Abstract
Purpose of review
This article argues that the time is approaching for data-driven disease modelling to take centre stage in the study and management of neurodegenerative disease. The snowstorm of data now available to the clinician defies qualitative evaluation; the heterogeneity of data types complicates integration through traditional statistical methods; and the large datasets becoming available remain far from the big-data sizes necessary for fully data-driven machine-learning approaches. The recent emergence of data-driven disease progression models provides a balance between imposed knowledge of disease features and patterns learned from data. The resulting models are both predictive of disease progression in individual patients and informative in terms of revealing underlying biological patterns.
Recent findings
Largely inspired by observational models, data-driven disease progression models have emerged in the last few years as a feasible means for understanding the development of neurodegenerative diseases. These models have revealed insights into frontotemporal dementia, Huntington's disease, multiple sclerosis, Parkinson's disease and other conditions. For example, event-based models have revealed finer graded understanding of progression patterns; self-modelling regression and differential equation models have provided data-driven biomarker trajectories; spatiotemporal models have shown that brain shape changes, for example of the hippocampus, can occur before detectable neurodegeneration; and network models have provided some support for prion-like mechanistic hypotheses of disease propagation. The most mature results are in sporadic Alzheimer's disease, in large part because of the availability of the Alzheimer's disease neuroimaging initiative dataset. Results generally support the prevailing amyloid-led hypothetical model of Alzheimer's disease, while revealing finer detail and insight into disease progression.
Summary
The emerging field of disease progression modelling provides a natural mechanism to integrate different kinds of information, for example from imaging, serum and cerebrospinal fluid markers and cognitive tests, to obtain new insights into progressive diseases. Such insights include fine-grained longitudinal patterns of neurodegeneration, from early stages, and the heterogeneity of these trajectories over the population. More pragmatically, such models enable finer precision in patient staging and stratification, prediction of progression rates and earlier and better identification of at-risk individuals. We argue that this will make disease progression modelling invaluable for recruitment and end-points in future clinical trials, potentially ameliorating the high failure rate in trials of, e.g., Alzheimer's disease therapies. We review the state of the art in these techniques and discuss the future steps required to translate the ideas to front-line application.
Keywords: ageing, Alzheimer, computational, disease progression, generative, modelling, multiple sclerosis, neurodegeneration, neurological, prion, statistical
INTRODUCTION
Neurodegenerative diseases present a global healthcare crisis. An estimated 47 million people suffer dementia worldwide and cost over $800 billion per year [1]. This will only intensify unless effective treatments are developed. Efforts in this regard currently face many challenges. Within a neurodegenerative disease, population and temporal heterogeneity couples with an oftentimes-protracted preclinical phase to render early diagnosis difficult. Across the spectrum of neurodegenerative diseases, differential diagnosis is complicated by overlapping phenotypes and common diseases. Tracking neurodegenerative disease progression has the same challenges, not to mention the prohibitive expense of longitudinal preclinical data acquisition at scale. Efforts are also confounded by comorbidities, environmental influences and ageing effects. Simplistic model of the complex, multifaceted nature of neurodegenerative diseases such as Alzheimer's disease has so far precluded the development of precision medicine for personalized treatment and care decisions, and is a likely factor underlying the glut of failed clinical trials.
Recent advances in computational approaches to the analysis of medical data are providing a powerful means to understand neurodegenerative diseases and to predict disease progression. By integrating a variety of clinical and biomedical data, including risk factors, biomarkers and interactions among them, these models give a uniquely holistic picture of disease progression from beginning to end. Such detailed understanding of the full disease time course provides new promise in overcoming the myriad challenges in managing neurodegenerative diseases.
Here, we review current progress in the field of neurodegenerative disease progression modelling. We start with an overview of the data upon which such models are built, continue to review the current technology and conclude with an outlook to the future.
Box 1.
no caption available
BIOMARKERS OF NEURODEGENERATIVE DISEASE
Accurate quantification of neurodegeneration in vivo is challenging. The multifactorial nature of neurodegenerative diseases necessitates considering a range of imaging and nonimaging biomarkers, which we review in this section. We pay specific attention to imaging biomarkers, which lend themselves particularly well to the kinds of model we discuss, but we mention also various other biomarkers that complement information from imaging.
Magnetic resonance imaging
MRI provides a unique noninvasive examination of tissue, in vivo[2]. MRI provides various markers of neurodegenerative disease including regional abnormalities in structure, microstructure, function and metabolism. Changes in these features can be subtle and occur at different disease stages.
Structural MRI reveals changes in brain structure. Abnormalities in the volume of a region of the brain typically reflect neurodegeneration in later stages of disease, although shape changes may occur earlier [3]. Higher specificity to volume loss is attained from short-term longitudinal data (atrophy) than from cross-sectional volumetric comparisons across individuals. Brain volume loss has been used for subject selection and as an endpoint in clinical trials for Alzheimer's disease [4].
Various quantitative or microstructural MRI techniques, for example using diffusion MRI [5,6], magnetization transfer [7], relaxometry [8], susceptibility imaging [9▪,10] or combinations thereof [11,12▪,13], can reveal changes at the cellular level that may precede macrostructural changes.
