Abstract
The microbial bioconversion of sterols can afford valuable steroid precursors, such as 4-androstene-3,17-dione (AD) and androsta-1,4-diene-3,17-dione (ADD). The Mycobacterium neoaurum MN4 mutant strain can produce AD in high yield and can tolerate a higher concentration of the substrate phytosterol than the parent strain M. neoaurum MN2. In order to further investigate the mechanisms underlying the enhanced substrate and product tolerance, we performed a genomic analysis of the MN2 and MN4 strains. The genomes were sequenced using a high-throughput approach and analyzed using software for genome assembly, gene prediction and functional annotation, KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation, COG (cluster of orthologous) group cluster analysis, GO cluster analysis, and SNP detection and annotation. Based on comparative genomics, 184 mutations were identified in MN4, the average variant rate of 1 variant every 27,249 bases, with a TS/TV value of 0.5877 and missense mutations in one key sterol degradation genes (ChoM1) and four side chain degradation genes that encode enzymes catalysing β-oxidation. The results suggest the high AD yield might be due to mutation of ChoM and genes encoding FadE, FadB and FadA β-oxidation enzymes. This study provides a theoretical basis for further functional genomics analysis and heterologous production of M. neoaurum MN2 secondary metabolites.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0818-2) contains supplementary material, which is available to authorized users.
Keywords: 4-androstene-3,17-dione(AD); Androsta-1,4-diene-3,17-dione(ADD); Comparative genomics; Mycobaterium neoaurum; Steroid
Introduction
3-hydroxy steroids such as cholesterol, phytosterols, and ergosterol are widely distributed in nature. Cholesterols are typically found in animals, while plant steroids include sitosterols, stigmasterol, campesterol, brassicasterol, and ergosterols are the major type in yeasts and other fungi (Hannich et al. 2011; Donova and Egorova 2012). As important components of cell membranes, they play a critical role in membrane fluidity, integrity, cell differentiation, and chemical tolerance, among many other processes (Beattie et al. 2005; Nicolaou et al. 2010).
Sterols are used by many mycobacteria as a natural source of carbon and energy. Steroid drugs are widely produced by microbial biotransformation or bioconversion of sterols into steroid precursors due to the low cost and abundance of sources. At present, more than 400 steroidal drugs are authorized for clinical use and are commercially available, with global annual sales of $400 million and an ever-increasing demand that is not far behind antibiotics (Tong and Dong 2009; Bureik and Bernhardt 2007). Steroid hormone drugs are important for both short-term use in the clinic and for long-term use to improve patient quality of life. Steroid-based drugs are widely used as anti-tumour, anti-inflammatory, anti-microbial, and anti-allergy agents; for the prevention and treatment of many severe diseases such as hormone-dependent forms of breast and prostate cancer, certain forms of colon cancer, obesity, diabetes, rheumatoid arthritis, hypertension, asthma, eczema, inflammation, metabolic disorders, neurodegenerative elderly diseases, central nervous system disorders, gynecopathy, anaphylactic shock, as replacement agents in the treatment of adrenal insufficiencies, in the inhibition of HIV integrase, in the prevention and treatment of infection by HIV, and in the treatment of declared AIDS (Bäckström et al. 2011; Douglas 2010; Garcia-Segura and Balthazart 2009; Suzuki et al. 1998; Finocchi and Ferrari 2011; Rugutt and Rugutt 2012).
Currently, the majority of pharmaceutically active steroids are produced through the selective degradation of sterol side chains, and 4-androstene-3,17-dione (AD) and androsta-1,4-diene-3,17-dione (ADD) serve as valuable sterol precursors that can be obtained from microbial bioconversion (Rodríguez et al. 2016). For this reason, a great number of non-pathogenic mycobacteria have been isolated and investigated for their ability to produce these high-value metabolites (Yao et al. 2013).
Mycobacterium neoaurum MN2 is a soil organism and well-characterized AD producer (Sun and Wang 2014). This strain has been mutated using UV and chemical approaches to generate the MN4 strain, which produces an even higher AD yield and displays increased tolerance to high concentrations of the substrate phytosterol than the parent strain. At a phytosterol concentration of 20 g/L, this strain is able to achieve an AD conversion rate of 40.9% during shake flask fermentation for 144 h, which is 15.1% higher than that of the original strain (Sun and Wang 2014). In the paper, we used M. neoaurum MN2 genome as a reference to allow for a comparative analysis to the previously sequenced mutated strain M. neoaurum MN4 which explored the high yield and substrate resistance mechanism of M. neoaurum MN4.
Materials and methods
Strains and culture conditions
The stock culture was maintained on an agar slant medium containing (per l) 3 g yeast extract, 5 g peptone, 20 g glucose, and 20 g agar (pH 7.0). M. neoaurum MN2 originally isolated from soil and characterized as an AD producer was cultivated in seed medium containing (per l) 6 g peptone, 8 g glucose, 10 g corn steep liquor, and 0.5 g K2HPO4 (pH 8.0) in a rotary shaker operating at 250 rpm and 31 °C for 4 days.
