Abstract
The nephron is the basic physiologic subunit of the mammalian kidney and is made up of several apicobasally polarized epithelial cell types. The process of apicobasal polarization in animal cells is controlled by the evolutionarily conserved Crumbs (CRB), Partitioning-defective, and Scribble protein complexes. Here, we investigated the role of protein associated with LIN-7 1 (Pals1, also known as Mpp5), a core component of the apical membrane–determining CRB complex in the nephron. Pals1 interacting proteins, including Crb3 and Wwtr1/Taz, have been linked to renal cyst formation in mice before. Immunohistologic analysis revealed Pals1 expression in renal tubular cells and podocytes of human kidneys. Mice lacking one Pals1 allele (functionally haploid for Pals1) in nephrons developed a fully penetrant phenotype, characterized by cyst formation and severe defects in renal barrier function, which led to death within 6–8 weeks. In Drosophila nephrocytes, deficiency of the Pals1 ortholog caused alterations in slit-diaphragm–like structures. Additional studies in epithelial cell culture models revealed that Pals1 functions as a dose-dependent upstream regulator of the crosstalk between Hippo- and TGF-β–mediated signaling. Furthermore, Pals1 haploinsufficiency in mouse kidneys associated with the upregulation of Hippo pathway target genes and marker genes of TGF-β signaling, including biomarkers of renal diseases. These findings support a link between apical polarity proteins and renal diseases, especially renal cyst diseases. Further investigation of the Pals1-linked networks is required to decipher the mechanisms underlying the pathogenesis of these diseases.
Keywords: Pals1, Mpp5, cell signaling, cell polarity, proteinuria, cystic kidney
During nephrogenesis, renal epithelial tissues undergo constant growth and differentiation, including functional diversification of the various segments of the tubule (tubulogenesis) and formation of the glomerular filtration barrier. The complex balance between proliferation and apoptosis that controls nephron maturation and kidney size is organized by several signal transduction systems, including those activated by Hippo and TGF-β.1–4
Apicobasal epithelial cell polarization begins early during the renal vesicle stage and continues during tubulo- and glomerulogenesis until mature nephrons are formed.1–3 Cell polarity is organized by evolutionarily conserved complexes assembled around the proteins CRB (Crumbs), PAR (Partitioning-defective), and SCRIB (Scribble), respectively. The CRB complex, which consists of Crumbs 3, Lin7c (lin-7 homolog C), Pals1 (protein associated with Lin7), and Patj (Pals1-associated tight junction protein), plays a central role in the determination of apical membranes in epithelial cells.5,6
Pals1 is a member of the evolutionarily conserved membrane-associated guanylate kinase (MAGUK) family.7 It is encoded by the gene Mpp5 (membrane protein, palmitoylated 5) and its orthologs in Drosophila and zebrafish are referred to as Stardust (Sdt) in Drosophila and Nagie Oko (Nok).8,9 Pals1 is the only apical polarity protein that interacts with all other components of the CRB complex.7,10,11 Moreover, Pals1 is also able to mediate binding between CRB and PAR complexes, because it interacts directly with the PAR component Par6.12 Knockdown studies in cell culture also revealed an essential role of Pals1 in the formation of tight and adherens junctions, indicating that cell junction assembly is closely linked to cell polarization in epithelial cells.13,14
More recent studies suggest that the CRB complex acts upstream of the Hippo signaling pathway, which controls tissue growth and organ size.15–18 Key proteins involved in the response to Hippo signaling are the transcriptional coactivators Yap (Yes-associated protein) and Taz (transcriptional coactivator with PDZ-binding motif, gene: Wwtr1). Active Hippo signaling results in the phosphorylation of Yap or Taz by the large tumor suppressor kinase (Lats kinase), which prevents their nuclear localization. By contrast, nonphosphorylated Yap/Taz can enter the nucleus to activate the expression of Hippo target genes.19,20
Because polarization and differentiation are highly coordinated during kidney development, we have investigated the role of Pals1 in these processes.
Results
Pals1 Haploinsufficiency in Nephrons Induces Early Cell Death, Proteinuria, and Renal Cyst Formation
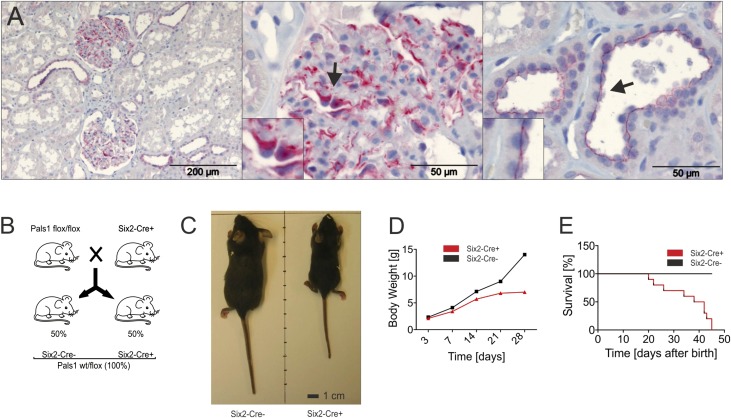
IHC staining of a human kidney specimen revealed a high level of expression of Pals1 in all renal epithelia, including tubulus cells and podocytes (Figure 1A). In order to address the role of Pals1 in the kidney, we examined the in vivo function of Pals1 in the entire nephron. Because Six2-positive cap-mesenchymal cells are precursors of all nephron epithelia, including podocytes as well as cells of the proximal and distal tubuli, we took advantage of a Cre-driver mouse line which expresses an EGFP-Cre fusion protein under the control of the Six2 promoter.2 Crosses between the Six2-Cre driver strain and homozygous conditional Pals1 knockout mice (Pals1fl/fl)21 resulted in heterozygous progeny (Pals1wt/fl), of which 50% were Cre-positive (Six2-Cre+) and 50% Cre-negative (Six2-Cre−) (Figure 1B). When we compared 4-week-old Six2-Cre+ mice with their Cre-negative littermate controls, we discovered that the Six2-Cre+ mice, lacking only one of the two Pals1 alleles in the nephron, were significantly smaller in size and weight (Figure 1, C and D) and died within the first 45 days (Figure 1E).
Figure 1.
Pals1 expression is important for the mammalian kidney. (A) Immunohistochemical analysis of human kidney sections revealed high levels of expression of Pals1 (red) in both tubular and glomerular regions of the organ. Left: Overview of Pals1 staining in a control biopsy specimen from a tumor-free human kidney. Scale bar, 200 µm. Middle: In glomeruli, Pals1 is predominantly expressed in podocytes. Right: Pals1 could also be localized in the apical membrane of renal tubular cells. Scale bar, 50 µm. (B) Scheme: Crosses between Six2-Cre strain and mice homozygous for a conditional Pals1 knockout allele (Pals1fl/fl) resulted in heterozygous F1 progeny (Pals1wt/fl), half of which are expected to carry the Cre recombinase gene (Six2-Cre+; Pals1wt/fl), whereas half are not (Six2-Cre−; Pals1wt/fl). These genotypes are hereafter referred to as Six2-Cre+ and Six2-Cre−, respectively. (C) A Six2-Cre+ mouse and a Six2-Cre− control photographed 4 weeks after birth. Note that Six2-Cre+ mice were smaller than their littermate controls. (D) Bodyweight analysis of Six2-Cre− and Six2-Cre+ mice. (E) Kaplan–Meyer analysis of Six2-Cre+ and Six2-Cre− mice.
Pals1 Haploinsufficiency Is Accompanied by the Formation of Glomerular and Tubular Cysts
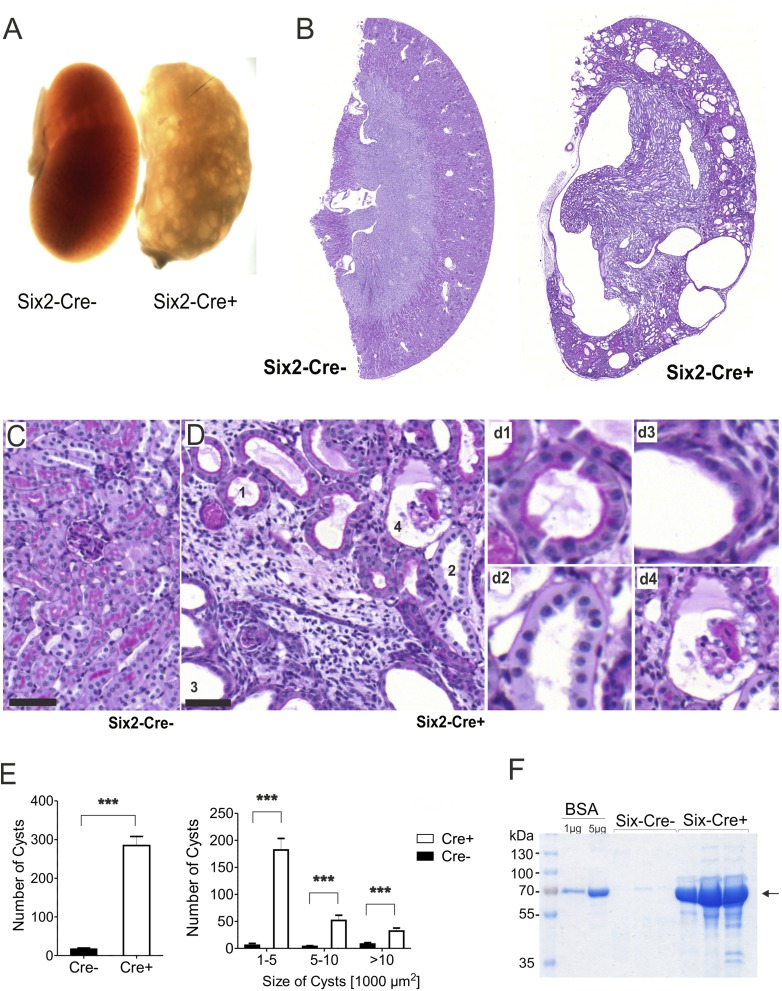
Kidneys derived from these mice were poorly perfused with blood (Figure 2A) and formed multiple cysts of varying size and morphology (Figure 2, B–E), which were completely absent in the littermate control (Figure 2, B and C). Moreover, Six2-Cre+ mice of both sexes developed proteinuria, starting 2 weeks after birth and increasing in severity over the following weeks (Figure 2F). This suggests that Pals1 expression is crucial for the formation and function of the renal filtration barrier (Supplemental Figure 1).
