Abstract
Background/Objectives
Genome-wide association studies (GWAS) have identified many loci associated with body mass index (BMI) in many different populations. Variants in the FTO locus are reported to be one of the strongest genetic predictors of obesity. Recent publications pointed also to a topologically associated domain (TAD) which is identified as a novel region affecting BMI. The TAD area encompasses the IRXB cluster (IRX3, IRX5, IRX6), FTO and RPGRIP1L genes.
Subjects/Methods
In this study, we investigated the relationship between variation of the FTO and IRX genes and obesity in Poles. We presented a case—control association analysis (normal versus overweight and/or obesity group) of Polish adult individuals (N = 5418). We determined whether or not the chromosomal region 16:53 500 000–55 500 000 contains polymorphic variants which are correlated with BMI in Polish population, including sex and age stratified analysis.
Results
The obtained results showed that the problem of weight-height abnormalities differently affects populations of Polish women and men (χ2 = 187.1; p<0.0001). From 353 SNPs enrolled to this study, 86 were statistically significant (highest χ2 = 15.72; p = 7.35E-05 observed for rs1558902). Linkage disequilibrium (LD) analysis revealed 61 blocks in the tested region of chromosome 16, with 24 SNPs located within the same block (block 8) of approximately 40 kb, in almost complete LD (|D’|>0.98, r2>0.80). We confirmed presence of the genetic susceptibility loci located in intron 1 of the FTO gene, which were correlated with BMI in our study group. For the first time, our analyses revealed strong association of FTO intronic variants (block 8) with overweight in group of men only. We have also identified association of the IRX region with overweight and/or obesity in Polish individuals.
Conclusion
Our study demonstrated how tested SNPs make differential contributions to obesity and overweight risk. We revealed sex dependent differences in the distribution of tested loci which are associated with BMI in the population of Poles.
Introduction
Obesity is one of the biggest health care problems worldwide. It is the most common nutritional disorder in developed countries, and it increases risk for hypertension, cardiovascular disease, and type 2 diabetes [1, 2]. It is a complex, multifactorial medical condition, affected by genetic and environmental risk factors [3–5]. According to the World Health Organization (2014) there are around 2 billion overweight adults, of those 670 million are considered to be affected by obesity (BMI ≥30 kg/m2) and 98 million severely affected by obesity (BMI ≥35 kg/m2). In Poland the current prevalence of overweight and obesity among the Polish population have been estimated to 36.54 and 13.34%, respectively (WHO website http://www.who.int/bmi).
Twin and adoption studies have revealed that genetic factors contribute to 40–70% variation of BMI within a population [6, 7]. Large-scale GWAS have identified many loci associated with BMI. In 2007 three independent studies identified FTO as obesity susceptibility gene [8–10]. A number of different obesity-susceptible loci have been reported by the GWAS approach (MC4R, MC3R, SLC6A14, TMEM18, POMC, BDNF, NEGR1) [11]. Among these findings, the biggest effect on obesity phenotype till date has the FTO. The FTO gene is located on the long arm of chromosome 16 (16q12.2) and encodes a protein with the activity of 2-oxoglutarate- and iron-dependent nucleic acid demethylase (II). The FTO is expressed in many tissues including hypothalamus, pituitary and adrenal glands [8, 12].
The BMI influencing variants of the FTO are associated with increased intake of energy [13], dietary fat [14], protein [15], increased appetite and reduced satiety [16] and loss of control over eating [17]. Fully functional FTO is critical for normal development and physiology [18]. Impact of obesity predisposing variants of FTO on BMI changes throughout life. They do not influence birth weight [19]; body weight starts to be affected in early childhood, and declines through adulthood [20–23]. Sex influence obesity through hormonal, genetic and environmental factors [24, 25]. This reflects in sex-specific body fat distribution and is associated with it genetic loci [26].
Numerous studies confirmed the association between FTO common variants and obesity phenotype in many different populations [9, 10, 27]. Up to date, the genetic variability of FTO in the Polish population was not explored well. There are few studies which focused only on distribution of rs9939609 AA [22, 23, 28] variant and its association with higher BMI in Polish children and adults. Wrzostek et al. found the association of the G allele of rs9930506 with higher BMI among Polish adult subjects [29]. Multiple studies have also focused on deciphering potential mechanisms, by which variants within a region of high LD in introns 1 and 2 of FTO confer the obesity risk [30]. Recent evidence from Landgraf and colleagues suggested that FTO variants directly affect adipocyte function through targeting IRX3 and IRX5 [30]. Presence of the FTO risk haplotype in lean children was found to be associated with increased adipocyte-specific expression of IRX3 and IRX5, whereas it was unaffected in a group of obese children.
The objective of the present study is to determine (by means of case-control association analysis) in what manner the polymorphic risk variants in FTO/IRXB region affect obesity and/or overweight in the Polish population. This is the first comprehensive study of cross-sectional individuals from the population at large, with representation from each geographical region of Poland. Presented case-control analysis comprising men and women subgroups gave a novel point of view on genetic association of FTO and IRXB cluster with obesity and overweight which depend on sex and age.
Materials and methods
Description of the participants
The participants were recruited in years 2010–2012 within research project TESTOPLEK and registered as a POPULOUS collection at the Biobank Lab of The Department of Molecular Biophysics of The University of Lodz [31]. Sampling was performed by a professional public opinion polling and survey-taking company (SMG/KRC Poland, a Millward Brown subsidiary). Each subject gave the written informed consent and fulfilled questionnaire. The saliva was collected into Oragene OG-500 DNA collection/storage receptacles (DNA Genotek, Kanata, Canada) from each individual. The approval for this study was obtained from The University of Lodz’s Review Board (KBBN-UL/II/2014). All procedures were performed in accordance with the Declaration of Helsinki (ethical principles for medical research involving human subjects).