DNA sequencing and data preprocessing
OMEGA bacterial genomic DNA extraction kit was used to genomic DNA extraction purchased from Feiyang BIOTECH Co., Ltd. (Guangzhou, China) according to the manufacturer’s instructions without modifications. The SDS method was used to isolate genomic DNA. Total DNA was subjected to quality control by agarose gel-electrophoresis and quantified by Qubit. Genome sequencing was performed using massively parallel sequencing (MPS) Illumina technology. Specifically, a paired-end DNA library was constructed with an insert size of 350 bp and sequenced using an Illumina HiSeq 4000 PE150 strategy at the Beijing Novogene Bioinformatics Technology Co., Ltd. Quality control of paired-end reads was performed using an in-house program. The NGS QC Toolkit (Patel and Jain 2012) was then used to filter low quality reads and details of the specific parameters are show in supporting information.
Prediction and functional annotation of encoded proteins
Velvet 1.2.10 assembler software (Zerbino and Birney 2008) was used to generate the whole-genome sequence by optimizing the Kmer value for the best assembly results.
Gene prediction software Glimmer 3.02 (Delcher et al. 2007) was used to predict open reading frames (ORFs), and BLASTP (Camacho et al. 2009) was used to align protein sequences against non-redundant (NR), Swiss-Prot (Boeckmann et al. 2003), InterPro (Mulder et al. 2002), Encyclopaedia of genes and genomes (KEGG) (Kanehisa and Goto 2000) Gene Ontology (GO) (Harris et al. 2004), and orthologous gene (COG) (Tatusov et al. 2000) databases (E values <1e-5) for functional annotation. Additionally, KEGG annotation genomic protein sequences were submitted to KAAS (Moriya et al. 2007) using the bidirectional hit (BDH) method.
Resequencing analysis
In order to explore the high yield and substrate resistance mechanism of M. neoaurum MN4. Differences between M. neoaurum MN4 and the reference M. neoaurum MN2 were visualised by aligning sample reads. The detail of the methods specific parameters are show in supporting information.
Results
Genome sequencing and general features
The genome of M. neoaurum MN2 was sequenced using a whole-genome shotgun sequencing strategy on the Illumina Hiseq 2500 platform, resulting in a 5.38 Mb genome sequence consisting of 43 contigs with an N50 length of 190,920 bp and an L50 of 10 (Table 1). the M. neoaurum MN4 also sequenced using a whole-genome shotgun sequencing strategy on the Illumina Hiseq 2500 platform, the sequencing information of the M. neoaurum MN2 and M. neoaurum MN4 are show in Table S1. It was reported that only 5 strains of mycobacteria neoaurum have been sequenced included M. neoaurum MN2 and M. neoaurum MN4. As we can see, five mycobacteria neoaurum are very similar in Genome size, GC content, CDS number, CDS mean length, and RNA gene number (Table 2).
Table 1.
General features of the Mycobacterium neoaurum MN2 genome
| Feature | No. |
|---|---|
| Number of contigs | 43 |
| Length of the genome assembly (Mb) | 5.38 |
| GC content (%) | 66.9% |
| tRNA number | 46 |
| Number of protein-coding genes | 4890 |
| Number of genes | 5049 |
| Average CDS size (bp) | 989 |
| Coding percentage (%) | 89.8% |
| Number of pseudo genes | 107 |
| GenBank No. | LQMX00000000 |
Table 2.
General features of several sequenced Mycobacterium neoaurum chromosomes
| Features | MN2 | VKMAc-1815D | ATCC25795 | DSM440744 |
|---|---|---|---|---|
| Length size (bp) | 5390,529 | 5421,267 | 5468,381 | 5504,703 |
| GC content (%) | 66.9 | 66.9 | 66.7 | 66.5 |
| CDS (no.) | 4890 | 4948 | 5059 | 5030 |
| Contigs (no.) | 43 | 1 | 42 | 45 |
| Average CDS size (bp) | 989 | 996 | 990 | 978 |
| CodingPercent (%) | 89.9 | 90.9 | 91.6 | 88.9 |
| tRNA number | 46 | 46 | 46 | 46 |
| Pseudogene (no.) | 105 | 25 | 22 | 82 |
| GenBank No. | LQMX00000000 | CP006936 | JMDW00000000 | CCDR000000000 |
Variant calling (SNPs and INDELs)
By mapping the reads for M. neoaurum MN4 against the genome of M. neoaurum MN2, differences were identified and comprised one point INS, two point deletions, with 181 SNPs variants (102 point missense variants, 60 synonymous variants, 18 intergenic regions, three stopped/lost reads, and one frameshift variant) (Tables S2 and S3).
All 184 variants were located in 36 contigs, with an average variant rate of 1 variant every 27,249 bases and a transitions/transversions (Ts/Tv) ratio of 0.5877 (Tables S4 and S5). Statistical analysis indicated that all single INSs, the three stop/lost variants, and the frameshift variant were located in coding regions and clustered in five genes, two of which were pseudogenes (Table S6). Furthermore, of all 102 missense variants, only 93 were in protein-coding genes, with 81 in genes and nine in pseudogenes. Details of missense variants and mutated genes are listed in Table S7.
