Abstract
Increased urine albumin excretion is highly prevalent in Hispanics/Latinos. Previous studies have found an association between urine albumin excretion and Amerindian ancestry in Hispanic/Latino populations. Admixture between racial/ethnic groups creates long-range linkage disequilibrium between variants with different allelic frequencies in the founding populations and it can be used to localize genes. Hispanic/Latino genomes are an admixture of European, African, and Amerindian ancestries. We leveraged this admixture to identify associations between urine albumin excretion (urine albumin-to-creatinine ratio [UACR]) and genomic regions harboring variants with highly differentiated allele frequencies among the ancestral populations. Admixture mapping analysis of 12,212 Hispanic Community Health Study/Study of Latinos participants, using a linear mixed model, identified three novel genome-wide significant signals on chromosomes 2, 11, and 16. The admixture mapping signal identified on chromosome 2, spanning q11.2–14.1 and not previously reported for UACR, is driven by a difference between Amerindian ancestry and the other two ancestries (P<5.7 × 10−5). Within this locus, two common variants located at the proapoptotic BCL2L11 gene associated with UACR: rs116907128 (allele frequency =0.14; P=1.5 × 10−7) and rs586283 (C allele frequency =0.35; P=4.2 × 10−7). In a secondary analysis, rs116907128 accounted for most of the admixture mapping signal observed in the region. The rs116907128 variant is common among full-heritage Pima Indians (A allele frequency =0.54) but is monomorphic in the 1000 Genomes European and African populations. In a replication analysis using a sample of full-heritage Pima Indians, rs116907128 significantly associated with UACR (P=0.01; n=1568). Our findings provide evidence for the presence of Amerindian-specific variants influencing the variation of urine albumin excretion in Hispanics/Latinos.
Keywords: human genetics, albuminuria, ethnic minority, genomics, ethnic groups, albumins
Increased urine albumin excretion is associated with a higher lifetime risk of ESRD and with increased cardiovascular disease risk.1,2 Both increased urine albumin excretion and ESRD differ by racial/ethnic groups in the United States, with the lowest and highest risks noted in European and Amerindian populations, respectively. Using sex-specific cut-points, increased urine albumin excretion prevalence in the United States is 10.3% in whites, 13.6% in blacks, 9.9% in Mexican Americans,3 >20% in American Indians,4 and 12%–14% in Hispanics/Latinos of the Hispanic Community Health Study/ Study of Latinos (HCHS/SOL).5 Hispanics/Latinos also have an approximately two-fold higher risk of ESRD than whites.6 However, Hispanics/Latinos are a heterogeneous group who show diversity in ancestry background including Amerindian, European, and West African.7 The percentages of African and Amerindian ancestry have previously been associated with increased urine albumin excretion prevalence in Hispanic/Latino populations.8,9
Despite the strong evidence for a role of ancestry in CKD susceptibility, few studies of kidney traits have leveraged the known genetic admixture in Hispanic/Latino populations to discover potential chromosomal regions that may harbor variants that confer risk for CKD traits such as urine albumin excretion. Among the genome-wide significant loci that have been identified and consistently replicated for urine albumin excretion, CUBN (chromosome 10) genetic variants are associated with urine albumin excretion in individuals of European ancestry and Hispanics/Latinos,10 and the HBB variant related to sickle cell trait (chromosome 11) is African-specific and associated with urine albumin excretion in Hispanics/Latinos with African admixture.11 An additional African-specific gene associated with increased urine albumin excretion and CKD is APOL1.12,13 Our recent work in the HCHS/SOL has confirmed a high proportion of Amerindian ancestry among Mainland Hispanics (Mexican, Central, and South American), who also had a low proportion of African ancestry.14 However, Mainland Hispanics have similar mean urine albumin excretion and frequency of increased urine albumin excretion compared with individuals of Caribbean background (Cuban, Dominican, Puerto-Rican), despite the absence of African-specific risk variants (APOL1 or HBB). This evidence suggests the presence of Amerindian ancestry variants in Hispanics/Latinos influencing urine albumin excretion.
There is great potential for genetic studies of CKD in Hispanics/Latinos to provide new insight into population-specific variants that confer risk for CKD traits but have not been uncovered through genome-wide association studies (GWAS). Admixture mapping leverages the known genomic heterogeneity of admixed individuals for improved genetic discovery, by identifying loci that contain genetic variants with allelic frequencies that are highly differentiated among ancestral populations and also significantly associated with a trait. It can use local ancestry at genomic regions to capture both common and rare variants. Prior research leveraging admixture has successfully identified the APOL1 alleles as a strong risk factor for clinically attributed hypertensive-associated CKD, FSGS, and HIV nephropathy in blacks.12,15
The HCHS/SOL study reported here used admixture mapping analysis in a large population sample of Hispanics/Latinos to identify loci that may harbor variants that increase urine albumin excretion. We identified a new locus at chromosome 2 that harbors an Amerindian-specific variant associated with urine albumin excretion and these findings were replicated in a cohort of Pima Indians.
Results
We analyzed a sample of 12,212 Hispanic/Latino individuals from the HCHS/SOL, with mean age of 46 years (SD=14 years), and with 41% men, 20% of patients with diabetes, 28% hypertensive, and 43% with obesity. The UACR was 6.54 mg/dl (interquartile range, 4.49–12.24). Three percent of individuals had an eGFR<60 ml/min per 1.73 m2 and 14.1% had increased UACR (≥17 mg/g in men and ≥25 mg/g in women).