Diffusion MRI and functional MRI also provide insight into brain connectivity. Diffusion MRI, via tractography, enables the study of structural connectivity, which can be affected in neurodegenerative diseases [14▪▪], and may act as a network supporting pathogen propagation [15,16]. Functional MRI based on blood-oxygen-level-dependent contrast imaging [17] is an indirect measure of neuronal activity based on blood flow. It can reveal regions active in cognitive tasks, which can be disrupted in disease [18]. Moreover, temporal correlations of this activity highlight functional connectivity and its disruption in disease [19,20]. Similar functional information can come from electroencephalography or magnetoencephalography [21].
PET uses a radioactive tracer bound to a metabolite to map out various processes across the brain. Notable pathogenic processes include glucose hypometabolism [22] and concentrations of β-amyloid [23] or tau [24▪▪]. Abnormalities of each are common markers of neurodegenerative diseases.
Cognitive and other clinical presentations
Cognitive and other clinical presentations play a key role in differentiating between neurodegenerative diseases. Tests that measure cognitive impairment are susceptible to learning effects [25▪▪] and are confounded by depression/anxiety [26]. In order to improve the sensitivity of cognitive assessments for early detection [27], cognitive composites have been developed [28,29].
Cerebrospinal fluid
Pathogenically abnormal levels of, for example, misfolded proteins (or neurofibrillary tangles) in the brain can be detected in cerebrospinal fluid (CSF) [30,31]. Changes are often detectable many years prior to the clinical presentation of symptoms. However, CSF markers suffer from considerable variability within and between laboratories and assays [31,32], require a highly invasive spinal tap and lack spatial specificity within the brain.
Fixed variables
Fixed-variables such as demographics, genetics, environmental or lifestyle factors are also influential on disease progression. For example, neurodegeneration differences exist between sexes [33], ApoE4 carriage is a genetic risk factor in Alzheimer's disease [34] and various environmental factors may influence neurodegeneration [35].
ADVANCED COMPUTATIONAL MODELS OF DISEASE PROGRESSION
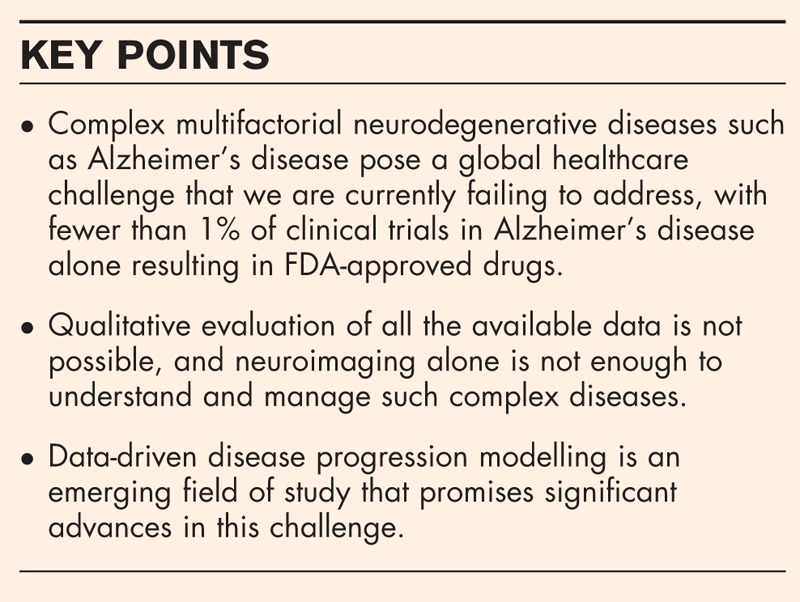
Data-driven disease models have emerged in recent years that provide uniquely rich disease signatures without relying on a-priori classification or staging of individuals. These models contrast with an earlier modelling paradigm (Fig. 1) including hypothetical models, such as those presented in [36–38], which sketch disease signatures like those in Fig. 1a but are not informed directly by measured data; traditional longitudinal models, for example [39–41] (Fig. 1b), which regress measured data against a prespecified variable that defines the disease stage; and pattern recognition approaches [42–44], which learn to classify cases from a labelled training database. Hypothetical models aid understanding, but lack the quantitative nature required for classification or prediction. Traditional models are inherently limited by, typically crude, a-priori staging and can model only variables that do not contribute to the stage assessment. Pattern recognition approaches are similarly limited by the a-priori labelling and provide no explicit disease signature to enhance understanding.
FIGURE 1.
Old paradigm disease progression models. (a) It shows the hypothetical model of [36], which illustrates qualitative sigmoid evolution in AD of scalar biomarkers such as CSF Aβ level, cognitive test scores and hippocampal volume or atrophy. The lack of quantitative information prevents direct diagnostic usage. (b) It shows a traditional longitudinal model of AD atrophy [39] by binning individuals a-priori into ‘mild’, ‘moderate’ and ‘severe’ classes based on cognitive test scores. The model can potentially match new individuals to the same stages using imaging data, but must exclude cognitive scores to avoid circularity. AD, Alzheimer's disease.
Discriminative models
Supervised machine-learning approaches use labelled data to discriminate between individuals who will/will not progress clinically, or do/do not have a disease [42–44]. They have shown the value of including both neuroimaging and nonimaging markers [44]. Straightforward classification approaches do not provide insight on the full disease time course, although many supervised machine-learning techniques also have the capability for regression so might be used to predict variables indicative of disease progression. However, such an approach still suffers from inherent limitations of the a-priori labelling.