COG clustering analysis
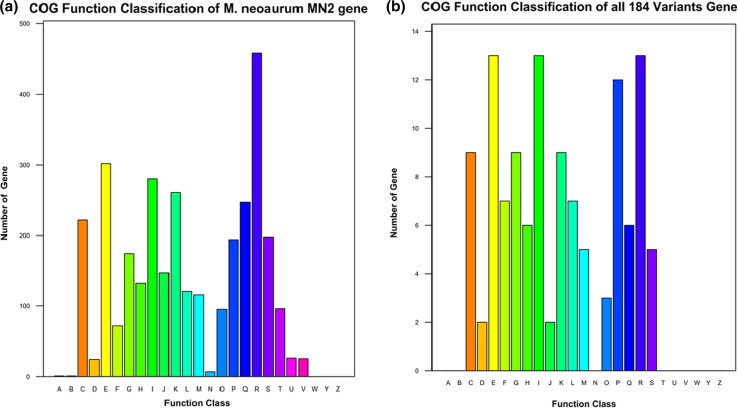
Based on the gene prediction and annotation results, M. neoaurum MN2 was predicted to contain 4891 protein-coding genes, and protein sequences were searched against the COG database using BLAST, with E value ≤1e-5 and identity ≥40% used to filter the results. Of all 184 variants, COG functional annotation was only successful for 121, and these were mainly in E (amino acid transport and metabolism, 10.7%), I (lipid transport and metabolism, 10.7%), R (general function prediction only, 10.7%), P (inorganic ion transport and metabolism, 9.9%) categories, suggesting variants were concentrated in metabolism and related functions (Fig. 1).
Fig. 1.
COG function classification of M. neoaurum MN2 gene (a) and 184 variants gene (b) (A RNA processing and modification, B chromatin structure and dynamics, C energy production and conversion, D cell cycle control, cell division, chromosome partitioning, E amino acid transport and metabolism, F nucleotide transport and metabolism, G carbohydrate transport and metabolism, H coenzyme transport and metabolism, I lipid transport and metabolism, J translation, ribosomal structure and biogenesis, K transcription, L replication, recombination and repair, M cell wall/membrane/envelope biogenesis, N cell motility, O posttranslational modification, protein turnover, chaperones, P inorganic ion transport and metabolism, Q secondary metabolites biosynthesis, transport and catabolism, R general function prediction only, S function unknown, T signal transduction mechanisms, U intracellular trafficking, secretion, and vesicular transport, V defense mechanisms, W extracellular structures, Y nuclear structure, Z cytoskeleton)
GO cluster analysis
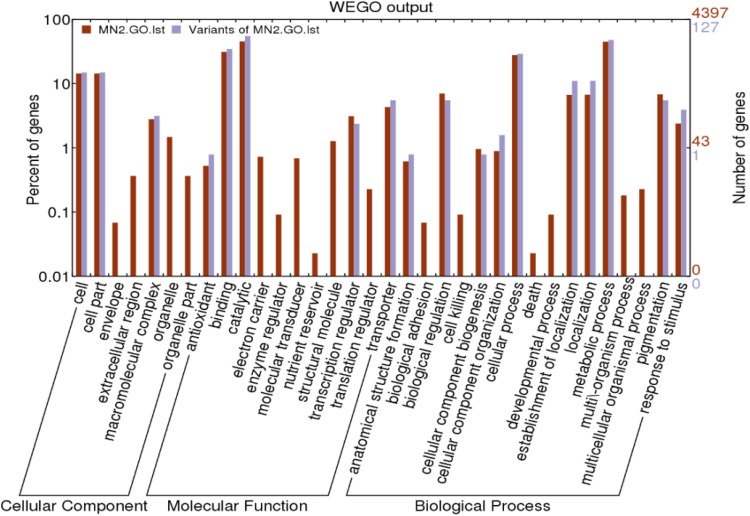
GO annotation classified most protein functions in the cellular components, molecular function, biological process categories. InterProScan alignment of MN2 GO annotation data with the WEGO online tools predicted 4891 protein-coding genes and 184 variants (Fig. 2).
Fig. 2.
Gene distribution based on Gene Ontology analysis (the Claret column is Gene Ontology analysis of MN2 and the gray column is Gene Ontology analysis about Variants of MN2)
As shown in Fig. 2, a large number of variants were found in cell, cell part, macromolecular complex, antioxidant, binding, catalytic, transcription regulator; transporter, anatomical structure formation, biological regulation, cellular component biogenesis, cellular component organization, cellular process, establishment of localization, localization, metabolic process, pigmentation, and response to stimulus subcategories, belonging to Cellular Component, Molecular Function, Biological Process categories. Because the variants were not concentrated in any particular genes or functional areas, we concluded that the mutation of MN2 was random in nature.