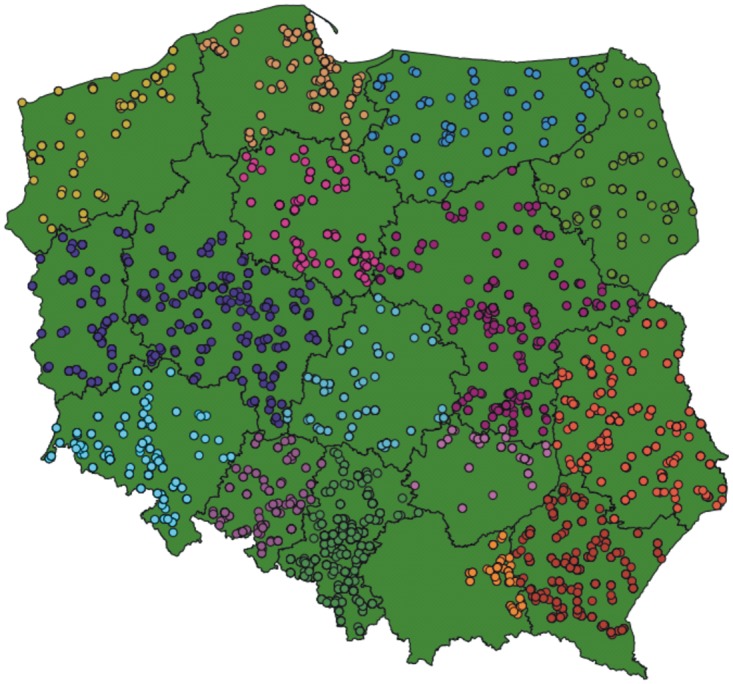
From over 10000 individuals throughout Poland, a total of 6047 participants were involved in creation a study group (Fig 1). The exclusion criteria were: diabetes, leukemia, bone marrow transplantation and cancer. Individuals who declared any of these diseases were excluded from the study (n = 488). A total of 5559 subjects declared themselves healthy. The study group consisted of 2812 females and 2747 males.
Fig 1. Geographic residence place of recruited participants included in study group.
BMI of each participant was calculated from the standard formula: BMI = weight/height2, based on self-assessment of height and weight controlled by trained pollsters.
DNA extraction and quantification
The saliva samples were stored in room temperature until initial processing. DNA was manually isolated from 500 μl of saliva using the manufacturer procedure (PrepitL2P, PD-PR-052, DNA Genotek, Kanata, Canada). The elution volume was 50 μl. DNA was quantified using broad range Quant-iT™ dsDNA Broad Range Assay Kit (Invitrogen™, Carlsbad, CA, USA). All DNA samples underwent PCR reaction for sex verification, which is a standard procedure of DNA quality control in our laboratory [32]. Only the DNA samples with proper concentration and quality were included and were diluted to 50 ng/μl in sterile DNase free water. The concentration was checked again using the same fluorometric method.
All laboratory methods related to the samples management were performed according to the standard operating procedures (SOPs) of Biobank Lab.
Genotyping
A total of 5559 DNA samples were genotyped for >550 000 SNPs using the 24x1 Infinium HTS Human Core Exome (Illumina Inc., San Diego, CA, USA) according to the protocol provided by the manufacturer. Briefly, DNA samples were amplified, then enzymatically fragmented and hybridized to the BeadChips. Afterwards, the BeadChips underwent an extension and X-staining processes. Next the BeadChips were scanned using iScan (Illumina Inc., San Diego, CA, USA). Raw fluorescence intensities were imported to the GenomeStudio with the Genotyping Module. All the data first underwent stringent quality control including sample exclusion (n = 141) if call rate was below 0.94 and 10% GenCall parameter was below 0.4. We filtered the data from the FTO and IRX region (NC_000016.9 (53 500 000–55 500 000)) which gave 353 SNPs for the analysis. All the SNPs reported in the presented study passed visual inspection for cluster quality. The results were exported from GenomeStudio using PLINK Input Report Plug-in v2.1.3 by forward strand. All sequence coordinates in this study are followed by GRCh37/hg19 reference sequence and were obtained from GenBank (http://www.ncbi.nlm.nih.gov).
Statistical analysis
Associations between SNPs and normal weight (control group) versus overweight and/or obesity (case group) were carried out using PLINK 2.050 [33] (http://pngu.mgh.harvard.edu/purcell/plink/).
All of the tested SNPs were first analyzed for consistency with the exact test for Hardy—Weinberg Equilibrium implemented by Wigginton et al. [34]. Differences in the distributions of allele frequencies between normal weight vs. overweight, normal weight vs. obese and normal weight vs. overweight & obese groups were evaluated under additive, dominant, and recessive inheritance models adjusted for sex and age. For these, significantly correlated with BMI categories, the odds ratio (OR) with 95% confidence interval (CI) was estimated. We have also applied the Cochran—Mantel—Haenszel (CMH) statistics to test association of SNPs with overweight and/or when assuming age stratification in group of female and male [35, 36].
Moreover, to investigate the impact of the tested polymorphisms on BMI, we have also performed a linear regression analysis, with BMI as a continuous trait and age and sex as covariates. Analysis of LD and calculation of haplotype frequencies were performed using the Haploview 4.2 software [37] (http://www.broadinstitute.org/haploview/haploview). Associations of estimated haplotypes were carried out using the case-control allelic chi-squared test.
Results
Characteristic of the study group
A total of 5559 unrelated Polish adult subjects were enrolled to the study group. According to the GWAS exclusion criteria described above we excluded 141 samples due to bad quality of the genotyping. After exclusion, there were 2758 women and 2660 men (N = 5418). Case (obesity and/or overweight individuals) and control (normal weight individuals) groups were established from the study group according to the BMI (kg/m2). The sample size for obese subjects (BMI ≥ 30 kg/m2) was 797 (male:female ratio 424:373, age 50.16 ± 13.55 years, BMI 33.15 ± 3.33 kg/m2), whereas for normal weight controls (BMI 18.5–24.99 kg/m2) was 2624 (male:female ratio 1127:1497, age 38.74 ± 14.13 years, BMI 22.27 ± 1.71 kg/m2). The underweight (BMI < 18.5 kg/m2) and overweight (BMI 25–29.99 kg/m2) groups consisting of 192 (male:female ratio 155:37, mean age 32.30 ± 12.31 years, BMI 17.49 ± 0.92 kg/m2) and 1805 individuals (male:female ratio 1072:733, with mean age 45.48 ± 14.25 years and BMI 27.19 ± 1.39 kg/m2), respectively, were also included to the analysis (Table 1).
