Abstract
Background and objectives
A new phenotype with overlapping characteristics between asthma and chronic obstructive pulmonary disease (COPD) called asthma–COPD overlap syndrome (ACOS) is emerging among inflammation diseases. To date, there is no agreement on specific criteria to define this syndrome, and the current guidelines are insufficient to classify the analogy and differences between overlap and COPD or asthma phenotypes. It would be necessary to identify new biomarkers able to identify these diseases clearly. Thus, the aim of this study was to identify a serum and supernatant of sputum microRNA (miRNA) expression profile of miRNA-145 and miRNA-338 in patients with asthma (n=13), COPD (n=31), and ACOS (n=8) and controls (n=7).
Methods
The expression was evaluated using quantitative real-time polymerase chain reaction (qRT-PCR). For statistical analysis, the ANOVA test, Kruskal–Wallis test, Mann–Whitney U-test, and Spearman’s rank correlation were used.
Results
The main finding of this work is that the expression of miRNA-338 is higher in the supernatant of different obstructive diseases than in peripheral blood, while miRNA-145 is higher only in the supernatant of asthma patients. The expression of both selected miRNAs is higher in the supernatant of asthma and COPD patients than in controls.
Conclusion
Differences in sputum miRNA expression profile were observed between patients with ACOS and asthma or COPD, which underline the potential role of miRNA as a biomarker that is able to discriminate patients with ACOS, asthma, and COPD.
Keywords: asthma–COPD overlap syndrome, ACOS, asthma, chronic obstructive pulmonary disease, miRNA, sputum
Introduction
Asthma and chronic obstructive pulmonary diseases (COPD), characterized by airway disorders associated with persistent inflammation, are classified as chronic conditions and are influenced by a combination of environmental, genetic, and epigenetic components.1 The combination of these two disorders has been recently defined as asthma–COPD overlap syndrome (ACOS). To date, there is no universal agreement about the definition of ACOS, but usually a combination of spirometric parameters and some clinical signs or symptoms are used to characterize a subset of patients who could be affected by this syndrome.2 According to Spanish guidelines, major and minor criteria were established to diagnose this mixed phenotype: two major criteria or two minor criteria should be met. On the other hand, according to Global Initiative for Asthma (GINA) guidelines, ACOS was defined as a syndrome characterized by persistent airflow limitation with several features usually associated with asthma and others associated with COPD.3,4 However, to better characterize this syndrome, it could be necessary to identify some biomarkers that can help physicians to diagnose this mixed phenotype in each patient, in order to improve diagnosis and therapy.
MicroRNAs (miRNAs) are assuming greater significance in research as novel regulators of gene expression, playing a central role in different pathophysiological processes. It has been shown that these classes of noncoding regulatory RNA are involved in several aspects of inflammation which is a determinant feature in many lung diseases such as asthma and COPD.5,6 Previous observations have suggested that the presence of miRNA in whole blood induced sputum, serum, and some other body fluids such as saliva.7–9
Few studies examined miRNA expression in asthma and COPD, but no studies have been conducted on the ACOS phenotype. We focused our attention on miRNA-145 and miRNA-338 which are involved in many processes of airway inflammation. miRNA-145 play a role in the regulation of airway smooth muscle (ASM) function, a component of airway wall, which contributes to the remodeling of airway in the presence of chronic inflammation in both asthma COPD diseases.10,11 While miRNA-338 seems to have a role in the control of cellular differentiation, apoptosis and tissue degeneration have been demonstrated in some types of human cancers, including non-small cell lung cancer (NSCLC).12
The aim of this study was not only to evaluate whether miRNAs are involved in the pathogenesis of main obstructive diseases such as asthma and COPD but also to define whether they could be useful to better characterize ACOS patients from a clinical point of view.