KEGG annotation of the M. neoaurum MN2 genome
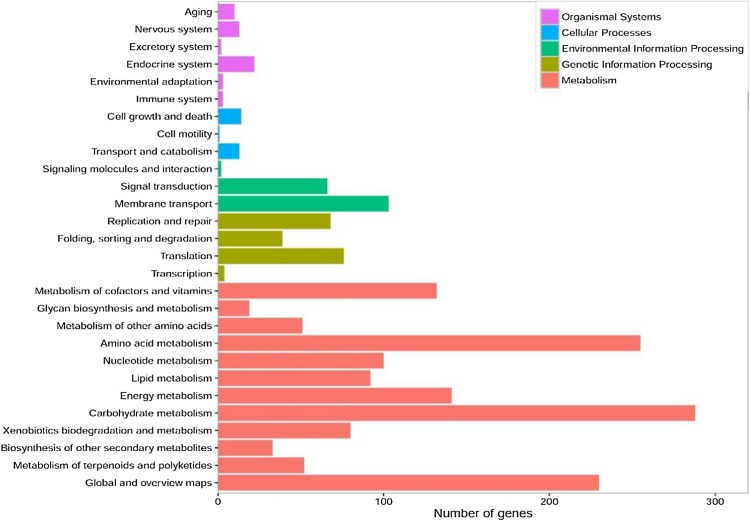
The results above identified 1991 protein-coding genes that were successfully annotated using the KEGG database (Fig. 3). A pathway map of steroid degradation is shown in Fig. 4.
Fig. 3.
Gene distribution based on KEGG Annotation of M. neoaurum MN2 (the Metabolism, genetic information Processing, Environmental Information Processing, Cellular Process, Organismal Systems accounted for 73.9, 9.3, 8.5, 1.4, 2.6% of genes, respectively)
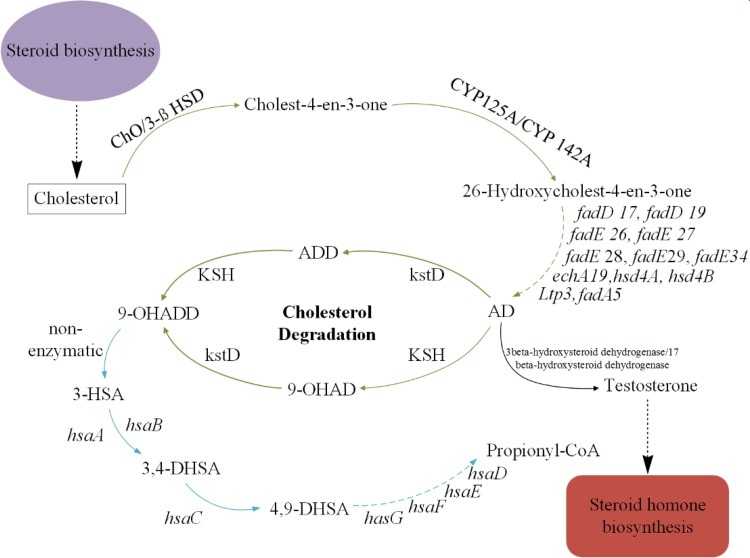
Fig. 4.
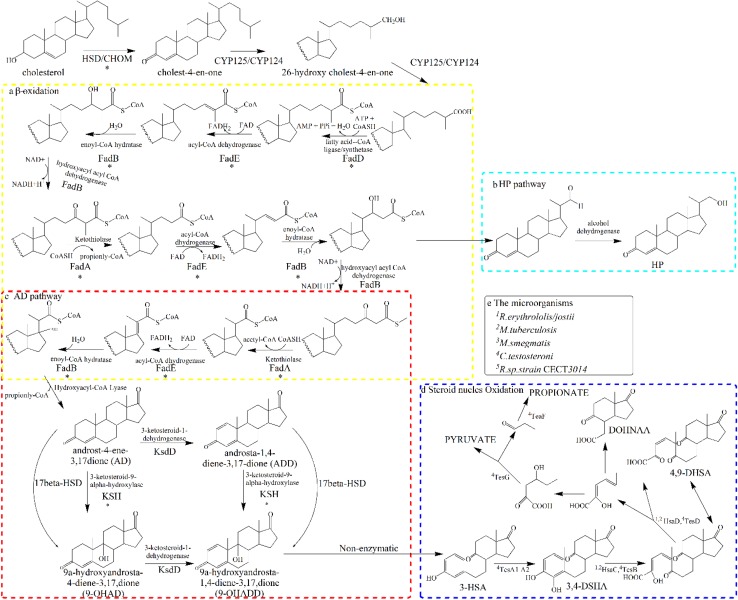
Pathway map of steroid degradation. The green arrow shows the degradation process of sterol side-chain, the blue arrow show the degradation of steroid nucleus. The dashed arrow indicate multiple enzymatic. AD 4-androstadiene-3,17-dione, 9-OHADD, 9α-hydroxy-1,4-androstadiene-3,17-dione, ChO cholesterol oxidase, 3β-HSD 3-beta-hydroxy-delta-5-steroid dehydrogenase, CYP125A cytochrome P450 125A1, FadD family putative fatty-acid-CoA ligase (fadD15,17,19), FadE family acyl-CoA dehydrogenase (fadE26,27,28,29,34), FadB family enoyl-CoA hydratase (ech19, hsd4B), FadA family acetoscetyl-CoA thiolase (fadA5, Ltp3), hsd4A Hydroxy-CoA dehydrogenase, KstD 3-Ketosteroid-1-dehydrogenase, KSH (KshA and KshB), 3-ketosteroid-9α-hydroxylase
Discussion
Steroid hormone production is a well-known example of the successful application of microbial biotransformation technology in large-scale industrial processes, as exemplified by the 1-dehydrogenation of steroids catalysed by Arthrobacter simplex, and the side-chain cleavage of phytosterol catalysed by Mycobacterium sp. (Mahato and Garai 1997; Fernandes et al. 2003). The current most marketed intermediates are AD and ADD, which are required for the commercial production of corticosteroids, mineralocorticoids, oral contraceptives, and other pharmaceutical steroids.
