Abstract
BACKGROUND: SIRT1 is a longevity gene that forestalls aging and age-related diseases including cancer, and has recently attracted widespread attention due to its overexpression in some cancers. We previously identified the overexpression of SIRT1 in ovarian carcinoma (OvCa) as a poor prognostic factor. However, mechanistic insights into the function of SIRT1 in OvCa have yet to be elucidated. METHODS: Quantitative real-time reverse PCR (qRT-PCR) and Western blotting were employed to examine the expression of SIRT1 in a panel of human OvCa cell lines. si-RNA or sh-RNA and cDNA technologies were utilized to knockdown or overexpress SIRT1, respectively. The effects of SIRT1 on proliferation and chemoresistance were examined using a WST-1 assay, and the underlying mechanisms were confirmed using an apoptotic assay, and the quantification of glutathione (GSH), and reactive oxygen species (ROS). The aggressiveness of SIRT1 was analyzed using in vitro invasion and migration assays. RESULTS: SIRT1 was more strongly expressed in OvCa cell lines than in the immortalized ovarian epithelium at the gene and protein levels. Stress up-regulated the expression of SIRT1 in dose- and time-dependent manners. SIRT1 significantly enhanced the proliferation (P < .05), chemoresistance (P < .05), and aggressiveness of OvCa cells by up-regulating multiple antioxidant pathways to inhibit oxidative stress. Further study into the overexpression of SIRT1 demonstrated the up-regulation of several stemness-associated genes and enrichment of CD44v9 via an as-yet-unidentified pathway. CONCLUSIONS: Our results suggest that SIRT1 plays a role in the acquisition of aggressiveness and chemoresistance by OvCa, and has potential as a therapeutic target for OvCa.
Introduction
Ovarian carcinoma (OvCa), primarily epithelial OvCa, is the eighth most common cause of cancer deaths in women worldwide [1]. In Japan, the incidence of epithelial OvCa, particularly endometriosis-associated OvCa such as clear cell carcinoma and endometrioid carcinoma, has markedly increased and continues to increase over that in Asian and Western countries [2].
Current treatments for OvCa include debulking surgery and adjuvant platinum-based chemotherapy. These treatment approaches have offered minimal survival benefits [1] due to increased recurrence and drug resistance, which lead to treatment failures [3]. The recurrence and drug resistance of OvCa have been linked to cancer stem cells (CSCs) [4], [5]. CSCs have been shown to possess a self-renewal ability, multi-lineage capabilities, and resistance to therapy by forming a significant residual of disease after therapy [6]. Among the proposed mechanisms responsible for CSC resistance, tolerance against oxidative stress has attracted a lot of attention [7]. Oxidative stress occurs once the production of reactive oxygen species (ROS) outweighs a cell's defense system comprising antioxidants and redox regulators [8]. Thus, the function-based mechanisms of CSCs need to be elucidated in more detail in order to identify novel therapeutic targets against chemoresistant/recurrent OvCa.
Sirtuins (SIRTs; SIRT1-SIRT7) are NAD (+) -dependent histone deacetylases that forestall aging and age-associated diseases in a broad range of organisms, from yeast to mammals [9]. SIRT1 has been reported to modulate the enzymatic activity of normal and diseased cells, including cancer cells [9]. Nevertheless, SIRT1 is a double-edged sword because it functions as an oncogene as well as a tumor suppressor [10]. SIRT1 deacetylates histone and non-histone targets (P53), thereby regulating cell cycle progression, apoptosis, cell senescence, and oxidative stress resistance, which allows cells to bypass cell-cycle control, leading to tumorigenesis [11], [12].
SIRT1 plays a crucial role in maintaining the proliferation/self-renewal abilities and pluripotency of embryonic stem cells [4], [5]. Previous studies reported that the associated stemness of SIRT1 was due to the control of p53 activity, which negatively modulates Nanog [13] or Oct4 expression [14].
Several studies have linked SIRT1 to cancer stemness, and CSCs have also been associated with resistance to conventional therapy. Therefore, SIRT1 is at a crossroads in the targeting of CSCs, recurrence, and drug resistance. A clearer understanding of the cellular survival mechanisms utilized by SIRT1 is important for developing novel treatment strategies to complement conventional therapies.
In the present study, using OvCa as a cancer model, we demonstrate the role of SIRT1 in the development of OvCa aggressiveness and chemoresistance.
Materials and Methods
Cell Lines and Culture Conditions
Human OvCa cell lines: IGROV-1, SKOV3, OVCAR3, ES2, and TOV112D, were purchased from ATCC (Rockville, MD), RMG1 was from Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan), and A2780 and its cisplatin-resistant derivative, A2780CDDP were kindly donated by Dr. Takashi Tsuruo (Cancer Chemotherapy Center, Tokyo, Japan). The immortalized ovarian surface epithelium cell line (OSE7E) was a kind gift from Dr. Hidetaka Katabuchi (Kumamoto University, Kumamoto, Japan) and was maintained in Dulbecco's modified Eagles/F12 medium (Gibco, St. Louis, MO). ES2 cells were maintained in McCoy 5A medium (Gibco, St. Louis, MO), RMG1 cells were maintained in F12 medium (Life Technologies, Carlsbad, CA), and A2780, A2780CDDP, OVCAR3, and IGROV-1 cells were maintained in RPM1 1640 medium (Gibco, St. Louis, MO). All cells were supplemented with 10% inactivated fetal bovine serum (Gibco, St. Louis, MO) and cultured at 37 °C in a humidified atmosphere containing 5% CO2. All cells were classified based on histology in Supplementary Table S1.
Cell Transfection and Selection
SIRT1-specific siRNA and scrambled siRNA (control), plasmids expressing SIRT1 short hairpin RNA (shRNA) or scrambled shRNA (control), and vectors expressing SIRT1 cDNA or an empty vector (control) (Origene, Rockville, MD) were used. Lipofectamine 2000 (Life Technologies, Carlsbad, CA) was used for plasmid transfection into cell lines as per the manufacturer's instructions. SIRT1-specific shRNA and cDNA colonies were selected by puromycin (Enzo Life Sciences, Farmingdale, NY, USA) or geneticin (EMD Millipore, Darmstadt, Germany), respectively. The sequences for SIRT1 shRNA (1, 2, and 3) or scrambled shRNA, and SIRT1 siRNA (A, B, and C) or scrambled siRNA are listed in Supplementary Table S2. Unless specified, SIRT1-siRNA (sequence C) named as “siSIRT1” and SIRT1-shRNA (sequence 1) named as “shSIRT1” were utilized.
Western Blotting
Protein was extracted from human OvCa cell lines following a previously described protocol [15]. Briefly, in Western blotting assays, equal amounts of protein extracts were subjected to 10% SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. After blocking for 1 hour with 5% skim milk, membranes were incubated with primary antibodies against SIRT1 (rabbit polyclonal; Cell Signaling, Danvers, MA, USA), HO-1 (rabbit polyclonal; Cell Signaling, Danvers, MA, USA), xCT (rabbit polyclonal; Abcam, USA), CD44v9 (rat monoclonal; Cosmo Bio, Tokyo, Japan), thioredoxin (rabbit polyclonal; Proteintech, USA), and beta-actin (ACTB) (mouse monoclonal; Sigma-Aldrich, St. Louis, MO, USA). Their corresponding peroxidase-labeled secondary antibodies were used for Western blotting. Detection was performed using ECL reagents (Amersham, Piscataway, NJ, USA) according to the manufacturer's guidelines.
