Abstract
Parallel detection approaches are of interest to many researchers interested in identifying multiple water and foodborne pathogens simultaneously. Availability and cost-effectiveness are two key factors determining the usefulness of such approaches for laboratories with limited resources. In this study, we developed and validated a high-density microarray for simultaneous screening of 14 bacterial pathogens using an approach that employs gold labeling with silver enhancement (GLS) protocol. In total, 8,887 probes (50-mer) were designed using an in-house database of virulence and marker genes (VMGs), and synthesized in quadruplicate on glass slides using an in-situ synthesis technology. Target VMG amplicons were obtained using multiplex polymerase chain reaction (PCR), labeled with biotin, and hybridized to the microarray. The signals generated after gold deposition and silver enhancement, were quantified using a flatbed scanner having 2-μm resolution. Data analysis indicated that reliable presence/absence calls could be made, if: i) over four probes were used per gene, ii) the signal-to-noise ratio (SNR) cutoff was greater than or equal to two, and iii) the positive fraction (PF), i.e., number of probes with SNR > 2 for a given VMG was greater than 0.75. Hybridization of the array with blind samples resulted in 100% correct calls, and no false positive. Because amplicons were obtained by multiplex PCR, sensitivity of this method is similar to PCR. This assay is an inexpensive and reliable technique for high throughput screening of multiple pathogens.
Keywords: Waterborne pathogens, Gold labeling and silver enhancement, Virulence and marker genes, Low cost microarray
Introduction
Hybridization-based screening or confirmation of multiple bacterial or viral pathogens often involves fluorescent labeling of the polymerase chain reaction (PCR)-based amplicons or extracted sample DNA, hybridization on a microarray with probes designed for the target genes, and signal quantification using a laser scanner (Cao et al., 2015; Miller et al., 2008; Stedtfeld et al., 2007). High cost of scanning equipment is a bottleneck for use in limited resource settings. Alternative strategies for labeling and signal quantification, e.g., gold nano-particles labeling with silver enhancement (GLS) followed by quantification with flatbed scanners can address this limitation (Grinev et al., 2017). Several studies have demonstrated its potential focusing on bacterial pathogens (Qi et al., 2010), swine viruses (Wang et al., 2013), and HIV-1 and Hep C (Tang et al., 2011), albeit using less than a dozen probes on the whole array (Table 1). Recently a microarray-based GLS approach demonstrated that it can also be used for detection of mutation in the epidermal growth factor receptor (Xue et al., 2014). Examples of antibody-based lateral flow assays using GLS are numerous (Ngom et al., 2010) but differ significantly in their performance compared to DNA-DNA hybridization.
Table 1.
Microarray-based studies using gold and silver labeling protocols with visual or flatbed-scanner-based signal quantification.
| Target Organisms | No. of probes | Scanning device | Max Grey Value | Reference |
|---|---|---|---|---|
| Six bacterial pathogensa | 6 | Flatbed scanner | 66±6 | Qi et al., 2010 |
| Seven swine viruses | 9 | Flatbed scanner | ~100 | Wang et al., 2013 |
| HIV-1 and Hep-C | 9 | Visual | NA | Tang et al., 2011 |
| EGFR gene mutationsb | 16 | Visual | ~4000 | Xue et al., 2014 |
Yersinia, Shigella, Salmonella, Brucella, E. coli O157:H7, and Cholera O139
epidermal growth factor receptor
From the studies conducted using GLS with DNA-DNA hybridization (Alexandre et al., 2001; Liang et al., 2004; Liu et al., 2006; Storhoff et al., 2004; Taton et al., 2000), it is evident that the range of signal intensity produced by the GLS approach is smaller than the range of signals obtained by fluorescent dyes and laser scanners (Wilson et al., 2002). For GLS using flatbed scanners, it is between 0 and 255 on the gray scale (with 0 being black and 255 being white) while for fluorescent dyes with laser scanners is between 0 and 65,000 arbitrary units. As signal-to-noise ratio (SNR), this may translate into 0 to 100 for GLS and 0 to 1,000 for fluorescent dyes. Previous studies have used gray scale (Liang et al., 2004; Taton et al., 2000) or SNR (Wan et al., 2005) when using GLS. Because microarrays designed to screen for multiple pathogens generally use multiplexed PCR amplification step prior to hybridization to enhance detection limit (Call, 2005; Call et al., 2003; Loy and Bodrossy, 2006), and do not provide quantitative information about the detected target, the numerical value of the SNR is not critical. However, to make reliable presence/absence calls, the SNR for target probes must be completely separated from the SNR for non-target probes. Thus, when a large number of bacterial targets must be screened using the GLS approach, it is critical to experimentally establish the cut-off for positive fraction (PF) defined as the number of probes having a SNR greater than a given value.
The objective of this study was to develop and validate a GLS-based approach for simultaneous detection of 14 bacterial pathogens using an in situ synthesized microarray and a flatbed scanner. Probes for an additional pathogens were present on the microarray but they were not validated with target VMGs and served only as a control set to evaluate non-specific hybridization. Reliable presence/absence calls were made based on a set of three filters related to the minimum number of probes per probe set, threshold SNR for positive signals, and a threshold for PF. Performance of the GLS approach was validated using blind samples containing mixtures of virulence and marker genes (VMGs) not known to the person hybridizing the array and performing data analysis. Because of the low-cost technologies used for scanning, the method represents a cost-effective approach (Table 2) to screen for multiple pathogens and provides a set of probes for many other pathogens that could be used after further validation. The bacterial pathogens chosen are common contaminants with a potential to be present in water and food globally (Logue et al., 2017).
Table 2.