However, the great majority of steroids are poorly soluble in water, with a solubility below 0.1 mM, which leads to a low bioconversion efficiency for hydrophobic substrates and products such as phytosterols that generally have a water solubility below 1 μM (Wang et al. 2011). The low solubility of phytosterols leads to inadequate mass transfer, which seriously hinders their bioavailability (Goetschel and Bar 1992; Phase and Patil 1994; Malaviya and Gomes 2008).
In this study, M. neoaurum MN4 was generated from the parent M. neoaurum MN2 strain by a combination of UV and chemical mutagenesis (DES). The ratio of AD to ADD in the transformed product of MN2 was 13:1, while the ratio of AD to ADD in the mutant strain MN4 was 20:1, MN4 displays an AD generation rate that is increased by 15% compared with MN2 (Sun and Wang 2014). The biotransformation ability of substrate-resistant strains and original strain show in Table 3 and the HPLC analysis of the transformed products are show in Fig. 5.
Table 3.
Biotransformation ability of substrate-resistant and original strain (Sun and Wang 2014)
| Strian | The AD Transformation rate (%) | The ADD transformation rate (%) | AD increase rate (%) |
|---|---|---|---|
| MN2 | 25.8 | 1.9 | – |
| MN4 | 40.9 | 2.1 | 15.1 |
Fig. 5.
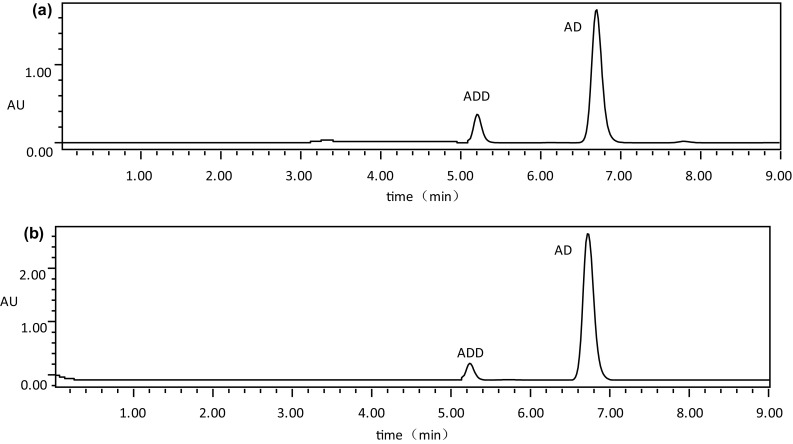
The HPLC analysis of transformation products by the strain. a The transformation of and original strain MN2 (the tansformation rate of AD and ADD are 25.8%, 40.9% in MN2. The ratio of AD to ADD in the transformed product of MN2 was 13:1). b The transformation of mutation strain MN4. (the tansformation rate of AD and ADD are 40.9%, 2.1% in MN4. The ratio of AD to ADD in the mutant strain MN4 was 20:1)
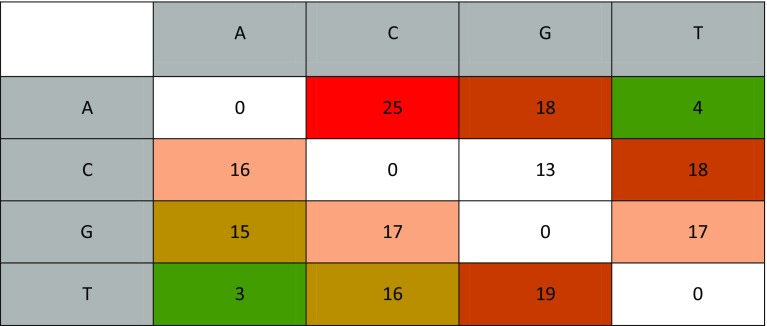
According to the results shown in Fig. 6, we can see that the number of G ⟺ A and C ⟺ T transitions, and the number of A ⟺ T and G ⟺ C transversions was 15, 18, 18, 16, 4, 3, 17, and 13, confirming the effectiveness of the combined UV and chemical mutagenesis. The high transversion of A > C and T > G indicates that UV mutagenesis was particularly effective.
Fig. 6.