SIRT1 Activity Assay
SIRT1 deacetylation activity was measured using a SIRT1 Activity Assay Kit (Fluorometric) (Abcam) as described previously [16] and analyzed in a microplate reader according to the manufacturer's guidelines. For this experiment, SIRT1 protein was concentrated by immunoprecipitation using protein A agarose beads (Santa Cruz, Dallas, TX, USA). OvCa cells with either endogenous low SIRT1 mRNA expression (ES2 cells), or high SIRT1 mRNA expression, with known cisplatin resistance (A2780CDDP cells) were selected for analyzing SIRT1 activity.
PCR Analysis
Total RNA was isolated using TRIzol reagent (Life Technologies), and complementary DNA (cDNA) synthesis and quantitative PCR (qPCR) were performed as previously described [17] using the PrimeScript RT-PCR kit (Takara Bio, Shiga, Japan) according to the manufacturer's instructions. The sequences of primers used were as follows: SIRT1 5’TCA GTG TCA TGG TTC CTT TGC-3′ (Forward), 5′-AAT CTG CTC CTT TGC CAC TCT-3′ (Reverse), ACTB 5′-GAC AGG ATG CAG AAG GAG ATT ACT-3′ (Forward), and 5′ –TGA TCC ACA TCT GCT GGA AGG T-3′ (Reverse) [16], and other primer sequences are listed in Supplementary Table S2 [8], [17], [18]. The Fold changes in target genes against the housekeeping gene (ACTB) were assessed using the ΔΔ cycle threshold (2-ΔΔCt) method. Data was representative of three independent experiments with eight replicates.
Drug Treatments
Anticancer drugs: paclitaxel (PTX) (Wako, Osaka, Japan) and cisplatin (CDDP) (Sigma-Aldrich, St. Louis, MO, USA), and a SIRT1 inhibitor (EX527) (Selleckchem, TX, USA) were dissolved in dimethylformamide (DMFA). In in vitro experiments, various concentrations of PTX and CDDP were added to cells for a fixed period between 0 to 72 hours and cytotoxicity was analyzed accordingly.
Cell Proliferation and Chemoresistance Assay (WST-1)
The WST-1 assay was performed to analyze the proliferation and chemoresistance of ovarian carcinoma cells. As previously described [19], cells at a density of 500–6000 cells/well were seeded on 96-well microplates. After confirmation of cells attaching to the bottom of wells, WST-1 assay was performed for 4 consecutive days in order to evaluate proliferation. To evaluate chemoresistance, the anti-cancer drugs or the selective SIRT1 inhibitor (EX527) were added into culture media, and then WST-1 assay was performed after 72 hours of incubation. These assays were done using WST-1 reagent (Roche Diagnostics, Basel, Switzerland) at 37 °C for 2.5 hours, optical density (at 450 nm) was measured using microplate reader (SYNERGY HT, Bio-Tek, Winooski, VT), and the viability rate was calculated. Data was representative of three independent experiments with 16 replicates.
Glutathione (GSH) and ROS Assays
Cells were seeded in dark-colored, flat-bottomed 96-well plates. In GSH assays, cells at a density of 5 x 103 cells/well were treated with GSH-Glo reagent (Promega, Madison, WI, USA) as stipulated by the manufacturer and luminescence was analyzed in a microplate reader. In ROS assays, cells at a density of 2 x 104 cells/well were stained with 10 μM of dichlorodihydrofluorescein diacetate (DCF-DA) (Sigma-Aldrich, St. Louis, MO, USA) for 45 minutes, treated with a ROS inducer (5 μM of CDDP) and/or ROS scavenger [5 or 10 mM of N-Acetyl-L-cysteine (NAC) (Wako, Osaka, Japan)] for 4 hours, and then analyzed in a microplate reader. We utilized a previously described protocol for GSH and ROS analyses [18]. Data was representative of three independent experiments with 4 replicates.
Soft Agar Colony Formation Assay
Agar was prepared as previously described [16]. Briefly, the bottom of each 60 mm dish (Corning, New York, NY, USA) was prepared by adding 3 ml of agar medium (1.5 ml of 1% agar and 1.5 ml of McCoy 5A with 20% FBS) with EX527 (10-6 M) or vehicle (0 M) and kept at room temperature to solidify. ES2-Con and ES2-SIRT1 cells pretreated with EX527 or vehicle for 24 hours were resuspended in McCoy with 10% FBS and EX527 or vehicle at a density of 1500 cells/ml, 1 ml of resuspended cells were mixed with 2 ml of agar medium containing EX527 or vehicle, and 2 ml of that mixture was layered on the top of the solidified bottom agar in each 60 mm-dish (1000 cells/dish) and maintained at 37 °C for 4 weeks. Then, the dishes were stained with crystal violet (0.04%), and the number of colonies was quantified. The data was representative of three independent experiments with 3 replicates.
In Vitro Migration and Invasion Assays
Migratory and invasive assays were performed as previously described [20]. Briefly, Matrigel inserts (Corning BioCoat Matrigel Invasion Chamber) were rehydrated as per the manufacturer's instructions, and a control membrane (Corning BioCoat Control Insert (No ECM)) was used for the migration assay. Cells (1 x 104) with serum-free medium were placed onto the upper chamber, medium with 10% FBS was placed into the lower chamber as a chemoattractant, and cells were incubated at 37 °C for 24 hours in a 5% CO2 incubator. Cells remaining on the upper side of the filter were wiped off with a cotton swab, and cells that had migrated to the underside of the membrane were fixed and stained (Diff-Quick, Sysmex, Kobe, Japan) as per the manufacturer's instructions and counted in four randomly selected microscopic fields. Migration and invasive activities were expressed as the mean number of migrated or invaded cells in four randomly selected high-power fields per chamber. The data was representative of three independent experiments.
Apoptosis Analysis
Cell apoptosis analyses were performed using the Annexin V Fluos staining kit (Roche Diagnostics, Basel, Switzerland) or Aposcreen Annexin V-PE (Southern Biotech, Birmingham, USA) for GFP-incorporated cell lines as per the manufacturer's protocol. Briefly, cells were collected and washed twice with cold PBS and resuspended in a 100 μl suspension of binding buffer with Annexin V and Propidium iodide (PI) or 7-Amino-Actinomycin D (7AAD) and incubated at room temperature for 15 min. Samples were then diluted with 400 μl of binding buffer and analyzed on a flow cytometer (BD FACS CANTO Becton, Dickinson, and Company). Data was representative of three independent experiments.
Statistical Analysis
Statistical analyses using SPSS Statistical software (IBM, Armonk, NY, USA) and the graphing software Excel (Microsoft, USA) were employed to analyze all data. We utilized the Student's t-test to compare mean values between two data sets and an ANOVA test to compare more than two data sets. All values were reported as the mean ± SD.