Cost and time comparison between GLS method and fluorescent labeling for microarray based detection.
| Fluorescent labeling | GLS method | |
|---|---|---|
| 1. Scanning equipment cost | $30,000–$60,000 | $100–$400 (Flatbed Scanner) |
| 2. Labeling cost per slide | $50–$100 | $10–$15 |
| 3. Time-to-result | ||
| PCR amplification | 1–2 hr | 1–2 hr |
| Fluorescent labeling | 2–3 hr | - |
| Biotin labeling | - | 1–2 hr |
| Hybridization | 2–10 hr | 2–10 hr |
| Silver enhancement | - | 1 hr |
| Scanning | 10–20 min | 10–20 min |
| Total | 5–15 hr | 5–15 hr |
Materials and Methods
Probe design and synthesis of VMG microarray
An in-house database containing 36 genera, 107 pathogens, 539 genes, and 3,183 VMG sequences obtained from GenBank was used to design 8,887 fifty-mer probes (Table S1–S3) using CommOligo (Li et al., 2005). The probe design criteria listed in Table S1 was similar to that reported in a previous study (He et al., 2005). These probes were synthesized in situ on a glass slide in quadruplicate (replicated four times) using a flexible in situ microarray technology described previously (Gao et al., 2004, 2001; LeProust et al., 2000). A 32-mer sequence (5′-CCTATAGTGAGTCGTATTAAGCAGCGCAGC-3′) was also synthesized in situ and replicated 1,214 times over the entire slide to serve as a control. A total of 551 probes targeting 40 VMGs associated with 14 pathogens served as the target probes either during initial development or with blind mixtures. Non-targeted probes were not validated further as part of this study and therefore only served for evaluated the specificity of the microarray.
Design of sample with known VMG mixture for method development
During method development, 22 VMGs targeted by 228 probes associated with 10 pathogens were used (Table 3; Known VMG mixture). Hybridization experiments to develop the protocol and data analysis scheme were carried out in triplicate. The 22 VMGs selected for method development were amplified from genomic DNA of the targeted pathogens by PCR. Target VMGs were amplified in five multiplexed reactions and the amplicons were labeled with biotin. It is worth noting that the overall VMG mixture also contained the amplicons for six additional VMGs that were filtered at the time of data analysis because only 1–2 probes per VMG were present on the array. A biotin labeled sequence complementary to the 32-mer in situ synthesized sequence was synthesized by Integrated DNA Technologies (Coralville, IA) and spiked in the hybridization mixture. This biotin labeled sequence served as a positive control for hybridization to the 32-mer in situ sequence mentioned above. The complete target mixture was hybridized to the microarray. Hybridized biotinylated targets were incubated with gold nano-particles using streptavidin-gold conjugate solution followed by silver enhancement to further amplify the generated signals. Thereafter, the VMG microarray was scanned using a conventional flatbed scanner and the data was analyzed using Genepix 5.0 software (Axon Instruments, Union City, CA).
Table 3.
List of targeted VMG and pathogens used in the known and blind validation mixture. The average signal to noise ratio (SNR), positive fraction of all probes for a given VMG (PF) and presence/absence calls are shown. SNR for method development was calculated as the average of median SNR values from each of three replicate set of experiments. “+” denotes spiked or present and “−” denotes not spiked or absent, a Unique probe, b Group-specific probe targeting tdh in (V. mimicus) and (V. parahaemolyticus)..
| S.No. | Organism | Gen Accession # | No. of Probes | Method Development | Method Validation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Known VMG mixture | Blind VMG mixture 1 | Blind VMG mixture 2 | ||||||||||
|
| ||||||||||||
| Spiked / Detected | SNR | PF | Spiked / Detected | SNR | PF | Spiked / Detected | SNR | PF | ||||
| 1. | A. hydrophila | ast AF419157a |
16 | +/+ | 3.3±0.42 | 0.95±0.08 | +/+ | 4.1 | 0.97 | −/− | 2.2 | 0.66 |
| 2. | C. jejuni | cadF AL139078 |
10 | +/+ | 3.5±0.68 | 0.90±0.01 | +/+ | 4.2 | 0.90 | +/+ | 3.0 | 0.93 |
| cdtA AF038283 |
11 | +/+ | 3.3±0.60 | 0.94±0.05 | +/+ | 4.0 | 0.98 | +/+ | 2.8 | 0.91 | ||
| cdtC AF038283 |
13 | +/+ | 3.3±0.66 | 0.90±0.02 | +/+ | 3.8 | 0.96 | −/− | 1.7 | 0.40 | ||
| hipO AL139076 |
9 | +/+ | 3.3±0.54 | 0.90±0.06 | +/+ | 3.8 | 0.83 | +/+ | 3.0 | 0.94 | ||
| 3. | C. perfringens | cpe AF416450 |