Patients and methods
Patients and clinical specimens
We enrolled 52 consecutive patients from the Clinic of Obstructive Pulmonary Diseases of the Ospedali Riuniti of Foggia, Foggia University, during the period of 3 months. Participants were classified into three groups based on their medical history: patients with asthma (n=13), patients with COPD (n=31), and patients with ACOS (n=8); seven healthy nonsmoker subjects were also enrolled as the control group. All the patients were non- or former smokers and had been clinically stable during the period of 6 months prior to testing. This study and all study protocols were approved by the Medical Ethics Committee of Ospedali Riuniti of Foggia (Ref 17/CE/2014) and written informed consent was obtained from all of the subjects.
Asthmatics were classified and treated according to GINA guidelines: we chose never-smoker patients with a classic reversibility of 12% and 200 mL increase in forced expiratory volume in 1 s (FEV1). COPD patients were classified according to Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines: former smokers with no reversible airflow limitation at spirometry. Furthermore, we selected ACOS patients according to Spanish guidelines using two major criteria: very positive bronchodilator test (increase in FEV1 ≥15% and ≥400 mL) and eosinophilia in sputum (>2%). Forced spirometry (before and after bronchodilation) was determined following international standards in all patients.13
Collection of blood and sputum
Spontaneous sputum was collected in a sterile cup before receiving any treatment and later treated with Sputolysin® (Calbiochem; EMD Millipore, Billerica, MA, USA) centrifuged at 1,000× g for 15 min to obtain the supernatant. At the same time, peripheral whole blood was collected in EDTA tubes and centrifuged at 3,000 rpm for 20 min to obtain serum. Both biological samples were stored at −80°C. All samples were analyzed within 6 months from collection.
The resuspended pellet was used to analyze the cell viability and to prepare the cytospin slides. The Dulbecco’s phosphate-buffered saline (D-PBS) cell suspension was centrifuged at 450 rpm (Shandon centrifuge) for 5 min.14 The slides were stained by Diff-Quick staining (Medion Diagnostic, Düdingen, Switzerland) for cell differentiation counts with 400 cells being counted from each slide. A value of <70% of contamination from squamous epithelial cells was accepted.
RNA isolation
Total RNA containing small RNA was extracted from serum and supernatant of sputum by using, respectively, the Trizol reagent and mirVana Paris Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA), in accordance with the manufacturer’s protocol. The concentration and quality of eluted RNA were measured using NanoDrop Spectrophotometer (Thermo Fisher Scientific). RNA purity was evaluated by the ratio of absorbance at OD260/OD280.
Reverse transcription and detection of miRNA expression by quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA was reverse transcribed using a TaqMan MicroRNA RT kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The identified miRNAs were evaluated in serum and supernatant by qRT-PCR with Taqman miRNA assay (Thermo Fisher Scientific) according to the manufacturer’s instructions. miRNA-16 (assay ID 000391) and RNU44 (assay ID 001094) were proven as internal control for miRNA quantification in serum and supernatant to normalize the Ct value of miRNA-145 (assay ID 002278) and miRNA-338 (assay ID 002658). Prior to purification, an exogenous miRNA (Cel-miR-39), a synthetic, nonhuman control, was added to each supernatant of sputum to monitor RNA isolation. Relative expression of targeted miRNAs was computed using the equation 2−ΔΔCt, where ΔCt = Ct (targeted miRNA) − Ct (internal control gene) with respect to the expression of miRNA in the control group.15 All of the assays were performed in triplicate and one no-template control (NTC) was carried out in each experiment.
Statistical analysis
Results are presented as mean ± SD. The ANOVA test was used to compare differences among groups. We used the Kruskal–Wallis test to compare the differential expression level of miRNAs among the three groups, while the Mann–Whitney U-test was employed to evaluate the differences between the expression of miRNA in serum and supernatant. Spearman’s rank correlation was used to assess the correlations between relative miRNA expression and clinical and biological parameters. A P-value <0.05 was considered statistically significant.