Unsupervised learning techniques, or clustering, offer potential in disease subtyping and staging [45,46,47▪▪]. Direct application of these ideas simply identifies groups of patients with similar appearance, but does not account for the temporal nature of progressive diseases where a single phenotype may appear very different at the beginning to the end of the full disease time course.
The recent rise of deep learning techniques potentially benefits discriminative models of disease progression, but such approaches typically need large amounts of data for both training and validation. Current medical data sets rarely approximate the data sets of billions of images that deep learning has used to show great success in, for example, computer vision tasks such as in [48], but the technology nevertheless may prove beneficial, particularly by exploiting transfer learning [49].
Generative models
Advanced generative models of neurodegenerative disease progression avoid the need for a-priori staging to construct a quantitative signature from all available biomarkers. They fall broadly into two categories: models for unstructured data and models for structured data.
Models for unstructured data
These models work with a vector of scalar biomarkers with no assumption of a particular structure or relationship among them. They are designed to fuse together information from the constellation of multimodal biomarkers typically available in studies of neurodegenerative diseases.
The event-based model (EBM) [50–52] (Fig. 2) learns an ordering and uncertainty in the ordering, in which a set of biomarkers becomes abnormal directly from a measured data set, possibly entirely cross-sectional, and requires no predefined staging variable. EBM uses the idea that more individuals from a cohort containing a spectrum of disease stages will show abnormality in biomarkers that change early in the progression. The EBM has been used to study familial Alzheimer's disease [50,52], Huntington's disease [50] and sporadic Alzheimer's disease [51,52].
FIGURE 2.
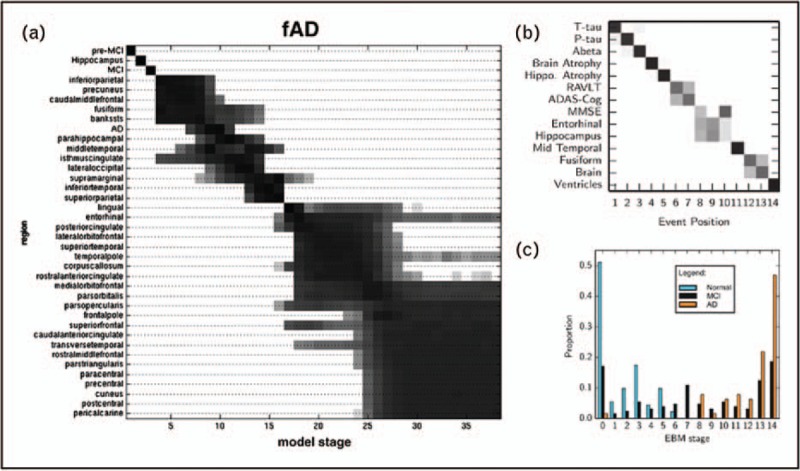
Event-based models of disease progression. (a) An EBM of familial AD from [49]. The set of biomarkers (vertical axis) includes atrophy rates in each cortical region. The positional variance diagram is a visualization of the uncertainty in the ordering: each row is a grayscale histogram of probabilities that the event occurs in each position of the ordered sequence. Groups of interchangeable events appear as large blocks (e.g. later in the progression, lower right), whereas strong confidence in the ordering of events appears as a thin diagonal (e.g. earlier in the progression, upper left). (b) An EBM of sporadic AD learned from the ADNI data set in [50] with more diverse biomarkers: CSF tau and Aβ; cognitive test scores; regional atrophy and volumes from MRI. The model defines a staging system based on the set of biomarkers showing abnormality. (c) Stage assignments of cognitively normal, mild cognitive impaired (MCI) and AD individuals showing classification accuracy similar to state-of-the-art pattern recognition approaches [44], but with an explicit generative model. AD, Alzheimer's disease; ADNI, Alzheimer's disease neuroimaging initiative; CSF, cerebrospinal fluid; EBM, event-based model.
Scalar trajectory models, derived from differential equation models [52–54] or self-modelling regression [55,56], reconstruct typical continuous trajectories of scalar biomarkers from a measured data set. As shown in Fig. 3, this gives a more complete temporal picture than the EBM's sequence of abnormality. Current implementations of differential equation models [52–54] treat each biomarker independently, whereas self-modelling regression pools information across biomarkers to estimate trajectories on a common time frame. Model estimation requires no staging variable, although the data set must contain at least short-term longitudinal information to support estimates of the temporal scale of the trajectories.
FIGURE 3.
