Abstract
Fluorescent pseudomonads protecting plant roots from phytopathogens by producing 2,4-diacetylphloroglucinol (DAPG) are considered to form a monophyletic lineage comprised of DAPG+ Pseudomonas strains in the “P. corrugata” and “P. protegens” subgroups of the “Pseudomonas fluorescens” group. However, DAPG production ability has not been investigated for many species of these two subgroups, and whether or not the DAPG+ Pseudomonas are truly monophyletic remained to be verified. Thus, the distribution of the DAPG biosynthetic operon (phlACBD genes) in the Pseudomonas spp. was investigated in sequenced genomes and type strains. Results showed that the DAPG+ Pseudomonas include species of the “P. fluorescens” group, i.e., P. protegens, P. brassicacearum, P. kilonensis, and P. thivervalensis, as expected, as well as P. gingeri in which it had not been documented. Surprisingly, they also include bacteria outside the “P. fluorescens” group, as exemplified by Pseudomonas sp. OT69, and even two Betaproteobacteria genera. The phl operon-based phylogenetic tree was substantially congruent with the one inferred from concatenated housekeeping genes rpoB, gyrB, and rrs. Contrariwise to current supposition, ancestral character reconstructions favored multiple independent acquisitions rather that one ancestral event followed by vertical inheritance. Indeed, based on synteny analyses, these acquisitions appeared to vary according to the Pseudomonas subgroup and even the phylogenetic groups within the subgroups. In conclusion, our study shows that the phl+ Pseudomonas populations form a polyphyletic group and suggests that DAPG biosynthesis might not be restricted to this genus. This is important to consider when assessing the ecological significance of phl+ bacterial populations in rhizosphere ecosystems.
Keywords: DAPG; 2,4-diacetylphloroglucinol; phl Operon; biocontrol agent; phlACBD genes
Introduction
About a century ago, the Pseudomonas genus was defined as a bacterial group gathering all aerobes, bacilli, and Gram-negative bacteria (Migula, 1894). The Pseudomonas classification has since considerably evolved through molecular phylogenetics, and phylogenies obtained using the rrs gene encoding 16S rRNA allowed the reclassification of many species into other bacterial genera (Brosch et al., 1996; Grimont et al., 1996; Anzai et al., 2000). Other molecular markers were also used to trace the evolutionary history of Pseudomonas, such as rpoB, rpoD, and gyrB genes (Ait Tayeb et al., 2005). In recent times, the use of concatenated phylogenetic markers or complete genome sequences has enabled a robust delimitation of the Pseudomonas genus (Yamamoto et al., 2000; Hilario et al., 2004; Frapolli et al., 2007; Mulet et al., 2010; Gomila et al., 2015; Garrido-Sanz et al., 2016), which encompasses several groups of closely-related species, such as the “P. fluorescens” group, “P. putida” group, “P. aeruginosa” group, “P. stuzeri” group, and “P. syringae” group.
Numerous Pseudomonas species or strains are presenting specific interactions with their eukaryotic hosts, such as humans (many strains belonging to the “P. aeruginosa” group), fungi (strains belonging to the species Pseudomonas tolaasii), or plants (many strains from the “P. fluorescens” group) (Munsch et al., 2000; Finnan et al., 2004; Mithani et al., 2010). Interaction with plants may involve (i) deleterious effects in the case of Pseudomonas plant pathogens, such as strains belonging to the species P. syringae, P. corrugata, or P. mediterranea (Catara et al., 2002; O'Brien et al., 2011; Trantas et al., 2015), or (ii) beneficial effects (Mirza et al., 2006) especially by protecting plants against a wide range of plant pathogens or pests (Mazurier et al., 2009; Almario et al., 2013a; Kupferschmied et al., 2013) through the production of many secondary metabolites such as 2,4-diacetylphloroglucinol (DAPG). This compound was also involved in the elicitation of plant defenses through induced systemic resistance (Iavicoli et al., 2003; Weller et al., 2012) and directly in the modulation of plant hormonal balance by acting as an auxin-mimetic compound (Brazelton et al., 2008).