5 | +/+ | 3.2±0.53 | 0.90±0.03 | −/− | −0.3 | 0.00 | −/− | 1.3 | 0.30 |
| plc AY277724 |
5 | +/+ | 3.5±0.66 | 0.80±0.03 | −/− | −0.4 | 0.05 | −/− | 0.5 | 0.05 | ||
| 4 | C. parvum | FGLN AAEE010000 11a |
20 | −/− | 0.3±0.54 | 0.20±0.12 | −/− | −0.1 | 0.04 | +/+ | 3.0 | 0.94 |
| gp40 AF022929 |
20 | −/− | 0.8±0.97 | 0.24±0.20 | −/− | 0.5 | 0.23 | +/+ | 2.8 | 0.81 | ||
| 5 | H. pylori | flaA AY319298 |
12 | +/+ | 3.5±0.58 | 0.96±0.02 | −/− | −0.2 | 0.00 | +/+ | 3.0 | 0.93 |
| ureB AY295085 |
19 | +/+ | 3.5±0.51 | 0.95±0.04 | −/− | −0.1 | 0.03 | −/− | 1.1 | 0.14 | ||
| glmM AY353251 |
4 | −/− | 0.5±0.47 | 0.23±0.11 | −/− | −0.2 | 0.03 | +/+ | 2.9 | 0.94 | ||
| 6 | L. pneumophila | mip AE017354 |
15 | +/+ | 3.6±0.57 | 0.96±0.03 | −/− | −0.2 | 0.03 | −/− | 1.2 | 0.15 |
| dotA AE017354 |
13 | −/− | 0.2±0.54 | 0.24±0.10 | −/− | −0.1 | 0.09 | +/+ | 2.8 | 0.83 | ||
| feoB CR628337 |
20 | −/− | 0.3±0.47 | 0.07±0.05 | −/− | −0.2 | 0.01 | +/+ | 2.7 | 0.89 | ||
| 7 | L monocytogenes | lisA M24199 | 5 | +/+ | 3.5±0.37 | 0.93±0.03 | +/+ | 4.2 | 0.95 | −/− | 0.9 | 0.10 |
| plcA AY367410 |
14 | +/+ | 3.5±0.66 | 0.97±0.03 | +/+ | 4.1 | 0.95 | +/+ | 2.9 | 0.93 | ||
| ami_AF0354 24 |
19 | −/− | 0.3±0.30 | 0.28±0.06 | −/− | 0.1 | 0.04 | +/+ | 3.0 | 0.93 | ||
| prfA AE017322 |
13 | −/− | 0.3±0.64 | 0.26±0.09 | −/− | −0.2 | 0.00 | +/+ | 2.9 | 0.98 | ||
| 8 | M. genitalium | P30 L43097 | 20 | −/− | 2.4±0.46 | 0.66±0.01 | −/− | 2.2 | 0.56 | +/+ | 2.7 | 0.99 |
| 9 | P. mirabilis | uca U28420 | 20 | −/− | 0.1±0.57 | 0.13±0.06 | −/− | 0.1 | 0.03 | +/+ | 2.9 | 0.93 |
| UmoA U66821a |
19 | −/− | 0.0±0.28 | 0.00±0.08 | −/− | −0.2 | 0.05 | +/+ | 2.9 | 0.82 | ||
| 10 | S. aureus | nuc BA000017 |
10 | +/+ | 3.7±0.46 | 0.93±0.04 | +/+ | 3.8 | 0.78 | +/+ | 3.1 | 0.94 |
| tsst1 AB084255 |
7 | +/+ | 3.4±0.62 | 0.96±0.04 | +/+ | 4.1 | 0.96 | −/− | 0.9 | 0.07 | ||
| arlR AF165314 |
20 | −/− | −0.5±0.16 | 0.01±0.02 | −/− | −0.3 | 0.03 | +/+ | 3.0 | 0.96 | ||
| lukE BA000017 |
20 | −/− | −0.2±0.29 | 0.05±0.03 | −/− | 0.0 | 0.00 | +/+ | 3.1 | 0.91 | ||
| 11 | S. agalactiae | covS CP000114 |
20 | −/− | −0.4±0.26 | 0.04±0.04 | +/+ | 4.2 | 0.96 | +/+ | 3.0 | 0.91 |
| Lmb AE014248 |
20 | −/− | 0.8±0.71 | 0.26±0.18 | +/+ | 4.0 | 0.96 | −/− | 1.5 | 0.35 | ||
| Cfb AE014283 |
20 | −/− | −0.2±0.37 | 0.03±0.06 | +/+ | 4.1 | 0.94 | +/+ | 2.9 | 0.91 | ||
| cylE AE014221 |
20 | −/− | 0.1±0.52 | 0.12±0.09 | +/+ | 4.1 | 0.93 | +/+ | 2.9 | 0.84 | ||
| 12 | V. cholerae | hlyA AF194418 |
4 | +/+ | 3.5±0.66 | 0.98±0.04 | +/+ | 4.0 | 1.00 | −/− | 1.8 | 0.25 |
| ompU AE004149 |
10 | +/+ | 3.4±0.51 | 1.00±0.01 | +/+ | 4.3 | 1.00 | −/− | 1.9 | 0.40 | ||
| toxR AE004179 |
15 | +/+ | 3.3±0.56 | 0.94±0.04 | +/+ | 4.1 | 0.90 | −/− | 1.4 | 0.30 | ||
| zot AF123249o1 39 |
15 | +/+ | 3.3±0.51 | 0.96±0.01 | +/+ | 4.0 | 0.93 | −/− | 1.7 | 0.37 | ||
| ace AF175708 |
15 | −/− | 0.2±0.54 | 0.12±0.91 | +/+ | 4.2 | 0.90 | −/− | 1.5 | 0.28 | ||
| tcpA AE004168 |
20 | −/− | 0.0±0.26 | 0.05±0.06 | +/+ | 4.1 | 0.93 | −/− | 1.4 | 0.16 | ||
| 13 | V. parahaemolyticus | tlh AB012596 | 13 | +/+ | 3.4±0.58 | 0.96±0.03 | +/+ | 4.4 | 0.94 | +/+ | 3.0 | 0.81 |
| toxR BA000031 |
11 | +/+ | 3.2±0.68 | 0.95±0.02 | +/+ | 4.1 | 1.00 | +/+ | 3.0 | 0.95 | ||
| tdh M64120b | 4 | +/+ | 3.5±0.56 | 0.90±0.10 | +/+ | 4.0 | 1.00 | −/− | 1.5 | 0.38 | ||
| 14 | Y. enterocolitica | ystsA U09235 | 5 | +/+ | 3.4±0.51 | 0.98±0.03 | −/− | 0.1 | 0.00 | −/− | 1.6 | 0.35 |
Strategy for blind samples with unknown VMGs for method validation
Two separate blind samples were prepared using amplicons generated by monoplex PCR of multiple VMGs including four additional pathogens that were not hybridized during development (Table 3: Blind mixtures 1 and 2). All PCR amplicons were cleaned using a QIAGEN PCR clean-up kit (QIAGEN, Valencia, CA), and quantified using the NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Approximately 2.5 x 10−14 mol of each amplicon was added to the blind sample mixture, and 250 ng of the final mixture was used for biotin labeling using Bioprime DNA labeling kit (Invitrogen, Carlsbad, CA). The blind samples targeted some VMGs that were included in the known VMG (Table 3, Blind VMG mixtures 1 and 2 marked as + for Spiked). The identity of the prepared samples in terms of pathogens targeted or VMGs mixture used was not revealed to the person carrying out the hybridization and data analysis for the blind samples until the after presence/absence calls were made.