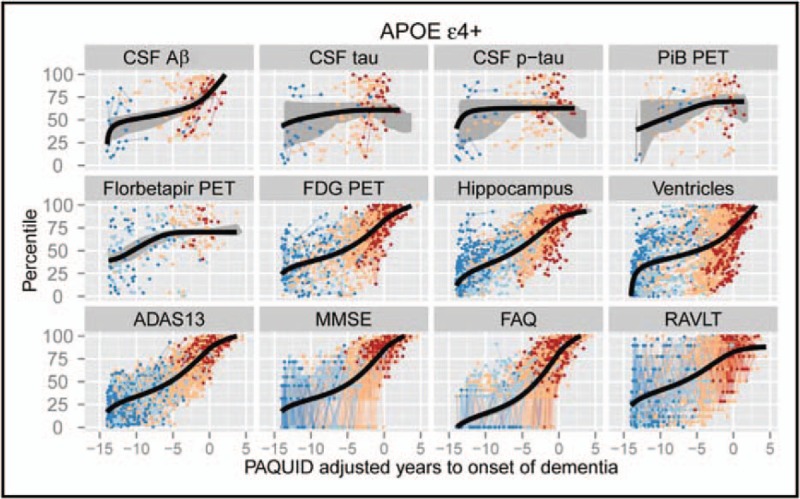
The temporally continuous self-modelling regression approach of [55]. The model shows the characteristic trajectories of a diverse set of biomarkers against a common continuous disease stage variable learned from the ADNI and PAQUID (Personnes Agées Quid) data sets. The model can potentially estimate the disease stage of a new patient by identifying the position along the trajectory set that best matches their data. ADNI, Alzheimer's disease neuroimaging initiative.
Models for structured data
These models work with structured data such as images, geometric meshes or networks in which data values have a well-defined spatial organization.
Spatiotemporal models [57,58,59▪▪] construct a four-dimensional picture of how an image or image region typically evolves in shape and/or appearance during a disease or developmental process. Traditional longitudinal models of this type are straightforward to construct by binning individuals according to a predefined variable, such as age or a cognitive test score, and computing statistics within each bin. Most current models [58,59▪▪] still rely on a-priori labelling. However, recent advances [57] avoid any predefined stage variable by simultaneously estimating a central image trajectory, a deformation of each time point of each individual to align with the central trajectory and a ‘timewarp’ for each individual and time point that defines the position along the central trajectory.
Network propagation models [60–64] attempt to explain disease progression in terms of spatial propagation through anatomical or functional networks in the brain, which thus induce measurement changes sequentially along the nodes of a network. The pattern of propagation depends on, and can thus reveal, mechanisms of propagation. In particular, Zhou et al.[62] translate various popular models for neurodegenerative disease propagation: ‘nodal stress’ – the most active regions or ‘hubs’ are most vulnerable [65,66]; ‘transneuronal spread’ – a prion-like agent propagates along network connections [15,16,67,68]; ‘trophic failure’ – opposite to ‘nodal stress’, the most isolated nodes are most vulnerable [69,70]; and ‘shared vulnerability’ – evenly distributed vulnerability over networks. Each model predicts different patterns of atrophy severity within networks, enabling comparison of the different propagation hypotheses in various neurological diseases. Dynamical system models [61,63] support the prion-like propagation hypothesis in a similar way. More recently, a multifactorial network model was proposed [64] that considers multiple brain networks and their interactions.
Although spatiotemporal models express the trajectory of much richer objects than scalar trajectory models, currently they typically model just one object (image or image region) derived from a single modality. Current formulations do extend naturally to vector-valued images, or multiple interacting regions, but the methods are memory and processor intensive so such extensions present practical difficulties. Network propagation models are typically used to make a single gross inference of propagation mechanism rather than a detailed disease signature so are somewhat distinct from the other models above and do not directly support diagnosis, staging or prognosis.
Current limitations
One of the most common limitations in disease progression models is the assumption of a common disease trajectory across individuals. Neurodegenerative diseases are highly heterogeneous and clearly violate this assumption. However, it does enable the models to elicit an average or canonical trajectory, which may often be sufficient to capture broad disease characteristics and discriminate different conditions. Various models naturally provide measures and visualizations of the uncertainty in that canonical trajectory, see for example the positional variance diagrams of the EBM in Fig. 2, although these cannot be interpreted directly as pictures of population heterogeneity. Various enhancements aim to estimate features of the population heterogeneity. For example, [71] estimates the distribution of the EBM ranking over Alzheimer's disease neuroimaging initiative individuals by replacing the single ordering with a Mallows model [72]; [47▪▪] identifies distinct components of Alzheimer's disease atrophy; and [73▪▪] uses a mixture of EBMs to identify subgroups of the population with distinct orderings. These ideas go some way toward ameliorating this limitation, but further work is required.
Another limitation in disease progression models is the assumption of biomarker independence. This may be valid for early vs. late biomarkers, but it is unlikely to be true for all biomarkers across the full disease time course. Some scalar trajectory models, in particular [55,56], avoid this assumption by explicitly pooling information across biomarkers in order to estimate disease progression. Recent spatially fine-grained models of disease accumulation across the brain use weak assumptions on the spatial correlation structure [74], or explicitly seek clusters with a correlated atrophy pattern [75]. The work in [75] further highlights, particularly for complex models such as spatiotemporal shape models of the brain, the need to balance parsimony and information content. This complexity control is necessary to make inference tractable in large data sets, while retaining the critical information about disease progression.
To date, little effort has gone into incorporating information from fixed variables into disease progression models. Simple approaches, such as constructing separate models for particular genotypes, for example as in [51], highlight that progression does depend on such information. However, lower level integration of such variables into the models may produce interesting new findings.