DAPG synthesis has been so far associated with the presence of the phl gene cluster, comprising the phlACBDE operon and other genes such as phlF, phlG, phlH, and phlI. The DAPG synthesis probably starts with the type III polyketide synthase (encoded by gene phlD), which allows the cyclization of malonyl-CoA to phloroglucinol (Achkar et al., 2005). Then, a monoacetylphloroglucinol (MAPG) acetyltransferase (encoded by phlACB) acetylates phloroglucinol to MAPG and then DAPG (Kidarsa et al., 2011). Besides the phlACBD genes directly involved in DAPG biosynthesis, the last gene of the phl operon (phlE) is involved in the efflux of DAPG (Abbas et al., 2004). Next to the phl operon, phlF and phlH code for transcriptional regulators, involved respectively in the repression and activation of DAPG production (Schnider-Keel et al., 2000). Gene phlG encodes a hydrolase allowing a return of DAPG to MAPG (Bottiglieri and Keel, 2006). The role of phlI (encoding a hypothetical protein) in DAPG production is still unknown (Hayashi et al., 2012).
DAPG+ Pseudomonas were isolated around the world and exhibited a cosmopolitan distribution (Wang et al., 2001; Almario et al., 2013b, 2014; Vacheron et al., 2016). They are ecologically important as they can control phytopathogens and contribute to disease suppressiveness (Sanguin et al., 2008; Almario et al., 2013a). Multilocus sequence analysis based on housekeeping genes classified them into six main phylogenetic groups noted A to F (Frapolli et al., 2007, 2012), belonging to the “P. corrugata” subgroup (multilocus phylogenetic groups A to E) and the “P. protegens” subgroup (multilocus phylogenetic group F) of the “P. fluorescens” group defined by Mulet et al. (2010). DAPG production is thus documented in some species of the “P. corrugata” subgroup (like P. brassicacearum) and “P. protegens” subgroup (like P. protegens), but it has not been investigated extensively in many species of these subgroups. Consequently, it is unclear whether or not phl+ Pseudomonas form a monophyletic group within the “P. fluorescens” group, as it was claimed (Moynihan et al., 2009). Similarly, the ability to produce DAPG has been proposed as a highly conserved trait (Moynihan et al., 2009), but again it was based on a rather limited number of species and genomes. Recent progress in the sequencing of Pseudomonas genomes, the phylogenetic assessment of phl+ Pseudomonas (Frapolli et al., 2012) and the description of the Pseudomonas genus (including new species and the recognition of “P. protegens” as a new subgroup; Gomila et al., 2015; Garrido-Sanz et al., 2016, 2017) make it now possible to conduct a more global assessment of this issue.
The aim of this study was to assess the distribution of the DAPG biosynthetic operon (phlACBD) among sequenced Pseudomonas genomes including some type strains. The occurrence of DAPG production ability in Pseudomonas spp. was determined, based on presence of the phlACBD operon and confirmatory HPLC analyses, and the phylogenies of phl+ and phl− Pseudomonas spp. were compared. In addition, the genomic context of the phlACBD operon in the phl+ Pseudomonas genomes available was further explored.
Materials and methods
Cultivation of Pseudomonas type strains and DNA extraction
The 12 Pseudomonas type strains (Table S1) from the “P. corrugata,” “P. chlororaphis,” and “P. fluorescens” subgroups were routinely grown on Luria-Bertani agar (Sambrook et al., 1989). For genomic DNA extraction, bacterial strains were grown overnight with shaking (150 rpm) in 20 mL of liquid LB medium, and DNA was extracted from 500 μL of bacterial culture using the NucleoSpin Tissue kit (Macherey-Nagel, Hoerdt, France), following the manufacturer's instructions. DNA was quantified spectrophotometrically and adjusted to 30 ng μL−1.
Search for phl genes in Pseudomonas type strains
The search for the phl operon (phlACBD genes) in genome-sequenced Pseudomonas type strains was carried out by BlastN using phlACBD genes sequences of P. kilonensis F113 as query. Blast parameters were fixed at 90% coverage and 75% identity in gene sequences (E < 10−20). In addition, confirmatory evidence for presence of absence of these genes in the 12 Pseudomonas type strains (Table S1) was sought by PCR targeting phlD (Figure S1). PCR was carried out in 50-μL volumes containing 3% DMSO, 1 × buffer (Roche Applied Science, Meylan, France), 1.5 mM MgCl2, 100 μM of each dNTP, 1 μM of primers B2BF and BPR4 (Almario et al., 2013a), 1.8 U of Taq Expand High Fidelity DNA polymerase (Roche Applied Science) and 1 μL of template genomic DNA. The cycling program included 3 min at 94°C, 30 amplification cycles of 1 min at 94°C, 1 min at 62°C and 1 min at 72°C, and an elongation step of 3 min at 72°C. In the absence of amplification, the phlD negative status of the strains was verified by conducting the PCR at lower primer hybridization temperatures (60, 58, 56, and 54°C instead of 62°C). PCR products were purified (MinElute PCR purification kit; Qiagen, Courtaboeuf, France) and both strands were sequenced (LGC Genomics, Berlin, Germany). The sequence pairs were trimmed and assembled with BioEdit v.7.0 (Hall, 1999), and their identity levels with phlD were determined using the BlastN algorithm and the nr Nucleotide Sequence Database.