DNA extraction from bacterial cultures
Target pathogens from which DNA was extracted for method development and validation included: Aeromonas hydrophila (ATCC 7966), Clostridium perfringens (ATTC 12916), Listeria monocytogenes (ATCC 15313), Vibrio parahaemolyticus (ATCC 43996), and Yersinia enterocolitica (ATCC 55075) -all obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were grown overnight according to the instructions provided by the ATCC for the respective organism. DNA was extracted using DNeasy Tissue Kit (QIAGEN, Valencia, CA) per the manufacturer’s instructions. For Staphylococcus aureus (ATCC 700699), Vibrio cholerae (ATCC 39315), Legionella pneumophila (ATCC 33152), Campylobacter jejuni (ATCC 700819), Helicobacter pylori (ATCC 700392), Cryptosporidium parvum (ATCC PRA-67D), Mycoplasma genetalium (ATCC 33530), Streptococcus agalactiae (ATCC BAA-611D), and Proteus mirabilis (ATCC 49565), only purified genomic DNA was obtained from the ATCC.
Primer selection and PCR amplification
Gene-specific primer reported earlier (Miller et al., 2008) were designed to amplify the regions of VMGs with a high probe density on the microarray. The designed primers were synthesized by Integrated DNA Technologies (Coralville, IA). A Blast search was used to screen the GenBank database and primer specificity was confirmed by finding mismatches to corresponding non-target sequences near the 3′ end of the primer. In multiplex PCR, the primers were separated so that each set contained 10 primer pairs segregated per their annealing temperature (53 °C to 58 °C). The primers were combined so that each pathogen was targeted by at least two multiplex reactions. Additional details regarding the primer design can be obtained elsewhere (Miller et al., 2008).
A PCR reaction mixture (25 μl) for amplification of each target was prepared by adding 2.5 μl of 10× PCR buffer, 2 μl of 2 mM MgCl2, 0.3 μl of AmpliTaq Gold (specified reagents purchased from Roche Molecular Systems, Pleasanton, CA), 2.5 μl of deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA), 10 μM of each primer (forward and reverse) for each VMG, 0.5 μl of BSA (New England Biolabs, Beverly, MA), and 1 μl of template DNA. Enzyme activation was carried out at 94 °C for 10 min, followed by PCR amplification for 40 reaction cycles. Each cycle included the following steps: denaturation at 94 °C for 60 s, annealing at 53 °C or 58 °C for 60 s, extension at 72 °C for 60 s and a final extension step at 72 °C for 7 min. The fragment size of PCR amplicons was determined by using an agarose gel stained with SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA) and visualized under a UV light source. The PCR products were purified using QIAquick PCR purification kit (QIAGEN, Valencia, CA) and quantified using NanoDrop ND-1000 spectrophotometer prior to hybridization.
Biotin labeling of VMG amplicons
The purified amplicons were labeled with biotin using Bioprime labeling kit (QIAGEN, Valencia, CA) as per the protocol described by the manufacturer with few modifications. Approximately 2.5 x 10−14 mol of each amplicon was added to prepare the known VMG mixture. To approximately 100 ng of this mixture in a microcentrifuge tube, 20 μl of 2.5× random primer solution (kept on ice) and water was added to obtain a total volume of 44 μl. This mixture was denatured by heating for 5 min at 95 °C and immediately cooled on ice for 5 min. Thereafter, 5 μl of 10× dNTP and 1 μl klenow fragment were added into the reaction vial, followed by an incubation step at 37 °C for 90 min. This step allowed the incorporation of biotin-labeled dUTP into the target DNA sequence. After incubation, 5 μl of stop buffer was added and the biotin-labeled amplicons were purified and quantified. The samples were dried and stored at −20 °C until needed for hybridization.
GLS hybridization protocol
Labeled PCR products were hybridized on the VMG microarray using the following protocol. The slide was washed twice with 0.2% sodium dodecyl sulphate followed by two additional washes with distilled water and dried using a dust remover can (Fisher Scientific, Pittsburgh, PA). The area on the glass slide containing in situ synthesized probes was covered with 6× SSPE (pH 6.7) for 1 min to help reduce background signal. Hybridization solution was prepared by adding 150 μl of 20× SSPE buffer, 50 μl of 10 mg/ml acetylated bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 0.5 μg of biotin labeled positive control, biotinylated target DNA (3,000 ng to 5,000 ng per slide) and distilled water to obtain a total volume of 500 μl. The solution was warmed at 65 °C for 5 min and target DNA was warmed at 95 °C for 5 min prior to hybridization.
