Abstract
Tea (Camellia sinensis) is one of the richest sources of flavan-3-ols, an important class of flavonoids. The expression level of gene-encoded key regulatory enzymes of flavan-3-ol/anthocyanin biosynthetic pathway, dihydroflavonol 4-reductase (DFR) and anthocyanidin reductase (ANR), has been highly correlated with the flavan-3-ol contents and antioxidant activity in tea plant. In the present study, pyramiding of CsDFR and CsANR in tobacco was achieved. However, single transgenic tobacco overexpressing either CsDFR or CsANR was documented earlier. In continuation, pyramided transgenic lines were evaluated for the possible, either same or beyond, effect on flavan-3-ol accumulation and protective ability against biotic and abiotic stresses. The pyramided transgenic lines showed early flowering and improved seed yield. The transcript levels of flavan-3-ol/anthocyanin biosynthetic pathway and related genes in pyramided transgenic lines were upregulated as compared to control tobacco plants. The accumulations of flavan-3-ols were also found to be higher in pyramided transgenic lines than control tobacco plants. In contrast, anthocyanin content was observed to be decreased in pyramided transgenic lines, while DPPH activity was higher in pyramided transgenic lines. In pyramided transgenic lines, strong protective ability against feeding by Spodoptera litura was documented. The seeds of pyramided transgenic lines were also found to have better germination rate under aluminum toxicity as compared to control tobacco plants. Interestingly, the synergistic effect of these two selected genes are not beyond from transgenic lines expressing either CsDFR and CsANR alone as published earlier in terms of flavan-3-ols accumulation. However, the unique flower color and better seed germination rate are some interestingly comparable differences that were reported in pyramided lines in relation to individual transgenic plants. In conclusion, the present results reveal an interesting dynamic between CsDFR and CsANR in modulating flavan-3-ol/anthocyanin levels and functional analysis of stacked CsDFR and CsANR transgenic tobacco lines.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0819-1) contains supplementary material, which is available to authorized users.
Keywords: Camellia sinensis, Flavan-3-ols, Transgenic tobacco, Early flowering, Antioxidants, Spodoptera litura, Aluminum toxicity
Introduction
Flavonoids represent the second largest group of plant secondary metabolites after alkaloids. Flavonoids are divided into several classes including anthocyanins and flavan-3-ols, and are abundant in different tissues of wide variety of plants (Kumar and Yadav 2013a; Ogo et al. 2013). The flavan-3-ols class of flavonoids is extensively investigated and involved in various biological processes including auxin transport regulation, seed development, pollen viability, and in defense responses under adverse conditions (Mahajan et al. 2011a, b, 2012; Falcone Ferreyra et al. 2012; Kumar and Yadav 2013b; Hammerbacher et al. 2014; Mahajan and Yadav 2014; Nakabayashi et al. 2014). The strong antioxidant activities exhibited by flavan-3-ols also contribute to tolerance against various environmental constraints (Mierziak et al. 2014; Nakabayashi et al. 2014). The flavan-3-ols are also well known for their interaction with proteins (Mierziak et al. 2014). In conclusion, flavan-3-ols, a class of flavonoids, are key metabolites for development and fitness of plants.
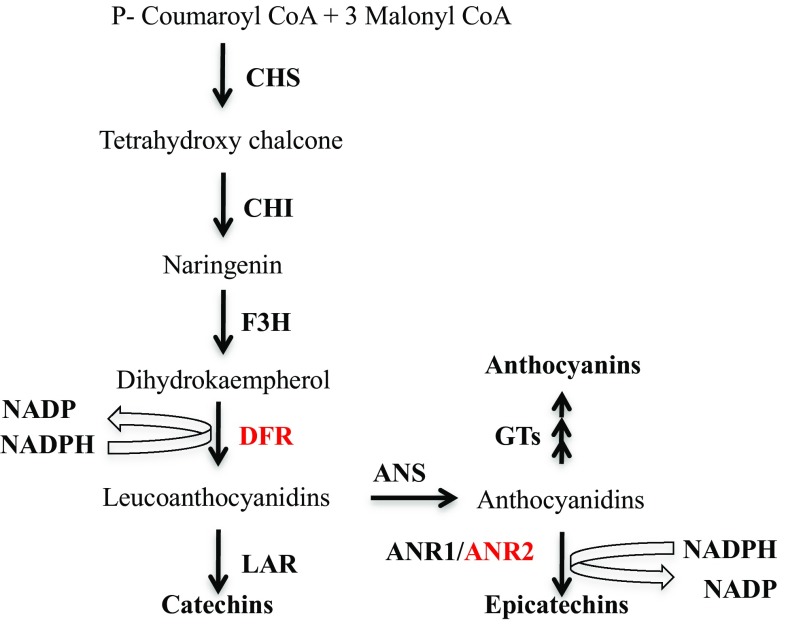
The biosynthesis of flavan-3-ols and anthocyanins is well investigated in terms of molecular genetics as well as biological mechanisms of the key regulatory enzymes in Arabidopsis (Dixon and Pasinetti 2010). The flavan-3-ol/anthocyanin biosynthetic pathway that leads to the production of flavan-3-ols and anthocyanins in plants is summarized in Fig. 1. The p-coumarate CoA, a product of phenylpropanoid pathway, condenses with three molecules of malonyl CoA to form dihydroflavonols using CHS (chalcone synthase), CHI (chalcone isomerase), and F3H (flavanone 3-hydroxylase) enzymes (Pang et al. 2013). Dihydroflavonol 4-reductase (DFR) catalyzes dihydroflavonol to leucoanthocyanidins which are further converted to anthocyanidins by ANS (anthocyanidin synthase) enzyme (Singh et al. 2009a, b). The dihydroflavonols can also be catalyzed by FLS (flavonol synthase) to produce flavonols which is a separate class of flavonoids. Therefore, DFR is considered as a branch key enzyme that controls the carbon flux direction toward biosynthesis of both flavan-3-ols and anthocyanins or either of them (Singh et al. 2009a, b). Other key branch enzymes of this pathway are identified as leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR), which catalyze the formation of catechins and epicatechins from leucoanthocyanidins and anthocyanidins, respectively (Pang et al. 2013). The anthocyanidins serve as the common substrate in the biosynthesis of flavan-3-ols as well as anthocyanins. The LAR and ANR enzymes divert the carbon flux towards biosynthesis of flavan-3-ols at the cost of anthocyanins. Among these regulatory enzymes, the evidence for LAR function is still unclear and less convincing than it is for DFR and ANR enzymes (Pang et al. 2013).
Fig. 1.
Basic representation of flavan-3-ol/anthocyanin biosynthetic pathway in plants. The enzymes are represented in abbreviated form: CHS chalcone synthase, CHI chalcone isomerase, DFR dihydroflavonol 4-reductase, F3H flavanone-3-hydroxylase, LAR leucoanthocyanidin reductase, ANR1 anthocyanin reductase1, ANR2 anthocyanin reductase 2, ANS anthocyanin synthase, GTs glucosyl transferase. The tea cDNA-encoded enzymes (red color) were used to be overexpressed simultaneously in transgenic tobacco plants
Flavan-3-ols/anthocyanins classes of flavonoids are well known for their higher antioxidant activity and are found to offer protection against major diseases in animals and plants. In addition, flavan-3-ols/anthocyanidins are also well-characterized nutrient ingredients in a number of fruits, which have attracted the attention of researchers to improve the antioxidant potential of plants by manipulations of flavan-3-ol contents (Wang et al. 2011; Kumar et al. 2013; Nakabayashi et al. 2014; Li et al. 2016; He et al. 2016; Rihani et al. 2017). The key rate-limiting enzymes of flavan-3-ol/anthocyanin biosynthetic pathway have been identified to allow their exploitation for metabolic engineering to improve the overall antioxidant potential of plants (Wang et al. 2011). A number of transcription factor genes have also been characterized for understanding the regulation mechanism of flavan-ol/anthocyanin biosynthetic pathway (Pang et al. 2013; Li et al. 2016; Pérez-Díaz et al. 2016; Rihani et al. 2017). Thus, this pathway is a special target for enhancement of antioxidant capacity via manipulation of flavan-3-ols in agricultural crops (Pang et al. 2013).
The metabolic engineering approach has highlighted two major strategies for the manipulations of flavan-3-ol contents in transgenic plants. The first strategy is to upregulate the pathway for flavan-3-ol production and second is to downregulate the competing branch pathways (Mahajan et al. 2011b, 2012; Yuan et al. 2012; Pang et al. 2013; Mierziak et al. 2014; Li et al. 2016; He et al. 2016; Lim et al. 2016; Pérez-Díaz et al. 2016; Rihani et al. 2017). The transgenic plants have been developed using either introduction of biosynthetic or regulatory genes from diverse plants to increase flavan-3-ol contents (Han et al. 2012; Hancock et al. 2012; Yuan et al. 2012; Kumar et al. 2013; Mahajan and Yadav 2014; Li et al. 2016; He et al. 2016; Pérez-Díaz et al. 2016; Rihani et al. 2017). The overexpression of DFR genes (DFR1 and DFR2) from Populus trichocarpa in tobacco and P. tomentosa has resulted in the accumulation of anthocyanins and monomeric and polymeric flavan-3-ol metabolites (Huang et al. 2012). Similarly, overexpression of wheat DFR gene increases anthocyanin accumulation in an Arabidopsis dfr mutant (Shin et al. 2016). The ANR from Malus domestica has also been investigated for flavan-3-ol content in transgenic tobacco (Han et al. 2012). Similarly, transgenic tobacco overexpressing Rosa rugosa ANR has been documented with improved tolerance against abiotic stress (Luo et al. 2016). The engineering of flavonoid biosynthetic pathway by co-expression of biosynthetic gene (ANR) and regulatory gene (PAP1) has also provided the evidence regarding the use of combinatorial approach for enhancing flavan-3-ol contents (Xie et al. 2006). On the other hand, different strategies including RNAi have been applied to downregulate the gene(s) encoding key competing enzyme(s) of biosynthetic pathway (Mahajan et al. 2011b; Jiang et al. 2013; Mahajan and Yadav 2014; Lim et al. 2016). The suppression of endogenous ANR genes (ANR1 and ANR2) has been documented with higher anthocyanin contents and lower polymeric flavan-3-ol content in soybean grain using redirection of metabolic flux (Kovinich et al. 2012). The strawberry fruits with enhanced flavan-3-ol level have been achieved through downregulation of anthocyanidin GT gene that also relies on the concept of redirection of metabolite flux toward biosynthesis of flavan-3-ols (Griesser et al. 2008). The suppression of DFR in tobacco has also been achieved with white flowers with altered flavan-3-ols/anthocyanins (Lim et al. 2016).