DAPG quantification by HPLC
DAPG production was measured by HPLC (Bonsall et al., 1997). Samples were analyzed with an Agilent 1200 series HPLC (Agilent Technologies, Santa Carla, USA) equipped with a degasser (G132A), a quaternary pump module (G1311A), an automatic sampler (G1329A), and a diode array detector (DAD G1315B). Separation of compounds was performed at room temperature with a C18 reverse-phase column. For each sample, 20 μL were injected and eluted at 1 mL min−1 using a step-by-step gradient increasing acetonitrile proportion in water: the gradient started at 40% of acetonitrile over 4 min and rose from 40 to 64% in 7.5 min, then reached 75% at 16.5 min, and ended at 100% at 18.5 min (100% maintained for 3 min before decreasing to 40% for 5 min). Chromatograms were recorded at 270 nm (maximum of absorbance of DAPG). The Chemstation Agilent software was used for integration of chromatograms, and quantitation of DAPG was done according to a standard curve with a chemical standard (Sigma-Aldrich). This experiment was done three times independently.
Phylogenetic analysis of Pseudomonas strains
The reconstruction of the Pseudomonas species phylogenetic tree was based on rpoB, gyrB, and rrs housekeeping genes. Nucleotide sequences were retrieved from Genbank (Table S1), then aligned using MUSCLE (Edgar, 2004) and informative positions were selected using Gblocks with relaxed parameters (Castresana, 2000). Alignments of the three genes were then concatenated and used to compute a Maximum Likelihood tree using PhyML (Guindon et al., 2010) with the following parameters: HKY85 model (Hasegawa et al., 1985), SPR topology search, estimation of invariants sites, and robustness assessment with 1,000 bootstraps. The phylogenetic reconstruction of the phlACBD operon was done using the same procedure. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified (e.g., strain F113) were renamed based on rrs-rpoD-gyrB phylogeny and on average nucleotide identity (ANI) data (Richter and Rosselló-Móra, 2009) (Figure 1A, Figure S2 and Tables S2–S7).
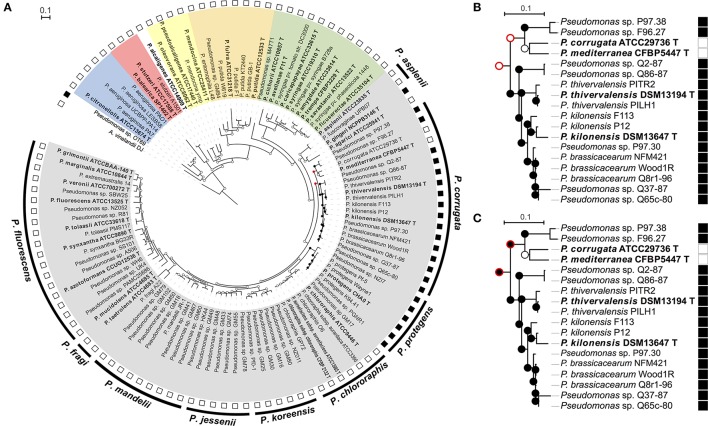
Figure 1.
Ancestral state character reconstruction by Maximum Parsimony of the gains and losses of the phl operon. (A) Phylogenetic rrs-gyrB-rpoB tree of selected Pseudomonas strains in which each node indicates presence (black circle) or absence of phl operon (phlACBD) (white circle). The red circle indicates multiple, equally-parsimonious scenarios that are shown in (B,C). The outer ring represents the subgroups defined in the “Pseudomonas fluorescens” group according to Mulet et al. (2010), the second ring correspond to the presence (black square) or the absence (white square) of the phl cluster. The different Pseudomonas groups are highlighted in color: Gray color, “P. chlororaphis” group; Green, “P. syringae” group; Purple, “P. putida” group; Yellow, “P. oleovorans” group; Red, “P. stutzeri” group; Blue, “P. aeruginosa” group. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7). (B,C) The two equally parsimonious scenarios inferred by ancestral reconstruction for the most recent common ancestor of the “P. corrugata” subgroup. Contradicting nodes are denoted by a red outer circle. Strains P97.30, Q37-87, and Q65c-80 likely belong to P. brassicacearum but are portrayed as Pseudomonas sp. in the absence of ANI data.