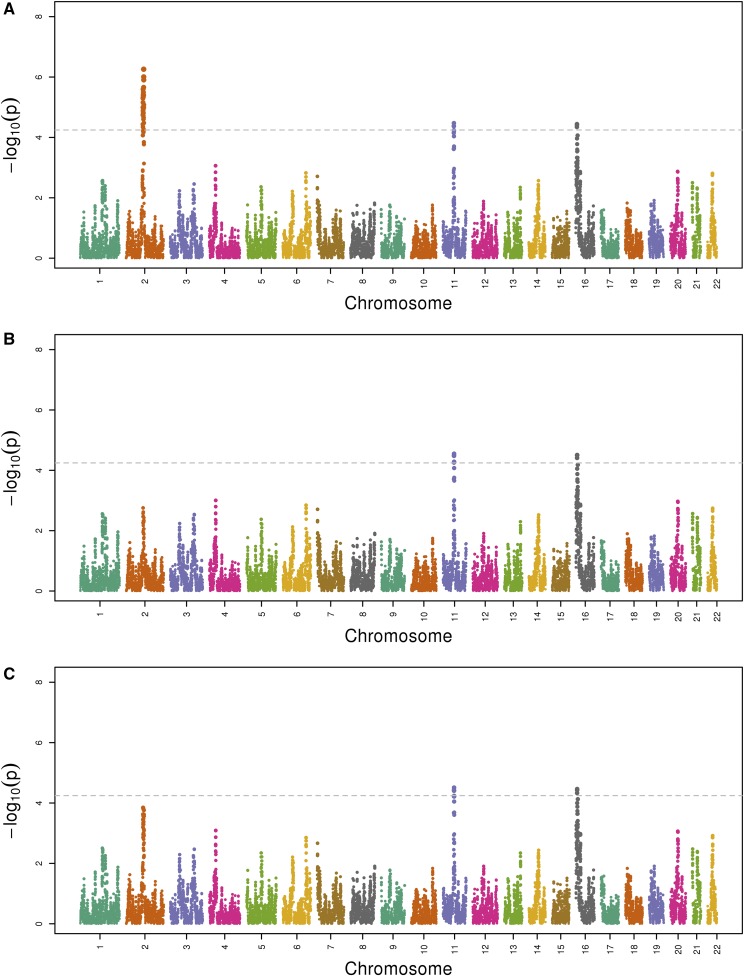
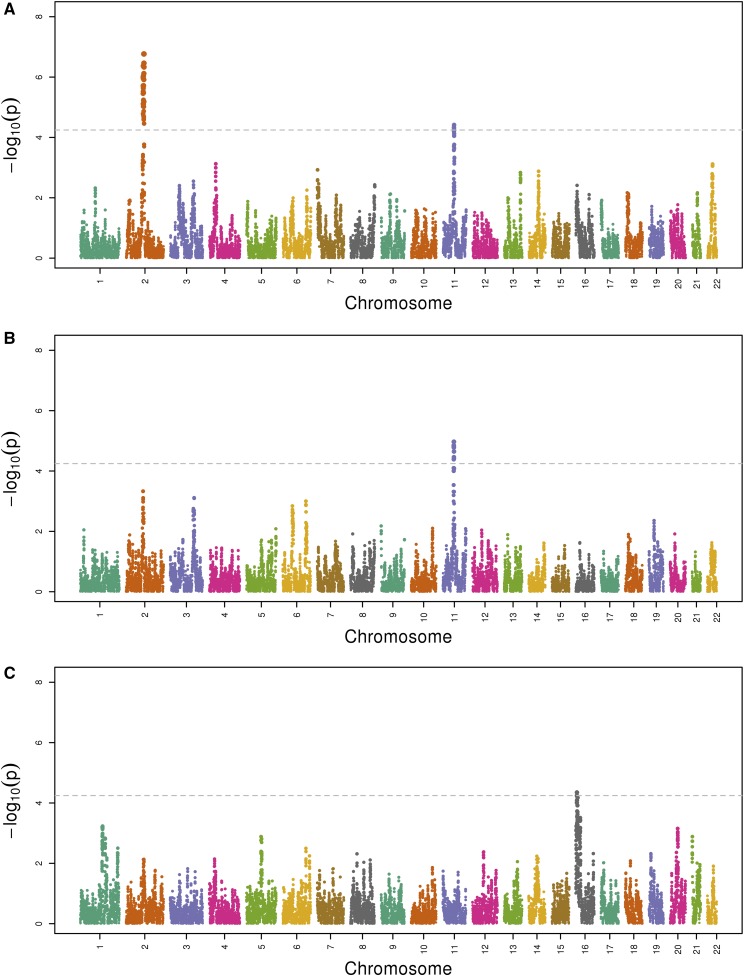
Local ancestry was estimated as European, African, or Amerindian at each of 419,645 genotyped SNPs across the autosomes.16 Admixture mapping consisted of a joint test of the effects of all three ancestry types on log-transformed UACR at each of the 419,645 SNP positions (using a linear mixed model with multiple fixed and random effects, as described in Concise Methods). This analysis identified three regions with P value(s) <5.7 × 10−5, indicating genome-wide significance (Figure 1A). The most significant peak spans 2q11.2–q14.1 with a minimum P value achieved (P=5.6 × 10−7) at 2q13. The second spans 11q13.2–q13.4 with a peak at 11q13.3 (P=3.4 × 10−5). The third peak is located in the 16p13.3 gene region (P=3.6 × 10−5). Secondary admixture mapping analyses that tested the effect of each local ancestry type separately revealed that the 2q13 locus was significantly associated with local Amerindian ancestry, whereas the 11q13 locus was associated with both local Amerindian and local European ancestries, and the 16p13 locus was associated with local African ancestry (Figure 2). The estimates for admixture mapping are shown in the Supplemental Table 1.