Other limitations arise from idiosyncrasies of medical data sets. Longitudinal data censoring because of participant dropout can lead to undersampling of later disease stages and, if not accounted for, can bias models towards earlier disease stages. Measurement error can bias even simple models, for example regression dilution, and so explicitly quantifying and expressing uncertainty within disease models is important [54]. Outliers can bias models and frequently arise, for example because of unrelated disease such as a brain tumour causing exaggerated brain volumes, or erroneous data entry. Missing data are also common and, although disease progression models often have formulations that handle missing data in theory [73▪▪], the effect on downstream inference remains to be quantified precisely.
FUTURE AND EMERGING TRENDS
Enhancing existing models
The combination of phenomenological models and mechanistic models offers powerful opportunities to compare hypotheses on disease propagation. Early explorations of mechanistic models [61,62] compare with only end-stage atrophy patterns. However, such models can predict the whole time course of neurodegeneration. Matching just the overall atrophy pattern is analogous to guessing the story of a movie from only the final scene. The temporal disease signatures uniquely provided by longitudinal data-driven disease progression models, such as the EBM or continuous trajectory models, open up the possibility of matching the whole pattern of appearance of disease – viewing the whole neurodegeneration movie as it unfolds for much stronger inferences on underlying mechanisms.
Recent advances include automatic determination of disease subtypes via data-driven mixture modelling [73▪▪]. The use of subtype models to identify the distinct phenotypes among patients provides fundamentally new knowledge that may reveal new subgroups in diseases that are known to be highly heterogeneous, or highlight common comorbidities. This has the potential to aid treatment development through identifying smaller and more homogeneous patient cohorts more likely to have coherent underlying biochemistry and disease, and thus to respond consistently to a single treatment. The subtype models further offer the radical possibility of challenging the traditional labels of dementia subtypes (Alzheimer's disease, dementia with Lewy bodies, frontotemporal dementia, posterior cortical atrophy and so on.) for the first time. Data-driven subtypes may prove more homogeneous and inherently easier to recognize from clinical data than the traditional labels. They may thus prove simpler targets for drug development thereby supporting precision medicine more readily, by matching individuals to treatments more precisely.
Enhancing these models to include causal inference will be an important advance. A recent multifactorial model [64] incorporated linear systems control theory into a model with causal structure1 to suggest necessary features of putative treatments that could have disease-modifying effects.
The complementary information provided by each type of disease progression model mentioned above can be combined into uniquely rich, quantitative signatures of disease that avoid the need for a-priori staging. This could be achieved through a single unified modelling framework. For example, there are natural links from the EBM to scalar trajectory models, to spatiotemporal trajectory models. Such a unified modelling framework potentially supports rigorous inference from the collection of disparate information available, balancing evidence from the multiple modalities appropriately.
Translational requirements to realize the potential of disease progression modelling
Currently, disease progression models remain largely in the technical research domain — being developed by computer scientists and statisticians in collaboration with clinical experts. They have great potential for application in the clinic and in clinical trials, but likely require several stages of development and proof of concept to reach the necessary level of maturity.
We must find ways to demonstrate consistent levels of performance. This requires robust and interpretable metrics of predictive performance, which include evaluation against known ground truth in simulations and well-phenotyped data sets (e.g. familial cohorts). Larger, less well-phenotyped data sets support performance evaluation in terms of model stability (e.g. cross-validation and intercentre generalizability), temporal resolution and staging self-consistency.
Clear performance metrics and yardsticks will provide developers of disease models with a clear understanding of how model assumptions bias model predictions and how to interpret and temper those predictions in practice. We must also find ways to transmit that understanding to end users to avoid abuse through misunderstanding. This involves the development of clear user interfaces to make the model output accessible and digestible by nonexpert users, and clear instructions and case studies demonstrating how to use the technology in practical situations.
Thus, key steps that remain for translation are:
-
(1)
External validation of predictive ability of developed models on research data sets. Several works already show this, but the community still has a need for unbiased evaluation in entirely unseen test sets. Community wide challenges such as [76,77] provide a valuable resource in this way.
-
(2)
Translation between data sets. Key questions remain on how well within-dataset performance will translate to other data sets, that is if we train models on a data set with a particular acquisition protocol, how well can we expect it to perform on unseen data from different protocols or acquired in different centres? Little exploration of this has been performed.
-
(3)
Demonstration in clinical trials. Initial steps to demonstrate potential to improve outcome via retrospective analysis of clinical trial data would support more ambitious work in demonstrating the potential of multimodal modelling in a prospective clinical trial.
-
(4)
Success in a small number of trials involving model developers can motivate adaptation of the technology for general usage without developer intervention and widespread deployment in treatment development. Such advances will require substantial efforts in interface design and user workflow modelling.
CONCLUSION
We have reviewed data-driven model-based analyses of neurodegenerative disease. We have argued the potential for generative data-driven models to take centre stage in the study and management of neurodegenerative diseases if we are to generate new avenues for disease understanding in the earliest, preclinical stages. This is necessitated by the challenges in monitoring any neurological disease over its full time course, coupled with overlapping phenotypes and lack of a single biomarker that is dynamic across the full disease time course. The main focus of development and application to date has been in Alzheimer's disease, but various efforts including the EuroPOND project are expanding the application to other dementias, multiple-sclerosis, prion diseases, normal ageing and development, and even non-brain applications. These techniques have the potential for widespread impact in realising precision medicine across many such domains.