Figure 2.
Pals1 is essential for the nephron function. (A) Bright-field microscopy of kidney sections from Pals wt/flox-Six2Cre+ (Six2-Cre+) and control mice (Six2-Cre−) shows that Six2-Cre+ kidneys are characterized by cyst-filled and bloodless tissues. (B–D) Periodic acid–Schiff staining of Six2-Cre+ and Six2-Cre− kidney sections shows the formation of multiple cysts in Six2-Cre+ kidneys. (C) Littermate control, higher magnification. (D) Pals1-depleted Six2-Cre+ kidney sections, higher magnification. Numbers mark different cysts, details on the right (d1–4). Scale bar, 50 µm. (E) Distribution of cyst numbers (left) and sizes (right) in Six2-Cre− and Six2-Cre+ kidneys. (F) SDS-PAGE analysis of urine from Six2 Cre− and Six2 Cre+ mice reveals proteinuria in Pals1-depleted mice.
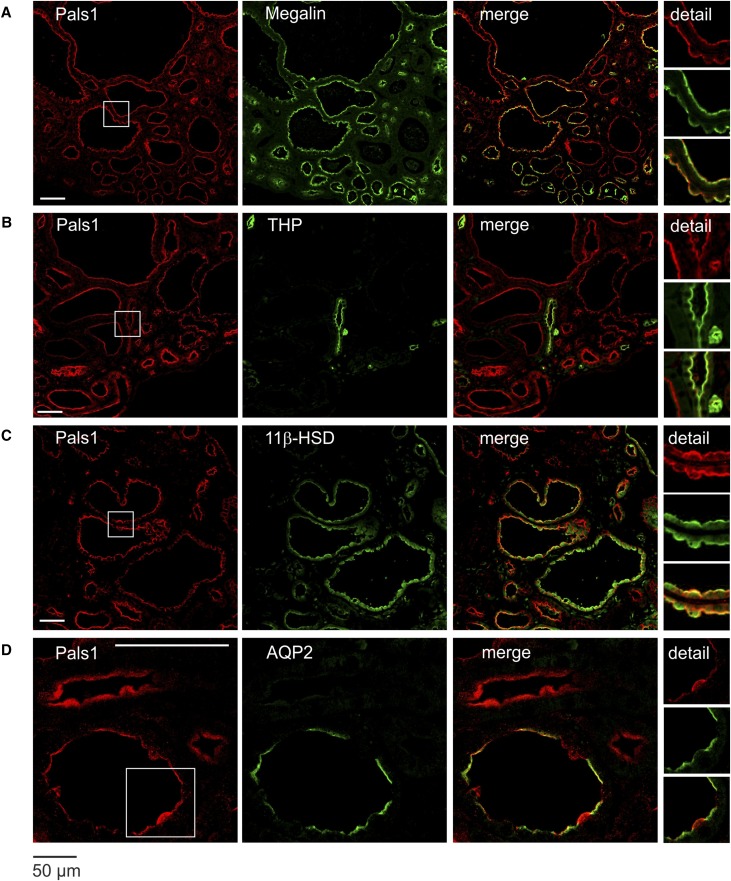
Pals1 is expressed along the entire nephron (Figure 3, Supplemental Figure 2). In tubular cysts, the overall apicobasal polarization of cyst-lining cells was preserved, as Pals1 was predominantly localized at the luminal side in the cysts, indicating that cyst formation was not necessarily linked to an unpolarized distribution of Pals1. Interestingly, staining for Pals1 of specimens obtained from patients with autosomal dominant polycystic kidney disease (ADPKD) confirmed that cyst-lining cells, even in strongly proliferating epithelia, maintained a predominantly apical Pals1 distribution (Supplemental Figure 3).
Figure 3.
Reduced Pals1 expression during nephrogenesis results in the formation of cysts. Immunofluorescence staining of Pals1 and costaining with diverse tubulus markers in Six2-Cre+ kidneys shows that remaining Pals1 is located to the apical membrane of all tubular cysts. Costaining of Pals1 with Megalin (proximal tubule marker, [A]), Tamm–Horsfall protein (thick ascending limb of the loop of Henle, [B]), 11β-HSD (distal tubules, [C]), and AQP2 (collecting duct, [D]). Scale bar, 50 µm.
Cysts are formed in various segments of the nephron (Figure 3, Supplemental Figures 2 and 4), namely in the glomeruli (glomerulocysts), in the proximal tubules (Megalin- and PNA-positive), in distal tubules (11β-HSD and PHA positive), and in the collecting duct (AQP2-positive). No cysts were observed in the thick ascending limb of the loop of Henle (Tamm-Horsfall–positive structures), although Pals1 is also expressed in this nephron segment. The presence of cysts even in regions that still contain both Pals1 alleles indicated the formation of secondary cysts, e.g., in the collecting duct (Figure 3D, Supplemental Figure 4, B–D).
Pals1 Reduction Leads to an Elevated Expression of Hippo Signaling Target Genes and Nuclear Accumulation of Taz and Yap in Cyst-Lining Cells
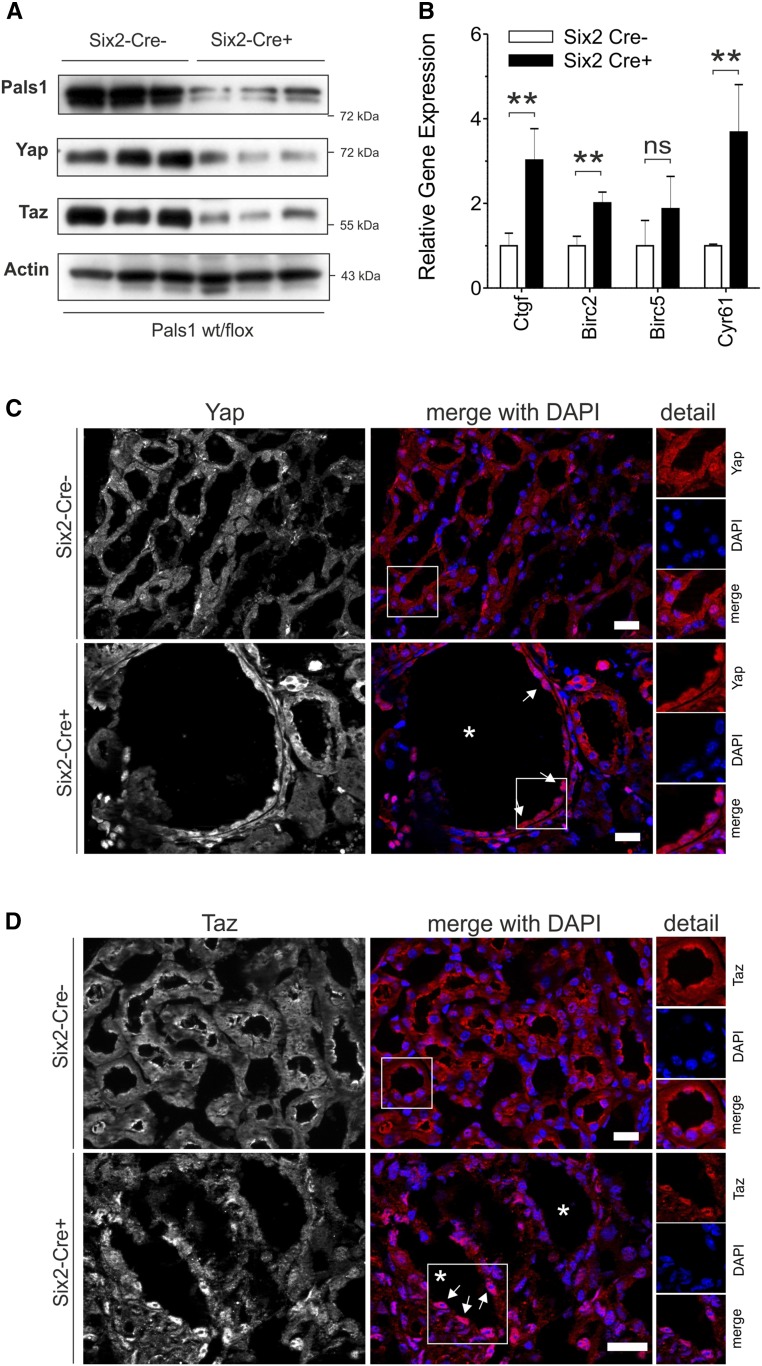
Mice lacking Taz develop a cystic-kidney–like phenotype,22–24 whereas Yap knockout mice die as embryos.25 In Pals1-deficient kidneys, levels of Yap and Taz were strongly reduced (Figure 4A). Real-time PCR analyses showed a significant increase in the expression of Hippo signaling target genes (Ctgf, Birc2, and Cyr61; Figure 4B) in Pals1-deficient kidneys, whereas basal expression of Hippo pathway genes (Yap, Taz/Wwtr1, members of the Amot and Lats families) and expression of apical polarity genes (with the exception of Pals1) remained unchanged (Supplemental Figure 5). Cryo-immunofluorescence staining using antibodies with high preference against Yap or Taz (see also Supplemental Figure 6) showed a predominant nuclear distribution of Yap and Taz in cyst-lining cells (Figure 4, C and D).