Figure 1.
Manhattan plot of admixture mapping of UACR in the HCHS/SOL population, n=12,212. (A) Three genome-wide loci reached the significance threshold (P<5.7 × 10−5, dashed lines). (B) Admixture mapping conditional on rs116907128. (C) Admixture mapping conditional on rs586283. In each plot, the x axis is chromosome position and the y axis is the −log10(P value) for the joint test of variation among the three local ancestry types.
Figure 2.
Manhattan plot of admixture mapping of UACR for each local ancestry type separately in the HCHS/SOL population, n=12,212: (A) Amerindian versus non-Amerindian; (B) European versus non-European; (C) African versus non-African. x axis is chromosome position and y axis is the −log10(P value) for admixture mapping analyses. The dashed line is the threshold for genome-wide significance (P<5.7 × 10−5).
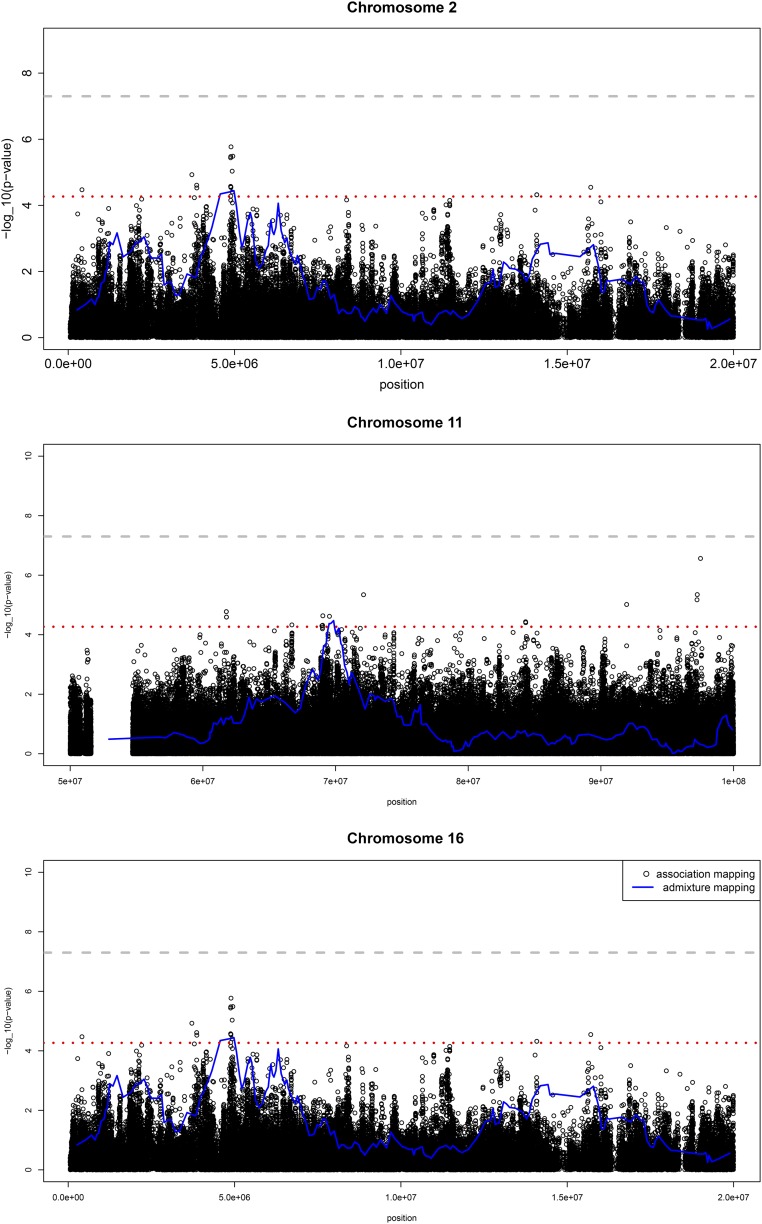
Single-variant association analyses performed using imputed genotypes within a 10 Mb window around 2q11.2–q14.1 yielded two SNPs significantly associated with UACR: rs116907128 (allele frequency for A allele =0.14; P=1.5 × 10−7) and rs586283 (allele frequency for C allele =0.35; P=4.2 × 10−7) (Figure 3A, Supplemental Material, Table 1). In analysis excluding diabetic individuals, the association of rs116907128 with UACR was unchanged (β=0.10; SEM=0.02; P=1.8 × 10−7), but the association of rs586283 with UACR was attenuated (β=0.05; SEM=0.01; P=2.1 × 10−4). Linkage disequilibrium between these two SNPs is low (r2=0.11), suggesting these are independent associations. There was no compelling evidence for associations within the regions containing the chromosomes 11 and 16 admixture signals (Figure 3, B and C).
Figure 3.
Single-variant and admixture association analyses within the regions on chromosomes 2, 11, and 16 showing admixture mapping signals in the HCHS/SOL population, n=12,212. For each plot, the x axis is chromosome position and the y axis is the −log10(P value) for admixture mapping analyses (blue lines) and single SNP association with UACR (black dots). The red dashed line is the threshold for the genome-wide admixture mapping (P<5.7 × 10−5). The gray dashed line is the threshold for association if the study was a GWAS.
Table 1.