Results
According to inclusion criteria, we selected 52 patients: 13 asthmatics, 31 COPD, and eight ACOS. Table 1 presents the main demographic, clinical, and functional characteristics of participants. The asthma patients were younger than the COPD subjects (52.46±13.19 vs 70.26±8.01 years), while the ACOS group had an intermediate age of 63.25±8.96 years. Baseline FEV1 % was lower in the COPD group than the others and, by definition, had poor improvement while both asthmatic and ACOS groups had higher improvement in FEV1 after administering salbutamol. As expected, there were many differences in sputum cell count: ACOS and asthmatic groups had the same percentage of eosinophils and both showed higher level compared to the COPD group (19.50±26.97%, 26.43±35.52% vs 1±1.58%, P=0.003). On the contrary, COPD patients had a higher percentage of neutrophils in the sputum compared to other groups (81.48±22.52% vs 52.09±31.79%, 52.71±35.54%, P=0.004; Table 1).
Table 1.
Clinical characteristics of population and sputum cellularity
| Characteristics | ACOS
|
Asthma
|
COPD
|
Control
|
P-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| Age (years) | 63.25 | 8.96 | 52.46 | 13.19 | 70.26 | 8.01 | 65.20 | 5.45 | 0.000 |
| BMI (kg/m2) | 27.73 | 3.02 | 29.16 | 7.66 | 28.52 | 5.03 | 28.10 | 4.13 | 0.852 |
| FEV1/FVC | 53.00 | 8.37 | 61.92 | 12.29 | 49.13 | 12.31 | 82.32 | 3.65 | 0.008 |
| FEV1 (%) | 67.00 | 16.83 | 75.77 | 22.45 | 52.68 | 19.40 | 87.27 | 4.76 | 0.003 |
| FVC (%) | 81.25 | 17.21 | 88.67 | 16.18 | 60.28 | 24.37 | 90.86 | 3.95 | 0.001 |
| PaCO2 (mmHg) | 37.44 | 2.41 | 41.67 | 7.02 | 44.64 | 6.11 | 41.27 | 2.01 | 0.050 |
| PaO2 (mmHg) | 74.58 | 5.00 | 72.67 | 10.12 | 69.96 | 10.08 | 80.36 | 4.24 | 0.588 |
| Eosinophils (%) | 26.43 | 35.52 | 19.50 | 26.97 | 1.00 | 1.58 | 1.1 | 0.3 | 0.003 |
| Neutrophils (%) | 52.71 | 35.54 | 52.09 | 31.79 | 81.48 | 22.52 | 15.76 | 5.38 | 0.004 |
| Macrophages (%) | 19.29 | 13.02 | 30.08 | 27.94 | 16.89 | 21.56 | 77.31 | 6.87 | 0.246 |
Note: Significant differences are marked in bold.
Abbreviations: ACOS, asthma–COPD overlap syndrome; BMI, body mass index; COPD, chronic obstructive pulmonary disease; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; SD, standard deviation.
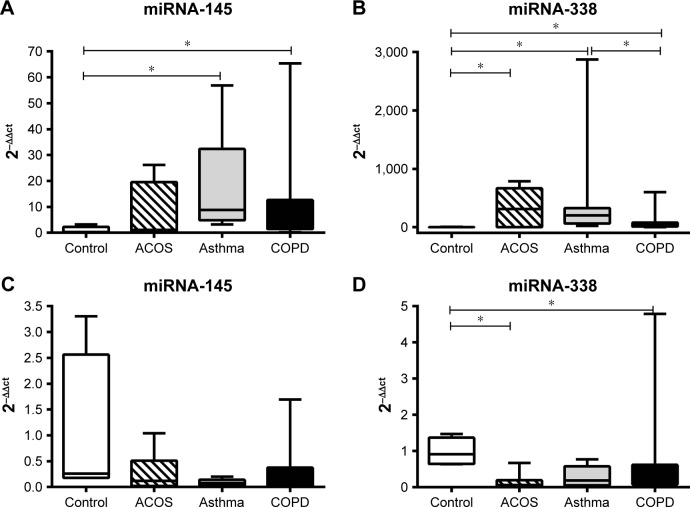
The expression of miRNA-145 in sputum was higher in asthmatic and COPD patients than in controls. The expression of miRNA-145 in ACOS patients was intermediate between two other groups, but no significant difference was detected. No difference was found in the expression of miRNA-145 in the serum among the three groups (Figure 1A and C). On the contrary, the expression level of miRNA-338 in the sputum was higher in all patient groups compared to controls; moreover, asthmatics showed a significantly higher miRNA-338 expression compared to COPD patients. Thus, in this case, ACOS patients also showed a higher level of miRNA-338 compared to controls (Figure 1B). Finally, the serum of ACOS and COPD patients showed a lower level of miRNA-338 expression compared to controls, while in asthma patients even if the level tends to be lower with respect to the control, any significant difference was demonstrated (Figure 1D).