Ancestral state reconstruction in Pseudomonas
Mesquite (Maddison and Maddison, 2010) was used to infer the presence or absence of the phlACBD operon at each node of the Pseudomonas species phylogenetic tree based on its distribution in terminal taxa. Analyses were done using the Maximum Parsimony method, which allows reconstructing ancestral states by minimizing character change events along a phylogenetic tree.
Genetic environment of the phl cluster in Pseudomonas genomes
Sequenced genomes of phl+ strains (Table S1) available in april 2016 were recovered and the 40-kb flanking genetic environment of the phl cluster (phlACBD operon and genes phlE, phlI, phlH, phlG, and phlF), encompassing the upstream and downstream regions, was analyzed using online JGI Integrated Microbial Genomes tools (https://img.jgi.doe.gov/).
The 40-kb flanking regions were screened for signs of genome instability based on G + C content analysis in Artemis (Rutherford et al., 2000), tRNA detection using tRNAscan-SE (Schattner et al., 2005), insertion sequence detection using ISfinder (Kichenaradja et al., 2010) and genomic island prediction using Island Viewer (Langille and Brinkman, 2009).
Results
Distribution of the phl operon among Pseudomonas strains
A total of 24 of 111 Pseudomonas strains displayed phlACBD genes. As expected, phl+ Pseudomonas strains clustered either in the “P. corrugata” or the “P. protegens” subgroup of the “P. fluorescens” group (Figure 1A and Figure S2). Within the “P. corrugata” subgroup, two clades could be distinguished. A first clade comprised phl+ strains from the multilocus phylogenetic groups A, B, C, and D defined in Frapolli et al. (2007), as well as phl+ type strains P. brassicacearumT, P. kilonensisT, and P. thivervalensisT. A second clade within the “P. corrugata” subgroup comprised phl+ strains (Pseudomonas sp. F96.27 and P97.38) from multilocus phylogenetic group E along with phl− P. mediterraneaT and P. corrugataT. Within the “P. protegens” subgroup, all the strains were harboring a phl operon and belonged to the multilocus phylogenetic group F defined in Frapolli et al. (2007). When the capacity to produce DAPG was tested in representative strains (including type strains) from the “P. chlororaphis” group, i.e., (i) P. fluorescensT, P. tolaasiiT, P. marginalisT (“P. fluorescens” subgroup), (ii) P. chlororaphis subsp. aurantiacaT, P. chlororaphis subsp. aureofaciensT, P. chlororaphisT (“P. chlororaphis” subgroup), (iii) P. brassicacearum T, P. kilonensisT, P. kilonensis F113 (formerly P. “fluorescens” F113), P. mediterraneaT, P. thievervalensisT, P. corrugata T (“P. corrugata” subgroup), (iv) P. protegens T, P. protegens Pf-5 (“P. protegens” subgroup), and (v) P. aspleniiT, P. fuscovaginaeT, P. rhodesiaeT, P. gingeriT (“P. asplenii” subgroup), only pseudomonads harboring the phlACBD genes could produce DAPG (Figure S3). Besides the “P. corrugata” and “P. protegens” subgroups, two other Pseudomonas strains possess a phl operon. The first one, P. gingeri NCPPB3146 T, is a member of the “P. fluorescens” group but does not belong to any defined subgroup (Mulet et al., 2010), whereas the second one, Pseudomonas sp. OT69 is a Pseudomonas strain clustering outside of the “P. fluorescens” group (Figure 1A).
Phylogeny of phl+ Pseudomonas
The phlACBD-based phylogenetic tree was substantially congruent with the one inferred from concatenated housekeeping genes rpoB, gyrB, and rrs since the differences between the two trees were restricted to lower-level branches and were not supported by high bootstrap scores (Figure S2). The phlACBD-based phylogenetic tree presented four clades. A first phlACBD clade included strains belonging to the “P. corrugata” subgroup (Figure S2), with the same separation found previously (Frapolli et al., 2007) between the multilocus phylogenetic groups A, B, C, and D on one side and group E on the other. A second phl clade comprised strains belonging to the “P. protegens” subgroup (multilocus group F). Finally, two clades harboring a single strain each, i.e., P. gingeri NCPPB3146 T and Pseudomonas sp. OT69, which were found in more ancestral branches of the phl operon tree (Figure S2).