Acknowledgements
The authors acknowledge their colleagues in the EuroPOND consortium (http://www.europond.eu), who contributed indirectly to this review and continue to contribute to the field of neurodegenerative disease progression modelling. The EuroPOND coinvestigators are Daniel C. Alexander, Neil P. Oxtoby, Sebastien Ourselin, Jonathan M. Schott, Olga Ciccarelli, Stanley Durrleman, Olivier Colliot, Meike W. Vernooij, Wiro J. Niessen, Stefan Klein, Frederik Barkhof, Giovanni B. Frisoni, Alberto Bizzi, Petra S. Hüppi and Wim Van Hecke.
Financial support and sponsorship
The authors receive funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 666992.
Conflicts of interest
The authors declare that they have no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Footnotes
Although the model structure (mathematics) is causal, the data-driven model (parameters estimated from the data) is not causal because it was built on cross-sectional data.
REFERENCES
- 1.Alzheimer's Disease International, Prince M, Comas-Herrera A, Knapp M, et al. World Alzheimer Report 2016 [Internet]. 2016; https://www.alz.co.uk/research/WorldAlzheimerReport2016.pdf [Google Scholar]
- 2.Bozzali M, Serra L, Cercignani M. Quantitative MRI to understand Alzheimer's disease pathophysiology. Curr Opin Neurol 2016; 29:437–444. [DOI] [PubMed] [Google Scholar]
- 3.Springer International Publishing, Cury C, Lorenzi M, Cash D. Reuter M, Wachinger C, Lombaert H, et al. Spatio-temporal shape analysis of cross-sectional data for detection of early changes in neurodegenerative disease. Spectral and Shape Analysis in Medical Imaging: First International Workshop, SeSAMI 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Revised Selected Papers 2016; 63–75. [Google Scholar]
- 4.Cash DM, Rohrer JD, Ryan NS, et al. Imaging endpoints for clinical trials in Alzheimer's disease. Alzheimer's Res Ther 2014; 6:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J 1994; 66:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assaf Y, Alexander DC, Jones DK, et al. The CONNECT project: combining macro- and micro-structure. Neuroimage 2013; 80:273–282. [DOI] [PubMed] [Google Scholar]
- 7.Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxationin vivo. Magn Reson Med 1989; 10:135–144. [DOI] [PubMed] [Google Scholar]
- 8.Laule C, Vavasour IM, Kolind SH, et al. Magnetic resonance imaging of myelin. Neurotherapeutics 2007; 4:460–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Deistung A, Schweser F, Reichenbach JR. Overview of quantitative susceptibility mapping. NMR Biomed 2017; 30:e3569. [DOI] [PubMed] [Google Scholar]; Review of both the background theory of quantitative susceptibility mapping and current applications in neuroimaging.
- 10.Wharton S, Bowtell R. Fiber orientation-dependent white matter contrast in gradient echo MRI. Proc Natl Acad Sci U S A 2012; 109:18559–18564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stikov N, Perry LM, Mezer A, et al. Bound pool fractions complement diffusion measures to describe white matter micro and macrostructure. Neuroimage 2011; 54:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪.Kim D, Doyle EK, Wisnowski JL, et al. Diffusion-relaxation correlation spectroscopic imaging: a multidimensional approach for probing microstructure. Magn Reson Med 2017; (in press, doi:10.1002/mrm.26629). [DOI] [PMC free article] [PubMed] [Google Scholar]; A new hybrid compartmental model for investigating white matter at the subvoxel scale.
- 13.Weiskopf N, Suckling J, Williams G, et al. Quantitative multiparameter mapping of R1, PD∗, MT, and R2∗ at 3T: a multicenter validation. Front Neurosci 2013; 7:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪▪.Iturria-Medina Y, Evans AC. On the central role of brain connectivity in neurodegenerative disease progression. Front Aging Neurosci 2015; 7:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]; Important review article for this emerging topic in mechanistic disease models.
- 15.Prusiner SB. Some speculations about prions, amyloid, and Alzheimer's disease. N Engl J Med 1984; 310:661–663. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner SB. A unifying role for prions in neurodegenerative diseases. Science 2012; 336:1511–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A 1990; 87:9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdulkadir A, Ronneberger O, Christian Wolf R, et al. Functional and structural MRI biomarkers to detect pre-clinical neurodegeneration. Curr Alzheimer Res 2013; 10:125–134. [DOI] [PubMed] [Google Scholar]
- 19.Pievani M, de Haan W, Wu T, et al. Functional network disruption in the degenerative dementias. Lancet Neurol 2011; 10:829–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pievani M, Filippini N, van den Heuvel MP, et al. Brain connectivity in neurodegenerative diseases: from phenotype to proteinopathy. Nat Rev Neurol 2014; 10:620–633. [DOI] [PubMed] [Google Scholar]
- 21.Hulbert S, Adeli H. EEG/MEG- and imaging-based diagnosis of Alzheimer's disease. Rev Neurosci 2013; 24:563–576. [DOI] [PubMed] [Google Scholar]
- 22.InTech, Teune LK, Bartels AL, Leenders KL. FDG- PET imaging in neurodegenerative brain diseases. Functional Brain Mapping and the Endeavor to Understand the Working Brain. 2013. [Google Scholar]
- 23.Jagust WJ. Amyloid imaging: coming to a PET scanner near you. Ann Neurol 2010; 68:277–278. [DOI] [PubMed] [Google Scholar]
- 24▪▪.Villemagne VL, Fodero-Tavoletti MT, Masters CL, Rowe CC. Tau imaging: early progress and future directions. Lancet Neurol 2015; 14:114–124. [DOI] [PubMed] [Google Scholar]; Tau imaging may become the weapon of choice for monitoring progression and treatment effect in diseases such as Alzheimer's disease. Authored by world leaders in PET imaging.