Nowadays, attention is being centralized on flavan-3-ol/anthocyanin biosynthetic pathway of tea plant (C. sinensis) because of extraordinary flavan-3-ol contents in their fresh foliar tissues that are being exploited for commercial purposes (Kumar et al. 2013). The genes encoding enzymes of flavan-3-ol biosynthetic pathway have been isolated and identified (Singh et al. 2009a, b; Pang et al. 2013). In continuation of these works, we also documented that the transgenic tobacco lines either CsDFR or CsANR cDNA from tea showed higher accumulation of flavan-3-ol content, early flowering, enhanced seed yield, improved total free radical scavenging activity, lesser feeding by S. litura, and better tolerance against Al toxicity (Kumar et al. 2013; Kumar and Yadav 2013b). In addition, we also correlated the relationship of CsANR overexpression in tobacco with higher accumulation of flavan-3-ols with lesser anthocyanin levels (Kumar and Yadav 2013a). Overall, these studies collectively help establish the potential role of CsANR and CsDFR in the accumulation of flavan-3-ol/anthocyanin content in tea plant.
A recombinant biosynthetic pathway carrying a cluster of CsF3H, CsDFR, and CsLAR genes has been designed and studied in relation to flavan-3-ol accumulation in E. coli (Umar et al. 2012). However, a number of studies have been conducted on the altered flavan-3-ol/anthocyanin contents in transgenic plants overexpressing individual transgene(s). Interestingly, there is a lack of studies on the combinatorial use of many genes simultaneously from plants including C. sinensis to analyze for a synchronized effect on manipulations of flavan-3-ols in plant systems. Thus, the idea of generation of pyramided transgenic lines overexpressing CsDFR and CsANR was implemented for evaluation purpose in relation to flavan-3-ol accumulation and for analyzing their combinatorial influence on the improvement of overall antioxidant potential. The pyramided transgenic tobacco lines were generated by cross-breeding approach and found to show improved flavan-3-ol contents, early flowering, better seed yield with slightly higher protection response against S. litura as compared to tobacco plants overexpressing CsDFR or CsANR alone. The pyramided transgenic lines were also documented for better germination rate under exposure of aluminum (Al) toxicity. To the best of our knowledge, the stacking of tea genes, especially genes encoding DFR and ANR, in tobacco through cross-pollination approach is the first documentation of its own kind. This work will also help to analyze the combinatorial effect of CsDFR and CSANR in pyramided transgenic lines over already published single transgenic lines, comparatively. This system also offers an opportunity to check the feasibility of pyramided transgenic approach for overaccumulation of flavan-3-ols over single transgenic lines. This report could be a step ahead toward establishing an alternative potential system for flavan-3-ol accumulation by genetic transformation approach in plant systems. Taking together, these findings indicate that elevated flavan-3-ol biosynthesis was mediated by simultaneous overexpression of CsDFR and CsANR, which thus improved the protective ability of transgenic tobacco plants under stress conditions.
Materials and methods
Plant materials, generation, and confirmation of pyramided transgenic tobaccos
Nicotiana tabacum vc. Xanthi was employed in all the experiments, because it is an excellent model system for transgenic generation and evaluation purposes. The transgenic tobacco plants overexpressing CsDFR and CsANR individually were generated by Agrobacterium tumefaciens-mediated transformation (Kumar and Yadav 2013a, b; Kumar et al. 2013). The stacking of CsDFR and CsANR in the same tobacco plant was developed using conventional breeding approach. The reciprocal crosses were obtained via fertilization of emasculated CsDFR (from collected seeds of D-15 homogenous transgenic lines)-overexpressing tobacco plants with pollens from CsANR (from collected seeds of A-05 homogenous transgenic lines)-overexpressing tobacco plants and vice versa. The D-15 (from CsDFR transgenic lines) and A-05 (from CsANR transgenic lines) lines were selected for crosses because they showed overall better response than the other generated transgenic tobacco lines (Kumar et al. 2013).
The research material was utilized from F2 generation. Pyramided transgenic F1 and F2 generation lines were selected by germination and growth on MS media with hygromycin antibiotic (50 mg/ml). Genomic DNA and semiquantitative PCR methods were used to confirm the integration and expression of CsDFR and CsANR transgenes in leaf portion of pyramided transgenic tobacco plants, respectively. The two confirmed positive pyramided transgenic lines of ♂CsDFR × ♀CsANR cross (DA-01 and DA-02) and ♂CsANR × ♀CsDFR cross (AD-01 and AD-02) were evaluated for various experiments.
RNA isolation and semiquantitative PCR
Total RNA was extracted from the leaf tissues of transgenic and control tobacco plants and was used for cDNA synthesis. RT-PCR experiments were performed (Kumar et al. 2013). The sequences of selected primers of NtPAL, NtC4H, Nt4CL, NtCHS, NtCHI, NtF3H, NtDFR, NtFLS, NtANR1, NtANR2, NtANS, NtTT1, NtTT2, and NtAN2 genes, and their respective annealing temperature are shown in Table S1. The primers corresponding to 26S rRNA-based internal control were used for semiquantitative expression analysis (Singh et al. 2004).
Evaluation of transgenic lines for morphological and yield parameters
The parameters such as days for flowering time, capsules number, seed yield, and thousand seed weight (grams) were analyzed as described previously in seven different plants with similar height from each selected line of both crosses (Kumar et al. 2013). Briefly, capsules numbers were counted at full maturation, and seed yield and thousand seed weight (grams) were measured after harvesting.
Estimation of flavan-3-ol content
Samples were prepared from the leaf tissues of selected transgenic tobacco lines and control tobacco plants and used for quantification of end products of flavan-3-ol biosynthetic pathway, flavan-3-ols by HPLC method as described previously (Kumar et al. 2013). The different flavan-3-ols, namely (+)-Cat, (−)-EC, (−)-EGC, and (−)-ECG, were used as references for estimation of respective constituent and purchased from Sigma-Aldrich, USA.
Anthocyanin content estimation
Total anthocyanin content was estimated from fresh fully opened flowers of transgenic tobacco lines and control tobacco plants (Kumar and Yadav 2013a). Briefly, anthocyanins were extracted in acidic HCl and absorbance of extracts was measured at 530 nm by UV spectrophotometer. The content was calculated as cyanidin equivalents using extinction coefficient 29.500 mol−1cm−1.
Estimation of antioxidant potential
The antioxidant activity of the methanolic extracts from leaves of pyramided transgenic tobacco plants and control tobacco plants was measured using DPPH (diphenylpicryl-hydrazyl) radical scavenging assay (Kumar et al. 2013).
Leaf disc non-choice experiment
The feeding behavior of Spodoptera litura (tobacco cutworms) was analyzed by preparing leaf discs as feeding material (6 cm diameter) from transgenic lines and control tobacco plants, and the experiment was performed as described earlier (Kumar et al. 2013).
Evaluation of seed germination under exposure of Aluminum toxicity
For germination study, control and transgenic tobacco seeds were surface sterilized, sown on half-strength Murashige and Skoog (MS) medium with 1 and 5 mM concentrations of Al in Petri dishes. Germination was recorded every day up to seven days to assess establishment of early seedlings under Al toxicity, and the plates were photographed.
Results
Generation and confirmation of pyramided transgenic tobacco plants overexpressing CsDFR and CsANR cDNAs
The homozygous transgenic individual tobacco lines of CsDFR (D-15) and CsANR (A-05) were selected for reciprocal crossing (Kumar et al. 2013). Seeds were harvested from mature pods of crossed lines of each combination (CsDFR ♂ × CsANR ♀ and CsANR ♀ × CsDFR ♂) and germinated on 0.8% agar medium. One-month-old 10–15 independent plants from two pyramided transgenic tobacco lines from both the combinations were acclimatized and shifted to greenhouse. There was no morphological difference between the CsDFR/CsANR overexpressing transgenic plants generated by reciprocal crosses with each other. Thus, there was no problem with transmission of either CsDFR or CsANR through male and female gametes. Three plants from each reciprocal cross and one plant from each of individual gene expressing transgenic line as well as control tobacco plants were used to confirm the integration and expression of CsDFR and CsANR transgenes.