Figure 1.
Evaluation of miRNA expression by qRT-PCR.
Notes: After normalization to miRNA-16 in each group, data were represented as mean ± SD and the obtained average values for each miRNA were used for statistics in ACOS, asthma, and COPD groups compared to healthy controls. Sputum miR-145 (A) was higher in asthma and COPD patients compared to healthy controls. At the same time, sputum miR-338 (B) was also present in higher abundance in all three groups analyzed, while there was no significant difference for serum miR-145 (C) in ACOS, asthma, and COPD patients compared to healthy controls. Serum miR-338 (D) was lower in ACOS, asthma, and COPD patients compared to healthy controls. The experiment was conducted in triplicate. *P<0.05.
Abbreviations: ACOS, asthma–COPD overlap syndrome; COPD, chronic obstructive pulmonary disease; qRT-PCR, quantitative real-time polymerase chain reaction.
There is no correlation between the expression level of miRNA both with main parameters of population such as age or FEV1 and with the cellularity of sputum.
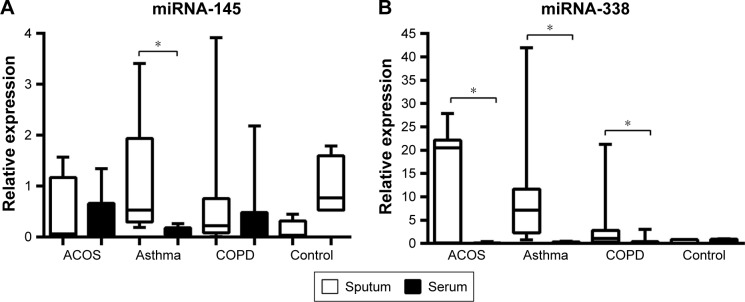
Interestingly, in all groups analyzed, the expression of miRNA-338 was higher in the sputum than in peripheral blood sample while no differences were found in healthy subjects. Moreover, asthma patients showed a higher level of miRNA-145 in sputum compared to serum, while in ACOS and COPD patients and controls the level of expression was almost the same in sputum and serum sample (Figure 2).
Figure 2.
Relative quantity of expression of miR-145 (A) and miR-338 (B) in sputum and serum of ACOS, asthma, and COPD patients and controls, calculated as 2−Δct.
Note: *P<0.05.
Abbreviations: ACOS, asthma–COPD overlap syndrome; COPD, chronic obstructive pulmonary disease.
Discussion
The main finding of this work is that the expression of miRNA-145 and miRNA-338 is higher in the supernatant of COPD and asthma patients than in controls. This result confirms that the lung environment participates in the pathogenesis of obstructive diseases and many biological factors are involved in the modulations of pathological mechanisms. For many years, much attention was given to biomarkers of inflammation and oxidative stress. Furthermore, the obtained results highlight that distinct patterns of miRNA expression in sputum may be involved in the pathogenesis of chronic obstructive diseases.
Different papers have recently demonstrated that some miRNAs are elevated in asthma and COPD patients.16 Moreover, the expression of miRNAs seems to be different in various asthma models. Wang et al17 identified different expressions of miR-145-5p, miR-636, miR-338-3p, miR-4485, miR-1229-3p, miR-4707-3p, and miR-3620-3p in the serum of patients with asthma, compared to patients with COPD. While in the exhaled breath condensate (EBC), Roff et al18 have analyzed the expression of 11 miRNAs, reporting decreased levels of miR-570-3p in EBC and in the serum from asthma patients. Altered miR-221 expression was documented in human primary ASM cells from patients with non-severe and severe asthma compared to healthy individuals.19
A pathophysiological role of miRNAs has been studied in asthma and in COPD and differential expression of miRNAs has been identified. Even if COPD and asthma are characterized by chronic airways inflammation, they have a distinct cellular composition of the airway inflammatory infiltrate. In fact, while in COPD, the main important role was usually played by neutrophils, macrophages, and lymphocytes, whereas eosinophils are the most prominent inflammatory cells in asthma. In addition, the inflammatory pattern is substantially different, and the miRNA expression seems to be different in these diseases.