Results
SIRT1 was More Strongly Expressed in OvCa Cell Lines Than in OSE7E Cells
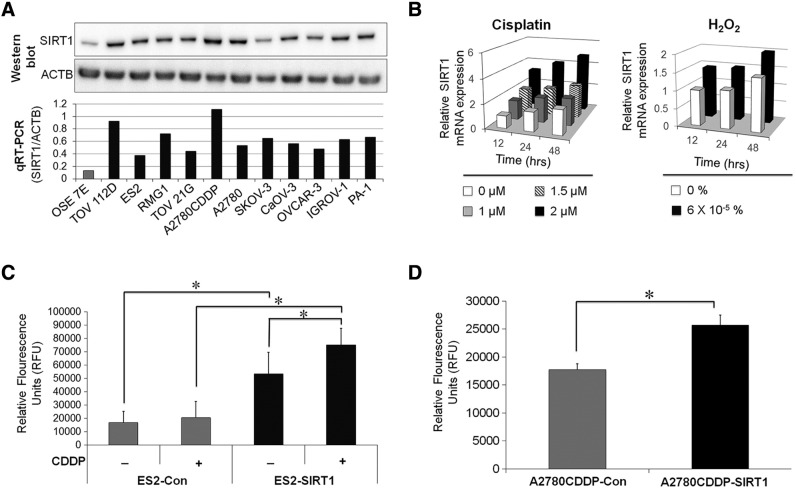
The expression of SIRT1 was assessed in a panel of OvCa cell lines and OSE7E cells. Western blotting and PCR results showed that the expression of SIRT1 was markedly stronger in OvCa cells than in OSE7E cells (Figure 1A). The expression of SIRT1 was up-regulated after exposure to various stressors such as cisplatin and hydrogen peroxide (Figure 1B) in dose- and time-dependent manners. Furthermore, the deacetylation activity of SIRT1 was up-regulated by the forced expression of SIRT1 (Figure 1, C and D) and cellular exposure to stress (Figure 1C). These results indicated that the elevated expression of SIRT1 by stresses or cDNA transfection enhanced the deacetylation activity of SIRT1 in OvCa cells.
Figure 1.
A: The expression of SIRT1 protein and mRNA was evaluated by Western blot and real-time quantitative RT-PCR (qRT-PCR) respectively. SIRT1 was more strongly expressed in OvCa cell lines than in an immortalized ovarian surface epithelium. B: The expression of SIRT1 mRNA in ES2 cells treated with cisplatin or hydrogen peroxide (H2O2) was evaluated by real-time qRT-PCR. Cytotoxic stresses such as cisplatin and H2O2 enhanced SIRT1 expression. C and D: The results of SIRT1 deacetylation activity assay of ES2 and A2780CDDP cells transfected with either SIRT1 cDNA (ES2-SIRT1/A2780CDDP-SIRT1) or the corresponding empty vector (ES2-Con/A2780CDDP-Con) as a control. SIRT1-overexpressing cells exhibited greater SIRT1 activity than control cells. CDDP treatment (5 μM for 24 hours) elevated SIRT1 activity in ES2 cells. Significance: * P < .05, significantly different from the controls.
SIRT1 Knockdown Decreased the Proliferation of OvCa Cells
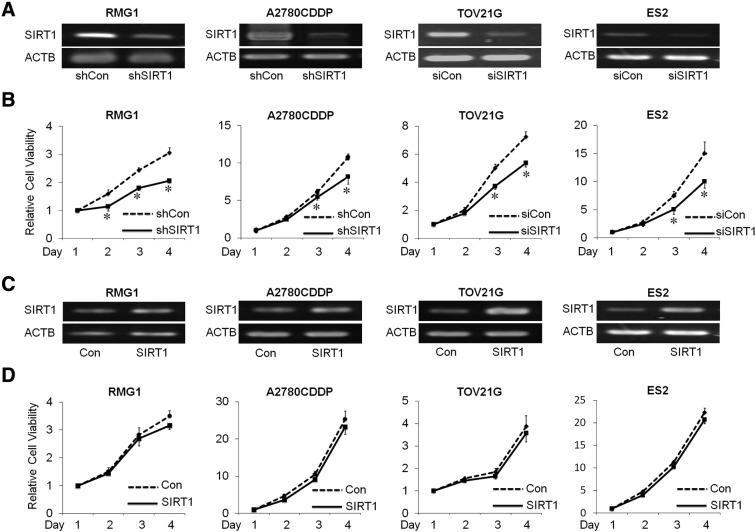
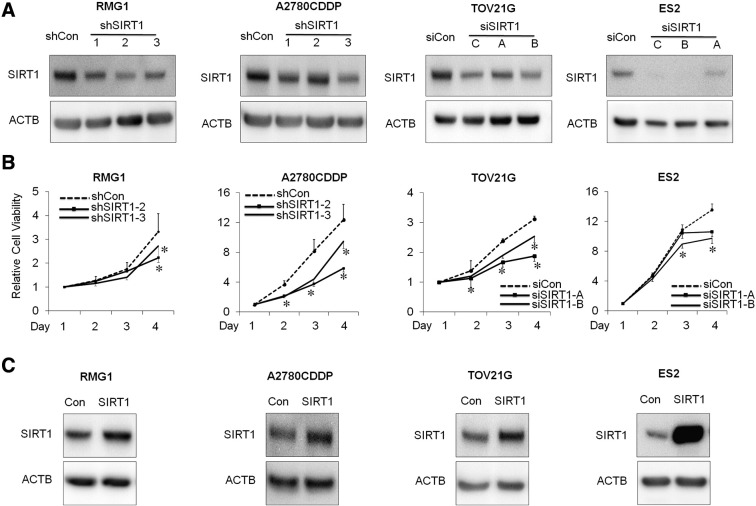
In order to analyze the function of SIRT1 in OvCa, the expression of SIRT1 was knocked down by either SIRT1-specific siRNA or shRNA (Figure 2A and Supplementary Figure S3A). The effects of SIRT1 on proliferation were measured by the WST-1 assay, and the results obtained revealed that the proliferation of SIRT1-silenced cells (si-SIRT1 and sh-SIRT1) was significantly lower than that in control cells (si-Con and sh-Con) (RMG1, A2780CDDP, and TOV21G, ES2 cells, P < .05; Figure 2B) (Supplementary Figure S3B). In an attempt to confirm these results, we overexpressed SIRT1 using SIRT1 cDNA to generate stable SIRT1-overexpressing RMG1, A2780CDDP, TOV21G and ES2 cells (−SIRT1) or corresponding empty vector (−Con) (Figure 2C and Supplementary Figure S3C). In contrast to our expectations, the overexpression of SIRT1 had no effect on the proliferation of OvCa cells until 72 hours. (Figure 2D).
Figure 2.
A: The expression of SIRT1 mRNA in RMG1, A2780CDDP, TOV21G, and ES2 cells transfected with either shRNA, siRNA sequences to knock down SIRT1 (shSIRT1 and siSIRT1), or scramble sequences as control (siCon and shCon). B: The effect of SIRT1 knockdown on cell proliferation was assessed using the WST-1 assay. Results were independently normalized by day 1. The knockdown of SIRT1 significantly decreased proliferation in OvCa cells (RMG1, A2780CDDP, TOV21G, and ES2). C: The expression of SIRT1 mRNA in RMG1, A2780CDDP, TOV21G, and ES2 cells transfected with SIRT1 cDNA (SIRT1) to overexpress SIRT1, or empty vector as control (Con) D: The effect of SIRT1 overexpression on cell proliferation were assessed using the WST-1 assay. Results were independently normalized by day 1. The overexpression of SIRT1 had no effect on the proliferation of OvCa cells. Significance: * P < .05, significantly different from the controls.
Supplementary Figure S3.