Figure 4.
Reduced Pals1 expression results in altered Hippo signaling. (A) Western blot analysis revealed that Pals1, Taz, and Yap, but not actin (loading control), are downregulated in Pals1-depleted (Six2-Cre+) mice (n=3). (B) Real-time PCR analysis showed increased expression of the Hippo target genes Ctgf, Birc2, Birc5, and Cyr61 in Six2-Cre+ mice (data are shown as mean±SD of at least three independent experiments). (C–D) Colocalization of Yap or Taz with the DNA-specific dye DAPI confirmed the nuclear accumulation of Yap and Taz in Six2-Cre+ cyst-lining cells. Asterisks mark cysts and arrows cyst-lining cells. All data shown in (A–D) were obtained from 4-week-old mice.
Stardust Knockdown Causes Dysfunctional Nephrocytes in Drosophila
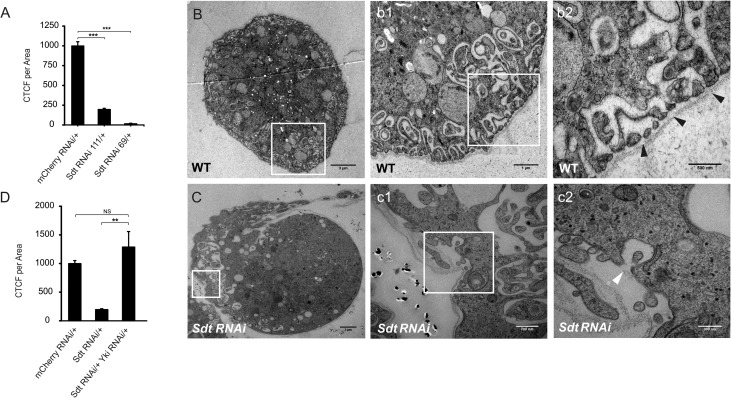
Drosophila nephrocytes control the filtration of the hemolymph by means of a podocyte-like slit-diaphragm26,27 and show high reabsorption and endocytotic activity,28 thus exhibiting remarkable functional similarities with mammalian nephrons. Indeed, selective RNAi knockdown of Stardust/Sdt (by Sdt RNAi 111/+ or Sdt RNAi 69/+), the Drosophila ortholog of Pals1, reduced the uptake efficiency of nephrocytes (Figure 5A) and was accompanied by the appearance of malformed slit-diaphragm–like structures (Figure 5, B and C and details). These uptake defects were rescued by simultaneous downregulation of Yorkie (Yki RNAi), which is the Drosophila counterpart of both Yap and Taz (Figure 5D). This finding indicated that defects in Sdt-depleted nephrocytes were linked to increased activation of Yki-dependent gene expression. Taken together, our data indicate that an imbalance in the relative levels of Sdt and Yki cause defects in the architecture of slit-diaphragm–like structures and function in endocytosis of fly nephrocytes.
Figure 5.
Stardust (Sdt) knockdown in Drosophila results in dysfunctional nephrocytes. (A) In comparison to the control strain (mCherry RNAi/+), depletion of Stardust (Sdt RNAi 111/+ and Sdt RNAi 69/+) led to strongly reduced filtration rates in RFP accumulation assays. CTCF, corrected total cell fluorescence. (B–C) Transmission electron microscopy analysis of wildtype (B) and Sdt-depleted nephrocytes (C) demonstrated that the defects in RFP endocytosis are accompanied by malformation of slit-diaphragm–like structures in Sdt RNAi nephrocytes ([C] and details in c1–c2; scale bars, 3 µm, 700 nm, and 300 nm, respectively) compared with the wildtype control ([B], details in b1–2; scale bars, 3 µm, 1 µm, and 500 nm, respectively). (D) Parallel downregulation of Yki in Sdt RNAi nephrocytes (Std RNAi/+ Yki RNAi/+) completely rescued the defect in RFP filtration shown in (A). For each genotype, >50 nephrocytes from at least six individuals were scored.
Reduced Pals1 Expression Results in an Inactivation of Hippo Signaling In Vitro
To explore the functional consequences of relative loss of Pals1 in more detail, we generated stable Pals1 MDCK and HEK293T knockdown cell lines. Introduction of the short hairpin RNAs sh1 and sh2 led to a 50%–70% reduction of endogenous Pals1 expression (Figure 6, Supplemental Figure 8).
Figure 6.
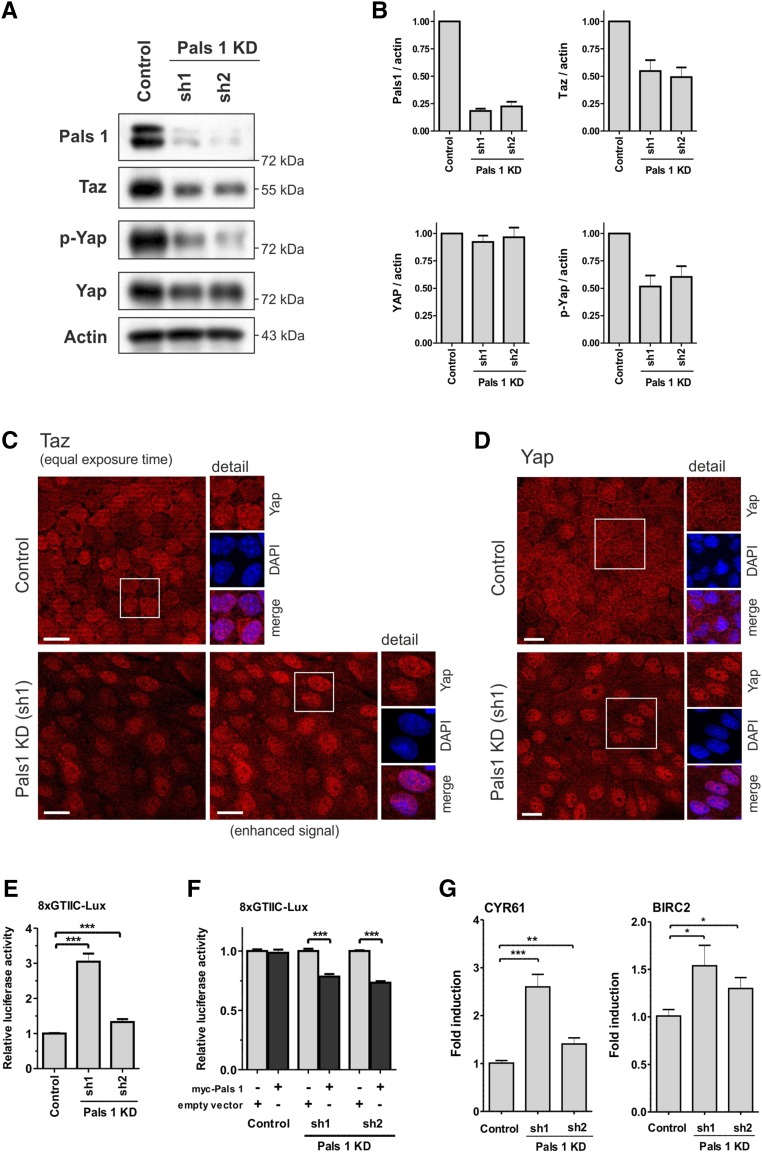
Pals1 controls Hippo signaling in vitro. (A) Knockdown of Pals1 in MDCK cells (sh1 and sh2) decreased expression of Taz and phosphorylation of Yap, relative to control cells. Expression level of Yap was not affected. Actin served as loading control. (B) Quantification of (A). (C) Analysis of confluent cells by immunofluorescence microscopy revealed a reduction in the expression and an increased nuclear localization of Taz (red) in Pals1 KD cells. (C–E) We used the 8xGTIIC reporter gene system, which expresses luciferase only when Yap/Taz is present in the nucleus, to analyze inactivation of Hippo signaling. The results of the 8xGTIIC-Lux reporter gene assay showed that Pals1 knockdown significantly enhances Yap/Taz-dependent transcriptional activity (data are presented as mean±SD and normalized to lane 1). (F) Conversely, Pals1 overexpression suppressed the enhanced Yap/Taz-dependent transcriptional activity in Pals1 KD cells. MDCK cells were transfected with myc-Pals1 or the empty vector. The data were normalized to the respective lane without Pals1 overexpression. (G) Expression of the Hippo target genes CYR61 and BIRC2 is increased in Pals1 KD cells (RT-PCR data are shown as mean+SD of at least three independent experiments).