Cao et al20 suggests a role of miR-183, miR-200b, and miR-200c in the regulation of BKCaβ1 expression, which in turn might be correlated with the severity of the disease. Donaldson et al21 found that plasma levels of muscle-specific miR-499 are associated with NF-κB p50 in mild/moderate COPD, whereas in severe and very severe disease, miR-206 and miR-133 are associated with circulating cytokines. Puig-Vilanova et al22 analyzed muscle-specific miRNAs (miR-1, miR-133, and miR-206), which were downregulated in COPD and hypothesized an adaptive mechanism to better overcome the continuous inspiratory loads of the respiratory system in COPD. However, to date, relatively very few miRNAs have been studied in detail and, hence, the precise biological relevance remains yet to be determined.
We quantified the expression of two miRNAs for which the published data indicate important parts on regulating interferon-gamma-inducible protein 30 and tumor necrosis factor receptor superfamily, miRNA-145 and miRNA-338, respectively.
A large number of miRNAs are differentially expressed in asthma in a predominantly cell-specific manner. These include miRNA-145 that has regulatory effects on the smooth muscle cell differentiation, plasticity, and phenotype differentiation and participates in the modulation of eosinophilic inflammation, mucus hypersecretion, Th2 cytokine production, and airway hyperresponsiveness in allergic airway diseases.23–25 Solberg et al26 examined 217 miRNAs in bronchial epithelium of asthma patients who have a very different level of expression compared to healthy subjects. In addition, the use of corticosteroids does not alter the expression of miRNA in the asthmatic population. O’Leary et al11 helped to identify miRNA-145 as a potential key regulator of ASM function in COPD. Despite it is highly expressed in the healthy lungs and in healthy ASM cells specifically, human and animal models showed that miRNA-145 could also contribute to the development of COPD.27–30
On the other hand, recent studies have indicated the important role of miRNA-338 in epithelial cell differentiation by facilitating the translocalization of β1-integrin to the basolateral membrane. Kos et al12 have shown that miRNA-338 regulates differentiation, apoptosis, and probably tissue degeneration, by modulation of the AATK mRNA. miRNA-338 is also downregulated in some types of human cancers, including NSCLC, where it has a role in cancer differentiation, pathological stage, and lymph node metastasis. This regulation might be explained by targeting the Ras-related protein 14 (RAB14) gene.31 Despite this evidence, the role of miRNA-338 in pulmonary functions has not been clearly identified. Our results seem to show that it could be involved in the pathogenesis of obstructive diseases. To date, we can only speculate that inflammation and cell proliferation at the base of the remodeling processes can be promoted by the activation of miRNA-338.
Another purpose of this study was to identify a novel possible biomarker that can be helpful in discriminating patients affected by ACOS. This is a common condition in which subjects present a commixture of clinical and spirometric characteristics presented in both COPD and asthma. There is a fervent debate around ACOS that seems to be a particular phenotype of COPD characterized by a younger age, less smoking history, and frequent and severe respiratory exacerbations. However, it is not always easy to distinguish ACOS subjects from COPD only using spirometry and bronchodilatator response. A biomarker able to discriminate these two entities can be useful in clinical practice. Recently, Iwamoto et al32 has shown that neutrophil gelatinase-associated lipocalin (NGAL), a protein associated with inflammation and airway injury, was higher in the sputum of ACOS patients than in COPD patients. The same paper confirmed that the sputum of COPD patients is characterized by neutrophils, while eosinophils are higher in asthma and ACOS patients. Our group studied mitochondrial DNA that seems to be altered in ACOS with respect to asthma or COPD, but it is indisputable that we are far from the routine use of these molecules in clinical practice.33
The results of the present study do not show differences between the expression of miRNA-145 and miRNA-338 in ACOS patients compared to asthma or COPD patients, and this confirms that this syndrome presents some characteristics overlapping between both diseases. However, the presence of different expression levels of miRNA in the sputum of all obstructive diseases compared to controls underlines a possible role of miRNA in the pathogenesis and development of obstructive diseases even if their role is not fully clarified because many confounding factors can influence their expression. Thus, a large population should be analyzed to have more information about their functions.