The integration of CsDFR and CsANR transgenes in pyramided transgenic plants was confirmed with genomic DNA PCR using gene-specific primers. Initially, PCR with primers specific to CsDFR transgene confirmed the presence of CsDFR transgene in all selected pyramided transgenic tobacco lines as well as in selected CsDFR overexpressing transgenic tobacco plant. The CsDFR transgene was not present in selected CsANR overexpressing tobacco plant (Fig. 2a). Another PCR with primers specific to CsANR transgene confirmed the presence of CsANR transgene in all selected pyramided transgenic tobacco lines as well as in selected CsANR overexpressing transgenic tobacco plant. The CsANR transgene was not present in selected CsDFR overexpressing tobacco plant (Fig. 2a). Thus, all selected tobacco plants from both crosses were found to be PCR positive for both CsDFR and CsANR transgenes.
Fig. 2.
Confirmation of pyramided transgenic tobacco plants by Genomic DNA and semiquantitative PCR and evaluation of confirmed lines for flowering time and yield parameters. a Genomic DNA PCR was used to confirm the integration of CsDFR and CsANR genes in genome of pyramided transgenic tobacco plants. b The transcript expression levels of both (CsDFR and CsANR) in transgenic tobacco lines were carried out using semiquantitative PCR. These experiments were carried out in three technical replicates. c The pyramided transgenic lines and control were kept in transgenic containment facility. d The days to flowering (days), capsule yield, seed yield, and seed weight per thousand seed (grams) in pyramided transgenic lines generated from reciprocal crosses in relation to control tobacco plants. Mean ± SD from three replications are represented
Similarly, expressions of both CsDFR and CsANR transgenes in all selected pyramided transgenic lines were also confirmed using semiquantitative PCR. Both the transgenes were observed with their expressions in pyramided transgenic tobacco plants (Fig. 2b).
Pyramided transgenic tobacco lines overexpressing CsDFR-CsANR documented for early flowering and improved yields
The pyramided transgenic tobacco lines and control plants were morphologically analyzed for height, stem diameter, and days to flowering with significant changes. However, no significant changes in growth pattern were observed in pyramided transgenic tobacco lines as compared to control tobacco plants (Fig. 2c, d). The pyramided transgenic lines were observed for early flowering after an average of 95 (CsDFR ♂ × CsANR ♀) and 87 (CsANR ♀ × CsDFR ♂) days compared to 105 days (for control tobacco plants) after seed germination (Fig. 2d). Both reciprocal crosses did not show significant change in plant height, stem diameter, and number of leaves as compared to control tobacco plants, which were measured at the flowering time. However, the pyramided transgenic lines were documented for higher number of fruits/capsules per plant, increased seed yield per plant, and weight of seed (grams) compared to control tobacco plants. The number of fruits per plant on an average were 51.5 (CsDFR ♂ × CsANR ♀) and 55.2 (CsANR ♀ × CsDFR ♂) as compared to 40.55 in control tobacco plants (Fig. 2d). The seed yield per plant was measured as 890 (CsDFR ♂ × CsANR ♀) and 1140 (CsANR ♀ × CsDFR ♂), compared to 747 mg in control tobacco plants. Weight of thousand seeds was 94 and 88 in pyramided transgenic lines, in contrast to only 70.5 mg calculated in control tobacco plants (Fig. 2d).
Of the confirmed pyramided transgenic tobacco lines, two transgenic lines DA-01 and DA-02 (from CsDFR ♂ × CsANR ♀ cross) and AD-01 and AD-02 (from CsANR ♀ × CsDFR ♂ cross) along with control tobacco plants were subsequently used for further analysis.
Increased transcript expression of flavan-3-ol/anthocyanin biosynthetic pathway genes in pyramided transgenic tobacco plants
To check the influence of simultaneous overexpression of both CsDFR and CsANR transgenes on the transcript level of various endogenous genes of flavan-3-ol/anthocyanin biosynthetic pathway, expression analysis of the leaf tissues of DA-01, DA-02, AD-01, and AD-02 transgenic lines vis-à-vis control tobacco plants was conducted. The expression of selected flavan-3-ol/anthocyanin biosynthetic endogenous genes (NtPAL, NtCHS, NtCHI, NtF3H, NtDFR, NtFLS, NtANR1, NtANR2, NtLAR, and NtANS) was found to be upregulated in pyramided transgenic tobacco plants as compared to control tobacco plants.
The transcript expression of NtPAL gene was observed to be increased by 95, 117, 119, and 112% in DA-01, DA-02, AD-01, and AD-02 pyramided transgenic lines as compared to control tobacco plants (Fig. 3a). In pyramided transgenic tobacco lines, the transcript expression of NtCHS gene was upregulated by approximately 52% (Fig. 3b). Similarly, transcript expression of NtCHI gene was also increased by 20–49% in all selected pyramided transgenic lines (Fig. 3c). The transcript expression of NtF3H gene was increased by 97, 110, 99, and 54% in DA-01, DA-02, AD-01, and AD-02 as compared to control tobacco plants (Fig. 3d). The transcript expression of NtDFR gene was observed to be upregulated in the range of 20–40% in all selected pyramided transgenic tobaccos as compared to control tobacco plants (Fig. 3e). Similarly, NtFLS gene was observed to be increased by 26–38% in all selected pyramided transgenic lines (Fig. 3f). The transcript expression of NtANR1 gene was upregulated by 20, 28, 29, and 79% in selected pyramided lines with respect to control tobacco plants (Fig. 3g). The transcript expression of NtANR2 gene was observed to be upregulated by 12–28% in pyramided transgenic lines (Fig. 3h). Transcript expression of NtLAR gene was upregulated by 13–21% in pyramided transgenic lines as compared to control tobacco plants (Fig. 3i). Similarly, NtANS gene was observed to be upregulated in pyramided transgenic lines by 12–41% (Fig. 3j). Hence, the simultaneous overexpression of both CsDFR and CsANR transgenes in pyramided transgenic tobacco plants has led to an increase in the endogenous expression of all the flavan-3-ol/anthocyanin biosynthetic pathway genes. However, individual transgenic tobacco overexpressing either CsDFR or CsANR transgene was observed to increase the expression of only NtCHS and NtANR2 genes (Kumar et al. 2013).
Fig. 3.
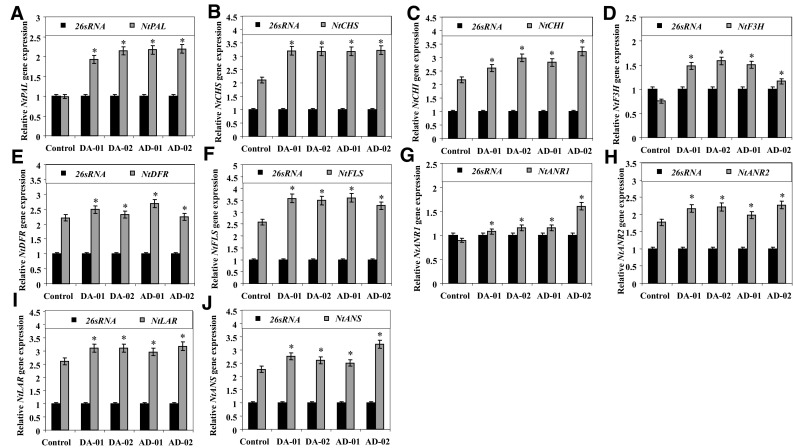
Relative transcript expression profiles of endogenous flavan-3-ol/anthocyanin biosynthetic pathway genes (a NtPAL, b NtCHS, c NtCHI, d NtF3H, e NtDFR, f NtFLS, g NtANR1, h NtANR2, i NtLAR, j NtANS) in pyramided transgenic tobacco lines vis-à-vis control tobacco plants. The expression level of 26S rRNA was used as reference control for semiquantitative expression analysis, and these experiments were repeated three times. Error bars indicate SD (n = 3), while asterisks indicate statistical significance (P < 0.05)
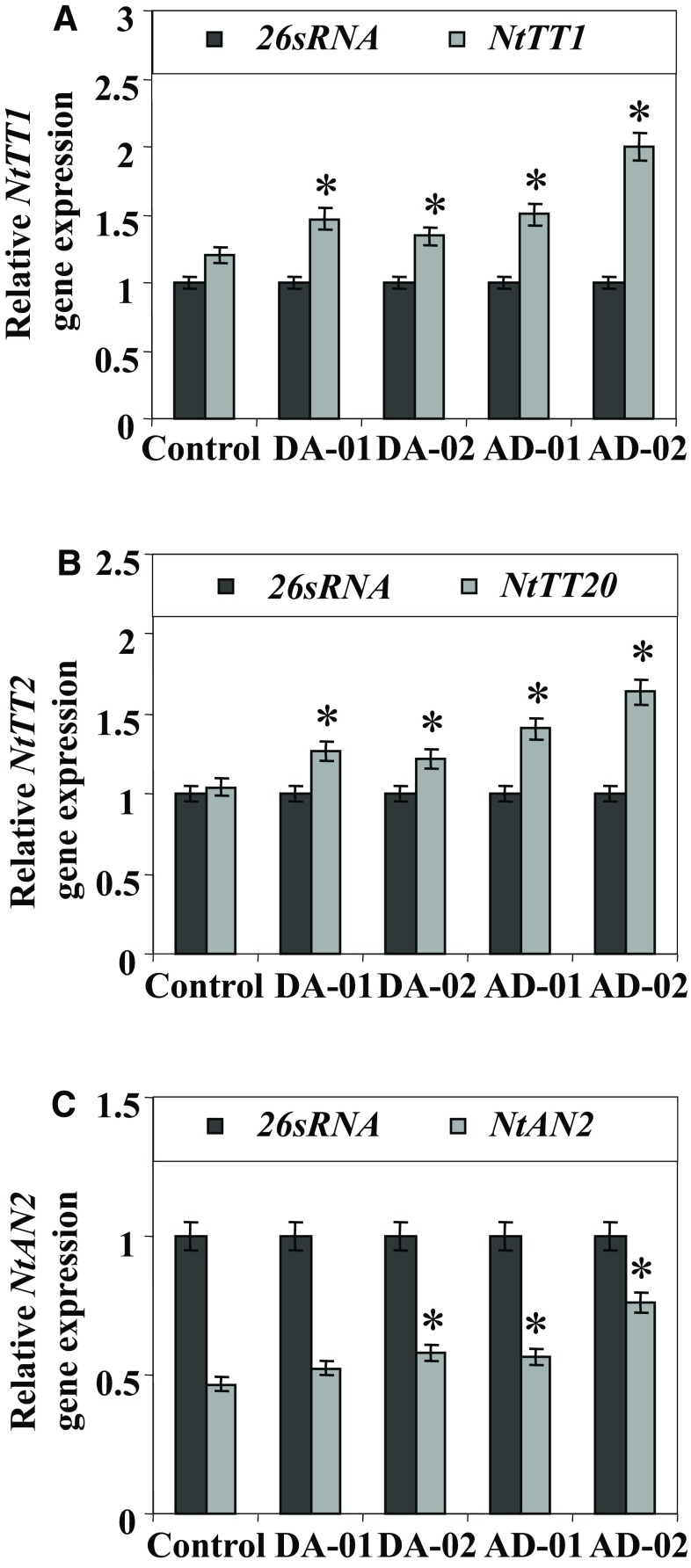
Similarly, the transcript expressions of gene-encoded flavan-3-ol/anthocyanin-related important transcription factors (NtTT1, NtTT20, and NtAN2) are also analyzed in all selected pyramided transgenic lines vis-à-vis control tobacco plants. The transcript expression of NtTT1 gene was observed to be increased by 122, 112, 125, and 167% in DA-01, DA-02, AD-01, and AD-02 as compared to control tobacco plants (Fig. 4a). The transcript expression of NtTT20 gene was upregulated by 117–157% in pyramided transgenic lines as compared to control tobacco plants (Fig. 4b). The transcript expression of NtAN2 was found to be upregulated by 106–154% in selected lines as compared to control tobacco plant (Fig. 4c).