In MDCK Pals1 KD cell lines, Taz expression and Yap phosphorylation (p-Yap S127) was significantly reduced, whereas Yap levels remained nearly unchanged (Figure 6, A and B) indicating that Hippo signaling, which triggers phosphorylation and cytoplasmic retention of Yap/Taz, is at least partially inactivated in the knockdown lines. This is supported by immunofluorescence analysis with Taz and Yap showing a predominant nuclear distribution in Pals1 KD cells (Figure 6, C and D). Increasing confluence of MDCK cells resulted in a stepwise export of Yap into the cytoplasm and cell junctions. By contrast, in Pals1 KD cells Taz/Yap kept a predominant nuclear localization even in confluent cells (Supplemental Figure 7).
We further tested whether the altered Yap/Taz localization in Pals1 KD cells was linked to enhanced expression of Yap/Taz target genes. We took advantage of the 8xGTIIC reporter gene system, which expresses luciferase only when Yap/Taz is present in the nucleus, to analyze inactivation of Hippo signaling.29 We found that reduced Pals1 expression resulted in a significant increase of luciferase activity in MDCK and HEK293T Pals1 KD cell lines (Figure 6E, Supplemental Figure 8B). Ectopically expressed Pals1, which compensates the reduced endogenous Pals1 expression in MDCK Pals1 KD cell lines, was able to reduce the 8xGTIIC reporter gene activity (Figure 6F). Real-time PCR revealed significant upregulation of Hippo target genes CYR61 and BIRC2 in Pals1 KD cell lines, indicating again (see also Figure 4B) that decreased Pals1 expression levels correlate with an activation of Yap/Taz-dependent gene expression (Figure 6G, Supplemental Figure 8C).
Cell Junction Dissociation Leads to Inactivation of Hippo Signaling In Vitro
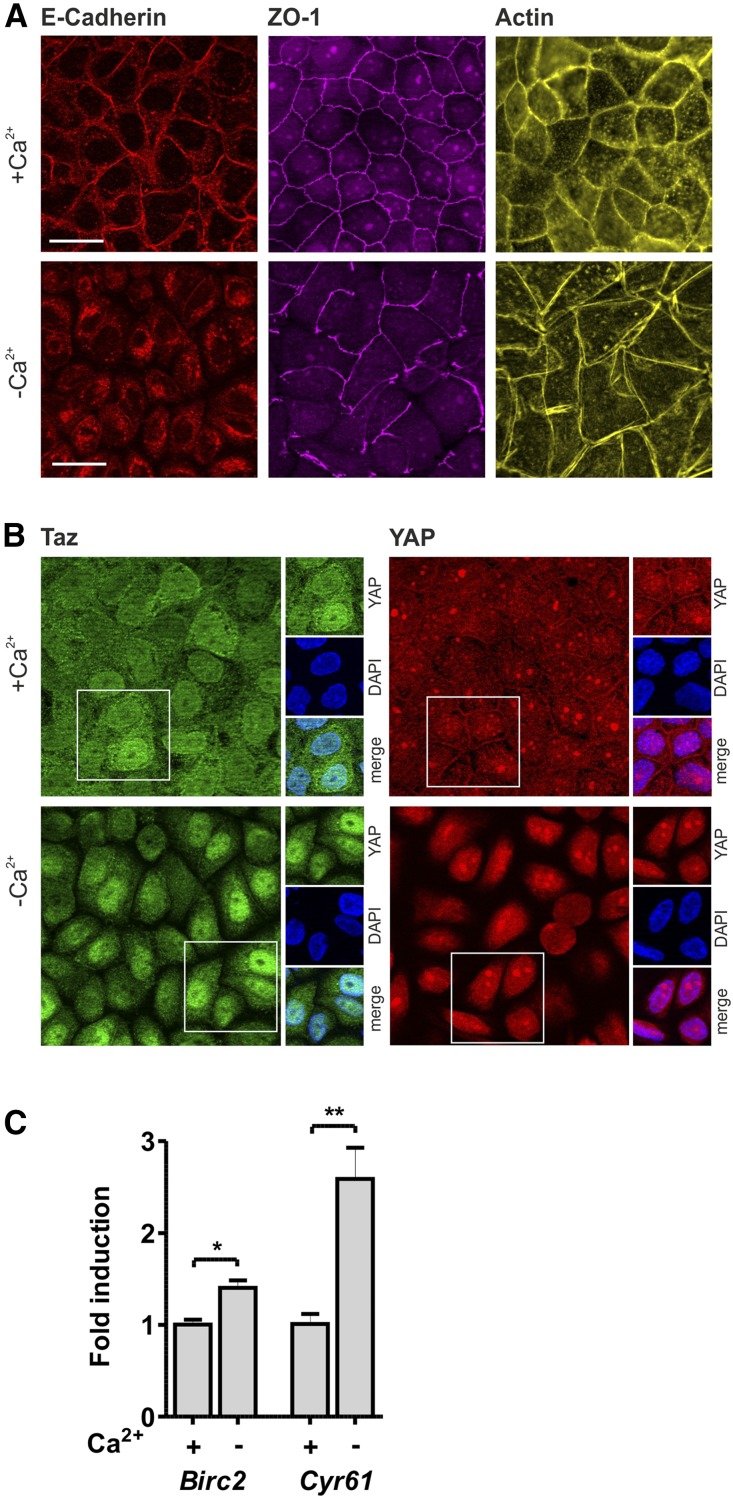
Knockdown studies in cell culture revealed an essential role of Pals1 in the formation of tight and adherens junctions, indicating that junction assembly is closely linked to cell polarization in epithelial cells.13,14 In order to analyze if the dissociation of cell-cell contacts influences Hippo signaling, monolayers of MDCK wildtype cells were cultivated under low Ca2+ conditions. Ca2+ depletion leads to disassembly of E-Cadherin–positive adherens junctions and ZO-1–positive tight junctions (Figure 7A), which results in a change of Yap and Taz localization (Figure 7B) accompanied by an increased expression of Yap/Taz target genes (Figure 7C).
Figure 7.
Disassembly of cell-cell contacts inactivates Hippo signaling. (A) Disassembly of cell-cell contacts in MDCK cells (wildtype) cultivated with Ca2+ (upper panel) and without Ca2+ (lower panel) stained for E-Cadherin (adherens junctions), ZO-1 (tight junctions), and actin cytoskeleton (Phalloidin). Scale bar, 20 µm. (B) Disassembly of cell-cell contacts by Ca2+ depletion in confluent wildtype MDCK cells increased nuclear localization of Taz (green) and Yap (red). Scale bars, 20 µm. (C) Ca2+ depletion resulted in an increased expression of Yap/Taz target genes CYR61 and BIRC2.