The main limitation of this study is that we selected only two miRNAs, while high numbers of them may be involved in the same diseases. Thus, a large panel should be evaluated to collect more details about their role in the pathogenesis of diseases. Nevertheless, our results can encourage the research in this field even if a large-scale validation study across multiple centers is required.
Conclusion
This pilot study suggests that miRNA could be important in obstructive diseases, because it may modulate the expression of different genes responsible for many biological processes such as development of airway epithelial cells, stress responses, formation of pulmonary surfactant, and inflammation, involved in the pathogenesis and evolution of obstructive lung diseases. Moreover, the study of miRNA may become a novel approach to evaluate new therapeutic target or the development of new miRNA-related inhaled medications.
Acknowledgments
This study was funded by tax allocated to the University of Foggia research fund in memory of Gianluca Montel.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10(3):281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson PG, Simpson JL. The overlap syndrome of asthma and COPD: what are its features and how important is it? Thorax. 2009;64(8):728–735. doi: 10.1136/thx.2008.108027. [DOI] [PubMed] [Google Scholar]
- 3.Soler-Cataluña JJ, Cosío B, Izquierdo JL, et al. Consensus document on the overlap phenotype COPD-asthma in COPD. Arch Bronconeumol. 2012;48(9):331–337. doi: 10.1016/j.arbres.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Global Initiative for Asthma Global Strategy for Asthma Management and Prevention; updated 2015. [Accessed June 5, 2017]. Available from: http://ginasthma.org/wp-content/uploads/2016/01/GINA_Report_2015_Aug11-1.pdf.
- 5.Hassan T, McKiernan PJ, McElvaney NG, Cryan SA, Greene CM. Therapeutic modulation of miRNA for the treatment of proinflammatory lung diseases. Expert Rev Anti Infect Ther. 2012;10(3):359–368. doi: 10.1586/eri.11.175. [DOI] [PubMed] [Google Scholar]
- 6.Pagdin T, Lavender P. MicroRNAs in lung diseases. Thorax. 2012;67(2):183–184. doi: 10.1136/thoraxjnl-2011-200532. [DOI] [PubMed] [Google Scholar]
- 7.Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15(17):5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Sun X, Wu Y, et al. Effects of miRNA-145 on airway smooth muscle cells function. Mol Cell Biochem. 2015;409(1–2):135–143. doi: 10.1007/s11010-015-2519-7. [DOI] [PubMed] [Google Scholar]
- 11.O’Leary L, Sevinç K, Papazoglou IM, et al. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett. 2016;590(9):1324–1334. doi: 10.1002/1873-3468.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kos A, OldeLoohuis NF, Wieczorek ML, et al. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One. 2012;7(2):e31022. doi: 10.1371/journal.pone.0031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller MR, Hankinson J, Brusasco V, et al. ATS/ERS Task Force Standardisation of spirometry. Eur Respir J. 2005;26(2):319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 14.Louhelainen N, Stark H, Mazur W, Rytilä P, Djukanovic R, Kinnula VL. Elevation of sputum matrix metalloproteinase-9 persists up to 6 months after smoking cessation: a research study. BMC Pulm Med. 2010;10:13. doi: 10.1186/1471-2466-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffl MW, Lange IG, Daxenberger A, Meyer HH. Tissue-specific expression pattern of estrogen receptors (ER): quantification of ER alpha and ER beta mRNA with real-time RT-PCR. APMIS. 2001;109(5):345–355. doi: 10.1034/j.1600-0463.2001.090503.x. [DOI] [PubMed] [Google Scholar]