A: Western blotting of SIRT1 and ACTB. The expression of SIRT1 protein in RMG1, A2780CDDP, TOV21G, and ES2 cells transfected with either shRNAs (shSIRT1–1 [shSIRT1], shSIRT1–2, and shSIRT1–3), siRNAs (siSIRT1-A, siSIRT1-B, and siSIRT1-C [siSIRT1]) sequences was suppressed compared with control cells transfected with the scrambled sequences (siCon and shCon). B: The effect of SIRT1 knockdown on cell proliferation was assessed using the WST-1 assay. Results were independently normalized by day 1. The knockdown of SIRT1 significantly decreased proliferation in OvCa cells (RMG1, A2780CDDP, TOV21G, and ES2). Results of shSIRT1–1 (shSIRT1) in RMG1 and A2780CDDP, and siSIRT1-C (siSIRT1) in TOV21G, and ES2 were shown in Fig. 2B. C: Western blotting of SIRT1 and ACTB. The expression of SIRT1 protein in RMG1, A2780CDDP, TOV21G, and ES2 cells transfected with SIRT1 cDNA (SIRT1) was enhanced compared with control cells transfected with empty vector (Con). Significance: * P < .05, significantly different from the controls.
Effects of SIRT1 on Tumor Formation Ability and The Expression of Stemness-Associated Genes
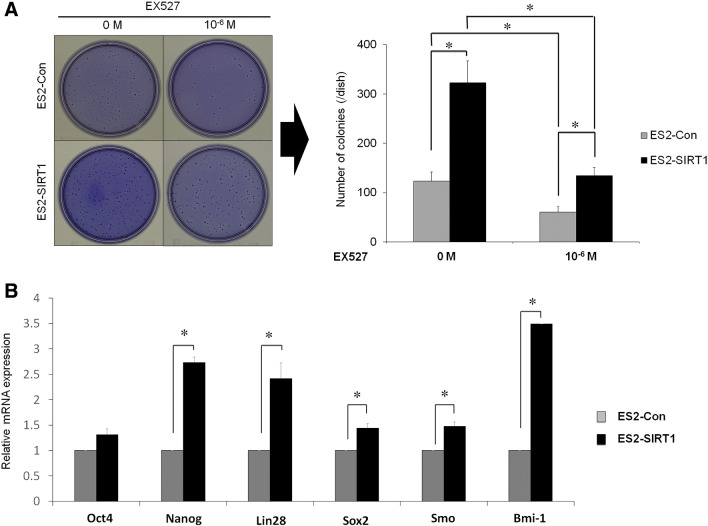
We performed soft-agar colony formation assays to further examine the effects of SIRT1 overexpression on proliferation. In ES2 cells, the overexpression of SIRT1 significantly increased colony formation abilities over those of the controls (Figure 3A, P < .05), and this effect was canceled out by the addition of the selective SIRT1 inhibitor, EX527. Then we analyzed the effect of SIRT1 on the expression of several stemness-associated genes (Oct4, Nanog, Lin28, Sox2, Smo, and Bmi-1) in ES2 cells. The qRT-PCR showed a significant increase in the mRNA levels of these genes except for Oct4 following the overexpression of SIRT1 (Figure 3B, P < .05). These results suggest that SIRT1 enhanced the tumor formation ability and increased the expression of several stemness-associated genes in OvCa cells.
Figure 3.
A: Effects of SIRT1 overexpression on the tumor formation ability. Representative photographs and graphic illustrations of ES2 cells with the overexpression of SIRT1 by SIRT1 cDNA (ES2-SIRT1) or an empty vector as control (ES2-Con). Tumor formation ability was assessed using the soft agar colony formation assay. The overexpression of SIRT1 (ES2-SIRT1) significantly increased the colony formation of ES2-SIRT1 cells over that of the control (ES2-Con), and the number of colonies was significantly decreased by the SIRT1 inhibitor (EX 527) in both ES2-SIRT1 and ES2-Con. B: Effects of SIRT1 overexpression on the expression of stemness associated genes. The mRNA levels for ES2-SIRT1 and ES2-Con were analyzed for the association with stemness-associated genes (Oct4, Nanog, Lin28, Sox2, Smo and Bmi-1) on a quantitative real-time PCR analysis (qRT-PCR). The qRT-PCR results show a significant increase in the mRNA levels of Nanog, Lin28, Sox2, Smo and Bmi-1 following the overexpression of SIRT1. Significance: * P < .05.
SIRT1 Enhanced the Chemoresistance of OvCa Cells
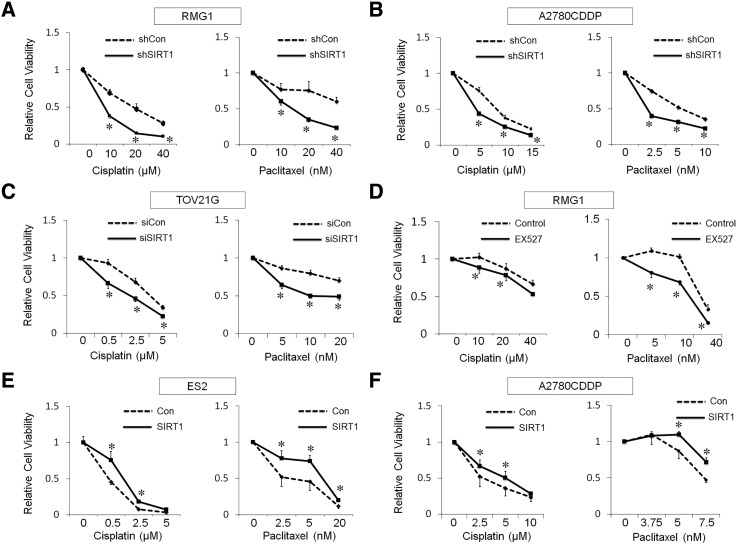
In order to investigate, the effects of SIRT1 on sensitivity against anti-cancer drugs, cell viability following a treatment with cisplatin or paclitaxel was measured using the WST-1 assay. The chemosensitivity of SIRT1-silenced cells (si- or sh-SIRT1) towards cisplatin and paclitaxel was greater than that of control cells (si- or sh-Con) (Figure 4, A–C, P < .05) in a dose-dependent manner. The inhibition of SIRT1 by EX527 (1 and 2 μM) significantly increased the chemosensitivity of RMG1 cells to cisplatin and paclitaxel, respectively (Figure 4D, P < .05).
Figure 4.
A–C: The effects of SIRT1 knockdown. SIRT1 knockdown (sh/siSIRT1) significantly attenuated chemoresistance of both cisplatin and paclitaxel compared with control (sh/siCon) in RMG1 (A), A2780CDDP (B) and TOV21G (C). D: The effect of SIRT1 inhibitor, EX527. EX527 attenuated cisplatin- and paclitaxel-resistance in RMG1 cells. E and F: The effect of SIRT1 overexpression. SIRT1 overexpression (SIRT1) significantly enhanced the chemoresistance compared with control (Con) of ES2 (E) and A2780CDDP (F). Significance: * P < .05, significantly different from the controls.
In addition, SIRT1-overexpressing cells (ES2-SIRT1 and A2780CDDP-SIRT1) exhibited weaker sensitivity to cisplatin (P < .05) and paclitaxel (P < .05) than their controls (ES2-Con and A2780CDDP-Con) (Figure 4, E and F). Taken together, these results suggest that SIRT1 enhanced the chemoresistance of OvCa cells.