Pals1 Depletion Results in an Increase in Responsiveness to TGF-β
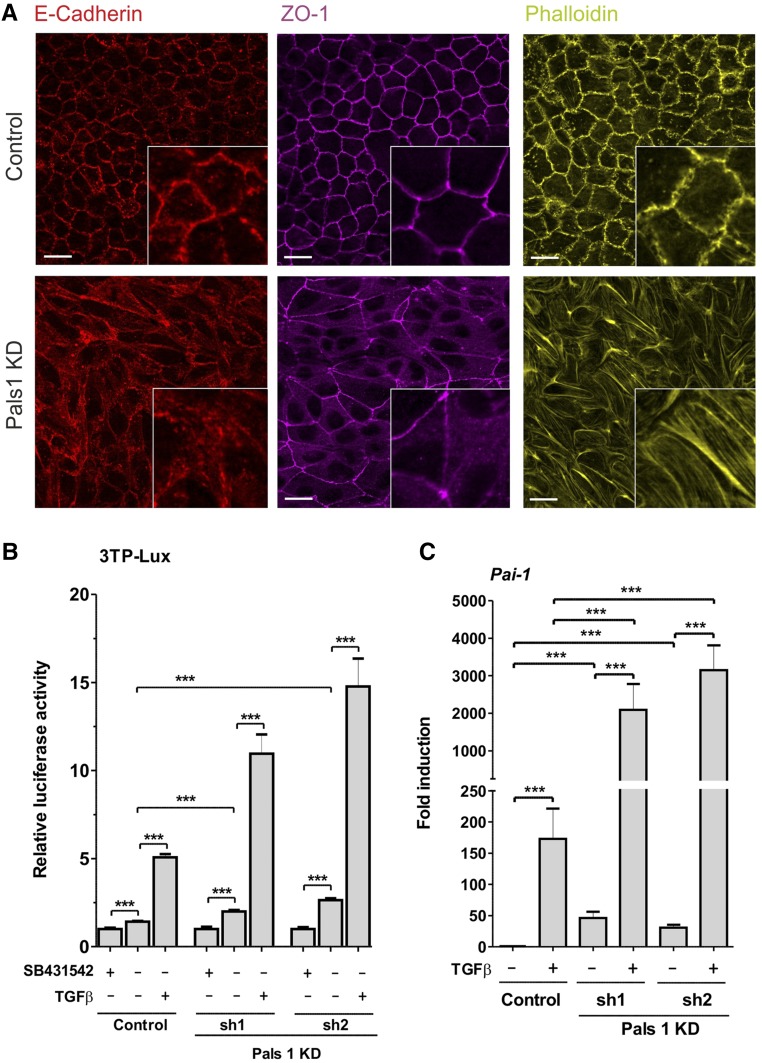
Similar to Ca2+-depleted MDCK cells, Pals1 KD cells showed a reduced formation of cell-cell contacts (Figure 8A, Supplemental Figure 7). Previous studies have pointed to a link between Hippo- and TGF-β–mediated signaling where disruption of the CRB complex enhances signaling by TGF-β and promotes EMT.18 In order to test whether Pals1 depletion in MDCK cells has an effect on TGF-β signaling, or TGF-β responsiveness, we used the 3TP-lux reporter gene system.30 We treated Pals1 KD and control cell lines with SB431542, an inhibitor of the TGF-β receptor, to completely block the endogenous basal activity of the receptor. Untreated Pals1 KD cells showed increased reporter gene activity relative to the untreated control cells. Administration of TGF-β (100 pM) led to a five-fold increase in the control, but to an 11- to 15-fold increase in reporter gene activity in Pals1 KD cells (Figure 8B), indicating that Pals1 serves as negative regulator of TGF-β–linked signaling. To confirm this, we analyzed expression levels of the TGF-β target gene Pai-1 (Figure 8C). The basal level of Pai-1 expression was found to be higher in Pals1 KD cell lines. After treatment with TGF-β, Pai-1 expression increased much more strongly than in the Pals1 KD cell lines. Thus, Pals1 depletion results in elevated responsiveness to TGF-β.
Figure 8.
Pals1 depletion increases responsiveness to TGF-β. (A) In Pals1 KD cells, the junction proteins E-cadherin and ZO-1 are more diffusely distributed and larger fractions of both remain in the cytoplasm. Instead of showing the normal cortical localization of actin, the knockdown cells form many stress fibers. MDCK cells were grown to high density, and E-cadherin, ZO-1, and actin were detected by immunofluorescence microscopy. Scale bars, 20 µm. (B) Pals1 knockdown resulted in an increased luciferase activity in the TGF-β–responsive 3TP-Lux luciferase reporter gene assay. Twenty-six hours after transfection with p3TP-Lux, control and Pals1 KD cells were treated with the TGF-β receptor antagonist SB431542 (10 µM), TGF-β (100 pM), or the corresponding solvent controls for 20 hours. 3TP-Lux reporter activity is presented as mean±SD and normalized to the value for the respective inhibitor-treated sample. (C) MDCK cells were grown to high density and treated with TGF-β (500 pM) or control solvent for 2 hours. Real-time PCR analysis revealed increased expression of the TGF-β target gene Pai-1 even under basal conditions, and the gene was highly induced upon TGF-β treatment.
Target Genes of the Hippo Pathway and Marker Genes of TGF-β Signaling Are Upregulated in Pals1-Depleted Kidneys
Next, we performed microarray analysis and compared the gene expression profile of kidneys of 4-week-old Six2-Cre+ mice with their corresponding Six2-Cre− littermate controls. The analyses showed, as expected, a 50% reduction in Pals1 expression in Six2-Cre+ kidneys.
The data set for each group (each n=3, Supplemental Figure 10A) was homogenous, therefore permitting the gene expression profiles of control (Six2-Cre−) and Pals1-depleted (Six2-Cre+) kidneys to be compared. By using the Pals1 regulation value (±1.5-fold) as threshold for further analysis, we found 1412 transcripts to be differentially expressed (Six2-Cre+ versus Six2-Cre− littermate controls, see Supplemental Material), of which 624 were up- and 788 were downregulated.
Similar to our in vitro results (Figures 6 and 8), target genes of the Hippo pathway, but also marker genes of TGF-β signaling, were significantly upregulated in Pals1-depleted kidneys (Supplemental Figure 10, Table 1). Moreover, many of the most highly upregulated genes were biomarkers of renal diseases, including marker genes for tubular-intestinal (e.g., Kim-1/Havcr1, Ngal/Lcn2, End1) and glomerular injury (Tagln1/2; Nphs1/2, Podxl), as well as for fibrosis and inflammation (Ccl2/Mcp-1, Gpnmb/Osteoactivin; Supplemental Material, Supplemental Table 1).
Table 1.
Expression of Hippo pathway–associated genes in Pals1-deficient kidneys compared with the littermate controls
| Gene | Name | Signaling Pathway | Six Cre2- versus Six2-Cre+ | |
|---|---|---|---|---|
| Fold Change | P Value | |||
| Myc | myelocytomatosis oncogene | Hippo | 3.94 | 1.31E-04 |
| Ctgf | connective tissue growth factor | Hippo | 2.52 | 4.01E-04 |
| Cyr61 | cysteine rich protein 61 | Hippo | 2.48 | 5.28E-03 |
| Axl | AXL receptor tyrosine kinase | Hippo | 2.04 | 6.85E-03 |
| Birc2 | baculoviral IAP repeat-containing 2 | Hippo | 1.87 | 3.86E-03 |
| Runx2 | Runt-related transcription factor 2 | Hippo | 1.54 | 9.33E-03 |
| Sox4 | SRY (sex-determining region Y)-box 4 | Hippo | 2.33 | 6.34E-05 |
| Tead1 | TEA domain family member 1 | Hippo | 1.59 | 9.94E-04 |
| Serpine1 | Serine/cysteine peptidase inhibitor, clade E, member 1 (Pai-1) | Hippo/TGF-β | 8.60 | 3.56E-04 |
| Runx1 | Runt-related transcription factor 1 | Hippo/TGF-β | 4.72 | 1.95E-04 |
| Cdkn1a | cyclin-dependent kinase inhibitor 1A | Hippo/TGF-β | 2.98 | 6.59E-05 |
| Col1a1 | collagen, type I, alpha 1 | Hippo/TGF-β | 2.81 | 6.20E-03 |
| Tgfbi | TGF, β induced | Hippo/TGF-β | 2.61 | 3.55E-03 |
| Col1a2 | collagen, type I, α 2 | Hippo/TGF-β | 2.47 | 7.88E-03 |
| Tgfb1 | TGF, β 1 | Hippo/TGF-β | 2.20 | 3.59E-03 |
| Fos | FBJ osteosarcoma oncogene | Hippo/TGF-β | 2.04 | 1.25E-02 |
| Junb | jun B proto-oncogene | Hippo/TGF-β | 2.30 | 2.34E-05 |
| Thbs1 | thrombospondin 1 | Hippo/TGF-β | 2.06 | 1.17E-02 |
| Tgfb3 | TGF, β 3 | Hippo/TGF-β | 1.90 | 2.96E-02 |
| Tsc22d1 | TSC22 domain family, member 1 | Hippo/TGF-β | 1.78 | 2.60E-03 |
| Dcn | decorin | Hippo/TGF-β | 1.92 | 2.86E-02 |
| Ifrd1 | IFN-related developmental regulator 1 | Hippo/TGF-β | 1.63 | 3.29E-03 |
Target genes of the Hippo signaling pathway and of genes that are affected by crosstalk between the Hippo and Wnt or Hippo and TGF-β are differentially regulated in Six2-Cre+ kidneys. The table lists a selection of genes that are significantly (see P value) upregulated in Pals1-deficient kidneys (Six2-Cre+) and are given in fold-change compared with the littermate controls (Six2-Cre−). Details of the microarray analysis are summarized in the Supplemental Material.
Discussion
In this study, we identified Pals1 as an essential regulator of renal epithelial morphogenesis. Intriguingly, heterozygous mice lacking one Pals1 (Mpp5) allele in the nephron developed a severe proteinuria, showed glomerular and tubular cysts, and died within 2 months of birth. The fact that even a partial reduction in Pals1 expression was sufficient to cause this complex renal phenotype is particularly interesting, given that other murine models lacking apical polarity proteins, such as Crb331–33 or Lin7c knockout mice,34 showed milder phenotype in the kidney, which moreover requires the loss of both alleles. These findings strongly support an essential, specific, and dose-dependent function for Pals1 in the nephron, especially during nephrogenesis.