- 16.Szymczak I, Wieczfinska J, Pawliczak R. Molecular background of miRNA Role in asthma and COPD: an updated insight. Biomed Res Int. 2016;2016:7802521. doi: 10.1155/2016/7802521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Huang Y, Liang Z, et al. Plasma miRNAs might be promising biomarkers of chronic obstructive pulmonary disease. Clin Respir J. 2016;10(1):104–111. doi: 10.1111/crj.12194. [DOI] [PubMed] [Google Scholar]
- 18.Roff AN, Craig TJ, August A, Stellato C, Ishmael FT. MicroRNA-570-3p regulates HuR and cytokine expression in airway epithelial cells. Am J Clin Exp Immunol. 2014;3(2):68–83. [PMC free article] [PubMed] [Google Scholar]
- 19.Perry MM, Baker JE, Gibeon DS, Adcock IM, Chung KF. Airway smooth muscle hyperproliferation is regulated by MicroRNA-221 in severe asthma. Am J Respir Cell Mol Biol. 2014;50(1):7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Z, Zhang N, Lou T, et al. microRNA-183 down-regulates the expression of BKCaβ1 protein that is related to the severity of chronic obstructive pulmonary disease. Hippokratia. 2014;18(4):328–332. [PMC free article] [PubMed] [Google Scholar]
- 21.Donaldson A, Natanek SA, Lewis A, et al. Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 2013;68(12):1140–1149. doi: 10.1136/thoraxjnl-2012-203129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puig-Vilanova E, Aguiló R, Rodríguez-Fuster A, Martínez-Llorens J, Gea J, Barreiro E. Epigenetic mechanisms in respiratory muscle dysfunction of patients with chronic obstructive pulmonary disease. PLoS One. 2014;9(11):e111514. doi: 10.1371/journal.pone.0111514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahan D, Ekman M, Larsson-Callerfelt AK, et al. Induction of angiotensin-converting enzyme after miR-143/145 deletion is critical for impaired smooth muscle contractility. Am J Physiol Cell Physiol. 2014;307(12):C1093–C1101. doi: 10.1152/ajpcell.00250.2014. [DOI] [PubMed] [Google Scholar]
- 24.Joshi SR, Comer BS, McLendon JM, Gerthoffer WT. MicroRNA regulation of smooth muscle phenotype. Mol Cell Pharmacol. 2012;4(1):1–16. [PMC free article] [PubMed] [Google Scholar]
- 25.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128(1):160–167. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Solberg OD, Ostrin EJ, Love MI, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186(10):965–974. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams AE, Larner-Svensson H, Perry MM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One. 2009;4(6):e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry MM, Tsitsiou E, Austin PJ, et al. Role of non-coding RNAs in maintaining primary airway smooth muscle cells. Respir Res. 2014;15:58. doi: 10.1186/1465-9921-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry MM, Adcock IM, Chung KF. Role of microRNAs in allergic asthma: present and future. Curr Opin Allergy Clin Immunol. 2015;15(2):156–162. doi: 10.1097/ACI.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 30.Brook PO, Perry MM, Adcock IM, Durham AL. Epigenome-modifying tools in asthma. Epigenomics. 2015;7(6):1017–1032. doi: 10.2217/epi.15.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Chen P, Zu L, Liu B, Wang M, Zhou Q. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am J Cancer Res. 2016;6(2):127–140. [PMC free article] [PubMed] [Google Scholar]
- 32.Iwamoto H, Gao J, Koskela J, et al. Differences in plasma and sputum biomarkers between COPD and COPD-asthma overlap. Eur Respir J. 2014;43(2):421–429. doi: 10.1183/09031936.00024313. [DOI] [PubMed] [Google Scholar]
- 33.Carpagnano GE, Lacedonia D, Carone M, et al. Study of mitochondrial DNA alteration in the exhaled breath condensate of patients affected by obstructive lung diseases. J Breath Res. 2016;10(2):026005. doi: 10.1088/1752-7155/10/2/026005. [DOI] [PubMed] [Google Scholar]