Table 1. Characterization of the study group (N = 5418).
| Group | BMI [kg/m2] | N = 5418 | Male:Female Ratio | Mean BMI [kg/m2] | Mean Age [years] |
|---|---|---|---|---|---|
| UNDERWEIGHT | <18.5 | 192 | 155: 37 | 17.5 ± 0.9 | 32.3 ± 12.3 |
| NORMAL WEIGHT | 18.5–24.99 | 2624 | 1127: 1497 | 22.3 ± 1.7 | 38.7 ± 14.1 |
| OVERWEIGHT | 25–29.99 | 1805 | 1072: 733 | 27.2 ± 1.4 | 45.5 ± 14.3 |
| OBESITY | ≥30 | 797 | 424: 373 | 33.1 ± 3.3 | 50.2 ± 13.6 |
Mean BMI ±SD; Mean Age ±SD
In our study we successfully genotyped SNPs spanning the 2 Mb region, encompassing the RPGRIP1L, FTO, IRX3, IRX5, IRX6 genes. This region was selected according to the Hunt et al. [38] to encompass a TAD defined in embryonic stem cells [39]. We found 353 SNPs in the tested area. We estimated the minor allele frequency (MAF) of all mentioned SNPs and realized that 89 of them had a MAF under 0.01 (S1 Table) so we excluded them from the further analysis. In the case and control groups (S2 Table), two SNPs, rs4784382 and rs1211435, showed evidence for deviations from Hardy-Weinberg equilibrium (HWE; p<1E-04) and they were also excluded. Total genotyping rate in all individuals was 0.999 for all tested SNPs.
In summary, we conducted association analyses for 262 SNPs with an estimated MAFs > 1% in 5418 unrelated Polish adults, divided into four groups as described above and presented in Table 1. The ratio of women to men in our study group was 1.037 (50.9%: 49.1%).
Linkage disequilibrium and haplotypes association analysis
We investigated the role of haplotypes formed by tested SNPs (n = 262) in the risk for obesity in our study group. Haplotyping was performed with Haploview [37]. As shown in Fig 2, LD analysis revealed 61 blocks in the tested region of chromosome 16, with 24 SNPs located within the same block 8 of approximately 42 kb, in almost complete LD (|D’|>0.98). This previously identified region in the first intron of the FTO contains rs76804286, localized next to the rs9939609, which is in complete dependence with all the SNPs in this block and with six SNPs from the block 3, and also with SNPs from the blocks 4, 5, 6 and 7. Haplotypes are depicted at S1 Fig. The strongest obesity associating haplotype (χ2 = 11.474, p = 0.0007) was in block 8 and occurred at a frequency of 0.46 in the case group and 0.41 in controls (S3 Table). Two other haplotypes were also associated with higher BMI: first in block 30 which is localized 200 kb upstream IRX3 and second in 57 localized 8 kb upstream IRX6, (p = 0.0079 and 0.0097, respectively).
Fig 2. Linkage disequilibrium analysis of 262 SNPs from chromosomal region 16:53 500 000–55 500 000.
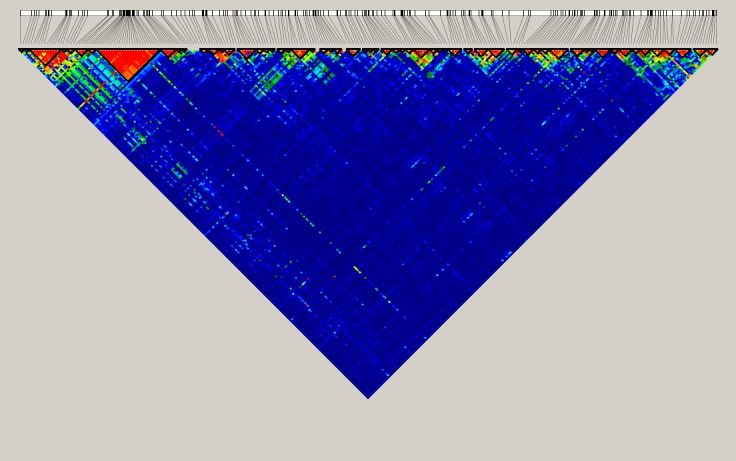
Pairwise linkage exhibited as LD coefficients (|D’|) between each pair of SNPs. Minor allele frequencies of all SNPs used in this analysis are ≥10%. Bolded triangles show haplotype blocks. Data were generated with Haploview version 4.2.
Case-control association studies
Using 262 selected SNPs, we performed tests of independence between BMI and alleles of each SNP using differently coded control and case phenotype group: normal vs. overweight, normal vs. obesity, normal vs. overweight and obesity. As the underweight group was represented by only 192 individuals we excluded it from association analysis.
We found that the association of alleles of tested SNPs with BMI was different in group of women and men (S4–S6 Tables). The most interesting finding from our analysis is that the FTO intronic region (47 kb block 8) was strongly associated with overweight only in males without impact of SNPs from this block on overweight in a group of females. Surprisingly, analysis of obesity phenotype in sex stratified manner did not revealed sex differences in this region (S2 and S3 Figs, respectively). A case-control analysis for obese female and male groups without sex stratification was performed and results are presented in supplementary materials (S7 Table and S4 Fig).
We have also performed analyses on different age groups in female and male. Six different age classes were set up in every 10 years interval (20–29, 30–39, 40–49, 50–59, 60–69, 70–79). Using CMH we tested the association of every SNP with overweight and/or obesity while assuming age class stratification. The observed effect of the SNPs impact on overweight and obesity was similar as described above but the estimated p values were lower while taking into account age stratification (S8 and S9 Tables). Differences in the distribution of estimated p values of the Chi square test between sexes in the normal weight versus obesity and normal weight and overweight association analysis with age stratification are depicted at Figs 3 and 4, respectively. This entirely confirmed the statistically significant association of block 8 SNPs with overweight group of male found in our analyses.
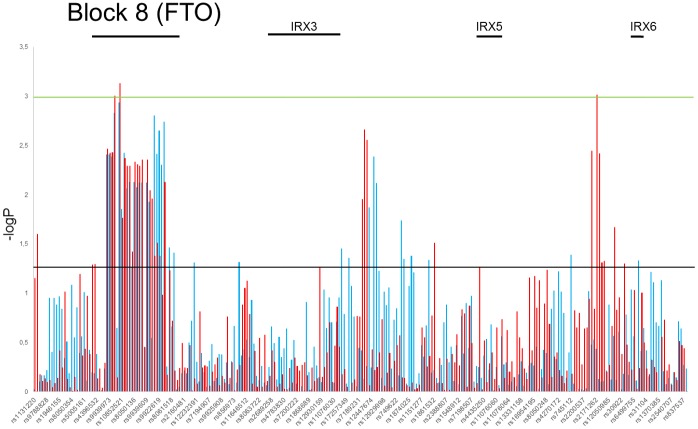
Fig 3. Distribution of estimated p values of the Chi square test in obesity phenotype stratified by age and sex.