Fig. 4.
Transcript expression levels of NtTT1 a, NtTT2, b and NtANR2 c in pyramided transgenic tobacco lines. The 26S rRNA was used as internal control for relative expression analysis. Error bars indicate SD (n = 3), while asterisks indicate statistical significance (P < 0.05)
Improved flavan-3-ol and altered anthocyanin contents in pyramided transgenic tobacco plants
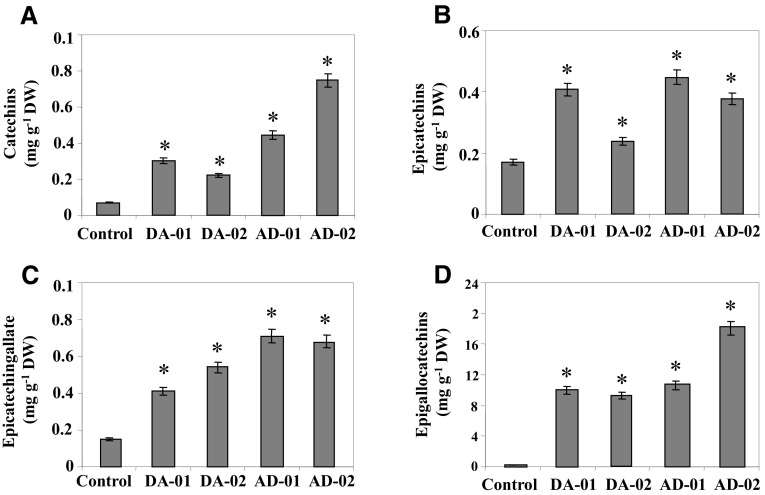
To check the influence of simultaneous expression of both CsDFR/CsANR transgenes on flavan-3-ol accumulation in pyramided transgenic tobacco plants, Cat, EC, ECG, and EGC contents were estimated by HPLC.
The Cat, EC, ECG, and EGC were found to be increased in all selected transgenic lines. The level of Cat was found to be 0.3 (DA-01), 0.22 (DA-02), 0.45 (AD-01), and 0.74 (AD-02) mg g−1 DW in pyramided transgenic lines, while it was detected at very low level (0.007 mg g−1 DW) in control tobacco plants (Fig. 5a). The content of EC was estimated to be 0.41 (DA-01), 0.24 (DA-02), 0.45 (AD-01), and 0.38 (AD-02) mg g−1 DW, while only 0.167 mg g−1 DW content was reported in control tobacco plants (Fig. 5b). The ECG content was measured as 0.41 (DA-01), 0.54 (DA-02), 0.71 (AD-01), and 0.68 (AD-02) mg g−1 DW as compared to 0.15 mg g−1 DW content in control tobacco plants (Fig. 5c). The EGC content was reported as 10 (DA-01), 9.4 (DA-02), 10.8 (AD-01), and 18.1 (AD-02) mg g−1 DW in pyramided transgenic lines, while it was reported to be only 0.022 mg g−1 DW in control tobacco plants (Fig. 5d). The HPLC chromatograms indicating identified peaks for different studied flavan-3-ols (Cat, EC, ECG, and EGC) with respect to their standards are documented in Figure S1.
Fig. 5.
Flavan-3-ol content in pyramided transgenic lines. Cat (a), EC (B), ECG (C), and EGC (D) content in pyramided transgenic lines vis-à-vis control tobacco plants. Error bars indicate SD (n = 3), while asterisks indicate statistical significance (P < 0.05)
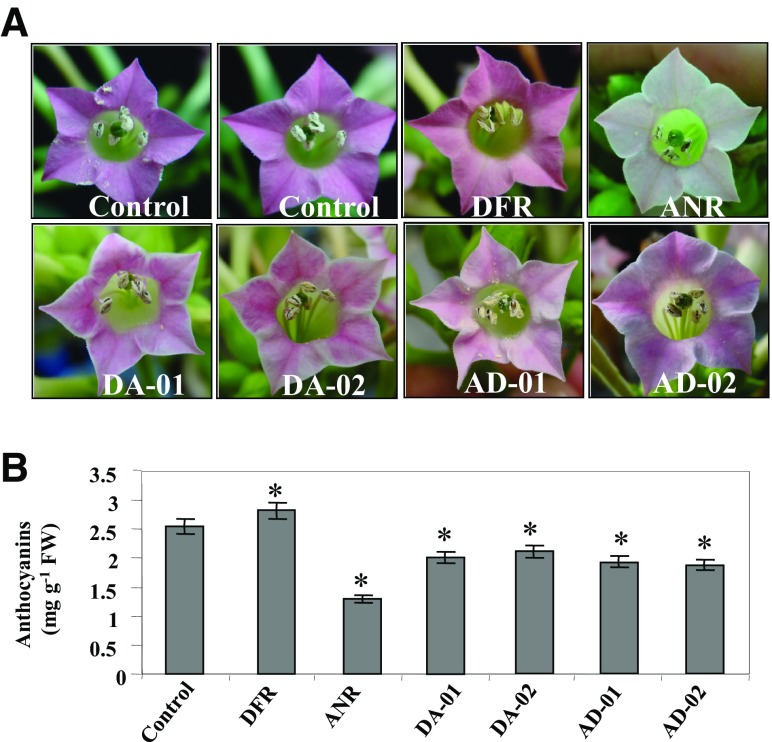
The difference in flower colors of pyramided transgenic lines (DA-01, DA-02, AD-01, and AD-02) from that of transgenic lines overexpressing either CsDFR or CsANR alone and control tobacco flower(s) is shown in Fig. 6. The flowers of pyramided transgenic lines showed altered pigmentation of pink and white patches. The colors of flowers of transgenic CsDFR and CsANR were dark pink and white, respectively. The flowers of control tobacco plant were reported to be light pink in color. On the basis of this phenotypic observation, anthocyanin contents of flowers of pyramided and single transgenic tobacco plants vis-à-vis control tobacco flowers were estimated. The anthocyanin contents were estimated as 2.1 (DA-01), 2.2 (DA-02), 1.9 (AD-01), and 1.85 (AD-02) per gram fresh weight (mg/g FW) vis-à-vis 2.8 (CsDFR) and 1.16 (CsANR) mg/g FW (Fig. 6). The anthocyanin content of wild tobacco flower was also estimated as 2.55 mg/g FW (Fig. 6).
Fig. 6.