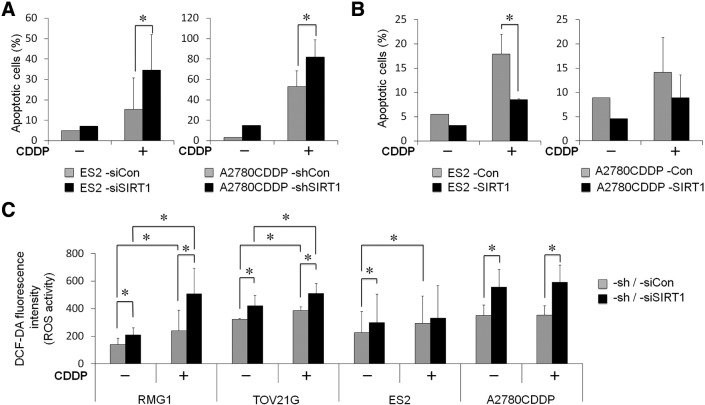
SIRT1 Enhanced the Chemoresistance of OvCa Cells by Inhibiting Apoptotic Cell Death
We were unable to demonstrate the effects of SIRT1 on well-known survival and apoptotic factors, including phospho-Akt, Bcl-2, BAX, or others (data not shown). Thus, we performed apoptotic assays utilizing Annexin V and PI/7AAD staining methods to evaluate the percentage of apoptotic cells. We found that cisplatin-induced apoptosis was greater in SIRT1-knockdown OvCa cells than in control cells (Figure 5A, P < .05). In order to confirm the above results, we utilized SIRT1-overexpressing ES2 and A2780CDDP cells. As expected, SIRT1-overexpressed cells had a significantly lower number of apoptotic cells than control cells (Figure 5B, P < .05).
Figure 5.
A: Effects of SIRT1 knockdown on cisplatin-induced apoptosis. The apoptosis analysis showed the percentage of apoptotic cells with (+) or without (−) the cisplatin (CDDP) treatment. The knockdown of SIRT1 significantly increased cisplatin-induced apoptosis in both ES2 and A2780CDDP cells. B: Effects of SIRT1 overexpression on cisplatin-induced apoptosis. The overexpression of SIRT1 (SIRT1) significantly decreased cisplatin-induced apoptosis compared with control (Con) in both ES2 and A2780CDDP cells. C: DCF-DA fluorescence levels indicating ROS activity in OvCa cells with (+) or without (−) CDDP. SIRT1 knockdown significantly increased ROS activity. Significance: * P < .05.
Effects of SIRT1 on Oxidative Stress
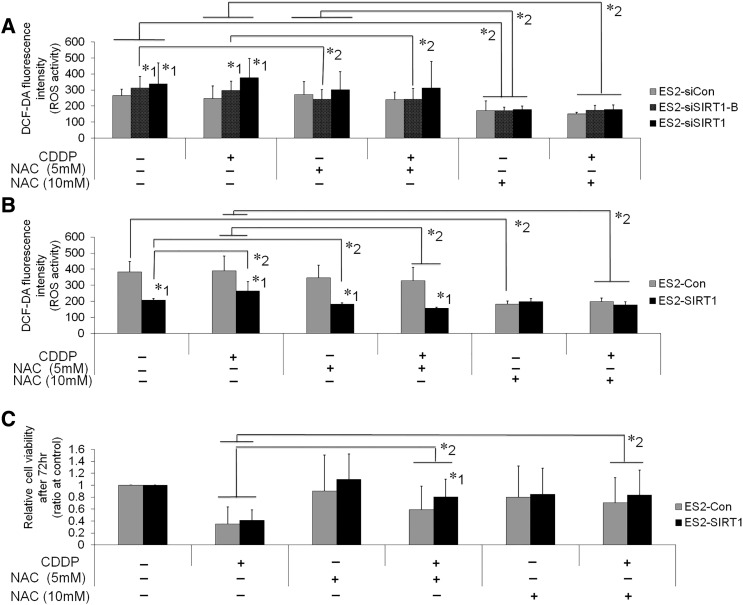
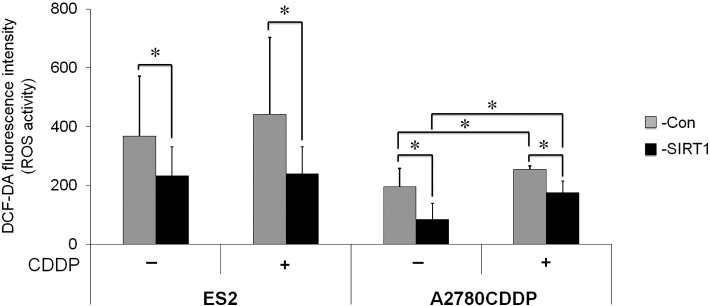
In order to investigate the underlying mechanisms for increases in SIRT1-associated cell growth and chemoresistance in more detail, we examined the effects of SIRT1 on oxidative stress. We analyzed ROS production by OvCa cells. DCF-DA fluorescence, indicating ROS production, was significantly stronger in SIRT1 knockdown cells than in control cells (Figure 5C, P < .05). This effect of SIRT1 knockdown on ROS production was canceled by the treatment with ROS scavenger, NAC (Supplementary Figure S5A). Furthermore, the CDDP treatment synergistically increased ROS production in RMG1 and TOV21G. Similarly, the overexpression of SIRT1 significantly decreased ROS production in ES2 and A2780CDDP cells (Supplementary Figure S4, P < .05). The treatment with 10 mM of NAC reduced ROS production of ES2-Con to the same level as ES2-SIRT1 (Supplementary Figure S5B). In addition, the treatment of NAC canceled the reduction of cell viability by CDDP (Supplementary Figure S5C).
Supplementary Figure S5.
Effects of the ROS scavenger, N-Acetyl-L-cysteine (NAC), in ES2 cells on ROS production (A and B) and cell viability (C). DCF-DA fluorescence intensity indicating ROS production. A: The knockdown of SIRT1 (ES2-siSIRT1, ES2-siSIRT1-B) significantly increased ROS production compared with control cells (ES2-siCon). NAC treatment (10 mM) significantly reduced ROS and canceled the increase of ROS by SIRT1 knockdown with or without CDDP treatment. B: The overexpression of SIRT1 (ES2-SIRT1) significantly decreased ROS production compared with control cells (ES2-Con). CDDP treatment increased ROS in ES2-SIRT1. NAC treatment significantly reduced ROS in both cells dose-dependently. The treatment with 10 mM of NAC significantly reduced ROS in ES2-Con to the same level as ES2-SIRT1 with or without CDDP treatment. C: NAC treatment significantly attenuated the cytotoxic effect of CDDP. Significance (P < .05), whereby *1: significantly different from control cells (ES2-siCon or ES2-Con), *2: significantly different between the cells indicated by bars.
Supplementary Figure S4.
Effects of SIRT1 overexpression on ROS production. DCF-DA fluorescence intensity indicating ROS activity in OvCa cells treated with (+) or without (−) CDDP. The overexpression of SIRT1 (−SIRT1) significantly decreased ROS activity compared with control (−Con) in both ES2 and A2780CDDP cells. Significance: * P < .05, significantly different from the controls.