P values were calculated in the test of independence between the obesity phenotype and MAFs at tested SNPs (S4 Table). The y axis shows the −log10 p values of 262 SNPs obtained in obesity versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
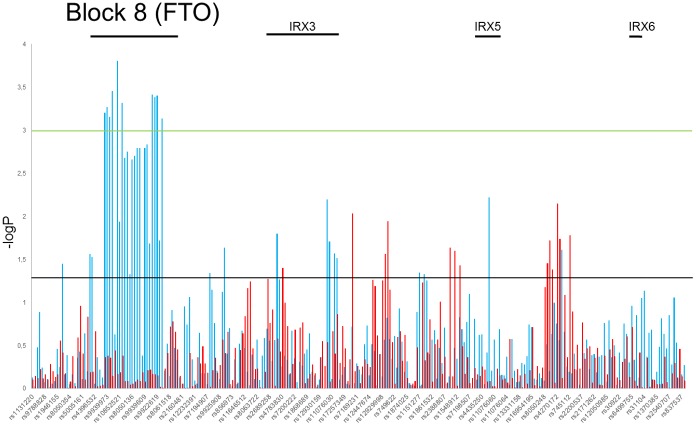
Fig 4. Distribution of estimated p values of the Chi square test in overweight phenotype stratified by age and sex.
P values were calculated in the test of independence between the overweight phenotype and MAFs at tested SNPs (S5 Table). The y axis shows the −log10 p values of 262 SNPs obtained in overweight versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
The FTO rs1558902 had the highest observed chi-square statistic (χ2 = 14.28, p = 0.0001572, OR = 1.265, SE = 0.06227, 95% CI = 1.12–1.43) in group of overweight men versus normal weight, whereas rs4270172, located 200 kb upstream IRX3, had the highest observed chi-square statistic (χ2 = 7.241, p = 0.007125, OR = 0.8063, SE = 0.08054, 95% CI = 0.6885–0.9442) in overweight females. When we analyzed the normal weight group versus group of obese women and obese men, the most statistically significant variants were located in FTO: rs1558902χ2 = 11.38, p = 0.000741, OR = 1.349, 95% CI = 1.133–1.605 and χ2 = 10.55, p = 0.001162, OR = 1.317, 95% CI = 1.115–1.556, respectively)) and rs12149832 (χ2 = 10.84, p = 0.000994, OR = 1.338, 95% CI 1.125–1.592 and χ2 = 10.09, p = 0.001493, OR = 1.309, 95% CI = 1.108–1.546, respectively).
We found additional association regions, in normal weight versus obesity analysis, which were located upstream IRX3 (block 29 and 30, highest observed p value for rs1420285: χ2 = 10.84, p = 0.000994, OR = 1.338, 95% CI = 1.125–1.592) and downstream IRX6 (blocks 53 and 54 with highest observed p value for rs2171262: χ2 = 10.87, p = 0.000975, OR = 1.431, 95% CI = 1.155–1.772) in females only (Fig 3). When we pulled together overweight and obesity groups we realized that in females group there are only 8 statistically significant SNPs compared with 27 in males (S6 Table).
We then searched for differences in the distributions of allele/genotype frequencies in case and control groups under additive, dominant, and recessive inheritance models. Stratification by BMI and sex revealed the influence of sex on the effect of tested SNPs on increased BMI in a genetic model-dependent manner (S10–S12 Tables). The strongest effect on obesity occurred again with FTO variants: rs1558902 and rs1421085 in females using a dominant model (p = 0.000463 and p = 0.0004744 respectively). In a group of males, two different FTO loci, rs12149832 and rs9930506, showed the strongest association with obesity under the recessive model (p = 0.003137 and 0.003741, respectively). Under the recessive inheritance model we also observed the strongest statistically significant association with BMI in range 25–29.99 kg/m2 for three loci in males and only one in females: rs4435250 (non-coding sequence located 100 kb downstream IRX5), rs9930506, rs12149832 (both FTO SNPs) and rs12444481 (non-coding sequence located 200 kb downstream IRX5), respectively.
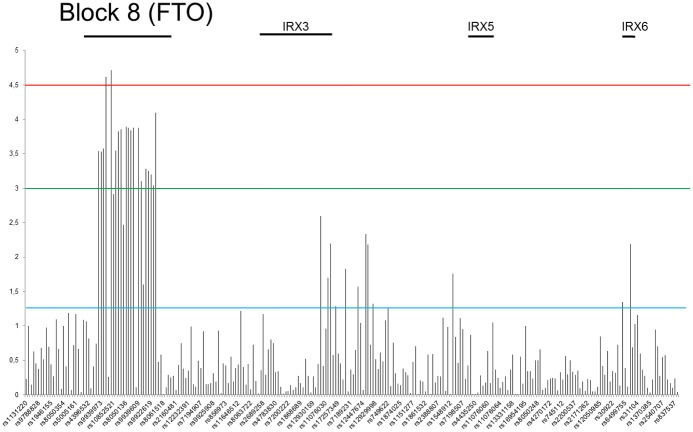
Previous studies suggested that genetic association to obesity at the FTO locus may be age dependent [40–42]. According to this, we examined the role of age in our age-diverse group (range from 20 to 77). Linear regression using BMI as a continuous trait (including underweight subjects) with sex and age as a covariates revealed differential contributions of SNPs to increased BMI. Obtained results are presented in Fig 5 and S13 Table. Age had bigger influence on BMI in group of women than in males. There were SNPs with positive and negative regression coefficient. The highest positive regression coefficient was observed only for three FTO variants: rs1558902, rs1421085 and rs9939609. The minor alleles of these SNPs were significantly more frequent in the obese group than in the normal-weight control group (0.5075 vs. 0.4509, 0.5069 vs. 0.4516 and 0.4812 vs. 0.4354, respectively) and carriers of the mentioned MAF had higher BMI than homozygous for the wild-type alleles (Beta = 0.3492, 95% CI = 0.1892–0.5093, p = 1.93E-05; Beta = 0.345, 95% CI = 0.185–0.5051, p = 2.42E-05; Beta = 0.3124, 95% CI = 0.1523–0.4725, p = 0.0001329, respectively).