- 25▪▪.Goldberg TE, Harvey PD, Wesnes KA, et al. Practice effects due to serial cognitive assessment: implications for preclinical Alzheimer's disease randomized controlled trials. Alzheimer's Dement Diagn Assess Dis Monit 2015; 1:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Practice effects are confounders in monitoring disease progression, particularly for clinical trials. This article focuses on this important issue and suggests ways around the problem.
- 26.Rohling ML, Green P, Allen LM, Iverson GL. Depressive symptoms and neurocognitive test scores in patients passing symptom validity tests. Arch Clin Neuropsychol 2002; 17:205–222. [PubMed] [Google Scholar]
- 27.Silverberg NB, Ryan LM, Carrillo MC, et al. Assessment of cognition in early dementia. Alzheimer's Demen 2011; 7:e60–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donohue MC, Sperling RA, Salmon DP, et al. The preclinical Alzheimer cognitive composite. JAMA Neurol 2014; 71:961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burnham SC, Raghavan N, Wilson W, et al. Novel statistically-derived composite measures for assessing the efficacy of disease-modifying therapies in prodromal Alzheimer's disease trials: an AIBL study. J Alzheimer's Dis 2015; 46:1079–1089. [DOI] [PubMed] [Google Scholar]
- 30.Blennow K, Hampel H. CSF markers for incipient Alzheimer's disease. Lancet Neurol 2003; 2:605–613. [DOI] [PubMed] [Google Scholar]
- 31.Blennow K, Dubois B, Fagan AM, et al. Clinical utility of cerebrospinal fluid biomarkers in the diagnosis of early Alzheimer's disease. Alzheimer's Demen 2015; 11:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos SJB, Visser PJ, Verhey F, et al. Variability of CSF Alzheimer's disease biomarkers: implications for clinical practice. PLoS One 2014; 9:e100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanamsagar R, Bilbo SD. Sex differences in neurodevelopmental and neurodegenerative disorders: focus on microglial function and neuroinflammation during development. J Steroid Biochem Mol Biol 2016; 160:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C-C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013; 9:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown RC, Lockwood AH, Sonawane BR. Neurodegenerative diseases: an overview of environmental risk factors. Environ Health Perspect 2005; 113:1250–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jack CR, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 2010; 9:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frisoni GB, Fox NC, Jack CR, et al. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 2010; 6:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aisen PS, Petersen RC, Donohue MC, et al. Clinical core of the Alzheimer's disease neuroimaging initiative: progress and plans. Alzheimer's Demen 2010; 6:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scahill RI, Schott JM, Stevens JM, et al. Mapping the evolution of regional atrophy in Alzheimer's disease: unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A 2002; 99:4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridha BH, Barnes J, Bartlett JW, et al. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol 2006; 5:828–834. [DOI] [PubMed] [Google Scholar]
- 41.Bateman RJ, Xiong C, Benzinger TLS, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med 2012; 367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klöppel S, Stonnington CM, Chu C, et al. Automatic classification of MR scans in Alzheimer's disease. Brain 2008; 131:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattila J, Koikkalainen J, Virkki A, et al. A Disease state fingerprint for evaluation of Alzheimer's disease. J Alzheimer's Dis 2011; 27:163–176. [DOI] [PubMed] [Google Scholar]
- 44.Young J, Modat M, Cardoso MJ, et al. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. Neuroimage Clin 2013; 2:735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitwell JL, Przybelski SA, Weigand SD, et al. Distinct anatomical subtypes of the behavioural variant of frontotemporal dementia: a cluster analysis study. Brain 2009; 132:2932–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nettiksimmons J, DeCarli C, Landau S, Beckett L. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimer's Demen 2014; 10:511–521. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47▪▪.Zhang X, Mormino EC, Sun N, et al. The Alzheimer's disease neuroimaging initiative: Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer's disease. Proc Natl Acad Sci U S A 2016; 113:E6535–E6544. [DOI] [PMC free article] [PubMed] [Google Scholar]; Example of a modern data-driven approach that takes into account the widely observed phenomenon of disease variability between people, such as multiple disease progression subtypes.