The VMG microarray was hydrated by soaking in 450 μl hybridization solution and placed inside a hybridization cassette (ArrayIt, Sunnyvale, CA). The hybridization cassette contained two small wells along the edges which were filled with 6× SSPE to ensure that the moisture content inside the chamber was 100%. Pre-warmed targets and biotin labeled positive control were added to the remaining hybridization solution (50 μl) and loaded on the slide under a cover slip without leaving any air bubbles. The setup was incubated in a water bath at 40 °C for 6 hr. After hybridization, the slide was washed with 6× SSPE at 40 °C for 5 min and quickly dipped in 4 °C water for 5 s and dried.
The hybridized slide was treated with a gold-labeling mixture containing 15 μl streptavidin-gold conjugate solution (Sigma-Aldrich, St. Louis, MO), 15 μl 20× SSPE buffer (Invitrogen, Carlsbad, CA), 5 μl acetylated bovine serum albumin, 0.5 μl of 5% tween 20 and 14.5 μl water. The solution was applied on the slide under a cover slip and incubated for 1 hr at room temperature. The slide was dipped two times each in 6× SSPE kept in 3 separate tubes. This was followed by fixing the slide with 2.5% gluteraldehyde solution (Sigma-Aldrich, St. Louis, MO) for 15 min under a fume hood. The slide was thoroughly washed with distilled water to avoid high background caused by buffer salts (24) and was kept submerged in it prior to the silver enhancement step.
The silver enhancement step was carried out in a darkroom (Newman and Jasani, 1998) since silver enhancer solutions are sensitive to light which leads to non-specific silver deposition (Alexandre et al., 2001). Equal volumes (12 ml each) of silver enhancer solution A containing silver salt and solution B used as the initiator (Sigma-Aldrich, St. Louis, MO) were mixed in a slide mailer (Fisher Scientific, Pittsburgh, PA) to obtain a total volume of 24 ml. Solution B initiated the reduction of silver salt in solution, thereby resulting in the precipitation of silver. Proportions of initiator and silver salt solutions were optimized to achieve high SNR. The VMG microarray was submerged in the silver solution and allowed to incubate for 25 min at room temperature. A fresh stock of silver solution was used to replace the original stock and the slide was allowed to incubate for an additional 10 min because a longer enhancement time with older solutions are known to cause self-nucleation and high background (Alexandre et al., 2001; Zhang et al., 2004). The array was washed with distilled water and dried. Signal preservation was achieved by quickly dipping the slide in archive solution (QIAGEN, Valencia, CA) as suggested by previous researchers (Lin et al., 2005; Reichert et al., 2000) and stored for scanning.
Data acquisition and analysis
Hybridized slides were scanned using Epson Perfection 4990 Flatbed Scanner (Epson, Long Beach, CA) set with a 16-bit gray scale. The images were scanned at the maximum resolution of 12,800 dpi (2 μm per pixel) and saved as Tiff files. The files were imported into Genepix 5.0 and were analyzed at a default wavelength of 600 nm. An overlay (unique for every microarray) was prepared and the SNR was quantified using Genepix 5.0. SNR was calculated as (signal of feature or spot - mean local background)/(standard deviation of mean local background). The data was analyzed using Microsoft Excel (Microsoft, Redmond, WA) and plotted using SigmaPlot 9.0 (Systat Software, Point Richmond, CA).
Each probe was assigned a unique name (genus_species_gene_accession number_sequence_group/unique probe_ replicate number) and sorted as per its accession number resulting in different probe set for each VMG. Each probe set consisted of a group of probes targeting a gene and organism for one or more strains. A set of three filters were developed and used to make reliable presence/absence calls. The PF of each probe set was calculated as the number of positive probes (SNR ≥ 2) in each probe set divided by the total number of probes within that set (Wilson et al., 2002).
Results and Discussion
Choice of data quality filters for the GLS protocol
For parallel screening of multiple pathogens using a large number of probes, it is critical to eliminate the potential for false negative and positive calls. Use of redundant probes for the same amplicons and computation of a “positive fraction” i.e., fraction of probes for the same amplicons giving positive signal is common for such diagnostic arrays (DeSantis et al., 2005; Miller et al., 2008; Wilson et al., 2002). Because GLS varies significantly in its labeling, hybridization, and data analysis methodology compared to the use of fluorescent labeling and laser scanner, the PF cut-off developed for the latter cannot be used directly for the GLS approach. These differences may be due to several reasons including: i) non-uniform silver deposition and manual hybridization, ii) use of local background signal to normalize the positive signals instead of using average background for the whole array or for selected spots as done in fluorescent-based techniques, and iii) cross-hybridization due to the presence of a large number of probes for closely related organisms.