Base changes (SNPs) following mutagenesis of MN2 to MN4. Columns are reference bases and rows are changed bases (e.g. column ‘A’ Row ‘T’ indicates how many ‘A’ bases have been replaced by ‘T’ bases). A red background indicates a higher number of base change
Whole-genome sequencing of MN2 revealed 43 contigs, and 5049 genes were predicted and annotated, including 4918 protein-coding genes. By comparative analysis, two cholesterol oxidase, three 3ß-HSD enzymes, two KstD enzymes, three KSH enzymes were identified in M. neoaurum MN2. In general, the number of KstD enzymes which catalyzed the steroid nucleus degradation are usually three at least but it is only two KstD enzymes in M. neoaurum MN2. (e.g. three KstD enzymes in ATCC2595 accession number are GQ411074, KF772209, KF772210; four KstD enzymes in DSM440744 accession number are CDQ47066, CDQ47402, CDQ44570, CDQ44573.1). Based on the above reason, it is can reduce the degradation of AD. Meanwhile, the number of enzymes associated with steroid uptake was up to five in M. neoaurum MN2 which increased the amount of steroid intake. (e.g. two cholesterol oxidase enzymes and one 3ß-HSD enzymes in ATCC2595 accession number are WP_030137457, WP_030134612, WP_030136274; two cholesterol oxidase enzymes and one 3ß-HSD enzymes in DSM440744 accession number are CDQ43574.1, CDQ46398.1, CDQ44698.) Cholesterol catabolic pathway genes in MN2 are shown in Table S8.
The degradation of sterols by mycobacterium involves three major process: sterols uptake, elimination of aliphatic side chain at C17 and steroid nucles oxidation (Donova and Egorova 2012). Research suggests that cholesterol side chain and ring degradation (steroid nucleus oxidation) were confirmed to be independent process, the ring degradation of sterols is initiated A-ring oxidation (Cholesterol oxidases), then 3-ketosteroid-1-dehydrogenase (KstD) and 3-ketosteroid-9α-hydroxylase (KshA) to open the steroid ring B (Fig. 7) (García et al. 2012; Jenna et al. 2011). Cholesterol oxidases (ChOs) are a class of interfacial enzymes involved in the steroid-transforming process that have been predicted to form part of the FMCM structure, thereby stimulating the transport of sterols (García et al. 2012). ChOs are flavoenzymes that catalyse the oxidation of sterols to sterones in the first step of steroid nucleus oxidation. Analysis of variants identified AVZ31_14310 (ChoM 1) as a missense variant (p.Ser408Phe). Since ChOs can access and extract sterols from the cell envelope, they play a prominent roles in initiating the metabolism of steroids, and a missense mutation of the gene encoding ChoM1 might alter the efficiency of steroid nucleus oxidation and increase the utilization of sterols.
Fig. 7.
The deduced cholesterol catabolic pathway of MN2. The side-chain degradation of cholesterol is shown in the dashed yellow box (a). The HP pathway is shown in the dashed light green box (b). The conversion of AD to ADD and AD to 9-OHAD are shown in the dashed red box (c). The Steroid Oxidation is shown in the blue box (d). Each step involved in the reaction of the enzyme (d) in front of the number representatives the different microorganisms show in the e (means that different steps of the biocatalytic reaction has different genes in different microorganisms). Reactions marked with asterisk refer to genes that were mutated following UV and chemical mutagenesis
The key step in microbial steroid catabolism is 9a-hydroxylation of 4-androstene-3,17-dione (AD) and 1,4-androstadiene-3,17-dione (ADD), which is catalysed by the terminal oxygenase (KshA) and ferredoxin reductase (KshB) two-component enzyme system. In Rhodococcus erythropolis SQ1, in which kshA and kshB genes were first identified, three additional kshA homologues are present (Geize et al. 2001; Van et al. 2008). KshB of Rhodococcus rhodochrous DSM43269 was shown to contain an FAD flavin cofactor and a plant-type Fe2S2 cluster (Capyk et al. 2009a, b; Petrusma et al. 2009). The KshB gene deletion strain RG4 can only transform phytosterol to sterone derivatives and cannot metabolize any further, indicating that side-chain degradation of sterols is blocked in this variant. Similarly, KshB gene deletion blocks phytosterol degradation (Petrusma et al. 2011). We identified two KshA genes (KshA-1 and KshA-2) and KshB genes in MN2 by BLASTP.
Resequencing analysis revealed 184 variants in MN2, and COG analysis showed that variants were concentrated in amino acid and lipid transport and metabolism processes, suggesting these were responsible for the improved performance of MN4 compared with MN2. Furthermore, the lipid transport and metabolism mutations were clustered in steroid uptake and metabolism genes, consistent with improved steroid uptake in MN4.