The four clades of phl+ Pseudomonas described above were clearly distinguished in the rrs-gyrB-rpoB tree (Figure 1A and Figure S2). Indeed, strains from multilocus groups A to D and type strains P. brassicacearumT, P. kilonensisT, and P. thivervalensisT formed one clade and were separated from group-E strains of the same “P. corrugata” subgroup, while strains from multilocus group F (i.e., “P. protegens” subgroup) clustered together, and P. gingeriT remained on a single branch forming an outgroup.
Ancestral state reconstruction for the phl operon
The polyphyletic distribution of phlACBD suggests that this operon could have undergone multiple losses and/or horizontal transfer events. Ancestral state reconstruction for the phl operon supported a scenario with multiple acquisitions. Indeed, this operon was independently acquired by Pseudomonas sp. OT69, P. gingeri NCPPB3146T and by the most recent common ancestor (MRCA) of the “P. protegens” subgroup (Figure 1A), and it was acquired possibly once or twice by the MRCA of the “P. corrugata” subgroup. Two parsimonious scenarios are possible to explain acquisition(s) of the phl operon by species of the “P. corrugata” subgroup. In the first scenario, two independent acquisitions can be proposed, i.e., by the MRCA of Pseudomonas strains P97.38 and F96.27 and the MRCA of the clade comprised of P. thivervalensis, P. kilonensis, P. brassicacearum, and Pseudomonas strains Q2-87 and Q65c-80 (Figure 1B). The second scenario is an ancestral acquisition by the MRCA of the “P. corrugata” subgroup and a subsequent loss in the MRCA of P. corrugata ATCC29736/P. mediterranea CFBP5447 (Figure 1C).
Seeking the nr database for phl homologs, the gene cluster containing phlFACBDE was also retrieved outside the Pseudomonas clade. This was the case for several Betaproteobacteria, i.e., Chromobacterium vaccinii MWU328, C. vaccinii MWU205, Chromobacterium piscinae ND17, and Pseudogulbenkiania ferrooxidans EGD-HP2 (Figure 2 and Figure S4).
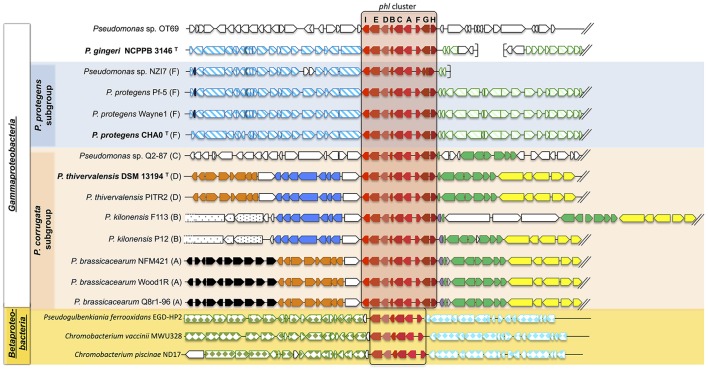
Figure 2.
Synteny of the genomic regions flanking the phl cluster among Pseudomonas genomes. The phl cluster is colored in red. Letters correspond to the name of each phl gene. Type strains are shown in bold, and letters following the strain name correspond to the multilocus phylogenetic groups defined in Frapolli et al. (2007). Each group of genes of homologous synteny downstream (at the left of the phl cluster) or upstream (at the right of the phl cluster) is presented using the same color/pattern, and strain-specific genes using white color. Brackets indicate the beginning or the end of a contig. The unclassified strains are positioned in a subgroup according to the phylogeny obtained in Figure 1. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7).
Genomic regions flanking the phl cluster in Pseudomonas
The genomic environment of the phl cluster (phlACBD operon and regulation/transport genes phlE, phlF, phlG, phlH, and phlI) was investigated 20 kb upstream and 20 kb downstream of the phl cluster (Figure 2). Apart from the phl cluster itself, no gene was common to all strains in the 40 kb surrounding region (Figure 2). However, the synteny showed higher conservation for strains belonging to the same Pseudomonas subgroup.