SIRT1 Knockdown Decreased the Ability of OvCa Cells to Counteract Oxidative Stress
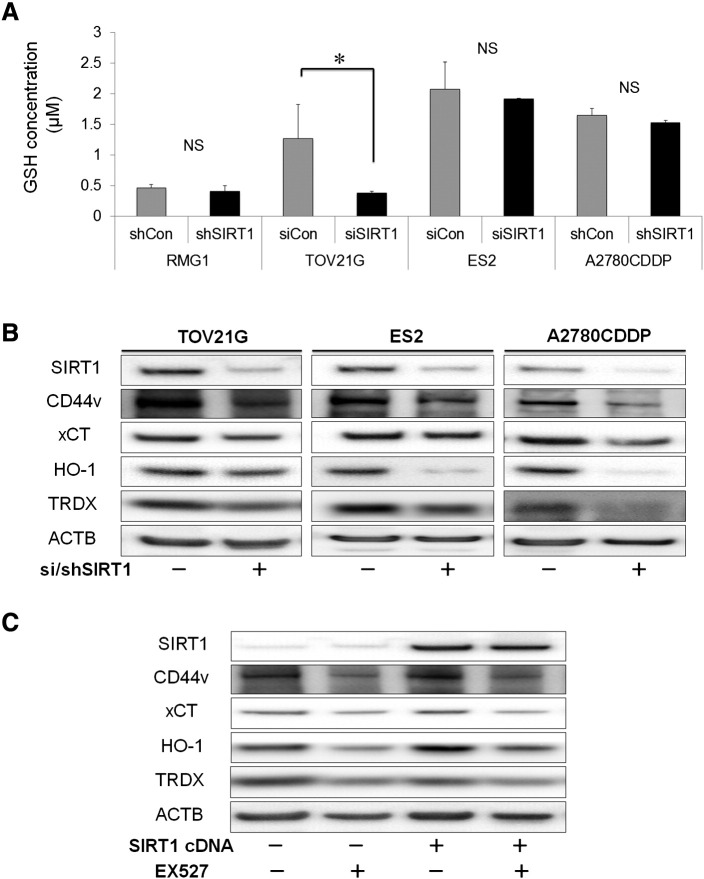
In order to gain mechanistic insights into how SIRT1 counteracts oxidative stress in OvCa, we analyzed cellular GSH levels and performed a Western blotting analysis for the cancer stem cell marker (CD44v9) and regulators of oxidative stress. The knockdown of SIRT1 significantly suppressed the cellular levels of GSH, a major antioxidant, compared with those in control cells in the TOV21G cell line (Figure 6A), suggesting that SIRT1 contributes to reductions in ROS by increasing cellular GSH levels in these cells. However, no significant difference was observed in other cell lines. Western blotting revealed that the knockdown of SIRT1 down-regulated the cancer stem cell marker CD44v9, a glutamate-cystine transporter system xCT, and the oxidative regulators: hemeoxygenase-1 (HO-1) and thioredoxin (TRDX) (Figure 6B). These results suggested that the knockdown of SIRT1 directly interfered with the CD44v9/xCT pathway, thereby regulating GSH levels inside cancer cells, which in turn, controlled ROS. Furthermore, the knockdown of SIRT1 also down-regulated the antioxidants HO-1 and TRDX, further emphasizing its role in breaking the synergistic defenses played by these antioxidants against ROS. Similar results were obtained using SIRT1 up-regulated cells (Figure 6C). The selective SIRT1 inhibitor (EX527) recapitulated the effects of the genetic knockdown of SIRT1 (Figure 6C). EX527 had effect on the activity of SIRT1 and not on the expression of SIRT1. In order to confirm these results, we performed qRT-PCR on CD44, xCT, and antioxidant genes (Supplementary Fig. S6A and B), and the results obtained were similar. It is important to note that the regulation of antioxidant genes and enrichment of CD44 by SIRT1 occurred at the protein and gene levels. These results were consistent with SIRT1 up-regulating multiple antioxidant pathways and enriching CD44v9, thereby enhancing the OvCa capacity for ROS defenses and, hence, tumor development.
Figure 6.
A: Glutathione (GSH) levels in OvCa cells. GSH is one of the major antioxidants. Among OvCa cell lines, intracellular GSH level was significantly decreased by SIRT1-knockdown (si/shSIRT1) compared with control (si/shCon) in TOV21G cells. B and C: Western blotting showed the expression of CD44v and xCT, involved in the synthesis of GSH, and other antioxidative enzymes such as heme oxygenase-1 (HO-1) and thioredoxin (TRDX) in OvCa cell lines. Knockdown of SIRT1 by siRNA or shRNA (B) and EX527 (a SIRT1 inhibitor) (C) decreased expression of these antioxidative proteins. In contrast, overexpression of SIRT1 by SIRT1 cDNA increased expression of antioxidative proteins in ES2, and these effects of SIRT1 were canceled out by EX527 (C). Significance: * P < .05.
Supplementary Figure S6.
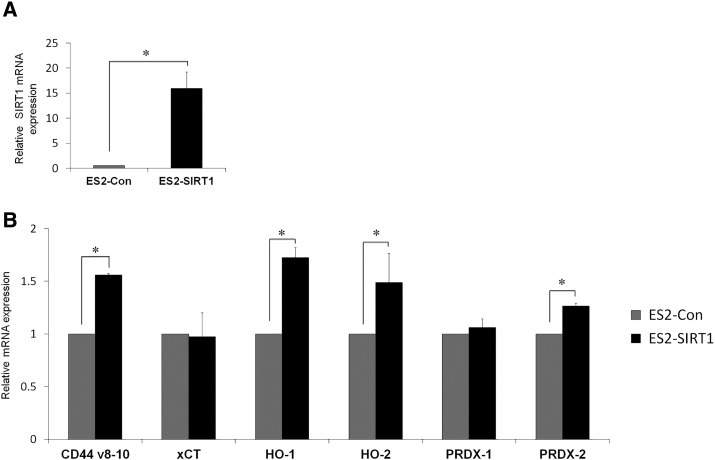
Effects of SIRT1 overexpression on the glutamate-cystine transporter system: CD44v8–10/xCT and antioxidative enzymes: heme oxygenase-1 (HO-1), heme oxygenase-2 (HO-2), Peroxiredoxin-1 (PRDX-1) and Peroxiredoxin-2 (PRDX-2). Quantitative real-time PCR analysis (qRT-PCR) of ES2 cells with the overexpression of SIRT1 by SIRT1 cDNA (ES2-SIRT1) or an empty vector as control (ES2-Con) (A and B). Quantitative mRNA levels showed effective overexpression of SIRT1 in ES2-SIRT1 cells compared with ES2-Con (A). The effects of SIRT1 overexpression in relation to mRNA levels of CD44v8–10, xCT, and antioxidative enzymes (HO-1, HO-2, PRDX-1, and PRDX-2) (B). SIRT1 overexpression (ES2-SIRT1) significantly up-regulated the expression of CD44v8–10, HO-1, HO-2, and PRDX-2) compared with control (ES2-Con) at the mRNA level. Significance: * P < .05
SIRT1 Overexpression Promoted the Aggressiveness of OvCa Cells
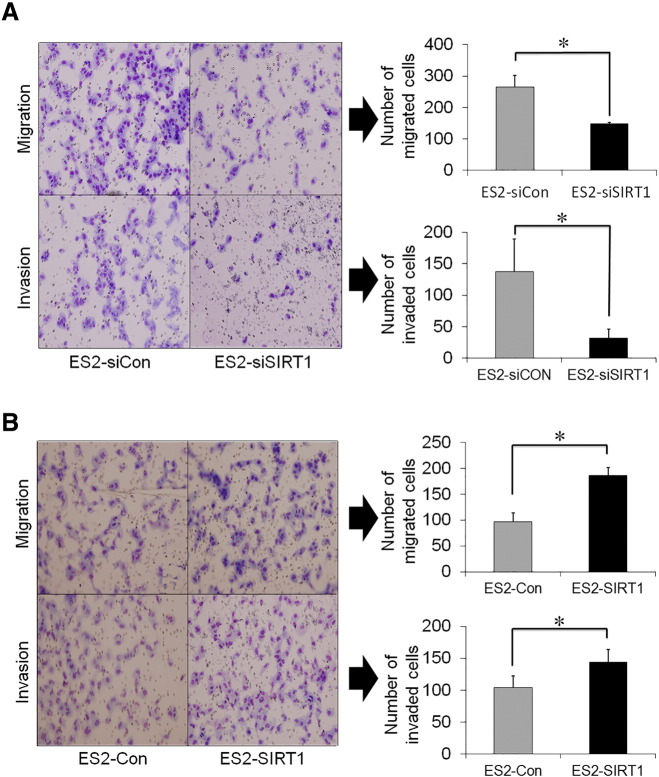
Cell migration and invasion assays demonstrated that the knockdown of SIRT1 (siRNA) significantly decreased the number of migrated cells on control inserts (without matrigel) and impaired cell invasion through matrigel more than the control (Figure 7A, P < .05). Similarly, SIRT1-overexpressing cells (ES2-SIRT1) had markedly higher the numbers of migrated cells and invaded cells (Figure 7B, P < .05) than control cells (ES2-Con). These results show that SIRT1 enhanced the invasiveness and migration ability of OvCa cells in vitro. Based on our in vitro experiments, we selected ES2 cell line, the most invasive and aggressive OvCa cell line, which was consistent with a previous study [21], and transduced it with vectors expressing SIRT1 (ES2-SIRT1) or an empty vector (ES2-Con). These results show that SIRT1 enhanced the aggressiveness of OvCa cells.