Anthocyanin content in flowers of transgenic lines vis-à-vis control tobacco plants. The upper panel represents the colors of flowers in transgenic vis-à-vis control tobacco plants. The petals of flowers from CsDFR were more pinkish in color, in contrast to flowers of control tobacco plants. The petals of CsANR flowers were documented white in color. The petal part of all flowers from selected pyramided transgenic lines showed altered pigmentation of white and pink patches. The lower panel documents the anthocyanin content in transgenic lines vis-à-vis control tobacco plants. Error bars indicate SD (n = 3), while asterisks indicate statistical significance (P < 0.05)
Improved antioxidant potential under the influence of CsDFR and CsANR overexpression in pyramided transgenic tobacco plants
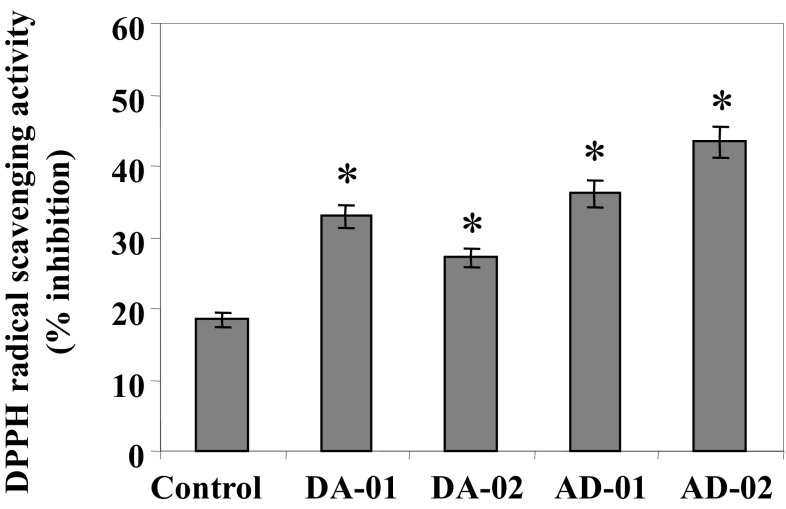
To check the influence of higher flavan-3-ol accumulation on antioxidant activity of pyramided transgenic tobacco plants, free radical scavenging activity was measured using DPPH assay. The methanolic extracts of the lyophilized leaf samples from pyramided transgenic lines showed 79% (DA-01), 47% (DA-02), 96% (AD-01), and 136% (AD-02) increase in DPPH free radical scavenging activity with respect to control tobacco plants (Fig. 7).
Fig. 7.
DPPH free radical scavenging activity in pyramided transgenic lines vis-à-vis control tobacco plants. Error bars indicate SD (n = 3), while asterisks indicate statistical significance (P < 0.05)
Increased protection ability of pyramided transgenic tobacco plants against S. litura attack
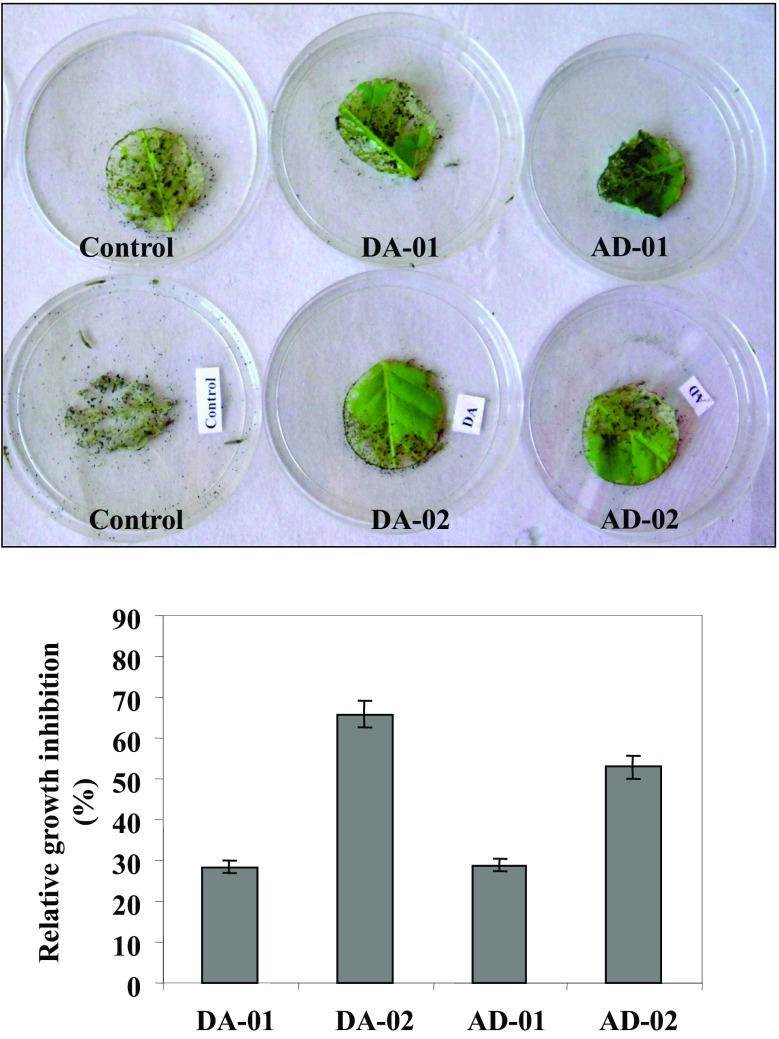
To check the anti-feeding effect of S. litura on pyramided transgenic tobacco plants, the non-choice method was used as adopted previously (kumar et al. 2013). The leaf disc of pyramided transgenic lines showed lesser feeding by larvae of S. litura as compared to leaf discs of control tobacco plants (Fig. 8). The lesser feeding response could be due to deterrent effect of accumulated flavan-3-ols in leaf discs as offered by non-choice method.
Fig. 8.
The anti-herbivore effect of pyramided transgenic tobacco lines against S. litura. The upper panel shows pictures of lesser feeding on leaf materials of pyramided transgenic lines by S. litura relative to control plants. The lower panel represents the improved relative percentage growth inhibition of S. litura after feeding on leaf materials from pyramided transgenic tobacco lines as compared to control. Error bars indicate SD (n = 3)
Hence, the growth retardation and lesser survival of S. litura larvae were also observed when fed on leaf discs of pyramided transgenic lines as compared to control tobacco plants. Percentage growth inhibition of larvae of S. litura that randomly fed on leaf discs of DA-01, DA-02, AD-01, AD-02 transgenic lines were 28, 65, 28, and 52%, respectively, as compared to the growth of other larvae that non-choicely fed on leaf discs of control tobacco plants (Fig. 8). The retarded growth of S. litura larvae feeding on leaf discs of pyramided transgenic lines has also indicated the decreased vigor of S. litura larvae.
Pyramided transgenic tobacco plants showed better seed germination rate under toxic Al stress condition
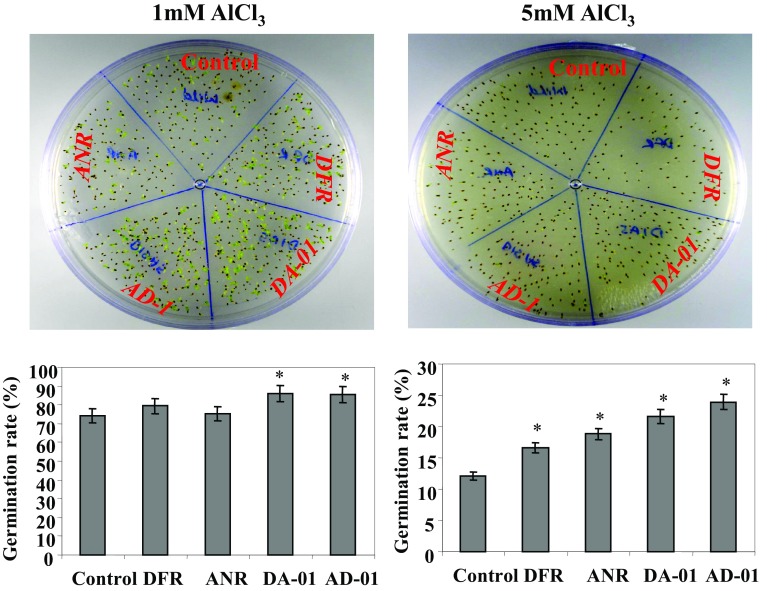
To check the influence of overexpression of transgenes CsDFR and CsANR on the germination behavior of seeds under Al exposure (1 mM and 5 mM), the germination rates of the seeds of selected pyramided transgenic lines (AD-01 and DA-01), individual transgenic lines (CsDFR and CsANR), and control tobaccos were calculated 4 days after initiating imbibition of seeds. The seed germination rates are 85.8% (DA-01) and 85.7% (AD-01) compared to 79.5% (CsDFR), 75.2% (CsANR), and 74.4% in control tobaccos under exposure of 1 mM Al toxicity. At 5 mM exposure of toxic Al, the rates of seed germination were 21.6% (DA-01) and 23.0% (AD-01), 16.6% (CsDFR), 18.8% (CsANR), and 12.11% in control tobaccos (Fig. 9). Significant differences in germination were observed in seeds of pyramided transgenic lines compared to individual transgenic lines as well as control tobaccos and a combinatorial role of CsDFR/CsANR was confirmed in the tolerance phenotype under toxic Al exposure.
Fig. 9.