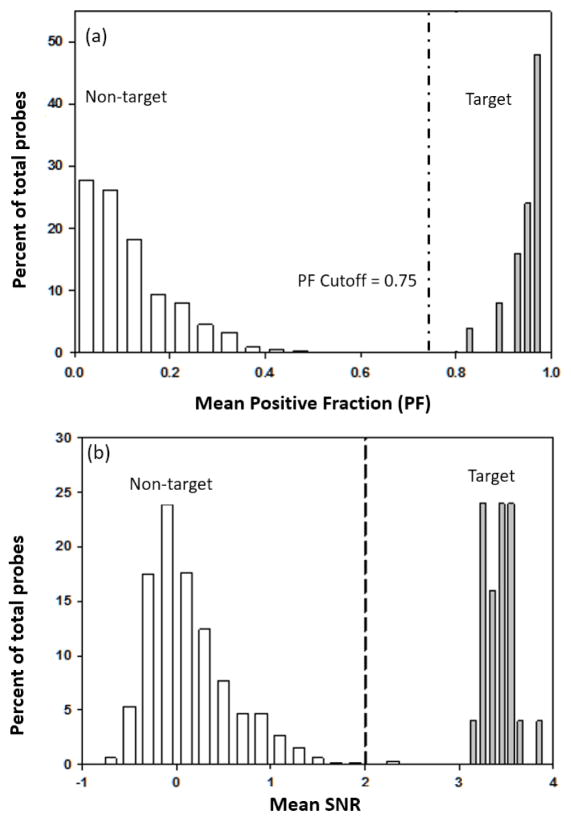
A known VMG mixture containing 43 amplicons targeting a total of 22 VMGs was used to develop the GLS protocol. The targeted amplicons were hybridized to a glass array followed by gold labeling with a silver enhancement step (Fig 1). There were three filters used to make reliable presence/absence calls. The first filter was related to the minimum number of probes and required that only those VMGs that had four or more probe be considered for further data analysis. The second filter was related to SNR and required that each probe in a probe set had an SNR ≥ 2.0 for the probe signal to be considered positive. The third filter was related to PF and required that each probe set must have a PF ≥ 0.75 for the VMG to be considered as present. For the first filter, a threshold value of four was chosen because it clearly separated the true positive signals from the true negative signals (Fig S1 a & b). All targeted VMGs with less than 4 probes per set resulted in positive fractions lower than 0.75 (Fig S1 c). All non-targeted VMGs had at least four or more probes per probe set. The PF of each probe set was used as an index to make final presence/absence calls. A clear separation between the targeted and non-targeted probe sets was achieved at a PF of 0.75 (Fig 2a). A similar PF cut-off value of 1.0 (DeSantis et al., 2005) and 0.8 (Wilson et al., 2002) were used in previous studies, but a lower PF cut-off value of 0.5 was also used (Miller et al., 2008).
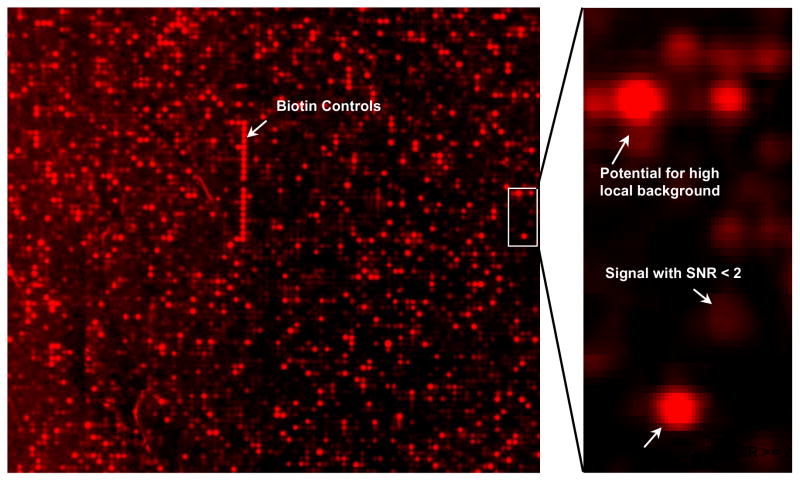
Figure 1.
Close-up of ~600 spots out of 8,887 probes analyzed using Genepix 5.0. Spots are approximately 50 μm in diameter. The continuous vertical stretch of bright red spots is the biotin labeled positive control spots. These controls were distributed throughout the entire slide to assess the hybridization uniformity.
Figure 2.
Frequency distribution of PF and SNR observed for hybridization with known VMG mixture. A total of 228 probes targeting 10 pathogens and 8,659 non-target probes containing four or more probes per set were plotted. The targets and non-targets are indicated by grey and white bars respectively. (a) Distribution based on mean PF for target and non-target probe sets. (b) Distribution based on mean SNR for target and non-target probe sets. Mean SNR was calculated as the average of median SNR values from each of three replicate set of experiments.
Although, the GLS approach exhibited a low range of positive SNR between 3.2 and 3.7, compared to the range of SNR obtained using fluorescent-based techniques, which was between 3 and 1000 (Miller et al., 2008), a clear separation between the targets and non-targets was achieved with the GLS approach using an SNR cutoff value of two (Fig 2b). This low range of SNR may be due to several reasons including i) non-uniform silver deposition and manual hybridization, ii) use of local background signal to normalize the positive signals instead of using average background for the whole array or for selected spots as done in fluorescent-based techniques, and iii) cross-hybridization due to the presence of a large number of probes for closely related organisms.
Performance of targeted and non-targeted probes for known VMG mixture
Of the 22 VMGs targeted by 4 to 20 probes per VMG, 21 had a mean PF between 0.9 and 1, while one VMG had a mean PF of 0.8 (Fig 2a). All probe sets targeting VMGs exhibited a PF much higher than the cut-off of 0.75. Considered at the probe level after applying the filters, 95% of the total targeted probes generated signals with a mean SNR between 3.2 and 3.7 (Fig 3b). Less than 0.3 % of non-targeted probes had a mean SNR greater than two (Fig 2b). In studies related to GLS approach, the signal intensity using flatbed scanner may also be quantified on the gray scale (Alexandre et al., 2001; Liang et al., 2004), using 0 for black and 255 for white. Previous studies using gray scale reported a range of signal intensity ranging between from 0 to 100 (Liang et al., 2004) and 50 to 150 (Taton et al., 2000). In this study, signals were quantified based on SNR between 0 and 5 which was a similar approach adopted by Wan and coauthors (Wan et al., 2005). They used a sandwich hybridization assay to detect hepatitis A virus and observed a maximum SNR of 2.7. A low range of SNR is rather normal for the GLS approach.