Previous studies in the cerebral cortex and the retina have shown that partial removal of Pals1 causes severe cortical malformations in the brain and in retinal precursor cells, respectively.21,35 This is in line with our data, as it emphasizes that the level of Pals1 expression is crucial for the proper development of complex tissues. The defects seen at the renal filtration barrier in mice indicate that podocyte injury is a main factor for the severe proteinuria, suggesting that Pals1-deficiency affects podocytes during glomerulogenesis. The loss of the slit-diaphragm–like structures in Sdt-depleted nephrocytes, which are the fly model for glomerular podocytes, confirmed the important role of Pals1 at the renal filtration barrier. Moreover, our studies in nephrocytes confirmed the role of Pals1 as an upstream regulator of Hippo signaling, as the defects caused by knockdown of the Pals1 ortholog Sdt were rescued by a simultaneous knockdown of Yki. These data extend recent findings that have pointed to a tight link between polarity proteins and Hippo signaling.15–18
Despite the evolutionarily conserved role of Pals1 and Sdt in controlling Hippo signaling, the presence of two Yki orthologs in mammals, Yap and Taz, argues for tissue-specific roles of the two proteins in mammalian systems. Indeed, loss of Taz is linked to pronounced formation of glomerular22 and tubular cysts23,24 in several mice models, whereas Yap knockout mice die at early embryonic stages.25 Distinct roles for Yap and Taz in the kidney have also been shown by Reginensi et al.36 These authors generated mice lacking either Taz, Yap, or both in the nephron and found that only Taz depletion is linked to a cystic-kidney–like phenotype.36 Pals1 KD cells might therefore—at least in part—reflect the kidney phenotype seen in Taz knockout mice.22–24,36
Although the overall expression of the cotranscriptional factors Yap and Taz in the nephron was downregulated in Pals1-deficient kidneys, we observed an increase in the expression of Yap/Taz target genes. This change is associated with a shift of Yap/Taz from the cytoplasm into the nucleus in cyst-lining cells. This is also seen in our in vitro model, in which Taz and Yap show mainly nuclear localization. Consequently, partial loss of Pals1 leads to incomplete inactivation of the Hippo pathway. Our data are in agreement with findings of Happé et al. who investigated the localization of Yap in mice models of polycystic kidney disease, as well as in renal tissues from patients with ADPKD and autosomal recessive polycystic kidney disease.37 They did not only observe an upregulation of Yap target genes in an ADPKD mouse model, but also found increased nuclear localization of Yap in cystic epithelia of renal tissues from patients with ADPKD and autosomal recessive polycystic kidney disease.37
Margolis and colleagues have shown that a loss of Pals1 delays the formation of tight junctions, and is accompanied by a decrease in the transepithelial electrical resistance.13,14 These data in turn imply that defects in the formation of cell-cell contacts prevent the establishment of a tight lateral diffusion barrier in epithelial cells, which is essential for the separation of apical and basolateral transmembrane proteins within the plasma membranes. In this study, we observed that both Ca2+ as well as Pals1 depletion triggers Yap/Taz-dependent gene expression, suggesting that the tightness of junctions is a crucial factor in the control of Hippo signaling. Thus, our in vitro data probably supports the hypothesis that a reduced Pals1 level in vivo results in a reduced tightness of the various renal epithelia of the nephron, leading to a more inactivated Hippo signaling pathway in Pals1 knockout animals compared with control animals. However, these conclusions need further confirmation by further in vivo and in vitro studies.
Intriguingly, it was recently shown that failure to establish mature cell-cell contacts prevents correct basolateral distribution of TGF-β receptors, and leads to the accumulation of TGF-β receptors at the apical membrane accompanied by a concomitant increase in the expression of TGF-β target genes. This observation suggests that delays in the formation of cell-cell junctions make epithelial cells more susceptible to TGF-β–linked signaling processes.18,38 Indeed, gene expression analyses of Pals1-deficient kidneys and Pals1 KD cell lines revealed an upregulation of target genes of the TGF-β pathway. Thus, our data are compatible with a model in which Pals1, beyond its function as a crucial polarity protein, plays a key role as upstream regulator of Hippo- and TGF-β–mediated signaling in the nephron. Whether altered TGF-β signaling is linked to CRB complex–dependent cell-density sensing in a Hippo signaling–dependent18,38 or –independent manner39 is currently a matter of debate. In any case, imbalances between Hippo and TGF-β signaling may result in a “signaling catastrophe,” triggering cyst formation due to incomplete inactivation of the Hippo pathway accompanied by highly increased susceptibility to pathogenic TGF-β signaling. Numerous studies showed the contribution of impaired TGF-β signaling on glomerular diseases.40–42 Moreover, it has been reported that TGF-β signaling contributes to cyst progression and fibrogenesis.43,44 Thus, both processes, inactivation of Hippo and activation of TGF-β signaling, contribute to the injury of glomerular and tubular nephron epithelia and cyst progression in the nephron.
So far, no mutations in Pals1 gene (MPP5) have been associated with renal, and in particular renal-cystic, diseases in humans. The severity of the phenotypes observed in Pals1-deficient mice emphasizes the central role of this protein in the brain and the kidney. The fact that Pals1 haploinsufficiency causes lethal phenotypes in mice underlines the importance of Pals1, and makes it unlikely that Pals1/MPP5 itself could be a target gene for rare inherited diseases. However, Pals1-interacting proteins have been linked to renal cyst formation in mice (Crb3, Lin7c, and Taz)22–24,31–34,36 and to certain kinds of nephronophthisis (nephrocystin-1 and -4), a human inherited cystic disease of childhood.45 These findings support the hypothesis that apical polarity complexes (CRB and PAR) are directly or indirectly linked to renal diseases, especially renal cyst diseases.
In summary, our data provide initial insights that Pals1 is a negative regulator of the Hippo and TGF-β signaling pathways, and is thus directly involved in the process of nephrogenesis. Future studies should therefore target the Pals1-linked networks, as well as factors that might control the intracellular turnover and stability of these proteins, in order to decipher the mechanisms underlying pathogenesis.
Concise Methods
Mouse Strains and Genotyping
The Pals1/Mpp5 cKO (conditional knockout, in a C56Bl/6 background) mouse was described earlier.21 Transgenic Six2-Cre mice were purchased from JAX.2 To provide for EGFP-labeling of Cre-expressing cells and quantification of Cre penetrance in the Pals1 cKO mice, we included the double fluorescence mTomato/mGFP (mT/mG) reporter mouse in our studies.46 For DNA isolation and genotyping, tissue was incubated in lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris/HCl pH8.3, 0.45% NP-40 0,45% Tween 20) containing 0.05 µg/µl proteinase K (Peqlab Biotechnology, Erlangen, Germany) at 55°C overnight. The proteinase K was then inactivated by incubation at 95°C for 10 minutes, the sample was centrifuged, and the DNA-containing supernatant was used for genotyping. Cre-positive mice were identified by PCR, using the primers Cre for (5′ GCATTACCGTCGATGCAACGAGTGATGAG 3′) and Cre rev (5′ GAGTGAACGAACCTGGTCGAAATCAGTGCG 3′), which results in a 450-bp PCR product. Pals1 mice were genotyped as previously described.21
Tissue Preparation and Histologic Analyses
Blocks of cortical tissue were processed by standard procedures for paraffin sections (2 µm) and stained with periodic acid–Schiff.
Immunohistologic Analyses
Characterization of Pals1 protein expression was done in kidneys from Six-Cre2− (wildtype) and Six2-Cre+ mice (n=2–3 each) using immunofluorescence microscopy. Paraffin-embedded formalin-fixed sections (2 µm) were deparaffinized and rehydrated using standard protocols. Kidney sections were blocked for 45 minutes with 1% (wt/vol) skim milk (Bio-Rad Laboratories, Munich, Germany) followed by incubation with primary antibodies (see Table 1) overnight at room temperature and subsequent washing in Tris buffer (50 mM Tris pH 7.4 supplemented with 0.05% [vol/vol] Tween 20; three times 5 minutes). Afterwards, the secondary fluorescence-labeled antibodies (see Supplemental Table 2) were applied for 30 minutes followed by subsequent washing. For double staining of Pals1 with Aquaporin 2 the protocol was modified. Sections were stained for Pals1 followed by blocking first with 10% (vol/vol) rabbit serum diluted in Tris buffer and second with donkey anti-rabbit Fab fragments (1:50 in Tris buffer) for 30 minutes each and interrupted by intensive washing in Tris buffer. DAPI was used to stain nuclei (1:1000 in distilled water for 5 minutes) followed by rinsing in Tris buffer (three times 5 minutes). Finally, sections were covered with mowiol mounting medium (Calbiochem, La Jolla) and analyzed using laser scanning confocal microscopy (LSM Zeiss 710 and Zen software; Zeiss GmbH, Jena, Germany).