In the “P. corrugata” subgroup, comparing the available genomes, the 40 kb genomic region surrounding the phl cluster showed high plasticity since only seven genes were conserved amongst all the strains analyzed (on the right of the phl cluster in Figure 2). Five different groups of genes presented synteny between “P. corrugata” subgroup strains. However, the distribution and position of these gene clusters were identical only within each multilocus group. Indeed, bacteria from the multilocus group A encompassing P. brassicacearum NFM421, Wood1R, and Q8r1-96 shared the exact same gene order in the vicinity of the phl cluster (Figure 2). Only multilocus group B (P. kilonensis P12 and F113) presented internal dissimilarity upstream of the phl cluster. P. kilonensis F113 showed an insertion of three genes for synthesis of an insecticidal toxin complex protein (cable pili-associated adhesin protein, virulence plasmid A protein, and an insecticidal toxin).
In the “P. protegens” subgroup (corresponding to multilocus group F), the region surrounding the phl cluster showed complete synteny (Figure 2). This was also the case for the downstream region of P. gingeri even though this strain falls outside the “P. protegens” subgroup.
Only Pseudomonas sp. OT69 harbored a specific phl genomic environment sharing no similarity with any other phl+ strain (Figure 2).
Insertion sequences, genes coding for tRNA, integrases or transposases, or other genes associated with genome instability were not found in the vicinity of the phl cluster in the genomes of the “P. fluorescens” group. In addition, there was no marked difference in G + C content when comparing the phl cluster to the rest of the genome (data not shown). Outside the “P. fluorescens” group, Pseudomonas sp. OT69 displayed a 29,375-bp putative genomic island of 28 genes (including the phl cluster) with a codon usage different from the rest of the genome, based on Hidden Markov Model analysis, suggesting recent horizontal transfer of this gene cluster.
Discussion
So far, analysis of the evolutionary history of the phl cluster has focused mainly on the phylogenetic relations among established phl+ pseudomonads, omitting closely-related Pseudomonas species of uncertain phl status. Here, we investigated the distribution of the phl cluster among a broader taxonomic range of Pseudomonas species, then established phylogenetic relations between phl+ and phl− Pseudomonas, and explored the genomic context of the phl cluster.
Within the “P. corrugata” subgroup, besides P. protegens and P. brassicacearum (for which DAPG production was documented; Ortet et al., 2011; Ramette et al., 2011; Bruto et al., 2014), the phl operon was also found in P. kilonensis and P. thivervalensis types strains, a possibility already suggested by the observation that closely-related pseudomonads displayed phlD (Frapolli et al., 2012). The phl operon is absent from the P. fluorescens species, which conflicts with the many literature reports indicating otherwise, but in which P. fluorescens was used as a convenient generic term for ill-defined fluorescent pseudomonads with biocontrol properties rather than an established taxonomic status per se (Couillerot et al., 2009). In the same way, an incorrect species assignment was detected for the strain P. “alcaligenes” OT69, which appears far from the P. alcaligenes type strain in the phylogenetic tree (Figure 1). Overall, the phl+ Pseudomonas did not form a monophyletic group, as the phl operon was found (i) in some but not all species of the “P. corrugata” subgroup, (ii) in all strains of the “P. protegens” subgroup, and (iii) in P. gingeri and Pseudomonas sp. OT69 but not in pseudomonads closely related to any of the latter.