As mentioned before, there were six VMGs that targeted 1–2 probes per gene. Due to the smaller number of probes per VMG, PF calculation was erroneous and therefore these VMGs were filtered out from further analysis. These included plcB, inlA and inlB (L. monocytogenes), seC (S. aureus), ctxB (V. cholerae) and ail (Y. enterocolitica) (Table S4). This filter indicates that during probe design, care must be taken to design at least four distinct probes per VMG for reliable presence/absence calls by a combination of PF and SNR.
The performance of non-targeted probes sets was also evaluated to assess cross-hybridization with the applied VMG mixture. All non-targeted probe sets had a PF below 0.7. And only two probe sets had a mean PF between 0.61 and 0.7, the highest PF range for a non-target probe set. As such, the cut-off value of PF for making presence calls was set at 0.75.
Presence/absence calls for blind samples
Validation of the GLS approach was accomplished by testing the microarray with blind samples containing spiked VMGs not known to the person performing hybridization experiments, processing data, and making calls. Presence/absence calls were made after applying the three filters described above. For the first blind sample, al spiked VMGs were found present with a SNR between 3.6 and 4.4 and a PF between 0.75 and 1 (Table 3). The lowest PF for a positive target was observed for nuc gene of S. aureus with a PF of 0.78 (Table 3). Further measurement of amplicon DNA concentration indicated that this low PF was due to unintentionally spiking gDNA at a lower concentration than was initially intended. PCR results of the blind sample confirmed a lower DNA concentration of the gene, resulting in a PF that was lower than others. Previous studies have shown that lower target abundance may result in lower positive fractions due to variation in signal intensity for different probes (Miller et al., 2008). For the second blind sample, all spiked VMGs were detected with a SNR between 2.7 and 3.3, and a PF between 0.81 and 0.99 (Table 3). There were no false positive calls made in any sample hybridized in this study. This is one of the key strengths of this approach in part due to the use of multiple VMGs and 4 to 20 probes per VMG.
A cost comparison shows differences in initial and repetitive costs in terms of scanners and labeling, respectively (Table 2), while time to positive results with both methods is similar. Since the GLS microarray was used to confirm presence of amplicons following PCR, sensitivity of the method in terms of absolute and relative abundance or tests with gDNA spiked into realistic samples (e.g. food and water matrix) was not evaluated. However both sensitivity and spikes into water matrices was described in our previous studies using fluorescent labeling (Miller et al., 2008). In detail, the analytical sensitivity was between 0.1% and 0.01% (relative abundance), depending on the pathogen and the marker gene, tested with river water, drinking water and tertiary effluent from a waste water treatment facility.
Conclusions
The GLS approach was evaluated using a VMG microarray with a flatbed scanner for simultaneous detection of 14 pathogens. For quantifying signals using the GLS approach, the flatbed scanner with a similar resolution to the one described in this paper may be available at a fraction of the cost compared to more expensive laser scanner employing fluorescent dyes. Reliable presence/absence calls were made based on a set of three filters; i) four or more probes per probe set, ii) SNR of each probe greater than or equal to two and iii) a PF cut-off of 0.75 per probe set. Assay specificity can be achieved through reliable probe design criteria and increased redundancy in probe sets. The range of SNR for the GLS approach was found to be lower as compared to fluorescent-based techniques. However, a clear separation between the target and non-targets can be achieved. The main limitation of the approach is that it can only be used for making presence/absence calls of amplicons and not for obtaining expression ratios. In addition, there is a potential for making false negative calls when the amplicon abundance is low. Although there were 8,887 probes on the VMG microarray, no false positive calls were made, indicating that there was minimal cross-hybridization on the array. Therefore, the GLS approach with VMG microarrays and a flatbed scanner can be used as a reliable and cost-effective tool for parallel screening of pathogens. The validated as well as theoretically designed probes can also be synthesized and used separately without the need for using a high density in situ synthesized array.
Supplementary Material
Highlights.
A gold-silver labeling technique was validated for the presence of 14 pathogens.
The labeling method allows use of a low cost flatbed scanner for presence/absence.
The method is extendable to detection of other pathogens in limited resource settings.
Hybridization of the 8,000 probe array with blind samples resulted in 100% correct calls.