Fig 5. Distribution of calculated p values in linear regression analysis.
P values are expressed as negative logarithm of the p values obtained in linear regression analysis with age and sex as a covariates. The y axis shows the −log10 p values of 262 SNPs, and the x axis shows their rs numbers. Horizontal blue, green and red lines represent the thresholds of p = 0.05 for significance without multiple correction, p = 8.2E-04 for LD block significance and p = 1.9E -04 for Bonferroni correction, respectively.
Discussion
In our study, we tested the genetic variation of 2 Mb region of chromosome 16, encompassing region of FTO, RPGRIP1L, AKTIP, RBL2, IRX3, IRX5 and, IRX6 genes. It was the first study with representation of individuals from each geographical region of Poland (N = 5418) focusing on this topic. Case—control association analysis was performed for 262 SNPs located in the tested region. Obtained results revealed that FTO SNPs rs1558902, rs1421085, rs12149832, rs9930506, rs9939609 had the strongest influence on obesity phenotype in our study group which is consistent with other studies [8–10, 27, 43–50]. These SNPs were in almost perfect LD (|D’|>0.98) with other 19 SNPs in block 8 located in non-coding sequence of intron 1. The haplotype analysis exhibited that the strongest obesity associated haplotype (χ2 = 11.474, p = 0.0007) which contained the minor alleles for SNPs distributed across a 45-kb stretch of intron 1 of FTO, was in block 8 and occurred at a frequency of 0.46 in the case group and 0.41 in controls. This region identified in our cohort was previously described by Hotta et al. as the 45 kb obesity associated region of high LD encompassing introns 1 and 2 of the FTO [48].
The most important discovery in our analysis was gender dependent manner of this association. SNPs in block 8 were statistically significant only in male group when we analyzed normal group versus overweight group. We did not observe this association in females. This unexpected finding was not observed so far in any previous studies. Thus, we concluded that overweight might be regulated by different genetic manner dependent on sex and in females is probably controlled by other region(s) in genome.
What is worth emphasizing, we have also noticed that SNPs included to block 8 were associated with obesity in normal versus obese group in females when assuming dominant inheritance model and in males when assuming recessive inheritance model. This genetic model-dependent sexual dimorphism for obesity susceptibility was also found in Mexicans but in different manner [51]. FTO SNPs were associated with obesity in recessive model and dominant model for females and males, respectively. Previous studies which concerned about different populations, reported that increased BMI heritability has sex-based differences and obesity trait is mediated through FTO and occurs independently in both sexes [51–53]. Meta-analysis of GWA studies of waist-hip ratio revealed evidence of gene-by-sex interactions involvement in body fat distribution [54]. Our study clearly demonstrates the importance of sex-stratification when analyzing complex traits. Sex can be perceived as risk factor which incorporates behavioral, physiological, and anatomical differences [55].
As the genetic association to obesity at the FTO locus was reported to be age dependent [40–42], we also examined the role of age in our age-diverse group (range from 20 to 77). Linear regression using BMI as a continuous trait and age and sex as a covariates revealed differential intronic FTO SNP contributions and disclosed that both age and sex had strong influence on BMI in our cohort (p = 3.06 × 10−145 and p = 9.05 × 10−34, respectively). We observed stronger association of intronic FTO region with BMI also when taking into account age class stratification in allelic association analysis.
The intronic region of FTO with high LD is packed with cis-regulatory elements, transcription factors binding sites, and DNase hypersensitive sites. These non-coding variants within FTO are connected with IRX3. In our Polish study group, we found one locus rs1126960, located in coding sequence of IRX3 and associated with obesity in males only when assuming additive and dominant models. This SNP was also associated with overweight under the recessive model in non sex-stratified analysis. Recent evidence suggests that FTO variants directly affect adipocyte function through targeting IRX3 and IRX5. Presence of the FTO risk haplotype in lean children was found to be associated with increased adipocyte-specific expression of IRX3 and IRX5, whereas it was unaffected in a group of obese children. IRX3 expression was elevated in adipose tissues and isolated adipocytes of lean compared to obese children and negatively correlated with BMI [30]. This new mechanism conferring genetic obesity risk whereby variants in FTO directly affect adipocyte function through the activation of IRX3 and IRX5 was previously discovered in lean adults by Claussnitzer et al. [56]. Although the functional interpretation of mechanism by which FTO variants interact with IRX3 and IRX5 remain clearly described, the genetic variation of IRXB cluster was not featured well. Hunt et al. sequenced the region of chromosome 16 encompassing the TAD area that includes the IRXB cluster in 284 Danish men [38]. Across tested area they identified 13373 simple (bi-allelic) variants, of which 12392 were SNPs and 981 were indels.
The haplotype analysis revealed that SNPs included in block 30, localized 200 kb upstream IRX3, were also associated with higher BMI (p = 0.0079). What is very interesting, we found that two SNPs, FTO rs1421084 and rs4238775 located downstream IRX5, in distance 1.3 Mb were in perfect LD. Near the location of IRX3 and IRX5, we also detected association between BMI and eleven SNPs located in non-coding region of chromosome 16 between IRX3 and IRX5 (16: 54 359 850–54 780 370) when assuming linear regression with age as a covariate. Hunt and colleagues [38] observed similar region upstream IRX5 which was independent of FTO but had, like FTO, an age dependent aspect, and its effect on BMI was similar.
Our next important finding is association between BMI and another haplotype (p = 0.0097) which contains SNPs from block 57, and localized 8 kb downstream from IRX6. These SNPs from 57 block (16: 55 326 804–55 349 837) were in high LD (|D’|>0.98) with SNPs located in coding region of IRX6 and included to block 58.
Few limitations of this research must be considered. All participants are Caucasians, therefore obtained results may not be extendable to other groups. The mean age of the study group (50.16±13.55 for individuals with obesity, and 45.48±14.25 for individuals with overweight) suggests that hormonal decline might be partially responsible for observed associations. We were unable to include to our statistical analysis many of significant factors such as level of physical activity, nutrient intake, socioeconomic status or other environmental covariates that might influence results, as they were not included in questionnaires.