Of the 184 variants, four missense variants (AVZ31_16285 p.Leu290Ile, AVZ31_22040 p.Gly241Asp, AVZ31_18165 p.Val231Glu, AVZ31_24580 p.Asn55Lys) were linked to side-chain degradation, specifically to FadE (AVZ1_16285, AVZ31_22040), FadB (AVZ31_18165), and FadA (AVZ31_24580) family enzymes. The initial step in the oxidation of sterol side-chains (and other C27 steroids) involves hydroxylation at carbon C-26, yielding 26-hydroxycholest-4-en-3-one, in a reaction catalysed by cytochrome P450 monooxygenases such as CYP125 and CYP142 (Capyk et al. 2009a, b; Rostoniec et al. 2009). Cholesterol side-chain β-oxidation is catalysed by an ATP-dependent sterol/steroid CoA ligase that catalyses the CoA activation of cholest-4-en-3-one-26-oic acid, examples of which include FadD19 of Rhodococcus rhodochrous DSM43269 (Sih et al. 1968a, b; Wilbrink et al. 2011). This reaction results in the formation of 17-ketosteroid following cleavage of the side-chain, and propionyl-CoA and acetyl-CoA are also formed during this process (β-oxidation) which involves four enzymes; acyl-CoA dehydrogenases (FadE family), enoyl-CoA-hydratases (FadB family), hydroxyacyl-CoA dehydrogenases, and thiolases (FadA family). Due to the presence of the cyclopentane D ring, propionyl-CoA and androst-4-ene-3,17-dione(AD) formation requires the participation of a hydroxyacyl-CoA lyase similar to other CoA-lyases instead of a conventional β-oxidation process (Atrat et al. 1991). Variants in genes encoding these enzymes may be responsible for the improved efficiency of β-oxidation and consequent AD/ADD production in MN4.
Not only did we find variants in cholesterol side chain degradation; we also identified ring degradation mutants. The active lipid transport and metabolism variant of MN2 and the missense ChoM1 (p.Ser408Phe) mutation possibly increased the absorption and utilization of sterol. Meanwhile, the missense mutations in FadE, FadB, and FadA family enzymes potentially improved the efficiency of β-oxidation. Mutations in all three processes could explain why MN4 displayed superior AD and ADD production capabilities than the parent MN2 strain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Jiangxi Provincial Science and Technology Department Support Program (Grant No. 20142BBG70016), the Natural Science Foundation of Jiangxi Province (Grant No. 20151BAB204003) and by the Development Foundation of the Key Lab of Protection and Utilization of Subtropical Plant Resources (Grant No. YRD201405).
Compliance with ethical standards
Conflict of interest
We declare that we have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0818-2) contains supplementary material, which is available to authorized users.
Ling-xia Xu and Hui-lin Yang contributed equally to this work.
References
- Atrat SP, Hösel P, Hörhold C. Interactions of Mycobacterium fortuitum with solid sterol substrate particles. J Basic Microbiol. 1991;31:413–422. doi: 10.1002/jobm.3620310605. [DOI] [Google Scholar]
- Bäckström T, Haage D, Löfgren M, Johansson IM, Strömberg J, Nyberg S, Andréen L, Ossewaarde L, Wingen GAV, Turkmen S. Paradoxical effects of GABA-A modulators may explain sex steroid induced negative mood symptoms in some persons. Neuroscience. 2011;191:46–54. doi: 10.1016/j.neuroscience.2011.03.061. [DOI] [PubMed] [Google Scholar]
- Beattie ME, Veatch SL, Stottrup BL, Keller SL. Sterol structure determines miscibility versus melting transitions in lipid vesicles. Biophys J. 2005;89:1760–1768. doi: 10.1529/biophysj.104.049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureik M, Bernhardt R (2007) Steroid hydroxylation: microbial steroid biotransformations using cytochrome P450 enzymes. In: Schmid RD, Urlacher VB (eds) Modern biooxidation—enzymes, reactions, and applications. Wiley-VCH, pp 155–176
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+:architecture and applications. BMC Bioinform. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capyk JK, Kalscheuer R, Stewart GR. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. J Biol Chem. 2009;284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capyk JK, D’Angelo I, Strynadka NC, Eltis LD. Characterization of 3-ketosteroid 9alpha-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem. 2009;284:9937–9946. doi: 10.1074/jbc.M900719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Gong YH, Cai YL, Bian YB. Genome sequence of the edible cultivated mushroomLentinula edodes(Shiitake) reveals insights into lignocellulose degradation. PLoS One. 2016;11(8):e0160336. doi: 10.1371/journal.pone.0160336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donova MV, Egorova OV. Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol. 2012;94:1423–1447. doi: 10.1007/s00253-012-4078-0. [DOI] [PubMed] [Google Scholar]
- Douglas M. Neurology of endocrine disease. Clin Med. 2010;10:387–390. doi: 10.7861/clinmedicine.10-4-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS. Microbial conversion of steroid compounds: recent developments. Enzyme Microbial Technol. 2003;32:688–705. doi: 10.1016/S0141-0229(03)00029-2. [DOI] [Google Scholar]
- Finocchi C, Ferrari M. Female reproductive steroids and neuronal excitability. Neurol Sci. 2011;32:31–35. doi: 10.1007/s10072-011-0532-5. [DOI] [PubMed] [Google Scholar]