Acknowledgments
This project was partially supported with funds from the National Institute of Health (5 R01 RR018625-03), and the Michigan Economic Development Corporation (GR-476 PO 085P3200517).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandre I, Hamels S, Dufour S, Collet J, Zammatteo N, Longueville F, Gala JL, Remacle J. Colorimetric silver detection of DNA microarrays. Anal Biochem. 2001;295:1–8. doi: 10.1006/abio.2001.5176. [DOI] [PubMed] [Google Scholar]
- Call D. Challenges and opportunities for pathogen detection using DNA microarrays. Crit Rev Microbiol. 2005;31:91–99. doi: 10.1080/10408410590921736. [DOI] [PubMed] [Google Scholar]
- Call D, Borucki M, Loge F. Detection of bacterial pathogens in environmental samples using DNA microarrays. J Microbiol Methods. 2003;53:235–243. doi: 10.1016/s0167-7012(03)00027-7. [DOI] [PubMed] [Google Scholar]
- Cao B, Wang S, Tian Z, Hu P, Feng L, Wang L. DNA Microarray Characterization of Pathogens Associated with Sexually Transmitted Diseases. PLoS One. 2015;10:e0133927. doi: 10.1371/journal.pone.0133927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T, Stone C, Murray S, Moberg J, Andersen G. Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol Lett. 2005;245:271–278. doi: 10.1016/j.femsle.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Gao X, Gulari E, Zhou X. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–596. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- Gao X, LeProust E, Zhang H, Srivannavit O, Gulari E, Yu P, Nishiguchi C, Xiang Q, Zhou X. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001;29:4744–4750. doi: 10.1093/nar/29.22.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinev A, Chancey C, Volkova E, Chizhikov V, Rios M. Development of a microarray-based assay for rapid monitoring of genetic variants of West Nile virus circulating in the United States. J Virol Methods. 2017;239:17–25. doi: 10.1016/j.jviromet.2016.10.011. [DOI] [PubMed] [Google Scholar]
- He Z, Wu L, Li X, Fields M, Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl Env Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeProust E, Pellois J, Yu P, Zhang H, Gao X. Digital light-directed synthesis. A microarray platform that permits rapid reaction optimization on a combinatorial basis. Comb Chem. 2000;2:349–354. doi: 10.1021/cc000009x. [DOI] [PubMed] [Google Scholar]
- Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang RQ, Tan C, Ruan KC. Colorimetric detection of protein microarrays based on nanogold probe coupled with silver enhancement. Immunol Methods. 2004;285:157–163. doi: 10.1016/j.jim.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Lin F, Sabri M, Alirezaie J, Li D, Sherman P. Development of a nanoparticle-labeled microfluidic immunoassay for detection of pathogenic microorganisms. Clin Diagn Lab Immunol. 2005;12:418–425. doi: 10.1128/CDLI.12.3.418-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cao X, Yang Y, Liu M-G, Wang Y-F. Array-based nano-amplification technique was applied in detection of hepatitis E virus. Biochem Mol Biol. 2006;39:247–252. doi: 10.5483/bmbrep.2006.39.3.247. [DOI] [PubMed] [Google Scholar]
- Logue CM, Barbieri NL, Nielsen DW. Advances in Food and Nutrition Research. 1. Elsevier Inc; 2017. Pathogens of food animals: sources, characteristics, human risk, and methods of detection. [DOI] [PubMed] [Google Scholar]
- Loy A, Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Acta. 2006;363:106–119. doi: 10.1016/j.cccn.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Miller S, Tourlousse D, Stedtfeld R, Baushke S, Herzog A, Wick L, Rouillard J, Gulari E, Tiedje J, Hashsham S. An in-situ synthesized virulence and marker gene (VMG) biochip for the detection of bacterial pathogens in water. Appl Env Microbiol. 2008;74:2200–2209. doi: 10.1128/AEM.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman G, Jasani B. Silver development in microscopy and bioanalysis: past and present. Pathology. 1998;186:119–125. doi: 10.1002/(SICI)1096-9896(1998100)186:2<119::AID-PATH160>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Ngom B, Guo Y, Wang X, Bi D. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: a review. Anal Bioanal Chem. 2010;397:1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- Qi H, Chen S, Zhang M, Shi H, Wang SQ. DNA microarrays for visual detection of human pathogenic microorganisms based on tyramine signal amplification coupled with gold label silver stain. Analy Bioanal Chem. 2010;398:2745–2750. doi: 10.1007/s00216-010-4189-3. [DOI] [PubMed] [Google Scholar]
- Reichert J, Csáki A, Köhler J, WF Chip-based optical detection of DNA hybridization by means of nanobead labeling. Anal Chem. 2000;72:6025–6029. doi: 10.1021/ac000567y. [DOI] [PubMed] [Google Scholar]
- Stedtfeld R, Wick L, Baushke S, Tourlousse D, Herzog A, Xia Y, Rouillard J, Klappenbach J, Cole J, Gulari E, Tiedje J, Hashsham S. Influence of dangling ends and surface-proximal tails of targets on probe-target duplex formation in 16S rRNA gene-based diagnostic arrays. Appl Env Microbiol. 2007;73:380–389. doi: 10.1128/AEM.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storhoff J, Marla S, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, Cork W, Müller U. Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron. 2004;19:875–883. doi: 10.1016/j.bios.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Tang J, Zhou L, Duan L, Feng Y, Cao X, Wang Y. Development and evaluation of a new detection tool — visual DNA microarray for simultaneous and specific detection of human immunodeficiency virus type-1 and hepatitis C virus. Mol Biol Rep. 2011;38:5341–5348. doi: 10.1007/s11033-011-0685-6. [DOI] [PubMed] [Google Scholar]
- Taton T, Mirkin C, Letsinger R. Scanometric DNA array Detection with Nanoparticle Probes. Science (80- ) 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Wan Z, Wang S, Li S-C, Duan L, Zhai J. Development of array-based technology for detection of HAV using gold-DNA probes. Biochem Mol Biol. 2005;38:399–406. doi: 10.5483/bmbrep.2005.38.4.399. [DOI] [PubMed] [Google Scholar]
- Wang X, Dang E, Gao J, Guo S, Li Z. Development of a gold nanoparticle-based oligonucleotide microarray for simultaneous detection of seven swine viruses. J Virol Methods. 2013;191:9–15. doi: 10.1016/j.jviromet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Wilson W, Strout C, DeSantis T, Stilwell J, Carrano A, Andersen G. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol Cell Probes. 2002;16:119–127. doi: 10.1006/mcpr.2001.0397. [DOI] [PubMed] [Google Scholar]
- Xue L, Fei J, Song Y, Xu R, Bai Y. Visual DNA microarray for detection of epidermal growth factor receptor (EGFR) gene mutations. Scand J Clin Lab Invest. 2014;74:693–699. doi: 10.3109/00365513.2014.951680. [DOI] [PubMed] [Google Scholar]
- Zhang G, Möller R, Kretschmer R, Csáki A. Microstructured arrays with pre-synthesized capture probes for DNA detection based on metal nano-particles and silver enhancement. Fluorescence. 2004;14:369–375. doi: 10.1023/b:jofl.0000031818.39925.4c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.