Influence of toxic Al exposure on seed germination from seeds of transgenic tobacco lines vis-à-vis control tobacco plant. The upper panel documents the pictures of a 4-day-old seedlings of CsDFR and CsANR transgenic lines vis-à-vis control tobaccos under 1 and 5 mM exposure of Al toxicity. The lower panel represents the rate of seed germination from transgenic lines vis-à-vis control tobacco plant after 4 days of initiating imbibition of seeds
Discussion
The anthocyanin and flavan-3-ol contents share the same metabolic intermediate and represent the extensively investigated biosynthetic pathway in plants. An effective transformation system for higher accumulation of flavan-3-ol content should rely on redirection of carbon flux toward biosynthesis of flavan-3-ols with higher accumulation of intermediate precursors. This could be achieved by either overexpression of genes encoding for DFR, ANR, LAR, F3H proteins, and silencing of gene encoding for FLS and ANS proteins at the point of carbon flux redirection due to their central position in the flavan-3-ol/anthocyanin biosynthetic pathway. Manipulation of flavan-3-ol/anthocyanin biosynthetic pathway has been shown to have the potential for improving flavan-3-ol content in plants (Mahajan et al. 2011b; Kumar et al. 2013; Mahajan and Yadav 2014: Hammerbacher et al. 2014; Li et al. 2016; He et al. 2016; Lim et al. 2016; Pérez-Díaz et al. 2016; Rihani et al. 2017). The transgenic flax plants overexpressing genes encoding CHS, CHI, and DFR have been crossed with transgenic flax line(s) that overexpressed the glucosyltransferase (GT) gene to improve protective ability against pathogen infection (Kostyn et al. 2012; Mierziak et al. 2014). The genes encoding DFR and ANR proteins were chosen at the point of redirection based upon their key regulatory roles in flavan-3-ol/anthocyanin biosynthetic pathway (Singh et al. 2009a, b, Kumar et al. 2013; Kumar and Yadav 2013a, b; Shin et al. 2016; Lim et al. 2016). The overexpression of CsDFR and CsANR has been documented with improved flavan-3-ol content in transgenic tobacco plants (Kumar et al. 2013; Kumar and Yadav 2013a, b).
After growing body of knowledge, it was our goal to achieve the optimum level of flavan-3-ol accumulation with improved tolerance against biotic and abiotic stresses through pyramiding of CsDFR and CsANR in transgenic tobacco plants relative to either transgene alone. The reciprocal crossing of CsDFR and CsANR overexpressing transgenic tobacco plants generated these pyramided transgenic tobacco lines. In continuation, the pyramided transgenic tobacco plants also showed early flowering, improved capsule numbers, and thousand seed weight compared with control tobacco plants. Mahajan et al. (2011b) have reported the correlation of NtFLS silencing in transgenic tobacco with retarded height, delayed flowering, and lesser yield compared to control tobacco plants. In continuation, different mutants from the same biosynthetic pathway have also been documented for their role in the overall architecture of plant, numbers of shoot/flower, and seed weight (Buer et al. 2010). Thus, the present study provides strong evidence to establish the correlation between higher accumulation of flavan-3-ol content with early flowering and improved seed yield. However, early flowering is generally considered to have negative effect on yield but overexpression of CsDFR and CsANR affects the flowering timing that can also have significant impact on post-flowering processes affecting fertility and pod development (Buer et al. 2010; Mahajan et al. 2011b; Kumar and Yadav 2013a). The other possible reason could be the modulation of phytohormone levels as experimentally proved earlier (Buer and Djordjevic 2009; Kumar et al. 2013). However, there is no significant difference in the above-said phenotypic results (early flowering, capsule numbers, and seed weight) of pyramided transgenic tobacco with respect to transgenic plants overexpressing CsDFR or CsANR alone. The reason could be that an optimal level of favan-3-ols that is induced in individual transgenic lines alone is enough for early flowering and other changes as documented earlier. However, the flower color phenotype of pyramided transgenic lines is significantly changed as compared to transgenic plants overexpressing CsDFR or CsANR alone and control tobacco plants.
Metabolic engineering for higher accumulation of flavan-3-ols is highly coordinated with significant changes in expression levels of genes associated with the flavan-3-ols/anthocyanin biosynthetic pathway under normal and stress conditions (Han et al. 2012; Kumar et al. 2013; Mahajan and Yadav 2014; Hammerbacher et al. 2014; Li et al. 2016; Luo et al. 2016; Pérez-Díaz et al. 2016). The overexpression of apple ANR gene has been observed to upregulate the expressions of only NtCHI and NtDFR genes of flavan-3-ol/anthocyanin biosynthetic pathway (Han et al. 2012). The overexpression of either CsDFR or CsANR has also been reported to upregulate the expression of only NtCHS and NtANR genes (Kumar et al. 2013). Thus, these reports have suggested that overexpression of a specific transgene is only responsible for change in expression of a set of related endogenous genes. On the other hand, the most striking results of the present study are that overexpressions of CsDFR and CsANR genes have triggered significant changes in the transcript levels of endogenous flavan-3-ol/anthocyanin biosynthetic pathway genes encoding NtPAL, NtCHS, NtCHI, NtF3H, NtDFR, NtFLS, NtANR1, NtANR2, NtLAR, and NtANS proteins as well as genes encoding NtTT1 (WIP1/Zn finger), NtTT2 (MYB), and NtAN2 (MYB) transcription factors. In general, like other regulators, these regulators (TT1, TT2, and AN2) could be positive target for induction of signals for higher accumulation of flavan-3-ols (Hichri et al. 2011; Xu et al. 2015). These regulators activate the expression levels of genes encoding enzymes of flavan-3-ol/anthocyanin biosynthetic pathway by binding to their respective promoter sites directly or indirectly (Hichri et al. 2011; Xu et al. 2015). In advance, the expression profiles of flavan-3-ol/anthocyanin biosynthetic pathway-related gene(s) associated with either overexpression or silencing of transcription regulator(s) is a strong evidence for transcriptional regulation of flavan-3-ol/anthocyanin biosynthetic pathway (Hichri et al. 2011; Xu et al. 2015). The constitutive simultaneous expressions of CsDFR and CsANR under 35S promoter resulted in accumulation of flavan-3-ols that could worked as a part of positive feedback loop and further upregulated the expression levels of flavan-3-ol/anthocyanin biosynthetic pathway genes. The tea-specific metabolites of flavan-3-ols, Cat, EC, ECG, and EGC have been reported to be higher in transgenic lines compared to control tobacco plants. The manipulation of flavan-3-ol content has been proved to be linked with modulation of biosynthesis of anthocyanins. The anthocyanin content has been found to be altered in flowers of pyramided transgenic lines as compared to control as well as transgenic plants overexpressing single gene alone (Kumar and Yadav 2013a). Tobacco flowers have been extensively studied as models for flavan-3-ol/anthocyanin biosynthesis and petal color is the most important indicator of anthocyanin content (Nakatsuka et al. 2012; Kumar and Yadav 2013a; Zhou et al. 2013). Altered flower color has also been reported earlier via manipulation of flavan-3-ol/anthocyanin biosynthetic pathway (Tanaka et al. 2010; Nishihara and Nakatsuka 2011; Nakatsuka et al. 2012; Kumar and Yadav 2013a; Zhou et al. 2013). The downregulation of Torenia NtF3′H as well as NtFLS genes and simultaneous overexpression of Gerbera DFR gene in tobacco has been reported to increase only anthocyanin biosynthesis (Nakatsuka et al. 2007). Similarly, knockdown of F3′H and F3′5′H genes with overexpression of rose DFR gene in Torenia violet cultivar resulted in flowers with darker pink petals with elevated level of anthocyanins (Nakamura et al. 2010). This report suggested that DFR encoded protein might be regulatory to redirect the flavan-3-ol/anthocyanin biosynthetic pathway towards biosynthesis of anthocyanins and their precursors. The inverse correlation between anthocyanins and flavan-3-ol levels in the tobacco flowers probably reflects fine-tuning between these two branches of flavonoid biosynthetic pathway. The other important enzyme ANR protein which is also a key rate-limiting enzyme catalyzes the biosynthesis of EC (flavan-3-ols) from anthocyanidin substrate (Zhang et al. 2012). The overexpression of ANR encoding protein in tobacco has diverted the carbon flux toward biosynthesis of EC, monomeric flavan-3-ols instead of anthocyanin biosynthesis. So, ANR catalyzed reaction plays a crucial role in determining the levels of anthocyanins and EC in flower (Xie et al. 2004; Han et al. 2012; Kumar and Yadav 2013a). The white flowers with decreased anthocyanin contents have been reported in CsANR transgenic tobacco plants (Kumar and Yadav 2013a; Han et al. 2012). So these studies have suggested that CsANR overexpression diverted the utilization of anthocyanidins toward biosynthesis of flavan-3-ols, thereby decreasing anthocyanin content. In the present study, the altered pigmentation has also been documented in flowers of transgenic lines. The anthocyanin contents in pyramided transgenic lines have also been measured at intermediate level in-between individual transgenic tobacco lines overexpressing either CsDFR (high) or CsANR (low) alone. The alterations of flower pigmentations and decrease in anthocyanin content are highly correlated with co-expression of CsDFR and CsANR in pyramided transgenic tobacco plants.