- 48.Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems (NIPS) 2012; pp 1097–1105. URL is http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. [Google Scholar]
- 49.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans Med Imaging. 2016; 35:1299–1312. [DOI] [PubMed] [Google Scholar]
- 50.Fonteijn HM, Modat M, Clarkson MJ, et al. An event-based model for disease progression and its application in familial Alzheimer's disease and Huntington's disease. Neuroimage 2012; 60:1880–1889. [DOI] [PubMed] [Google Scholar]
- 51.Young AL, Oxtoby NP, Daga P, et al. A data-driven model of biomarker changes in sporadic Alzheimer's disease. Brain 2014; 137:2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oxtoby NP, Young AL, Lorenzi M, et al. Model-based comparison of autosomal-dominant and late-onset Alzheimer's disease progression in the DIAN and ADNI studies. Alzheimer's Demen 2016; 12:668–669. [Google Scholar]
- 53.Villemagne VL, Burnham S, Bourgeat P, et al. Amyloid deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol 2013; 12:357–367. [DOI] [PubMed] [Google Scholar]
- 54.Oxtoby NP, Young AL, Fox NC, et al. Initiative TADN. Learning imaging biomarker trajectories from noisy Alzheimer's disease data using a Bayesian multilevel model. In BAMBI 2014. Lecture Notes in Computer Science 8677:85–94. [Google Scholar]
- 55.Jedynak BM, Lang A, Liu B, et al. A computational neurodegenerative disease progression score: method and results with the Alzheimer's disease neuroimaging initiative cohort. Neuroimage 2012; 63:1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donohue MC, Jacqmin-Gadda H’E, Le Goff M’, et al. Estimating long-term multivariate progression from short-term data. Alzheimer's Demen 2014; 10:S400–S410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durrleman S, Pennec X, Trouve A, et al. Toward a comprehensive framework for the spatiotemporal statistical analysis of longitudinal shape data. Int J Comp Vis 2013; 103:22–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huizinga W, Poot DHIJ, Roshchupkin G, et al. Modeling the brain morphology distribution in the general aging population. In Proc. SPIE 9788, Medical Imaging 2016: Biomedical Applications in Molecular, Structural, and Functional Imaging, 2016. URL is http://proceedings.spiedigitallibrary.org/proceeding.aspx?articleid=2511101. [Google Scholar]
- 59▪▪.Lorenzi M, Pennec X, Frisoni GB, Ayache N. Disentangling normal aging from Alzheimer's disease in structural magnetic resonance images. Neurobiol Aging 2015; 36 Supplement 1:S42–S52. [DOI] [PubMed] [Google Scholar]; Age is the primary risk factor in most neurodegenerative diseases and can have a confounding effect on disease progression monitoring if not disentangled from pathological loss of brain tissue.
- 60.Seeley WW, Crawford RK, Zhou J, et al. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009; 62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron 2012; 73:1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou J, Gennatas ED, Kramer JH, et al. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 2012; 73:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iturria-Medina Y, Sotero RC, Toussaint PJ, Evans AC. and the Alzheimer's Disease Neuroimaging Initiative. Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput Biol 2014; 10:e1003956–e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iturria-Medina Y, Carbonell FM, Sotero RC, et al. Multifactorial causal model of brain (dis)organization and therapeutic intervention – application to Alzheimer's disease. Neuroimage 2017; 152:60–77. [DOI] [PubMed] [Google Scholar]
- 65.Buckner RL, Sepulcre J, Talukdar T, et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 2009; 29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 2011; 71:35–48. [DOI] [PubMed] [Google Scholar]
- 67.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci 2010; 11:155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013; 501:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Appel SH. A unifying hypothesis for the cause of amyotrophic lateral sclerosis, Parkinsonism, and Alzheimer disease. Ann Neurol 1981; 10:499–505. [DOI] [PubMed] [Google Scholar]
- 70.Salehi A, Delcroix J-D, Belichenko PV, et al. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron 2006; 51:29–42. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, Alexander D: Probabilistic event cascades for Alzheimer's disease. In Advances in Neural Information Processing Systems (NIPS), 2012:3104–3112. URL is http://papers.nips.cc/paper/4856-probabilistic-event-cascades-for-alzheimers-disease. [Google Scholar]
- 72.Beckett LA: Maximum likelihood estimation in Mallows's model using partially ranked data. In Probability models and statistical analyses for ranking data. Papers presented at the AMS-IMS-SIAM conference, Amherst, MA, USA, June 1990. New York: Springer-Verlag; 1993:92–107. [Google Scholar]
- 73▪▪.Young AL, Oxtoby NP, Huang J, et al. Multiple orderings of events in disease progression. In Information Processing in Medical Imaging (IPMI) 2015. Lecture Notes in Computer Science 9123:711–722. [DOI] [PubMed] [Google Scholar]; One of the first examples of an advanced computational model of disease progression that incorporates automated subtyping.
- 74.Bilgel M, Prince JL, Wong DF, et al. A multivariate nonlinear mixed effects model for longitudinal image analysis: Application to amyloid imaging. Neuroimage 2016; 134:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marinescu RV, Eshaghi A, Lorenzi M, et al. A vertex clustering model for disease progression: application to cortical thickness images. In Information Processing in Medical Imaging (IPMI) 2017, (in press). to appear in http://www.springer.com/gb/book/9783319590493. [Google Scholar]
- 76.Bron EE, Smits M, van der Flier WM, et al. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: the CADDementia challenge. Neuroimage 2015; 111:562–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Allen GI, Amoroso N, Anghel C, et al. Crowdsourced estimation of cognitive decline and resilience in Alzheimer's disease. Alzheimer's Demen 2016; 12:645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]