Moynihan et al. (2009) proposed that the phl cluster was present in the last common ancestor of the “P. fluorescens” group but was only retained in one branch, giving rise to a divide between a phl+ and a phl− group of fluorescent Pseudomonas and thus a paraphyletic distribution. However, our results based on a higher number of species (type strains) and related strains do not support this hypothesis. On the contrary, ancestral character reconstructions favored multiple independent acquisitions rather than one ancestral event followed by vertical inheritance. First, ancestral state reconstruction (Figure 1) indicates that the phl cluster was absent from the last common ancestor of the “P. fluorescens” group, and was probably acquired three times in this group, i.e., by the MRCA of the “P. corrugata” subgroup, by the MRCA of the “P. protegens” subgroup, and by the species P. gingeri. Second, phl+ Pseudomonas form a polyphyletic group, in that the phl cluster is not present in all descendants of the last common ancestor of phl+ Pseudomonas, suggesting that the phl cluster may have undergone several loss and/or horizontal transfer events. Thus, the only uncertainty in the reconstruction was found in the “P. corrugata” subgroup where two equally-likely scenarios may be proposed. However, the scenario suggesting an acquisition by the MRCA of the “P. corrugata” subgroup and a subsequent loss by the MRCA of P. corrugata ATCC29736T/P. mediterranea CFBP5447T can be favored on the basis of two arguments. First, large scale genomic analysis suggests that gene loss is more common and likely than genomic acquisitions (Wolf and Koonin, 2013). Second, P. corrugata ATCC29736T and P. mediterranea CFBP5447T are the only species in the “P. corrugata” subgroup known to be plant pathogens, causing pith necrosis in tomato, eggplant, pepper, and tobacco (Catara et al., 2002). Since DAPG is able to elicit plant defenses via ISR pathways (Iavicoli et al., 2003), the phl cluster may have been counter-selected and lost in these species during their adaptation to a phytopathogenic lifestyle. In the case of P. syringae, gene loss is a major mechanism of adaptation to pathogenicity (Mithani et al., 2010), and indeed it was recently shown that P. corrugata and P. mediterranea genomes specialized into pathogenicity via gene loss events (Trantas et al., 2015).
While signs of recent horizontal transfers were only found in the distant strain Pseudomonas sp. OT69, no signature was identified in the other Pseudomonas strains, supporting the ancestral acquisition by each clade. Recent literature proposed that the phlACBD operon was presumably formed in Pseudomonas through the acquisition of phlACB from Archaea and phlD from another unclear origin, possibly Streptomyces, Vibrio (Kidarsa et al., 2011), or even plants (Cook et al., 1995; Ramette et al., 2001). The current study showed that the phlACBD operon (and genes phlE and phlF) is also present in the Betaproteobacteria C. vaccinii and Pseudogulbenkiania ferrooxidans EGD-HP2, suggesting a more complex evolutionary history (Figures 2 and Figure S4).
Thus, we have shown that the phl+ Pseudomonas form a polyphyletic group. This is an important finding to take into consideration when assessing the ecological significance of DAPG+ populations, which have been proposed as soil health bioindicators because of their contribution to the suppression of plant diseases (Sanguin et al., 2008; Almario et al., 2013a). First, it means that “DAPG-producing bacteria” corresponds to a broader phylogenetic range than usually thought (Sanguin et al., 2008). Second, it raises the possibility of more diversified regulation patterns for DAPG production than in Pseudomonas strains investigated so far (Duffy and Défago, 1999; Schnider-Keel et al., 2000). Third, a larger range of DAPG-producing bacteria implies more varied combinations of ecological traits in addition to DAPG production, which are likely to influence the response of these bacteria to environmental factors and their contribution to ecological processes such as plant-disease suppression (Sanguin et al., 2008; Frapolli et al., 2010; Schreiner et al., 2010).
Author contributions
JA, MB, YM, and DM conceived and designed the experiments. JA performed the experiment. MB performed the ancestral state reconstruction, JV and CP the DAPG HPLC analyses. JV, MB, JA, and DM analyzed the data and JA, JV, YM, and DM wrote the manuscript; all authors contributed to the discussion and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JA and MB were supported by a Ph.D. grant by the Ministère Français de la Recherche. JV was supported by a Ph.D. grant from Academic Research Cluster 3 of Rhône-Alpes Region. This study was supported by the ANR project SymbioMaize (ANR-12-JSV7-0014-01). We thank Wafa Achouak (CEA, Cadarache, France) and Christelle Gruffaz (Université de Strasbourg, France) for gift of Pseudomonas type strains, and Audrey Dubost for iBio platform expertise.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01218/full#supplementary-material
Presence/absence of phlD gene in the 12 Pseudomonas type strains sought by PCR targeting phlD. MW, molecular weight; 1 and 6, negative control; 2, P. brassicacearum T; 3, P. kilonensis T; 4, P. thivervalensis T; 5, P. mediterranea T; 7, P. corrugata T; 8, P. protegens T; 9, 10, and 15, P. kilonensis F113 (positive control); 11, P. chlororaphis subsp. aurantiaca T; 12, P. chlororaphis subsp. aureofaciens T; 13, P. chlororaphis subsp. chlororaphis T; 14, P. tolaasii T; 16, P. marginalis T; 17, P. chlororaphis T.