Association findings for SNP rs116907128, which lies within the 2q11.2–q14.1 locus identified by admixture mapping
| Study | Alleles | A allele frequency | UACRa | eGFR P Value | Type 2 Diabetes | Diabetic Nephropathyb | |||
|---|---|---|---|---|---|---|---|---|---|
| β (SEM) | P Value | OR | P Value | OR (95% CI) | P Value | ||||
| HCHS/SOL study | A/C | 0.14 | 0.11 (0.02) | 1.5 × 10−7 | 0.97 | 0.99 | 0.79 | NA | |
| Full-heritage Pima Indiansc | A/C | 0.54 | 0.32 (0.13) | 9.6 × 10−3 | 0.85 | 1.07 | 0.33 | 1.22 (1.01 to 1.48) | 0.04 |
OR, odds ratio; 95% CI, 95% confidence interval; NA, not available.
Test for heterogeneity estimated from HCHS/SOL Hispanics/Latinos and Pima Indians not significant (Q=2.55 on 1 degrees of freedom; P=0.11).
Diabetic nephropathy defined as case (UACR≥300 mg/g or ESRD; n=398) versus control (UACR<300 mg/g; n=1251) for a total of n=1649.
UACR (n=1568); eGFR (n=1552); type 2 diabetes (total n=3699; 1724 cases; and 1975 controls).
To determine if the two identified SNPs on chromosome 2 account for the admixture mapping findings, we repeated the admixture mapping analyses using these SNPs as covariates. Adjusting for rs116907128 markedly reduced the admixture mapping signal (P for admixture =0.002) at the chromosome 2 locus (Figure 1B), whereas conditioning on rs586283 slightly attenuated the association findings (P for admixture =1.4 × 10−4) (Figure 1C, Supplemental Table 1). Analyses including both SNPs showed similar results to conditional analysis on rs116907128 (data not shown). Neither SNP was associated with eGFR in Hispanics/Latinos (rs116907128, P=0.97; and rs586283, P=0.15) or diabetes (P=0.81 and 0.46, respectively). SNP rs116907128 is monomorphic in 1000 Genome Project European and African samples, but it is a common variant among full-heritage Pima Indians (A allele frequency =0.54). In a replication analysis conducted in full-heritage Pima Indians, rs116907128 associated with UACR as a continuous trait (n=1568; β=0.32; P=0.01; P for heterogeneity Hispanics/Latinos and Pima Indians =0.11) and nominally with diabetic nephropathy in a case (UACR≥300 mg/g or ESRD) versus control (UACR<300 mg/g) analysis (n=1649; P=0.04; odds ratio=1.22; 95% confidence interval, 1.01 to 1.48). There was a nonsignificant increase in prevalence of diabetic nephropathy by duration of diabetes among Pima Indians with AA/AC genotypes compared with CC genotypes but the genotype-duration of diabetes interaction was not significant (P=0.12; Supplemental Figure 1). The rs116907128 SNP was not associated with eGFR (n=1552; P=0.85) or type 2 diabetes (n=3699; P=0.33) in full-heritage Pima Indians.
We used Haploreg17 (Epigenome Roadmap Project and the Encyclopedia Of DNA Elements [ENCODE] Project) to query the evidence for regulatory function of SNPs in the chromosome 2 region with significant admixture mapping signal. The rs116907128 position, located 5′ upstream of BCL2L11, overlaps enriched regulatory annotations for DNase I hypersensitivity sites in multiple tissues including fetal kidney and immune cells. This variant also overlaps transcription factor binding sites, including histone H3K4me1, H3K4me3, H3K27ac, and H3K9ac in several cell lines.
Discussion
This study used admixture mapping analyses to identify novel loci that may harbor variants affecting UACR and conferring risk for UACR. The main findings of our study are the identification of three novel genomic regions on chromosomes 2, 11, and 16 for which UACR is associated with local ancestry in a large and diverse population sample of Hispanics/Latinos. This approach identified an Amerindian-specific locus on chromosome 2, driven by a variant located 5′ of BCL2L11, common in Amerindians but rare in European and African populations. Amerindian genetic ancestry has been significantly associated with urine albumin excretion among Hispanics/Latinos,8,9 and Amerindian variants have been previously shown to associate with type 2 diabetes susceptibility in Hispanics/Latinos.18,19 The admixture mapping approach is on the basis of local ancestry—i.e., the number of alleles at a specific variant position that derive from each member of a given set of ancestral populations (in this case Amerindian, West African, and European). This approach also accounts for global ancestry (across all autosomes) as a fixed covariate in the linear mixed-model regression. In contrast, although GWAS typically adjust for global ancestry, they do not directly account for local ancestry in detecting genotype-phenotype associations. Our findings provide important information on the underlying genetic architecture of ancestry-specific genetic risk for UACR. Here, we have identified three specific loci that may contribute to associations between global ancestry and UACR or increased UACR. These results support further investigation of Amerindian ancestry to better understand Hispanic CKD risk.
To date, the only other validated genome-wide significant loci for UACR are CUBN and HBB (which is related to sickle cell trait10,11). Two additional UACR loci located at 2q21 (HS6ST1) and 11q14 (near RAB38/CTSC) were recently described in a GWAS of diabetic individuals (n=5509–5825) of European ancestry.20 These regions do not overlap our identified loci. We previously have shown significant genome-wide associations of CUBN and HBB with UACR in the HCHS/SOL study.21 The most significant CUBN variant has similar allele frequency in West African (AFR=0.17), European (EUR=0.12), and Hispanic (0.14) populations using 1000 Genomes data. Therefore, this variant will not be expected to be identified in admixture mapping.