- García JL, Uhía I, Galán B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb Biotechnol. 2012;5:679–699. doi: 10.1111/j.1751-7915.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Segura LM, Balthazart J. Steroids and neuroprotection: new advances. Front Neuroendocrinol. 2009;30(2):V–IX. doi: 10.1016/j.yfrne.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geize RVD, Hessels GI, Gerwen RV, Meijden PVD, Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ 1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol Lett. 2001;205:197–202. doi: 10.1111/j.1574-6968.2001.tb10947.x. [DOI] [PubMed] [Google Scholar]
- Goetschel R, Bar R. Formation of mixed crystals in microbial conversion of sterols and steroids. Enzyme Microbial Technol. 1992;14(6):462–469. doi: 10.1016/0141-0229(92)90138-E. [DOI] [Google Scholar]
- Hannich JT, Umebayashi K, Riezman H. Distribution and functions of sterols and sphingolipids. Cold Spring Harb Persp Biol. 2011;3:725–738. doi: 10.1101/cshperspect.a004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:258–261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenna K, Capyk IC, Gruninger Robert, Strynadka Natalie C, Eltis Lindsay D. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J Biol Chem. 2011;286:40717–40724. doi: 10.1074/jbc.M111.289975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahato SB, Garai S. Advances in microbial steroid biotransformation. Steroids. 1997;62:332–34532. doi: 10.1016/S0039-128X(96)00251-6. [DOI] [PubMed] [Google Scholar]
- Malaviya A, Gomes J. Androstenedione production by biotransformation of phytosterols. Biores Technol. 2008;99:6725–6737. doi: 10.1016/j.biortech.2008.01.039. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A. The genome analysis toolkit: a MapReduce framework for analyzing next generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P. InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3:225–235. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusma M, Dijkhuizen L, Geize RVD. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron–sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl Environ Microbiol. 2009;75:5300–5307. doi: 10.1128/AEM.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusma M, Hessels G, Dijkhuizen L, Robert VDG. Multiplicity of 3-ketosteroid-9 alpha-hydroxylase enzymes in Rhodococcus rhodochrous DSM 43269 for Specific degradation of different classes of steroids. J Bacteriol. 2011;193:3931–3940. doi: 10.1128/JB.00274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phase N, Patil S. Natural oils are better than organic solvents for the conversion of soybean sterols to 17-ketosteroids by Mycobacterium fortuitum. World J Microbiol Biotechnol. 1994;10:228–229. doi: 10.1007/BF00360894. [DOI] [PubMed] [Google Scholar]
- Rodríguez García A, Fernánde Alegre E, Morales A, Sola Landa A, Lorraine J, Macdonald S, Dovbnya D, Smith MCM, Donova M, Barreiro C. Complete genome sequence of ‘Mycobacterium neoaurum’ NRRL B-3805, an androstenedione (AD) producer for industrial biotransformation of sterols. J Biotechnol. 2016;224:64–65. doi: 10.1016/j.jbiotec.2016.03.021. [DOI] [PubMed] [Google Scholar]
- Rostoniec KZ, Wilbrink M, Capyk JK, Mohn WW, Ostendorf M, van der Geize R. Cytochrome P450125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii, RHA1. Mol Microbiol. 2009;74:1031–1043. doi: 10.1111/j.1365-2958.2009.06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugutt JK, Rugutt KJ. Antimycobacterial activity of steroids, long-chain alcohols and lytic peptides. Nat Prod Res. 2012;26:1004–1011. doi: 10.1080/14786419.2010.539977. [DOI] [PubMed] [Google Scholar]
- Sih CJ, Tai HH, Yun YT, Sang SL, Coombe RG. Mechanisms of steroid oxidation by microorgansism. XIV. Pathway of cholesterol side-chain degradation. Biochemistry. 1968;7:808–818. doi: 10.1021/bi00842a039. [DOI] [PubMed] [Google Scholar]
- Sih CJ, Wang KC, Tai HH. Mechanisms of steroid oxidation by microorganisms. 13. C22 acid intermediates in the degradation of the cholesterol side chain. Biochemistry. 1968;7:796–807. doi: 10.1021/bi00842a038. [DOI] [PubMed] [Google Scholar]
- Sun WJ, Wang XL. Screening of substrate-resistant mutant strains producing Androst-4-ene-3,17-dione and optimization of Biotransformation medium. Food Sci. 2014;35:158–163. [Google Scholar]
- Suzuki Y, Doukyu N, Aono R. Lithocholic acid side-chain cleavage to produce 17-keto or 22-aldehyde steroids by Pseudomonas putida strain ST-491 grown in the presence of an organic solvent. diphenyl ether. Agric Biol Chem. 1998;62:2182–2188. doi: 10.1271/bbb.62.2182. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W-Y, Dong X. Microbial biotransformation: recent developments on steroid drugs. Recent Pat Biotechnol. 2009;3:141–153. doi: 10.2174/187220809788700157. [DOI] [PubMed] [Google Scholar]
- Van DGR, Hessels GI, Nienhuis-Kuiper M, Dijkhuizen L. Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9alpha-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB. Appl Environ Microbiol. 2008;74:7197–7203. doi: 10.1128/AEM.00888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FQ, Yao K, Wei DZ (2011) From soybean phytosterols to steroid hormones. Soybean Health. InTech—Open Access Publisher, Rijeka, pp 232–252
- Wilbrink MH, Petrusma M, Dijkhuizen L. Geize R.V.D. FadD19 of DSM43269, a steroid-coenzyme a ligase essential for degradation of C-24 branched sterol side chains. Appl Environ Microbiol. 2011;77:4455–4464. doi: 10.1128/AEM.00380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, Chen S. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Wang FQ, Zhang HC, Wei DZ. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab Eng. 2013;15:75–87. doi: 10.1016/j.ymben.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.