The aim of this study was also to improve antioxidant potential by increasing the flavan-3-ol content with co-expression of CsDFR and CsANR transgene simultaneously over single transgenic lines. The monomeric and polymeric forms of flavan-3-ols are best known for their antioxidant properties and their potential protective benefits against biotic and abiotic stresses (Buer et al. 2010; Falcone Ferreyra et al. 2012; Mierziak et al. 2014; Nakabayashi et al. 2014; Luo et al. 2016). The transgenic tobaccos overexpressing CsDFR and CsANR individually have also been documented for higher antioxidant properties (Kumar et al. 2013). In this study, we estimated DPPH radical scavenging activity that was found to be slightly higher than transgenic tobacco overexpressing CsDFR or CsANR transgene alone. The improved flavan-3-ol content with higher in vitro antioxidant activity has been inversely correlated with reactive oxygen-mediated cell death in CsDFR and CsANR transgenic lines as experimentally proved earlier (Kumar et al. 2013). Similarly, overexpression of ANR from Rosa rugosa has also been documented with improved tolerance to abiotic stress through elevated ROS scavenging activity (Luo et al. 2016). So, the strategy to improve the antioxidant potential of plants via overaccumulation of flavan-3-ols could be an effective strategy for tolerance against biotic and abiotic stresses (Kumar et al. 2013; Nakabayashi et al. 2014; Meng et al. 2015; Mitsunami et al. 2014; Mahajan and Yadav 2014). The flavan-3-ols play an important role in plant herbivore interaction or against pathogen infections (Porth et al. 2011; Barbehenn and Constabel 2011; Kumar et al. 2013; Mahajan and Yadav 2014; Mitsunami et al. 2014; Hammerbacher et al. 2014). Transgenic tobaccos overexpressing gene-encoded key enzymes involved in flavan-3-ol/anthocyanin biosynthetic pathway have been documented with better tolerance to biotic stresses, including herbivore (Misra et al. 2010; Kumar et al. 2013; Mitsunami et al. 2014) and fungal infection (Mahajan and Yadav 2014; Mierziak et al. 2014). So, evaluation of pyramided transgenic lines for their protective ability against feeding by S. litura was done. The defensive ability of pyramided transgenic plants was found to be improved against the generalist herbivore S. litura. The combinatorial approach indicates that simultaneous overexpression of CsDFR and CsANR slightly improves the tolerance against feeding by S. litura relative to single transgene alone. The abiotic environmental constraints also induce higher accumulation of flavan-3-ols/anthocyanins in plants (Kusano et al. 2011; Yuan et al. 2012; Meng et al. 2015). The flavan-3-ol/anthocyanin metabolites scavenge the accumulated reactive oxygen species triggered by abiotic stresses. A few reports have also discussed the correlation between flavan-3-ol/anthocyanin accumulation with strong radical scavenging and tolerance to abiotic stresses such as UV-B irradiation, salt, drought, water stresses, chilling stress, and against Al toxicity (Kusano et al. 2011; Yuan et al. 2012; Kumar and Yadav 2013b; Mahajan and Yadav 2014; Nakabayashi et al. 2014; Meng et al. 2015).
In addition, extraordinary amount of flavan-3-ol content makes tea an Al-hyperaccumulator plant and encourages for assessment of aluminum toxicity as reported earlier (Osaha et al. 2011; Kumar and Yadav 2013a). In the present study, we also reported the germination rate of seeds from pyramided transgenic tobacco was found to be significantly higher under Al exposure. The higher amount of flavonoids in tea is well known for providing tolerance against Al though detoxified by potential antioxidant mechanism (Mukhopadyay et al. 2012). Seedlings of transgenic tobacco plants overexpressing CsDFR and CsANR transgene alone relative to control were documented for better growth under exposure of Al toxicity (Kumar and Yadav 2013b). These observations confirm the usefulness of flavan-3-ols for enhancing both biotic and abiotic stress tolerance in pyramided transgenic tobacco plants. Thus, we could demonstrate that gene stacking approach is promising for flavan-3-ol production in plants. However, the optimum level of flavan-3-ol accumulation with improved tolerance against biotic and abiotic stresses through simultaneous overexpression of CsDFR and CsANR in transgenic tobacco plants is not beyond the individual transgenic lines.
Conclusions
In conclusion, this report demonstrated the coordinated expression of CsDFR and CsANR in tobacco with early flowering, flavan-3-ol accumulation, and higher DPPH scavenging activity, which not only protected against feeding of S. litura but also showed better seed germination under Al toxicity. Since the single transgenic lines have been studied and the work was published, it would be important here to know the significance of CsDFR-CsANR-stacked tobacco transgenic over single transgenic lines. The overall results documented from pyramided transgenic lines were much better than those reported earlier from individual transgenic tobacco plants of either CsDFR or CsANR. In continuation, this work also emphasizes the potential role of CsDFR- and CsANR-encoded proteins in fine-tuning of anthocyanin accumulation in flower and germination rate of seeds under Al exposure. Further work is needed to investigate how the end product of flavan-3-ol/anthocyanin biosynthetic pathway is recognized by the plant cell and triggers subsequent mechanisms that control the optimum level of flavan-3-ols. In addition to stacking of CsDFR and CsANR, there is also need for identification of any potential regulatory gene(s), to design a novel strategy for maximum accumulation of antioxidant flavan-3-ol contents in agriculturally important crops including cotton, flax, maize, and rice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1 Sequences of primers used in the present study. Figure S1 HPLC spectra details of standard (Catechin, Cat; Epicatechin, EC; Epicatechin gallate, ECG; Epigallocatechin, EGC) and identification of similar peaks in pyramided transgenic tobacco lines as compared to control tobacco plant in left and right side, respectively (PPTX 150 kb)
Acknowledgements
The authors are thankful to the Director, CSIR-Institute of Himalayan Bioresource Technology, Palampur for providing the necessary facility to conduct the research and valuable suggestions during course of work. Thanks are due to Dr. Gireesh Nadda for expert technical assistances in anti-feeding experiment. The Council of Scientific and Industrial Research (CSIR), GOI sponsored this work under NMITLI program (TLP003). VK is also thankful to CSIR for award of SRF.
Abbreviations
- 4CL
4-coumarate: CoA ligase
- ABTS
2,2-azinobis (3-ethyl-benzothiazoline-6-sulfonic acid)
- AN2
Anthocyanin 2
- ANR1
Anthocyanidin reductase 1
- ANR2
Anthocyanidin reductase 2
- ANS
Anthocyanidin synthase
- C4H
Chalcone-4-hydrolase
- Cat
Catechin
- cDNA
Complementary deoxyribonucleic acid
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- DFR
Dihydroflavonol 4-reductase
- DPPH
2,2-diphenyl-1-picryl-hydrazyl
- EC
Epicatechin
- ECG
Epicatechin gallate
- EGC
Epigallocatechin
- F3H
Flavanone 3-hydroxylase
- FLS
Flavonol synthase
- MS
Murashige and Skoog medium
- Nt
Nicotiana tabacum
- PAL
Phenylalanine lyase
- PCR
Polymerase chain reaction
- RT
Reverse transcriptase
- ROS
Reactive oxygen species
- TT1
Transparent testa 1
- TT2
Transparent testa 2
Author contribution
VK and SKY conceived and designed present research. VK conducted experiments. VK and SKY analyzed data. VK wrote the manuscript. All authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0819-1) contains supplementary material, which is available to authorized users.
References
- Barbehenn RV, Constabel CP. Tannins in plant-herbivore interactions. Phytochemistry. 2011;72:1551–1565. doi: 10.1016/j.phytochem.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Buer CS, Djordjevic A. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot. 2009;60:751–763. doi: 10.1093/jxb/ern323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djoordjevic A. Flavonoids: new roles for old molecules. J Integr Plant Biol. 2010;52:98–111. doi: 10.1111/j.1744-7909.2010.00905.x. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Pasinetti GM. Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol. 2010;154:453–457. doi: 10.1104/pp.110.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra MN, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser M, Hoffmann T, Bellido ML, Rosati C, Fink B, Kurtzer R, Aharoni A, Munoz-Blanco J, Schwab W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008;146:1528–1539. doi: 10.1104/pp.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbacher A, Paetz C, Wright LP, Fischer TC, Bohlmann J, Davis AJ, Fenning TM, Gershenzon J, Schmidt Axel. Flavan-3-ols in Norway spruce: biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol. 2014;164:2107–2122. doi: 10.1104/pp.113.232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Vimolmangkang S, Soria-Guerra RE, Korban SS. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J Exp Bot. 2012;63:2437–2447. doi: 10.1093/jxb/err415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S. Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol. 2012;159:1204–1220. doi: 10.1104/pp.112.195420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Li Y, Lawson D, De-Yu Xie. Metabolic engineering of anthocyanins in dark tobacco varieties. Physiol Plant. 2016;159:2–12. doi: 10.1111/ppl.12475. [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gou J, Jia Z, Yang L, Sun Y, Xiao X, Song F, Luo K. Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS One. 2012;7:e30364. doi: 10.1371/journal.pone.0030364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Wang JY, Jia HF, Jia WS, Wang HQ, Xiao M. RNAi-mediated silencing of the flavanone 3-hydrooxylase gene and its effect on flavonoid biosynthesis in strawberry fruit. J Plant Growth Regul. 2013;32:182–190. doi: 10.1007/s00344-012-9289-1. [DOI] [Google Scholar]
- Kostyn K, Czemplik M, Kulma A, Bortniczuk M, Skala J, Szopa J. Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci. 2012;190:103–115. doi: 10.1016/j.plantsci.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Kovinich N, Saleem A, Rintoul TL, Brown DCW, Arnason JT, Miki B. Coloring genetically modified soybeans grains with anthocyanins by suppression of the proanthocyanidins genes ANR1 and ANR2. Transgenic Res. 2012;21:757–771. doi: 10.1007/s11248-011-9566-y. [DOI] [PubMed] [Google Scholar]