Our main admixture mapping finding is in a region containing BCL2L11, which encodes a proapoptotic protein that belongs to the BCL-2 protein family. Several lines of evidence suggest a role of this gene and its protein in kidney development and kidney disease states. MicroRNA (miRNA)–mediated regulation of Bcl1211 expression in mice plays an important role in nephron progenitor survival during kidney development.22 miRNAs are involved in post-transcriptional repression of target mRNAs. Loss of miRNAs in nephron progenitors increases apoptosis and elevates the expression of protein Bim (also known as BCL2L11) leading to a decrease in nephron number.22 Bim null mice manifest systemic autoimmune disease and immune complex GN.23 In an experimental study of diabetic nephropathy, increased advanced glycation end products promoted Bcl2l11 expression, leading to podocyte apoptosis.24 Establishing the functional role of this gene in CKD will require targeted in vivo studies using knockout and/or transgenic mouse models, in addition to longitudinal population studies. The variant we identified, rs116907128, located at 5′ of BCL2L11, overlaps enriched regulatory annotations for DNase I hypersensitivity sites and histone marks in fetal and adult kidney, and in several other cell lines including immune cells. This evidence suggests a regulatory function in these cells and tissues. Among full-heritage Pima Indians, this variant is associated with increased UACR (n=1568; P=0.01) and diabetic nephropathy (n=1649; P=0.04), but not with eGFR or type 2 diabetes. In HCHS/SOL Hispanics/Latinos, there is also no association with either type 2 diabetes or eGFR. Interestingly, the “A” allele for rs116907128 is more common in Pima Indians than admixed Hispanics/Latinos, and the association with UACR was stronger in full Pima Indians.
The resolution of admixture mapping depends on the lengths of local ancestry tracts (i.e., the contiguous positions on a given chromosome having the same local ancestry). In Hispanics/Latinos, local ancestry tracts are relatively large because the admixture among European, African, and Amerindian ancestors was relatively recent so recombination has not yet broken them down into small segments. Therefore, the regions with genome-wide significance in admixture mapping are relatively large. Here, we used single-variant association testing to further localize the genetic effect(s) within these broader regions. There is an SNP within the admixture mapping signal for UACR on chromosome 2 (rs116907128), which has a P value for association with UACR of 1.5 × 10−7. This P value is significant after multiple-testing adjustment within that region, but it does not meet the genome-wide significance level of 5 × 10−8. Therefore, it was not identified as in our previous GWAS for UACR in HCHS/SOL.21 This example illustrates how admixture mapping can be complementary to genome-wide association testing of single variants as well as localizing genetic factors contributing to global ancestry associations with diseases and their component traits.
In the regions detected on chromosome 11 and 16, we found no SNPs that passed the region-specific multiple-testing adjustment. This could be due to low power in association testing (e.g., due to relatively low combined allele frequencies of the tag SNPs), or due to lack of good proxies of the causal SNP(s). Further studies will require sequencing the chromosome 11 and 16 regions to identify variants (common and rare) accounting for these admixture signals in Hispanics/Latinos. In addition, fine-mapping of loci associated with Amerindian local ancestry are likely to be more successful when performed in American Indian populations.
Our study is limited by the available genomic markers (imputed and genotyped) for fine-mapping of regions. Although we found strong associations of rs116907128 (and rs586283) at the chromosome 2 locus, which accounted for most of the local ancestry association in the admixture mapping, there was still a fairly small admixture P value after adjusting for the SNPs, suggesting that there must be other variants in the region also contributing to the signal, or that the SNPs are only in partial LD with one or more causal variants. Admixed mapping approaches capture both rare and common variants, but association analyses are underpowered to detect low-frequency variants. Further studies using sequencing are needed to identify additional low-frequency and rare variants in the region that could account for the admixture mapping findings. The replication of our findings in Pima Indians supports the role of the locus on UACR variation. Future studies will examine longitudinal changes in kidney function and UACR to better characterize the relevance of BCL2L11 to increased UACR and CKD, which may provide important clues to the clinical effect of this gene in CKD. HCHS/SOL is currently examining participants in a second visit, from which data could be used for these follow-up studies.
In summary, we identified three novel loci for UACR using admixture mapping in Hispanics/Latinos, including a region on chromosome 2 within which the signal is driven by an Amerindian-specific variant near the BCL2L11 gene, which has a UACR association that replicated in Pima Indians. Our study provides important information on the presence of Amerindian-specific genetic variants associated with UACR in Hispanics/Latinos and American Indians.
Concise Methods
Study Population
HCHS/SOL is a community-based cohort study of 16,415 self-identified Hispanic/Latino adults aged 18–74 years from randomly selected households near four United States field centers (Chicago, Bronx, Miami, and San Diego) with baseline examination (2008–2011) and yearly telephone follow-up assessment for at least 3 years as previously described.25 Participants self-identified as having Hispanic/Latino background, with the largest groups being Central American (n=1730), Cuban (n=2348), Dominican (n=1460), Mexican (n=6471), Puerto-Rican (n=2728), and South American (n=1068). The clinical examination included physical measures, behavioral/lifestyle factors, and sociodemographic assessments,25 in addition to collection of fasting blood and spot urine samples. The study was approved by the Institutional Review Board at each participating institution, and all participants provided written informed consent. The final sample used for analyses was 12,212 individuals.