Cryo-Sections
After explantation, kidney tissue was directly embedded in Tissue-Tek (Sakura Finetek BV, Leiden, The Netherlands), and frozen sections (4–5 µm) were cut in a cryostat and mounted on slides. The slides were fixed with methanol/10% H2O2 at −20°C for 10 minutes, washed three times with PBS and incubated with 0.1% Triton-X-100 for 10 minutes at room temperature. For staining, the slides were first treated with blocking solution (5% BSA) for 20 minutes, then incubated with the first antibody solution (in 1% BSA) overnight at room temperature. After washing with PBS, the slides were incubated with the secondary antibody solution (in 1% BSA) for 45 minutes at room temperature. After further repeated washing the nuclei were stained for 5 minutes with DAPI and embedded in Fluoromount (Sigma-Aldrich) for microscopy.
We analyzed the number and size of glomeruli per kidney section as well as the distance between visceral and parietal epithelial cells in each glomerulus, and the size and number of cysts. Kidneys of six littermates (Six2-Cre+ versus Six2-Cre−) and one section of each kidney were used for quantification purposes. The number of glomeruli represents the total amount of glomeruli seen per kidney section (whole slides). The size of glomeruli and the distance between visceral and parietal cells were measured using panoramic viewer (3DHISTECH). We defined “cysts” as epithelium-lined fluid-filled cavities larger than approximately 35 µm in diameter and 1000 µm2 in area. The total number of cysts per kidney section was taken into account. To determine the mean area of cysts per section, the cysts were treated as circles (A = πr2). The diameter was measured with panoramic viewer, and the area of the cyst (A = πr2) was calculated.
Fly Stocks and RFP Accumulation Assay
Drosophila melanogaster stocks were cultured on standard cornmeal-agar food and maintained at 25°C. RFP filtration assays were performed at 29°C. Briefly, secreted RFP (ANP-RFP) is filtrated and taken up by garland nephrocytes, in which protein expression was downregulated using the nephrocyte-specific UAS/GAL4 driver line sns-GCN:GAL427 and the following responder lines: UAS::mCherry-RNAi (negative control, #35778; Bloomington Stock Center, Bloomington, BL), UAS::Sdt-RNAi-111 (#37510; BL), and UAS::Sdt-RNAi-69 (#15342R-2; National Institute of Genetics, Shizuoka, Japan). For rescue experiments, sns-GCN::GAL4, ANP-RFP was recombined with UAS::Sdt-RNAi-111 and crossed to UAS::Yki-RNAi (#40497; Vienna Drosophila Resource Center, Austria) and UAS::Yki-S168A.47 Garland nephrocytes were isolated from wandering third-instar larvae by microdissection in HL3.1 saline,48 fixed in 4% PFA in PBS for 1 hour, stained with DAPI for 20 minutes, washed with PBS, and mounted in Mowiol. Samples were imaged using an LSM-510 confocal microscope (ZEISS, Jena, Germany) and RFP accumulation in garland nephrocytes was subsequently analyzed using ImageJ. For each genotype, >50 nephrocytes from at least six individuals were scored.
Evaluation of nephrocyte ultrastructure was done by transmission electron microscopy. Briefly, garland nephrocytes were microdissected in HL3.1 saline, high-pressure frozen (EM-PACT2; Leica, Wetzlar, Germany), freeze-substituted in acetone/2% OsO4/5% H2O/0.25% uranyl acetate (AFS2; Leica, Wetzlar, Germany), and finally embedded in Epon. For transmission electron microscopy, 70-nm-thick sections were cut using a diamond knife (Diatome, Biel, Switzerland) with an ultramicrotome (Leica UC6 or UC7; Wetzlar, Germany). Samples were subsequently imaged with a TEM-902 transmission electron microscope (ZEISS, Jena, Germany).
Cell Culture and Generation of Stable Cell Lines
MDCK and HEK293T cells were grown in standard DMEM medium (Life Technologies) supplemented with glutamine and antibiotics (37°C and 5% CO2) as described previously.49,50 For Ca2+ depletion experiments, MDCK cells were cultivated in Ca2+-free DMEM (Life Technologies) for 16 hours. Stable knockdowns of endogenous canine Pals1 in MDCK cells were generated using pSuperior-Puro (Oligoengine) according to the manufacturer’s instructions. The short hairpin sequences used for silencing of endogenous canine Pals1 in MDCK cells were 5′-GCATGGTACGCTGACATTTGT-3′ (cPALS1-sh1) and 5′-AGAGGATAGTAGACAAGTTCT-3′ for (cPALS1-sh2), and for endogenous human Pals1 in HEK293T cells 5′-GCCAGTTCATCATAAGGAAGG-3′ (hPals1-sh1), and 5′-GCTACAGTTCGTAATGAAATG-3′ (hPals1-sh2), and 5′-CCTAAGGTTAAGTCGCCCTCG-3′ for the sh control. Selection of pSuperior-Puro–containing cells was done with 1 µg/ml puromycin.
Luciferase Assays
Luciferase assays were performed in MDCK cells to measure Yap/Taz-dependent (8xGTIIC-Lux system) or TGF-β signaling–dependent (3TP-Lux system) gene expression as described earlier.18,29 To normalize transfection, luciferase reporter genes were cotransfected with pCMV-β gal plasmids. All transfections were done with Lipofectamin 2000 (Life Technologies) according to the manufacturer’s instructions.
Preparation of Cell and Tissue Lysates and Western Blot
Kidneys were homogenized using an ULTRA-TURRAX (IKA) high-performance disperser in 1× Laemmli buffer (4% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol blue, 0.0625 M Tris-HCl; pH 6.8). Cells grown on dishes were lysed in 1× Laemmli. After boiling for 5 minutes, the sample was pushed through a 20-gauge needle and equal volumes of lysates were fractionated on 8%–15% SDS-PAGE gels (Biorad, Munich, Germany) and analyzed by western blot as described previously.49,50 Nonspecific binding of antibodies to unoccupied areas of the PVDF membrane was prevented by preincubation in blocking solution (5% BSA in TBS-T) for 1 hour. Afterwards, the membranes were incubated with primary antibodies overnight at 4°C. After washing the membrane several times in TBS-T, the secondary antibody was applied for 30 minutes at room temperature and the membrane was washed three times before incubating with Lumi-Light (Roche) to visualize the specific antigens.
Antibodies
The antibodies used for western blot analyses or immunofluorescence studies are listed in the Supplemental Table 2.
Real-Time PCR Analysis and Evaluation
Total RNA was isolated from cells and from mouse tissues using the Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). To verify the quality of RNA obtained from mouse tissue samples, the RNA integrity number (RIN) was determined using the RNA 6000 Nano LabChip technology (Agilent Technologies, Santa Clara). Only RNA with RIN≥6.7 was used for further analysis. RNA purity values (260/280 and 260/230) were above 1.99, and 1.8, respectively. The Agilent Bioanalyzer RIN values of the samples indicated no interfering degradation. Aliquots (1–3 µg) of total RNA were converted into cDNA using the SuperScript III Reverse Transcription Kit (Invitrogen, Darmstadt, Germany) according to the manufacturer’s instructions.
Real-time PCR was performed using the SYBR Green PCR Master Mix (Life Technologies), the Biorad CFX384 Touch (Bio-Rad Laboratories GmbH, Munich), and Bio-Rad CFX Manager v3.0 software. Relative expression levels of genes of interest were calculated as ΔCt values normalized to the GAPDH control. Sequences of the primers used are listed in the Supplemental Material.
DNA Microarrays
RNA concentration and purity were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). RNA integrity was checked using an Agilent Bioanalyzer (software version B.02.07.SI532) on an RNA Nano Chip (both Agilent Technologies Inc., Santa Clara, CA). Probing of Affymetrix GeneChip Mouse Gene 2.0 ST microarrays (Affymetrix UK Ltd., High Wycombe, UK) was performed as described in the manufacturer’s protocols. The probes were prepared from 100-ng aliquots of total RNA in accordance with the instructions supplied with the Affymetrix WT PLUS Reagent Kit. After hybridization, the arrays were washed and stained using the Affymetrix GeneChip Fluidics Station 450 and scanned using the Affymetrix GeneChip Scanner 3000 7G.
Microarray Data Analyses
Microarray data quality was checked as recommended by the manufacturer and quantified using the quality metrics in the Partek Genomics Suite software (Partek Inc., St. Louis, MO). Statistical analyses of microarray data were performed using the Partek Genomics Suite. CEL-files (containing raw expression measurements) were imported into Partek GS. The robust multiarray average algorithm was used for normalization. The array data were quantile-normalized and log-2–transformed. For each probe, a one-way ANOVA was performed: Yij = μ + Groupi + εij, where Yij represents the jth observation on the ith group and μ is the common effect for the whole experiment. εij represents the random error present in the jth observation on the ith group. The errors εij are assumed to be normally and independently distributed with mean 0 and SD δ for all measurements.51 For each probe, the Fisher least significant difference was tested to statistically compare the difference between the means of the groups’ expression measurements. An FDR value ≤0.05 was used as the threshold for significance.52 Only genes with a fold-change of ±1.4 were regarded as significantly changed in expression. Microarray raw data will be available through Gene Expression Omnibus accession number GSE77628.