Figure 7.
A: Effects of SIRT1 knockdown by siRNA transfection on the migration and invasion of ES2 cells. SIRT1 knockdown (ES2-siSIRT1) significantly decreased the invasion and migration compared with control (ES2-siCon). B: Effects of SIRT1 overexpression by cDNA transfection on the migration and invasion of ES2 cells. SIRT1 overexpression (ES2-SIRT1) significantly enhanced the invasion and migration compared with control (ES2-Con). Significance: * P < .05.
Discussion
Increases in the recurrence and drug resistance of OvCa maybe attribute to CSCs, forming a significant residual of disease after therapy [4]. Our results favor a model in which SIRT1 inhibits oxidative stress by maintaining the stemness of CSC-like cells, thereby driving ovarian tumorigenesis. The present study showed that SIRT1 is more strongly expressed in a panel of OvCa cell lines than in an immortalized ovarian surface epithelium at the gene and protein levels. These results were consistent with our clinical data analysis, in which SIRT1 was more strongly expressed in OvCa tissues than in the normal ovarian epithelium, and SIRT1 was an independent prognostic predictor of overall survival regardless of the tumor stage [22]. We also identified the expression of SIRT1 in endometrial carcinoma as a poor prognostic factor [16]. The overexpression of SIRT1 is not only limited to gynecological cancers, it has been previously reported in various cancer types [23], [24], [25], [26], [27], [28], and the function of SIRT1 (tumor promoter or suppressor) remains controversial [29], [30]. The function of SIRT1 was recently reported to be tissue-dependent [31], and our results herein demonstrate that SIRT1 functions as an oncogene in OvCa.
SIRT1 has been reported to increase the aggressiveness of various cancer cells by regulating pathways related to cell growth, genome integrity, and cell death [9]. Therefore, we dissected the mechanisms of SIRT1 mediating OvCa chemoresistance and aggressiveness. The expression of SIRT1 was enhanced following exposure to cytotoxic stress (Figure 1B), and the forced expression of SIRT1 further increased the deacetylation activity of SIRT1 (Figure 1, C–D). The significant increase of SIRT1 deacetylation activity was observed by the forced expression of SIRT1 but not observed by the endogenous increase of SIRT1 mRNA with CDDP (Figure 1, B and C), suggesting that SIRT1 may be mainly involved in the intrinsic drug resistance, rather than the acquired resistance. Furthermore, the inhibition of SIRT1 significantly reduced the proliferation of OvCa cells, as reported previously [27]. Cisplatin and paclitaxel sensitivities were both increased by SIRT1 knockdown and decreased by SIRT1 overexpression. These findings were consistent with previous findings [26], [32]. Taken together, these results demonstrate that SIRT1 is involved in OvCa cell growth and resistance to chemotherapy. The overexpression of SIRT1 (SIRT1 cDNA) did not affect the proliferation of OvCa cells (Figure 2D). We speculate that this was because the cellular level of SIRT1 in OvCa cells was sufficient for growth; hence, the forced expression of SIRT1 had no additive effect.
A literature search on the mechanisms underlying chemoresistance directed us to how a subset of CSCs evades chemotherapy by counteracting oxidative stress-induced apoptosis [34]. The present study revealed that the ROS scavenger, NAC, attenuated the cytotoxic effect of CDDP (Supplementary Fig. S5). As expected, SIRT1 inhibited apoptotic cell death by down-regulating oxidative stress (ROS). A large number of studies have reported that excessive ROS production is detrimental to cancer cells because it disrupts the cancer signaling pathways that promote proliferation, migration, and invasion [35], [36]. Therefore, cancer cells avoid the harmful effects of ROS by actively utilizing multiple antioxidant systems [35], for example, the most abundant antioxidant system (GSH). This finding has only been confirmed in one OvCa cell line (TOV21G). In contrast to previous findings our results failed to elucidate the functions of GSH in attenuating ROS in RMG1, ES2, and A2780CDDP cell lines; however, our results did show that SIRT1 positively up-regulated the CD44v9/xCT pathway, a glutamate-cystine transporter system. We speculate that another antioxidant pathway besides GSH may be involved, as was described by Harris et al., that cancer cells require GSH for the initial stages of cancer initiation, but not thereafter, partly due to utilizing another pathway: thioredoxin [35]. This finding that is consistent with our results. The present study is the first to propose a possible link between SIRT1, GSH, and CD44v9/xCT in OvCa; however, this requires further clarification in future studies.
Increasing evidence has suggested the SIRT1 family's stem cell-like abilities [4], [5]. Our results are consistent with this finding because SIRT1 promoted the formation of colonies and increased the expression of several stemness-associated genes. To date, accumulated evidence has also identified CD44 as a cell surface marker for CSCs derived from solid tumors, for example, colon, breast, ovary and pancreatic cancers [36], [37]. Furthermore, the expression of CD44, particularly the variant isoform (CD44v9), leads to defenses against ROS [8]. On the other hand, our Western blotting analysis showed that the knockdown of SIRT1 also down-regulated the oxidative regulators HO-1 and thioredoxin. Consistent with our results, several studies have shown that CSCs up-regulate antioxidant pathways, thereby controlling ROS generated from oxidative stress, leading to resistance and ultimately cancer cell growth [33], [34], [38], which further confirms the finding by Harris et al. that thioredoxin replaced the utilization of GSH in established tumors [34].
The present study revealed that SIRT1 facilitated the aggressiveness of OvCa cells through increases in the migration, invasion, and tumorigenicity. Several reports have shown that SIRT1 promoted EMT in several cancers through the transcription repression of epithelial genes including E-cadherin while increasing the expression of mesenchymal genes including N-cadherin and vimentin, resulted in promoting aggressiveness [39], [40]. Repression of E-cadherin by SIRT1 was observed in ES2 in our study (data not shown).