Genotypes and Imputation
HCHS/SOL participants were genotyped using a Custom Illumina array containing a total of 2,536,661 SNPs of which 2,427,090 are from a standard Illumina Omni2.5M array and the remaining 109,571 are custom SNPs. Quality control was completed using methods previously described.26 Genotype imputation was performed using the 1000 Genomes Project phase 1 cosmopolitan reference panel. We estimated kinship coefficients and principal components using methods previously described.14
Phenotypes and Covariates
Albumin (milligrams per deciliter) and creatinine (grams per deciliter) were measured in urine specimens using an immunoturbidometric method and a creatinase enzymatic method, respectively. Log-transformed UACR, defined as the ratio of albumin-to-creatinine, was the main phenotype. For variants associated with UACR within regions, we also examined the association with eGFR, estimated from the Chronic Kidney Disease Epidemiology Collaboration equation on the basis of the measured serum creatinine, sex, age,27 and diabetes, with cases defined as having fasting time >8 hours and fasting glucose levels ≥126 mg/dl; or fasting≤8 hours and fasting glucose ≥200 mg/dl; or postoral glucose tolerance test glucose ≥200 mg/dl; or hemoglobin A1C ≥6.5%; or if on current treatment with a hypoglycemic agent.
Statistical Analyses
The HCHS/SOL has approximately 2000 related individuals, and a complex sampling design. To account for survey design and for correlation structure due to relatedness, we fit variance components for kinship, household, and census block group using linear mixed models.14 All analyses were additionally adjusted for age, sex, center, and the five first principal components. Sampling weights were also included as a fixed effect to account for the sampling design.14
Admixture Mapping
We inferred local ancestry at a set of 236,456 SNPs in common between HCHS/SOL and reference-panel datasets (selected populations from the Human Genome Diversity Panel,28 HapMap 3,29 and 1000 Genome phase 130) used to detect and estimate European, West African, and Amerindian ancestry at genomic locations, as previously described.16 We ran an unsupervised analysis using ADMIXTURE software31 with K=3 populations, and retained individuals with >90% estimated ancestry from one of the inferred ancestral populations. We phased the HCHS/SOL data combined with the reference panels using Beagle 4.032 and inferred local ancestry using RFMix.33 Using these inferred local ancestries,16 we implemented a genome-wide admixture mapping approach for UACR utilizing a linear mixed model as described above with a joint test for the local ancestry count from all three ancestries (Amerindian, West African, European). As a secondary analysis, we performed admixture mapping, testing the local ancestry of each ancestral group separately. On the basis of previously reported simulation analyses in HCHS/SOL,34 a P value of 5.7 × 10−5 controls the family-wise error rate of admixture mapping at level 0.05.
Association Mapping within a Region
We used linear mixed models as described above within each identified region from the admixture mapping to test the association of each variant with UACR. P values were adjusted for the number of tests performed.
Functional Annotation of Variants
We used Haploreg to examine the overlap of two noncoding SNPs of interest on chromosome 2 with chromatin state and protein binding annotations. These annotations were obtained from the Roadmap Epigenomics and ENCODE projects for specific tissues and cells.17
Replication
Replication of rs116907128 was assessed in a population-based sample of Southwest United States American Indians who are full Pima Indian heritage and had participated in a longitudinal study conducted by the National Institute of Diabetes and Digestive and Kidney Diseases.35,36 This included n=1568 individuals for UACR; n=1552 for eGFR; n=3699 for diabetes case-control; and n=1649 for diabetic nephropathy case-control. Rs116907128 was previously genotyped using a Pima Indian custom Axiom array (Affymetrix). All genotypes analyzed on this array met quality control metrics (call rate ≥90%; discrepant rate ≤2 pairs among 100 blind duplicate pairs; and lack of deviation from Hardy–Weinberg equilibrium with a P>10−4). The SNP associations with UACR and diabetic nephropathy were performed using linear and logistic models, respectively, adjusted for age and sex. We tested the heterogeneity across Hispanics/Latinos and Pima Indian estimates using the Cochran Q test.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the Hispanic Community Health Study/ Study of Latinos (HCHS/SOL) for their important contributions (investigators website, http://www.cscc.unc.edu/hchs/).
The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following institutes/centers/offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, and National Institutes of Health Institution–Office of Dietary Supplements. The Genetic Analysis Center at Washington University was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by the NHLBI grant HL123677-01 and by NIDDK 1R56DK104806-01A1 (N.F.). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center grant DK063491.
Preliminary results from this study were presented at the American Society of Human Genetics meeting in Vancouver, Canada, in October 19, 2016.
This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016091010/-/DCSupplemental.