Statistical Analyses
The evaluation was done using GraphPad prism (Graph Pad software). All data show SD of at least three independent experiments and were analyzed using paired t test. * P<0.05; ** P<0.01; *** P<0.001.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Katja Brinkmann, Brunhilde Hähnel, Julia Schröer, Truc Van Le, and Karin Wacker for excellent technical assistance. In addition, we thank Prof. Xaralabos Varelas (Boston, MA) and Prof. Stefano Piccolo (Padua, Italy) for providing constructs, Prof. Thomas Benzing and Prof. Berhard Schermer (Cologne, Germany) for Taz−/− tissue, and Prof. Christopher A. Walsh (Boston, MA) for the conditional Pals1 knockout mice.
The work was supported by grants from the German Research Foundation to T.W. (DFG, SCHL 1845/2-1) and M.P.K. (SFB699, KR3901/1-2) and by a grant from the Else Kröner-Fresenius-Stiftung (EKFS 2014_A271) to T.W. and H.P., respectively.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016040474/-/DCSupplemental.
References
- 1.Costantini F, Kopan R: Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18: 698–712, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause M, Rak-Raszewska A, Pietilä I, Quaggin SE, Vainio S: Signaling during kidney development. Cells 4: 112–132, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernascone I, Martin-Belmonte F: Crossroads of Wnt and Hippo in epithelial tissues. Trends Cell Biol 23: 380–389, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Boulan E, Macara IG: Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 15: 225–242, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pieczynski J, Margolis B: Protein complexes that control renal epithelial polarity. Am J Physiol Renal Physiol 300: F589–F601, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamberov E, Makarova O, Roh M, Liu A, Karnak D, Straight S, Margolis B: Molecular cloning and characterization of Pals, proteins associated with mLin-7. J Biol Chem 275: 11425–11431, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Knust E, Tepass U, Wodarz A: crumbs and stardust, two genes of Drosophila required for the development of epithelial cell polarity. Dev Suppl: 261–268, 1993 [PubMed] [Google Scholar]
- 9.Wei X, Malicki J: nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet 31: 150–157, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B: The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of crumbs and discs lost. J Cell Biol 157: 161–172, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roh MH, Fan S, Liu C-J, Margolis B: The crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116: 2895–2906, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B: Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5: 137–142, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Straight SW, Shin K, Fogg VC, Fan S, Liu C-J, Roh M, Margolis B: Loss of PALS1 expression leads to tight junction and polarity defects. Mol Biol Cell 15: 1981–1990, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Chen XW, Margolis B: PALS1 regulates E-cadherin trafficking in mammalian epithelial cells. Mol Biol Cell 18: 874–885, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson BS, Huang J, Hong Y, Moberg KH: Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol 20: 582–590, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE: Lgl, aPKC, and crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol 20: 573–581, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Chen C-L, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G: The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A 107: 15810–15815, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL: The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell 19: 831–844, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Piccolo S, Dupont S, Cordenonsi M: The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev 94: 1287–1312, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Hansen CG, Moroishi T, Guan K-L: YAP and TAZ: A nexus for Hippo signaling and beyond. Trends Cell Biol 25: 499–513, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho SH, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ, Malicki J, LaMantia AS, Broccoli V, Walsh CA: The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron 66: 69–84, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W: Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci USA 104: 1631–1636, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Y, Kolb R, Hong J-H, Carroll J, Li D, You J, Bronson R, Yaffe MB, Zhou J, Benjamin T: TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol 27: 6383–6395, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, Nakagawa O, Small EV, Cobo-Stark P, Igarashi P, Murakami M, Tominaga J, Sato T, Asano T, Kurihara Y, Kurihara H: Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol 294: F542–F553, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL: Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weavers H, Prieto-Sánchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, Ruiz-Gómez M, Skaer H, Denholm B: The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM: Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136: 2335–2344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, Zhao Y, Chao Y, Muir K, Han Z: Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J Am Soc Nephrol 24: 209–216, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S: Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183, 2011 [DOI] [PubMed] [Google Scholar]
- 30.Wrana JL, Attisano L, Cárcamo J, Zentella A, Doody J, Laiho M, Wang X-F, Massagué J: TGF β signals through a heteromeric protein kinase receptor complex. Cell 71: 1003–1014, 1992 [DOI] [PubMed] [Google Scholar]
- 31.Whiteman EL, Fan S, Harder JL, Walton KD, Liu C-J, Soofi A, Fogg VC, Hershenson MB, Dressler GR, Deutsch GH, Gumucio DL, Margolis B: Crumbs3 is essential for proper epithelial development and viability. Mol Cell Biol 34: 43–56, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charrier LE, Loie E, Laprise P: Mouse Crumbs3 sustains epithelial tissue morphogenesis in vivo. Sci Rep 5: 17699, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szymaniak AD, Mahoney JE, Cardoso WV, Varelas X: Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector yap. Dev Cell 34: 283–296, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen O, Funke L, Long JF, Fukata M, Kazuta T, Trinidad JC, Moore KA, Misawa H, Welling PA, Burlingame AL, Zhang M, Bredt DS: Renal defects associated with improper polarization of the CRB and DLG polarity complexes in MALS-3 knockout mice. J Cell Biol 179: 151–164, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho SH, Kim JY, Simons DL, Song JY, Le JH, Swindell EC, Jamrich M, Wu SM, Kim S: Genetic ablation of Pals1 in retinal progenitor cells models the retinal pathology of Leber congenital amaurosis. Hum Mol Genet 21: 2663–2676, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim D-S, Pawson T, Wrana J, McNeill H: Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet 9: e1003380, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Happé H, van der Wal AM, Leonhard WN, Kunnen SJ, Breuning MH, de Heer E, Peters DJM: Altered Hippo signalling in polycystic kidney disease. J Pathol 224: 133–142, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Narimatsu M, Samavarchi-Tehrani P, Varelas X, Wrana JL: Distinct polarity cues direct Taz/Yap and TGFβ receptor localization to differentially control TGFβ-induced Smad signaling. Dev Cell 32: 652–656, 2015 [DOI] [PubMed] [Google Scholar]
- 39.Nallet-Staub F, Yin X, Gilbert C, Marsaud V, Ben Mimoun S, Javelaud D, Leof EB, Mauviel A: Cell density sensing alters TGF-β signaling in a cell-type-specific manner, independent from Hippo pathway activation. Dev Cell 32: 640–651, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K: Developmental signalling pathways in renal fibrosis: The roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol 12: 426–439, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tossidou I, Schiffer M: TGF-β/BMP pathways and the podocyte. Semin Nephrol 32: 368–376, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Herman-Edelstein M, Weinstein T, Gafter U: TGFβ1-dependent podocyte dysfunction. Curr Opin Nephrol Hypertens 22: 93–99, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Małgorzewicz S, Skrzypczak-Jankun E, Jankun J: Plasminogen activator inhibitor-1 in kidney pathology (Review). [Review] Int J Mol Med 31: 503–510, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Norman J: Fibrosis and progression of autosomal dominant polycystic kidney disease (ADPKD). Biochim Biophys Acta 1812: 1327–1336, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delous M, Hellman NE, Gaudé HM, Silbermann F, Le Bivic A, Salomon R, Antignac C, Saunier S: Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum Mol Genet 18: 4711–4723, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L: A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Oh H, Irvine KD: In vivo analysis of Yorkie phosphorylation sites. Oncogene 28: 1916–1927, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao X, Ding X, Guo Z, Zhou R, Wang F, Long F, Wu F, Bi F, Wang Q, Fan D, Forte JG, Teng M, Yao X: PALS1 specifies the localization of ezrin to the apical membrane of gastric parietal cells. J Biol Chem 280: 13584–13592, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Schulze U, Vollenbroeker B, Braun DA, Van Le T, Granado D, Kremerskothen J, Fraenzel B, Klosowski R, Barth J, Fufezan C, Wolters DA, Pavenstadt H, Weide T: The Vac14-interaction network is linked to regulators of the endolysosomal and autophagic pathway. Mol Cell Proteomics 2014. Available at http://www.ncbi.nlm.nih.gov/pubmed/24578385. Accessed March 21, 2014 [DOI] [PMC free article] [PubMed]
- 50.Wennmann DO, Vollenbroker B, Eckart AK, Bonse J, Erdmann F, Wolters DA, Schenk LK, Schulze U, Kremerskothen J, Weide T, Pavenstadt H: The Hippo pathway is controlled by Angiotensin II signaling and its reactivation induces apoptosis in podocytes. Cell DeathDis 5: e1519, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenhart C: The assumptions underlying the analysis of variance. Biometrics 3: 1–21, 1947 [PubMed] [Google Scholar]
- 52.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z: Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses 45: 486–490, 1995 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.