The p values presented in our study are not corrected for multiple testing. We did not aim in our study to discover new variants with genetic susceptibility to obesity. They have been previously identified by many of GWA studies in many populations. We mainly focused on characteristic of the Polish population in FTO and IRXB cluster regions and checked whether or not it is possible to observe any of sex specific differences in our sex stratified analyses. This sophisticated approach has enabled us to discover several novel and important findings described above.
The question of what significance threshold is appropriate for GWAS seems to be unresolved [57]. The usage of the GWA study correction (p<7.2E-08) in our particular study is overly as we only include to our analysis 2 Mb region of chromosome 16. A Bonferroni correction for 262 SNPs, would give a p value significance threshold of 1.9E-04. If we use the number of LD blocks (n = 61) identified by Haploview to estimate the corrected p value threshold, which becomes 0.05/61 = 8.2E-04, we would have considered only the FTO intron variants: rs1558902, rs1421085, rs12149832, rs9940128, rs9930333 and rs9939973 as the statistically significant corresponding to obesity in normal versus obese group without adjustment for sex. We have also controlled our statistical significance by performing 100 000 permutation tests in Haploview. Only two variants rs1558902 and rs1421085 passed permutation p value threshold (0.0185 and 0.0218, respectively). The rs12149832 showed suggestive evidence (p = 0.0675) of association with obesity in tested population of Poles after permutation tests (S14 and S15 Tables).
In conclusion, we have comprehensively analyzed the association of genetic variants and haplotypes located in the chromosomal FTO/IRXB region with increased and declined values of BMI in large and well characterized Polish cohort. This is the first study focusing on full characteristics of FTO and IRXB regions in the Polish population. Moreover, we performed comprehensive case-control analysis for different subgroups based on BMI including also significance of sex and age. Our greatest finding from this study was sex-dependent manner of FTO associations with obesity/overweight phenotype. Different variants of FTO were associated with BMI values in a group of women and men. We have also confirmed dependence of FTO intronic SNPs with other polymorphisms from the region of IRXB cluster. Our observations highlight the necessity, and challenge to reveal the genetic basis of obesity complex trait in population of Poles but also give a novel point of view for FTO genetic association with overweight and obesity as a more complex and not yet well-known issue. Thus, in the future we plan to extend our research of obesity and overweight phenotype by performing GWA study to further analyze these traits in Poles.
Supporting information
(TIF)
P values were calculated in the test of independence between the obesity phenotype and MAFs at tested SNPs (S4 Table). The y axis shows the −log10 p values of 262 SNPs obtained in obesity versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
P values were calculated in the test of independence between the overweight phenotype and MAFs at tested SNPs (S5 Table). The y axis shows the −log10 p values of 262 SNPs obtained in overweight versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
P values were calculated in the test of independence between the obesity phenotype and MAFs at tested SNPs (S7 Table). The y axis shows the −log10 p values of 262 SNPs obtained in obesity versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
The threshold was set up on MAF<0.01.
(XLSX)
SNP—SNP identifier, Group—Code indicating sample (ALL-all cohort, Case-obesity, Control-normal weight), A—Minor allele code, B—Major allele code, AA/AB/BB—Genotype counts, O(HET), E(HET)—Observed and Expected heterozygosity, respectively; p—HWE p-value. The cut off was set up (p<1E-04).
(XLSX)
Bold font denotes haplotypes with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of SNP on chromosome 16; A—minor allele; NMISS—Number of non-missing individuals included in analysis; BETA—regression coefficient; SE—standard error of regression coefficient; L95—Lower bound of 95% confidence interval; U95—Upper bound of 95% confidence interval; STAT—Coefficient t-statistic; P—Asymptotic p-value for t-statistic, bold font denotes markers with statistically significant p values and shaded cells indicate the meaning the additive effects of allele dosage when assuming age and sex as a covariates.
(XLSX)
1850 permutation exceed highest observed Chi-square (29.95). Best observed Chi-square 15.355 for rs1558902. Bold font denotes markers with statistically significant p values.
(XLSX)
15527 permutation exceed highest observed Chi-square (29.035). Best observed Chi-square 11.474 for Block 8. No statistically significant association.
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Polish POIG grant 01.01.02-10-005/08 TESTOPLEK from the European Regional Development Fund and through the Polish Ministry of Science and Higher Education Grant DIR/WK/2017/01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. Epub 2000/04/15. doi: 10.1038/35007508 . [DOI] [PubMed] [Google Scholar]
- 2.Dixon JB. The effect of obesity on health outcomes. Mol Cell Endocrinol. 2010;316(2):104–8. Epub 2009/07/25. doi: 10.1016/j.mce.2009.07.008 . [DOI] [PubMed] [Google Scholar]
- 3.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–51. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 4.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring, Md). 2006;14(4):529–644. Epub 2006/06/03. doi: 10.1038/oby.2006.71 . [DOI] [PubMed] [Google Scholar]