References
- 1.Gerstein HC, Mann JF, Yi Q, Zinman B, Dinneen SF, Hoogwerf B, Halle JP, Young J, Rashkow A, Joyce C, Nawaz S, Yusuf S, Investigators HS: Albuminuria and risk of cardiovascular events, death, and heart failure in diabetic and nondiabetic individuals. JAMA 286: 421–426, 2001 [DOI] [PubMed] [Google Scholar]
- 2.Astor BC, Matsushita K, Gansevoort RT, van der Velde M, Woodward M, Levey AS, Jong PE, Coresh J, Chronic Kidney Disease Prognosis Consortium, Astor BC, Matsushita K, Gansevoort RT, van der Velde M, Woodward M, Levey AS, de Jong PE, Coresh J, El-Nahas M, Eckardt KU, Kasiske BL, Wright J, Appel L, Greene T, Levin A, Djurdjev O, Wheeler DC, Landray MJ, Townend JN, Emberson J, Clark LE, Macleod A, Marks A, Ali T, Fluck N, Prescott G, Smith DH, Weinstein JR, Johnson ES, Thorp ML, Wetzels JF, Blankestijn PJ, van Zuilen AD, Menon V, Sarnak M, Beck G, Kronenberg F, Kollerits B, Froissart M, Stengel B, Metzger M, Remuzzi G, Ruggenenti P, Perna A, Heerspink HJ, Brenner B, de Zeeuw D, Rossing P, Parving HH, Auguste P, Veldhuis K, Wang Y, Camarata L, Thomas B, Manley T: Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int 79: 1331–1340, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattix HJ, Hsu CY, Shaykevich S, Curhan G: Use of the albumin/creatinine ratio to detect microalbuminuria: Implications of sex and race. J Am Soc Nephrol 13: 1034–1039, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Robbins DC, Knowler WC, Lee ET, Yeh J, Go OT, Welty T, Fabsitz R, Howard BV: Regional differences in albuminuria among American Indians: An epidemic of renal disease. Kidney Int 49: 557–563, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Ricardo AC, Flessner MF, Eckfeldt JH, Eggers PW, Franceschini N, Go AS, Gotman NM, Kramer HJ, Kusek JW, Loehr LR, Melamed ML, Peralta CA, Raij L, Rosas SE, Talavera GA, Lash JP: Prevalence and correlates of CKD in Hispanics/Latinos in the United States. Clin J Am Soc Nephrol 10: 1757–1766, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.USRD System : 2014 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States, Bethesda, MD, National Institute of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2014 [Google Scholar]
- 7.Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, Chen WM, Wong Q, Williams K, Kerr KF, Taylor KD, Tsai MY, Goodarzi MO, Sale MM, Diez-Roux AV, Rich SS, Rotter JI, Mychaleckyj JC: Population structure of Hispanics in the United States: The multi-ethnic study of atherosclerosis. PLoS Genet 8: e1002640, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peralta CA, Li Y, Wassel C, Choudhry S, Palmas W, Seldin MF, Risch N, Siscovick D, Arnett D, Psaty B, Shlipak MG: Differences in albuminuria between Hispanics and whites: An evaluation by genetic ancestry and country of origin: The multi-ethnic study of atherosclerosis. Circ Cardiovasc Genet 3: 240–247, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez Burchard E, Borrell LN, Choudhry S, Naqvi M, Tsai HJ, Rodriguez-Santana JR, Chapela R, Rogers SD, Mei R, Rodriguez-Cintron W, Arena JF, Kittles R, Perez-Stable EJ, Ziv E, Risch N: Latino populations: A unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health 95: 2161–2168, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boger CA, Chen MH, Tin A, Olden M, Kottgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tonjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarod K, Skorpen F, Syvanen AC, Illig T, Baumert J, Koenig W, Kramer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Volzke H, Stumvoll M, Magi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlov J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH: CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naik RP, Derebail VK, Grams ME, Franceschini N, Auer PL, Peloso GM, Young BA, Lettre G, Peralta CA, Katz R, Hyacinth HI, Quarells RC, Grove ML, Bick AG, Fontanillas P, Rich SS, Smith JD, Boerwinkle E, Rosamond WD, Ito K, Lanzkron S, Coresh J, Correa A, Sarto GE, Key NS, Jacobs DR, Kathiresan S, Bibbins-Domingo K, Kshirsagar AV, Wilson JG, Reiner AP: Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. JAMA 312: 2115–2125, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernandez-Rhodes L, Justice AE, Graff M, Young KL, Seyerle AA, Avery CL, Taylor KD, Rotter JI, Talavera GA, Daviglus ML, Wassertheil-Smoller S, Schneiderman N, Heiss G, Kaplan RC, Franceschini N, Reiner AP, Shaffer JR, Barr RG, Kerr KF, Browning SR, Browning BL, Weir BS, Aviles-Santa ML, Papanicolaou GJ, Lumley T, Szpiro AA, North KE, Rice K, Thornton TA, Laurie CC: Genetic diversity and association studies in US Hispanic/Latino populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 98: 165–184, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, Berns JS, Briggs W, Cho ME, Dart RA, Kimmel PL, Korbet SM, Michel DM, Mokrzycki MH, Schelling JR, Simon E, Trachtman H, Vlahov D, Winkler CA: MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet 40: 1175–1184, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning SR, Grinde K, Plantinga A, Gogarten SM, Stilp AM, Kaplan RC, Aviles-Santa ML, Browning BL, Laurie CC: Local ancestry inference in a large US-Based Hispanic/Latino Study: Hispanic Community Health Study/Study of Latinos (HCHS/SOL). G3 (Bethesda) 6: 1525–1534, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward LD, Kellis M: HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–D934, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estrada K, Aukrust I, Bjorkhaug L, Burtt NP, Mercader JM, Garcia-Ortiz H, Huerta-Chagoya A, Moreno-Macias H, Walford G, Flannick J, Williams AL, Gomez-Vazquez MJ, Fernandez-Lopez JC, Martinez-Hernandez A, Jimenez-Morales S, Centeno-Cruz