [Google Scholar]
- 45.Slominski A, Kim TK, Brozyna AA, Janjetovic Z, Brooks DL, Schwab LP, et al. The role of melanogenesis in regulation of melanoma behavior: melanogenesis leads to stimulation of HIF-1alpha expression and HIF-dependent attendant pathways. Archives of biochemistry and biophysics. 2014;563:79–93. doi: 10.1016/j.abb.2014.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brozyna AA, Jozwicki W, Carlson JA, Slominski AT. Melanogenesis affects overall and disease-free survival in patients with stage III and IV melanoma. Hum Pathol. 2013;44(10):2071–4. doi: 10.1016/j.humpath.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seregard S. Conjunctival melanoma. Surv Ophthalmol. 1998;42(4):321–50. [DOI] [PubMed] [Google Scholar]
- 48.Farber M, Schutzer P, Mihm MC Jr. Pigmented lesions of the conjunctiva. J Am Acad Dermatol. 1998;38(6 Pt 1):971–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan Meier curves demonstrating the correlation between SDHD protein expression and disease-free (a) and overall (b) survival of cutaneous melanoma patients. Patients with low expression of SDHD protein (solid line) display reduced disease-free and overall survival compared with patients high expression of SDHD protein (dashed line).
(TIF)
All the cell lines analysed express SDHD protein.
(TIF)
Data Availability Statement
All relevant data are within the paper.