- Kumar V, Yadav SK. Overexpression of CsANR increased flavan-3-ols and decreased anthocyanins in transgenic tobacco. Mol Biotechnol. 2013;54:426–435. doi: 10.1007/s12033-012-9580-1. [DOI] [PubMed] [Google Scholar]
- Kumar V, Yadav SK. Overexpression of CsDFR and CsANR enhanced root flavonoids and improved root architecture to provide tolerance against aluminum toxicity in tobacco. Plant Root. 2013;7:65–76. doi: 10.3117/plantroot.7.65. [DOI] [Google Scholar]
- Kumar V, Nadda G, Kumar S, Yadav SK. Transgenic tobacco overexpressing tea cDNA encoding dihydroflavonol 4-reductase and anthocyanidin reductase induces early flowering and provides biotic stress tolerance. PLoS One. 2013;8:e65535. doi: 10.1371/journal.pone.0065535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, Kondou Y, Goto H, Kawashima M, Matsuda F, Niida R, Matsui M, Saito K, Fernie AR. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 2011;67:354–369. doi: 10.1111/j.1365-313X.2011.04599.x. [DOI] [PubMed] [Google Scholar]
- Li P, Dong Q, Ge S, He X, Verdier J, Li D, Zhao J. Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanin precursor flux in legume. Plant Biotechnol J. 2016;14:1604–1618. doi: 10.1111/pbi.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SH, You MK, Kim DH, Kim JK, Lee JY, Ha SH. RNAi-mediated suppression of dihydroflavonol 4-reductase in tobacco allows fine-tuning of flower color and flux through the flavonoid biosynthetic pathway. Plant Physiol Biochem. 2016;109:482–490. doi: 10.1016/j.plaphy.2016.10.028. [DOI] [PubMed] [Google Scholar]
- Luo P, Shen Y, Jin S, Huang S, Cheng X, Wang Z, Li P, Zhao J, Bao M, Ning G. Overexpression of Rosa rugosa anthocyanin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016;245:35–49. doi: 10.1016/j.plantsci.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Mahajan M, Yadav SK. Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol Biol. 2014;85:551–573. doi: 10.1007/s11103-014-0203-z. [DOI] [PubMed] [Google Scholar]
- Mahajan M, Ahuja PS, Yadav SK. Post- transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS One. 2011;6:e28315. doi: 10.1371/journal.pone.0028315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M, Kumar V, Yadav SK. Effects of flavonoid-mediated free IAA regulation on growth and development of in vitro-grown tobacco seedlings. Int J Plant Dev Biol. 2011;5:42–48. [Google Scholar]
- Mahajan M, Joshi R, Gulati A, Yadav SK. Increase in flavan-3-ols by silencing flavonol synthase mRNA affects the transcript expression and activity levels of antioxidant enzymes in tobacco. Plant Biol. 2012;14:725–733. doi: 10.1111/j.1438-8677.2011.00550.x. [DOI] [PubMed] [Google Scholar]
- Meng C, Zhang S, Deng YS, Wang GD, Kong FY. Overexpression of a tomato flavanone 3-hydroxylase-like protein gene improves chilling tolerance in tobacco. Plant Physiol Biochem. 2015;96:388–400. doi: 10.1016/j.plaphy.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Mierziak J, Kostya K, Kulma A. Flavonoids as important molecules of plant interactions with the environment. Molecules. 2014;19:16240–16265. doi: 10.3390/molecules191016240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra P, Pandey A, Tiwari M, Chandrashekar K, Sidhu OP, Asif MH, Chakrabarty D, Singh PK, Trivedi PK, Nath P, Tuli R. Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 2010;152:2258–2268. doi: 10.1104/pp.109.150979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunami T, Nishihara M, Galis I, Alamgir KM, Hojo Y, Fujita K, Sasaki N, Nemoto K, Sawassaki T, Arimura G. Overexpression of the PAP1 transcription factor reveals a complex regulation of flavonoid and phenylpropanoid metabolism in Nicotiana tabacum plants attacked by Spodoptera litura. PLoS One. 2014;9:e108849. doi: 10.1371/journal.pone.0108849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadyay M, Bantawa P, Das A, Sarkar B, Bera B, Ghosh P, Mondal TK. Changes of growth, photosynthesis and alteration of leaf antioxidative defence system of tea [Camellia sinensis (L.) O. Kuntze] seedlings under aluminium stress. Biometals. 2012;25:1141–1154. doi: 10.1007/s10534-012-9576-0. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Fukuchi-Mizutani M, Fukui Y, Ishiguro K, Suzuki K, Suzuki H, Okazaki K, Shibata D, Tanaka Y. Generation of pink flower varieties from blue Torenia hybrida by redirecting the flavonoid biosynthesis pathway from delphinidin to pelargonidin. Plant Biotechnol. 2010;27:375–383. doi: 10.5511/plantbiotechnology.10.0610a. [DOI] [Google Scholar]
- Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M. Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthesis genes. Plant Cell Rep. 2007;26:1951–1959. doi: 10.1007/s00299-007-0401-0. [DOI] [PubMed] [Google Scholar]
- Nakatsuka T, Saito M, Yamada E, Fujita K, Kakizaki Y, Nishihara M. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologs of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot. 2012;63:6505–6517. doi: 10.1093/jxb/ers306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara M, Nakatsuka T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotech Lett. 2011;33:433–441. doi: 10.1007/s10529-010-0461-z. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Ozawa K, Ishimaru T, Murayama T, Takaiwa F. Transgenic rice seed synthesizing diverse flavonoids at high level: a new platform for flavonoid production with associated health benefits. Plant Biotechnol J. 2013;11:734–746. doi: 10.1111/pbi.12064. [DOI] [PubMed] [Google Scholar]
- Osaha H, Endo I, Hara Y, Matsushima Y, Tange T. Transient proliferation of proanthocyanidin-accumulating cells in the epidermal apex contributes to highly aluminum-resistant root elongation in camphor tree. Plant Physiol. 2011;155:433–446. doi: 10.1104/pp.110.166967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Abeysinghe ISB, He J, He X, Huhman D, Mewan KM, Sumner LW, Yun J, Dixon RA. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013;161:1103–1116. doi: 10.1104/pp.112.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Díaz JR, Pérez-Díaz J, Madrid-Espinoza J, González-Villanueva E, Moreno Y, Ruiz-Lara S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol Biol. 2016;90:63–76. doi: 10.1007/s11103-015-0394-y. [DOI] [PubMed] [Google Scholar]
- Porth I, Hamberger B, White R, Ritland K. Defense mechanisms against herbivory in Picea: sequence evolution and expression regulation of gene family members in the phenylpropanoid pathway. BMC Genom. 2011;12:608. doi: 10.1186/1471-2164-12-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihani KAL, Jaconsen H-J, Hofmaan T, Schwab W, Hassan F. Metabolic engineering of apple by overexpression of the MdMyb10 gene. J Genet Eng Biotechnol. 2017 doi: 10.1016/j.jgeb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Choi M-G, Kang C-S, Park C-S, Choi S-B, Park Y-L. Overexpressing the wheat dihydroflavonol 4-reductase gene TaDFR increases anthocyanin accumulation in an Arabidopsis dfr mutant. Genes Genom. 2016;38:333–340. doi: 10.1007/s13258-015-0373-3. [DOI] [Google Scholar]
- Singh K, Raizada J, Bhardwaj P, Ghawana S, Rani A, Singh H. 26S rRNA-based internal control gene primer pair for reverse transcription-polymerase chain reaction-based quantitative expression studies in diverse plant species. Anal Biochem. 2004;335:330–333. doi: 10.1016/j.ab.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Singh K, Kumar S, Yadav SK, Ahuja PS. Characterization of dihydroflavonol 4- reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze] Plant Biotechnol Rep. 2009;3:95–101. doi: 10.1007/s11816-008-0079-y. [DOI] [Google Scholar]
- Singh K, Rani A, Paul A, Dutt S, Joshi R, Gulati A, Ahuja PS, Kumar S. Differential display mediated cloning of anthocyanidin reductase gene from tea (Camellia sinensis) and its relationship with the concentration of epicatechin. Tree Physiol. 2009;29:837–846. doi: 10.1093/treephys/tpp022. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F, Kale G, Senior M, Dyson B, Nakamura N, Katsumoto Y, Chandler S. Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosci Biotechnol Biochem. 2010;74:1760–1769. doi: 10.1271/bbb.100358. [DOI] [PubMed] [Google Scholar]
- Umar KM, Abdulkarim SM, Radu S, Hamid AA, Saari N. Engineering the production of major catechins by Escherichia coli carrying metabolite genes of Camellia sinensis. Sci World J. 2012;2012(519031):1–7. doi: 10.1100/2012/529031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen S, Yu O. Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol. 2011;91:949–956. doi: 10.1007/s00253-011-3449-2. [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Dixon RA. Anthocyanidin reductase from Medicago truncatula and Arabidopsis thaliana. Achiev Biochem Biophys. 2004;422:91–102. doi: 10.1016/j.abb.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma B, Wright E, Wang ZY, Dixon RA. Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J. 2006;456:895–907. doi: 10.1111/j.1365-313X.2006.02655.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Liu Y, Wu C, Chen S, Wang Z, Yang Z, Shauanshuang Q, Huang L. Water deficit affected flavonoid accumulation by regulating hormone metabolism in Scutellaria baicalensis Georgi roots. PLoS One. 2012;7:e42946. doi: 10.1371/journal.pone.0042946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Y, Gao K, Zhao L, Wang Y, Sun M, Gao L, Xia T. Characterization of anthocyanidin reductase from shuchazao green tea. J Sci Food Agric. 2012;92:1533–1539. doi: 10.1002/jsfa.4739. [DOI] [PubMed] [Google Scholar]
- Zhou X-W, Fan Z-Q, Chen Y, Zhu Y-L, Li J-Y, Yin H-F. Functional analyses of a flavonol synthase–like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation. J Biosci. 2013;38:593–604. doi: 10.1007/s12038-013-9339-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Sequences of primers used in the present study. Figure S1 HPLC spectra details of standard (Catechin, Cat; Epicatechin, EC; Epicatechin gallate, ECG; Epigallocatechin, EGC) and identification of similar peaks in pyramided transgenic tobacco lines as compared to control tobacco plant in left and right side, respectively (PPTX 150 kb)