- 5.Lakshman R, Elks CE, Ong KK. Childhood obesity. Circulation. 2012;126(14):1770–9. Epub 2012/10/03. doi: 10.1161/CIRCULATIONAHA.111.047738 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hjelmborg J, Fagnani C, Silventoinen K, McGue M, Korkeila M, Christensen K, et al. Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity (Silver Spring, Md). 2008;16(4):847–52. Epub 2008/02/02 doi: 10.1038/oby.2007.135 . [DOI] [PubMed] [Google Scholar]
- 7.Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. The American journal of clinical nutrition. 2008;87(2):398–404. Epub 2008/02/09. . [DOI] [PubMed] [Google Scholar]
- 8.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–6. Epub 2007/05/15. doi: 10.1038/ng2048 . [DOI] [PubMed] [Google Scholar]
- 9.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. Epub 2007/04/17. doi: 10.1126/science.1141634 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115 Epub 2007/07/31. doi: 10.1371/journal.pgen.0030115 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh RK, Kumar P, Mahalingam K. Molecular genetics of human obesity: A comprehensive review. C R Biol. 340(2):87–108. Epub 2017/01/17. doi: 10.1016/j.crvi.2016.11.007 . [DOI] [PubMed] [Google Scholar]
- 12.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72. Epub 2007/11/10. doi: 10.1126/science.1151710 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. The New England journal of medicine. 2008;359(24):2558–66. Epub 2008/12/17. doi: 10.1056/NEJMoa0803839 . [DOI] [PubMed] [Google Scholar]
- 14.Park SL, Cheng I, Pendergrass SA, Kucharska-Newton AM, Lim U, Ambite JL, et al. Association of the FTO obesity risk variant rs8050136 with percentage of energy intake from fat in multiple racial/ethnic populations: the PAGE study. Am J Epidemiol. 2013;178(5):780–90. Epub 2013/07/04. doi: 10.1093/aje/kwt028 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad T, Lee IM, Pare G, Chasman DI, Rose L, Ridker PM, et al. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care. 2011;34(3):675–80. Epub 2011/01/27. doi: 10.2337/dc10-0948 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. International journal of obesity (2005). 2009;33(1):42–5. Epub 2008/10/08. doi: 10.1038/ijo.2008.174 . [DOI] [PubMed] [Google Scholar]
- 17.Tanofsky-Kraff M, Han JC, Anandalingam K, Shomaker LB, Columbo KM, Wolkoff LE, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. The American journal of clinical nutrition. 2009;90(6):1483–8. Epub 2009/10/16. doi: 10.3945/ajcn.2009.28439 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85(1):106–11. Epub 2009/06/30. doi: 10.1016/j.ajhg.2009.06.002 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilpelainen TO, den Hoed M, Ong KK, Grontved A, Brage S, Jameson K, et al. Obesity-susceptibility loci have a limited influence on birth weight: a meta-analysis of up to 28,219 individuals. The American journal of clinical nutrition. 2011;93(4):851–60. Epub 2011/01/21. doi: 10.3945/ajcn.110.000828 . [DOI] [PubMed] [Google Scholar]
- 20.Qi L, Kang K, Zhang C, van Dam RM, Kraft P, Hunter D, et al. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57(11):3145–51. Epub 2008/07/24. doi: 10.2337/db08-0006 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22(17):3597–607. Epub 2013/05/15. doi: 10.1093/hmg/ddt205 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitek A, Rosset I, Strapagiel D, Majewska M, Ostrowska-Nawarycz L, Żądzińska E. Association of FTO gene with obesity in Polish schoolchildren. Anthropol Rev. 2014;77(1):33–44. doi: 10.2478/anre-2014-0003 [Google Scholar]
- 23.Rosset I, Strapagiel D, Sitek A, Majewska M, Ostrowska-Nawarycz L, Żądzińska E. Association of FTO and TMEM18 polymorphisms with overweight and obesity in the population of Polish children. Anthropol Rev 2016;79(1):17–33. doi: 10.1515/anre-2016-0002 [Google Scholar]
- 24.Wisniewski AB, Chernausek SD. Gender in childhood obesity: family environment, hormones, and genes. Gend Med. 2009;6 Suppl 1:76–85. Epub 2009/06/12. doi: 10.1016/j.genm.2008.12.001 . [DOI] [PubMed] [Google Scholar]
- 25.Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011;7(8):e1002215 Epub 2011/08/20. doi: 10.1371/journal.pgen.1002215 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung YJ, Perusse L, Sarzynski MA, Fornage M, Sidney S, Sternfeld B, et al. Genome-wide association studies suggest sex-specific loci associated with abdominal and visceral fat. International journal of obesity (2005). 2016;40(4):662–74. Epub 2015/10/21. doi: 10.1038/ijo.2015.217 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PloS one. 2007;2(12):e1361 Epub 2007/12/27. doi: 10.1371/journal.pone.0001361 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luczynski W, Zalewski G, Bossowski A. The association of the FTO rs9939609 polymorphism with obesity and metabolic risk factors for cardiovascular diseases in Polish children. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2012;63(3):241–8. Epub 2012/07/14. . [PubMed] [Google Scholar]
- 29.Wrzostek M, Zakrzewska A, Ruczko L, Jabłonowska-Lietz B, Nowicka G. Association between rs9930506 polymorphism of the fat mass & obesity-associated (FTO) gene & onset of obesity in Polish adults. Indian J Med Res. 2016;143(3): 281–7. doi: 10.4103/0971-5916.182617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf K, Scholz M, Kovacs P, Kiess W, Korner A. FTO Obesity Risk Variants Are Linked to Adipocyte IRX3 Expression and BMI of Children—Relevance of FTO Variants to Defend Body Weight in Lean Children? PloS one. 2016;11(8):e0161739 Epub 2016/08/26. doi: 10.1371/journal.pone.0161739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strapagiel D. Sobalska-Kwapis M, Słomka M, Marciniak B. Biobank Lodz—DNA Based Biobank at the University of Lodz, Poland. Open J Bioresources. 2016;3(1):e6 http://doi.org/10.5334/ojb.22. [Google Scholar]
- 32.Strapagiel D, Majewska M, Słomka M, Janik K, Sobalska M, Bartosz G, inventors Method for determining sex. Poland 2015 2015-06-22.