F, Mendoza-Caamal E, Revilla-Monsalve C, Islas-Andrade S, Cordova EJ, Soberon X, Gonzalez-Villalpando ME, Henderson E, Wilkens LR, Le Marchand L, Arellano-Campos O, Ordonez-Sanchez ML, Rodriguez-Torres M, Rodriguez-Guillen R, Riba L, Najmi LA, Jacobs SB, Fennell T, Gabriel S, Fontanillas P, Hanis CL, Lehman DM, Jenkinson CP, Abboud HE, Bell GI, Cortes ML, Boehnke M, Gonzalez-Villalpando C, Orozco L, Haiman CA, Tusie-Luna T, Aguilar-Salinas CA, Altshuler D, Njolstad PR, Florez JC, MacArthur DG: Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 311: 2305–2314, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, Aguilar-Salinas CA, Gonzalez-Villalpando C, Florez JC, Orozco L, Haiman CA, Tusie-Luna T, Altshuler D: Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506: 97–101, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, Li M, Li Y, Mijatovic V, Ko YA, Taliun D, Luciani A, Chen MH, Yang Q, Foster MC, Olden M, Hiraki LT, Tayo BO, Fuchsberger C, Dieffenbach AK, Shuldiner AR, Smith AV, Zappa AM, Lupo A, Kollerits B, Ponte B, Stengel B, Kramer BK, Paulweber B, Mitchell BD, Hayward C, Helmer C, Meisinger C, Gieger C, Shaffer CM, Muller C, Langenberg C, Ackermann D, Siscovick D, Boerwinkle E, Kronenberg F, Ehret GB, Homuth G, Waeber G, Navis G, Gambaro G, Malerba G, Eiriksdottir G, Li G, Wichmann HE, Grallert H, Wallaschofski H, Volzke H, Brenner H, Kramer H, Mateo Leach I, Rudan I, Hillege HL, Beckmann JS, Lambert JC, Luan J, Zhao JH, Chalmers J, Coresh J, Denny JC, Butterbach K, Launer LJ, Ferrucci L, Kedenko L, Haun M, Metzger M, Woodward M, Hoffman MJ, Nauck M, Waldenberger M, Pruijm M, Bochud M, Rheinberger M, Verweij N, Wareham NJ, Endlich N, Soranzo N, Polasek O, van der Harst P, Pramstaller PP, Vollenweider P, Wild PS, Gansevoort RT, Rettig R, Biffar R, Carroll RJ, Katz R, Loos RJ, Hwang SJ, Coassin S, Bergmann S, Rosas SE, Stracke S, Harris TB, Corre T, Zeller T, Illig T, Aspelund T, Tanaka T, Lendeckel U, Volker U, Gudnason V, Chouraki V, Koenig W, Kutalik Z, O’Connell JR, Parsa A, Heid IM, Paterson AD, de Boer IH, Devuyst O, Lazar J, Endlich K, Susztak K, Tremblay J, Hamet P, Jacob HJ, Boger CA, Fox CS, Pattaro C, Kottgen A: Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes 65: 803–817, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer HJ, Stilp AM, Laurie CC, Reiner AP, Lash J, Daviglus ML, Rosas SE, Ricardo AC, Tayo BO, Flessner MF, Kerr KF, Peralta C, Durazo-Arvizu R, Conomos M, Thornton T, Rotter J, Taylor KD, Cai J, Eckfeldt J, Chen H, Papanicolau G, Franceschini N: African Ancestry-Specific Alleles and Kidney Disease Risk in Hispanics/Latinos [published online ahead of print Sep 20, 2016]. J Am Soc Nephrol doi:10.1681/ASN.2016030357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho J, Pandey P, Schatton T, Sims-Lucas S, Khalid M, Frank MH, Hartwig S, Kreidberg JA: The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J Am Soc Nephrol 22: 1053–1063, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A: Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Chuang PY, Dai Y, Liu R, He H, Kretzler M, Jim B, Cohen CD, He JC: Alteration of forkhead box O (foxo4) acetylation mediates apoptosis of podocytes in diabetes mellitus. PLoS One 6: e23566, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavange LM, Kalsbeek WD, Sorlie PD, Aviles-Santa LM, Kaplan RC, Barnhart J, Liu K, Giachello A, Lee DJ, Ryan J, Criqui MH, Elder JP: Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 20: 642–649, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS, Investigators G: Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34: 591–602, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, Kusek JW, Manzi J, Van Lente F, Zhang YL, Coresh J, Levey AS, Investigators C-E: Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 367: 20–29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalli-Sforza LL: The human genome diversity project: Past, present and future. Nat Rev Genet 6: 333–340, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE: Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR: A global reference for human genetic variation. Nature 526: 68–74, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander DH, Novembre J, Lange K: Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19: 1655–1664, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning SR, Browning BL: Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81: 1084–1097, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maples BK, Gravel S, Kenny EE, Bustamante CD: RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 93: 278–288, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schick UM, Jain D, Hodonsky CJ, Morrison JV, Davis JP, Brown L, Sofer T, Conomos MP, Schurmann C, McHugh CP, Nelson SC, Vadlamudi S, Stilp A, Plantinga A, Baier L, Bien SA, Gogarten SM, Laurie CA, Taylor KD, Liu Y, Auer PL, Franceschini N, Szpiro A, Rice K, Kerr KF, Rotter JI, Hanson RL, Papanicolaou G, Rich SS, Loos RJ, Browning BL, Browning SR, Weir BS, Laurie CC, Mohlke KL, North KE, Thornton TA, Reiner AP: Genome-wide association study of platelet count identifies ancestry- specific loci in Hispanic/Latino Americans. Am J Hum Genet 98: 229–242, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC: An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63: 1130–1138, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson RL, Muller YL, Kobes S, Guo T, Bian L, Ossowski V, Wiedrich K, Sutherland J, Wiedrich C, Mahkee D, Huang K, Abdussamad M, Traurig M, Weil EJ, Nelson RG, Bennett PH, Knowler WC, Bogardus C, Baier LJ: A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes 63: 369–376, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.