In conclusion, our results show that SIRT1 is more strongly expressed in OvCa cells than in the immortalized ovarian surface epithelium. Further analyses revealed that SIRT1 enriched the CSC pool and played a pertinent role in the chemoresistance of OvCa by counteracting oxidative stress. Taken together, our results indicate that the targeting of SIRT1 offers a novel therapeutic target against CSCs, ultimately reducing the chemoresistance burden of OvCa.
The following are the supplementary data related to this article.
Relative expressions of SIRT1 based on histology.
The Primer sequences for target genes and RNAi sequences used in this study.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Acknowledgments
The authors sincerely remain grateful for the excellent technical assistance by Fumi Tsunoda and Eiji Uchida (Research Assistants; Department of Obstetrics and Gynecology, Shinshu University School of Medicine).
Footnotes
This work was supported by JSPS KAKENHI Grant Number 15 K10711.
References
- 1.Ozols RF. Treatment goals in ovarian cancer. Int J Gynecol Cancer. 2005;15(Suppl 1):3–11. doi: 10.1111/j.1525-1438.2005.15351.x. [DOI] [PubMed] [Google Scholar]
- 2.Ushijima K. Current status of gynecologic cancer in Japan. J Gynecol Oncol. 2009;20:67–71. doi: 10.3802/jgo.2009.20.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA, Palle K. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One. 2014;9:e107142. doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 7.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 8.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(−) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 9.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 10.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013;61:95–110. doi: 10.1016/j.freeradbiomed.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, Su L, Chen WY. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther. 2013;6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang ZN, Chung SK, Xu Z, Xu Y. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells. 2014;32:157–165. doi: 10.1002/stem.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiozawa T, Horiuchi A, Kato K, Obinata M, Konishi I, Fujii S, Nikaido T. Up-regulation of p27Kip1 by progestins is involved in the growth suppression of the normal and malignant human endometrial glandular cells. Endocrinology. 2001;142:4182–4188. doi: 10.1210/endo.142.10.8455. [DOI] [PubMed] [Google Scholar]
- 16.Asaka R, Miyamoto T, Yamada Y, Ando H, Mvunta DH, Kobara H, Shiozawa T. Sirtuin 1 promotes the growth and cisplatin resistance of endometrial carcinoma cells: a novel therapeutic target. Lab Invest. 2015;95:1363–1373. doi: 10.1038/labinvest.2015.119. [DOI] [PubMed] [Google Scholar]
- 17.Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, Shabbir A, So JB, Chan SL. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630–2641. doi: 10.1158/0008-5472.CAN-13-2309. [DOI] [PubMed] [Google Scholar]
- 18.Yamada Y, Miyamoto T, Kashima H, Kobara H, Asaka R, Ando H, Higuchi S, Ida K, Shiozawa T. Lipocalin 2 attenuates iron-related oxidative stress and prolongs the survival of ovarian clear cell carcinoma cells by up-regulating the CD44 variant. Free Radic Res. 2016;50:414–425. doi: 10.3109/10715762.2015.1134795. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki A, Horiuchi A, Ashida T, Miyamoto T, Kashima H, Nikaido T, Konishi I, Shiozawa T. Cyclin A2 confers cisplatin resistance to endometrial carcinoma cells via up-regulation of an Akt-binding protein, periplakin. J Cell Mol Med. 2010;14:2305–2317. doi: 10.1111/j.1582-4934.2009.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto T, Kashima H, Suzuki A, Kikuchi N, Konishi I, Seki N, Shiozawa T. Laser-captured microdissection-microarray analysis of the genes involved in endometrial carcinogenesis: stepwise up-regulation of lipocalin2 expression in normal and neoplastic endometria and its functional relevance. Hum Pathol. 2011;42:1265–1274. doi: 10.1016/j.humpath.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 21.Shaw TJ, Senterman MK, Dawson K, Crane CA, Vanderhyden BC. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol Ther. 2004;10:1032–1042. doi: 10.1016/j.ymthe.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Mvunta DH, Miyamoto T, Asaka R, Yamada Y, Ando H, Higuchi S, Ida K, Kashima H, Shiozawa T. Overexpression of SIRT1 is Associated With Poor Outcomes in Patients With Ovarian Carcinoma. Appl Immunohistochem Mol Morphol. 2016 doi: 10.1097/PAI.0000000000000316. 10.1097/PAI.0000000000000316 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Sun K, Jiao S, Cai N, Zhao X, Zou H, Xie Y, Wang Z, Zhong M, Wei L. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci Rep. 2014;4:7481. doi: 10.1038/srep07481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elangovan S, Ramachandran S, Venkatesan N, Ananth S, Gnana-Prakasam JP, Martin PM, Browning DD, Schoenlein PV, Prasad PD, Ganapathy V. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor α in breast cancer. Cancer Res. 2011;71:6654–6664. doi: 10.1158/0008-5472.CAN-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 26.Cao B, Shi Q, Wang W. Higher expression of SIRT1 induced resistance of esophageal squamous cell carcinoma cells to cisplatin. J Thorac Dis. 2015;7:711–719. doi: 10.3978/j.issn.2072-1439.2015.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenzinger A, Endris V, Klauschen F, Sinn B, Lorenz K, Warth A, Goeppert B, Ehemann V, Muckenhuber A, Kamphues C. High SIRT1 expression is a negative prognosticator in pancreatic ductal adenocarcinoma. BMC Cancer. 2013;13:450. doi: 10.1186/1471-2407-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Y, Zhang J, Wu S, Li B, Liu S, Cheng J. SIRT1 promotes proliferation and inhibits apoptosis of human malignant glioma cell lines. Neurosci Lett. 2012;525:168–172. doi: 10.1016/j.neulet.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2:648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stünkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011;16:1153–1169. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- 32.Zhao G, Cui J, Zhang JG, Qin Q, Chen Q, Yin T, Deng SC, Liu Y, Liu L, Wang B. SIRT1 RNAi knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells. Gene Ther. 2011;18:920–928. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 33.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Tochhawng L, Deng S, Pervaiz S, Yap CT. Redox regulation of cancer cell migration and invasion. Mitochondrion. 2013;13:246–253. doi: 10.1016/j.mito.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Sacks JD, Barbolina MV. Expression and Function of CD44 in Epithelial Ovarian Carcinoma. Biomolecules. 2015;5:3051–3066. doi: 10.3390/biom5043051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basakran NS. CD44 as a potential diagnostic tumor marker. Saudi Med J. 2015;36:273–279. doi: 10.15537/smj.2015.3.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsuhiro N, Satoru K, Bo Z, Xiuzhi Z, Yasunara M, Masahiro T, Yoshiko M, Noriko M, Manabu H, Satoshi O. Prognostic impact of CD133 expression as a tumor-initiating cell marker in endometrial cancer. Hum Pathol. 2010;41:1516–1529. doi: 10.1016/j.humpath.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Hao C, Zhu P, Yang X, Han Z, Jiang J, Zong C, Zhang X, Liu W, Zhao Q, Fan T. Overexpression of SIRT1 promotes metatstasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Cancer. 2014;14:978. doi: 10.1186/1471-2407-14-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, Dai Y. SIRT1 induces EMT by cooperating with transcription factors and enhance prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative expressions of SIRT1 based on histology.
The Primer sequences for target genes and RNAi sequences used in this study.