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. doi: 10.1086/519795 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–93. Epub 2005/03/25. doi: 10.1086/429864 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huh I, Kwon MS, Park T. An Efficient Stepwise Statistical Test to Identify Multiple Linked Human Genetic Variants Associated with Specific Phenotypic Traits. PloS one. 2015;10(9):e0138700 Epub 2015/09/26. doi: 10.1371/journal.pone.0138700 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48. Epub 1959/04/01. . [PubMed] [Google Scholar]
- 37.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. Epub 2004/08/07. doi: 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 38.Hunt LE, Noyvert B, Bhaw-Rosun L, Sesay AK, Paternoster L, Nohr EA, et al. Complete re-sequencing of a 2Mb topological domain encompassing the FTO/IRXB genes identifies a novel obesity-associated region upstream of IRX5. Genome Med. 2015;7:126 Epub 2015/12/09. doi: 10.1186/s13073-015-0250-3 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. Epub 2012/04/13. doi: 10.1038/nature11082 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graff M, Gordon-Larsen P, Lim U, Fowke JH, Love SA, Fesinmeyer M, et al. The influence of obesity-related single nucleotide polymorphisms on BMI across the life course: the PAGE study. Diabetes. 2013;62(5):1763–7. Epub 2013/01/10. doi: 10.2337/db12-0863 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19(3):545–52. Epub 2009/11/03. doi: 10.1093/hmg/ddp504 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jess T, Zimmermann E, Kring SI, Berentzen T, Holst C, Toubro S, et al. Impact on weight dynamics and general growth of the common FTO rs9939609: a longitudinal Danish cohort study. International journal of obesity (2005). 2008;32(9):1388–94. Epub 2008/07/30. doi: 10.1038/ijo.2008.110 . [DOI] [PubMed] [Google Scholar]
- 43.Field SF, Howson JM, Walker NM, Dunger DB, Todd JA. Analysis of the obesity gene FTO in 14,803 type 1 diabetes cases and controls. Diabetologia. 2007;50(10):2218–20. Epub 2007/07/28. doi: 10.1007/s00125-007-0767-0 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. Epub 2007/10/19. doi: 10.2337/db07-0910 . [DOI] [PubMed] [Google Scholar]
- 45.Wahlen K, Sjolin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. Journal of lipid research. 2008;49(3):607–11. Epub 2007/12/01. doi: 10.1194/jlr.M700448-JLR200 . [DOI] [PubMed] [Google Scholar]
- 46.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab. 2008;93(4):481–4. Epub 2007/12/07. doi: 10.1016/j.ymgme.2007.10.011 . [DOI] [PubMed] [Google Scholar]
- 47.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. Epub 2010/10/12. doi: 10.1038/ng.686 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, Komatsu R, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. Journal of human genetics. 2008;53(6):546–53. Epub 2008/04/02. doi: 10.1007/s10038-008-0283-1 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Kilpelainen TO, Liu C, Zhu J, Liu Y, Hu C, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55(4):981–95. Epub 2011/11/24. doi: 10.1007/s00125-011-2370-7 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45(6):690–6. Epub 2013/04/16. doi: 10.1038/ng.2608 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saldana-Alvarez Y, Salas-Martinez MG, Garcia-Ortiz H, Luckie-Duque A, Garcia-Cardenas G, Vicenteno-Ayala H, et al. Gender-Dependent Association of FTO Polymorphisms with Body Mass Index in Mexicans. PloS one. 2016;11(1):e0145984 Epub 2016/01/05. doi: 10.1371/journal.pone.0145984 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobsson JA, Danielsson P, Svensson V, Klovins J, Gyllensten U, Marcus C, et al. Major gender difference in association of FTO gene variant among severely obese children with obesity and obesity related phenotypes. Biochem Biophys Res Commun. 2008;368(3):476–82. Epub 2008/02/06. doi: 10.1016/j.bbrc.2008.01.087 . [DOI] [PubMed] [Google Scholar]
- 53.Zhang M, Zhao X, Cheng H, Wang L, Xi B, Shen Y, et al. Age- and sex-dependent association between FTO rs9939609 and obesity-related traits in Chinese children and adolescents. PloS one. 2014;9(5):e97545 Epub 2014/05/16. doi: 10.1371/journal.pone.0097545 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. Epub 2010/10/12. doi: 10.1038/ng.685 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34(8):846–53. Epub 2010/11/26. doi: 10.1002/gepi.20540 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. The New England journal of medicine. 2015;373(10):895–907. Epub 2015/08/20. doi: 10.1056/NEJMoa1502214 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32(3):227–34. Epub 2008/02/27. doi: 10.1002/gepi.20297 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
P values were calculated in the test of independence between the obesity phenotype and MAFs at tested SNPs (S4 Table). The y axis shows the −log10 p values of 262 SNPs obtained in obesity versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
P values were calculated in the test of independence between the overweight phenotype and MAFs at tested SNPs (S5 Table). The y axis shows the −log10 p values of 262 SNPs obtained in overweight versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
P values were calculated in the test of independence between the obesity phenotype and MAFs at tested SNPs (S7 Table). The y axis shows the −log10 p values of 262 SNPs obtained in obesity versus normal weight association analysis, and the x axis shows their rs numbers. Horizontal black and green lines represent the thresholds of p = 0.05 for significance without multiple correction and p = 8.2E-04 for LD block significance, respectively. Red and blue columns represents females and males group, respectively.
(TIF)
The threshold was set up on MAF<0.01.
(XLSX)
SNP—SNP identifier, Group—Code indicating sample (ALL-all cohort, Case-obesity, Control-normal weight), A—Minor allele code, B—Major allele code, AA/AB/BB—Genotype counts, O(HET), E(HET)—Observed and Expected heterozygosity, respectively; p—HWE p-value. The cut off was set up (p<1E-04).
(XLSX)
Bold font denotes haplotypes with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of tested SNP on chromosome 16; A, B—Minor and major allele names, respectively (based on whole sample); F_A—Frequency of MAF in cases; F_U—Frequency of MAF allele in controls; CHISQ—Basic allelic test chi-square (1df); P—Asymptotic p-value for this test; OR—Estimated odds ratio (for A, i.e. B is reference); L95—Lower bound of 95% confidence interval for odds ratio; U95—Upper bound of 95% confidence interval for odds ratio. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
AFF—Genotypes/alleles in cases, UNAFF—Genotypes/alleles in controls, GENO—Genotypic model (AA versus AB versus BB); ADD—Additive model (AA versus BB); DOM—Dominant model (AA+AB versus BB); REC—Recessive model (AA versus AB+BB); CHISQ—Chi-squated statistic, P—Asymptotic p-value. By convention, in all models, A is the minor allele and B is the major allele. Bold font denotes markers with statistically significant p values.
(XLSX)
SNP—rs number of tested SNP; BP—Physical position of SNP on chromosome 16; A—minor allele; NMISS—Number of non-missing individuals included in analysis; BETA—regression coefficient; SE—standard error of regression coefficient; L95—Lower bound of 95% confidence interval; U95—Upper bound of 95% confidence interval; STAT—Coefficient t-statistic; P—Asymptotic p-value for t-statistic, bold font denotes markers with statistically significant p values and shaded cells indicate the meaning the additive effects of allele dosage when assuming age and sex as a covariates.
(XLSX)
1850 permutation exceed highest observed Chi-square (29.95). Best observed Chi-square 15.355 for rs1558902. Bold font denotes markers with statistically significant p values.
(XLSX)
15527 permutation exceed highest observed Chi-square (29.035). Best observed Chi-square 11.474 for Block 8. No statistically significant association.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.