Comparison of species tree of phl+ Pseudomonas strains and phlACBD tree for these bacteria. (A) Phylogenetic analysis based on concatenated housekeeping genes rpoB, gyrB, and rrs. When available, membership to the multilocus phylogenetic groups defined in Frapolli et al. (2007) is given in parenthesis. (B) Phylogenetic analysis of phl+ Pseudomonas based on nucleotide sequences for the operon phlACBD. In both panels, the Maximum Likelihood tree was inferred using PhyML model, and nodal robustness was assessed using 1,000 bootstrap replicates. Type strains are shown in bold and written in blue. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7).
DAPG production in Pseudomonas type strains. (A) HPLC chromatogram, UV, and mass spectra of DAPG standard. (B) HPLC detection of DAPG in Pseudomonas strain supernatants.
Phylogenetic analysis of the phl operon for the main Pseudomonas subgroups and betaproteobacterial species. When available, membership to the multilocus phylogenetic groups defined in Frapolli et al. (2007) is given in parenthesis. The Maximum Likelihood tree was inferred using PhyML model, and nodal robustness was assessed using 500 bootstrap replicates. The “P. corrugata” subgroup is highlighted in orange and the “P. protegens” subgroup in blue. Type strains are indicated in bold and red. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7).
Pseudomonas strains and accession numbers for nucleotidic sequences used in this study.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. protegens species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. protegens species.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. chlororaphis species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. chlororaphis species.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. brassicacearum, P. kilonensis, and P. thivervalensis species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. brassicacearum, P. kilonensis, and P. thivervalensis species.
References
- Abbas A., McGuire J. E., Crowley D., Baysse C., Dow M., O'Gara F. (2004). The putative permease PhlE of Pseudomonas fluorescens F113 has a role in 2,4-diacetylphloroglucinol resistance and in general stress tolerance. Microbiology 150, 2443–2450. 10.1099/mic.0.27033-0 [DOI] [PubMed] [Google Scholar]
- Achkar J., Xian M., Zhao H., Frost J. W. (2005). Biosynthesis of phloroglucinol. J. Am. Chem. Soc. 127, 5332–5333. 10.1021/ja042340g [DOI] [PubMed] [Google Scholar]
- Ait Tayeb L., Ageron E., Grimont F., Grimont P. A. D. (2005). Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res. Microbiol. 156, 763–773. 10.1016/j.resmic.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Almario J., Moënne-Loccoz Y., Muller D. (2013a). Monitoring of the relation between 2,4-diacetylphloroglucinol-producing Pseudomonas and Thielaviopsis basicola populations by real-time PCR in tobacco black root-rot suppressive and conducive soils. Soil Biol. Biochem. 57, 144–155. 10.1016/j.soilbio.2012.09.003 [DOI] [Google Scholar]
- Almario J., Muller D., Défago G., Moënne-Loccoz Y. (2014). Rhizosphere ecology and phytoprotection in soils naturally suppressive to Thielaviopsis black root rot of tobacco. Environ. Microbiol. 16, 1949–1960. 10.1111/1462-2920.12459 [DOI] [PubMed] [Google Scholar]
- Almario J., Prigent-Combaret C., Muller D., Moënne-Loccoz Y. (2013b). Effect of clay mineralogy on iron bioavailability and rhizosphere transcription of 2,4-diacetylphloroglucinol biosynthetic genes in biocontrol Pseudomonas protegens. Mol. Plant Microbe Interact. 26, 566–574. 10.1094/MPMI-11-12-0274-R [DOI] [PubMed] [Google Scholar]
- Anzai Y., Kim H., Park J. Y., Wakabayashi H., Oyaizu H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50, 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Bonsall R. F., Weller D. M., Thomashow L. S. (1997). Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63, 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottiglieri M., Keel C. (2006). Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2,4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 72, 418–427. 10.1128/AEM.72.1.418-427.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton J. N., Pfeufer E. E., Sweat T. A., McSpadden Gardener B. B., Coenen C. (2008). 2,4-diacetylphloroglucinol alters plant root development. Mol. Plant Microbe Interact. 21, 1349–1358. 10.1094/MPMI-21-10-1349 [DOI] [PubMed] [Google Scholar]
- Brosch R., Lefèvre M., Grimont F., Grimont P. A. D. (1996). Taxonomic diversity of pseudomonads revealed by computer-interpretation of ribotyping data. Syst. Appl. Microbiol. 19, 541–555. 10.1016/S0723-2020(96)80026-9 [DOI] [Google Scholar]
- Bruto M., Prigent-Combaret C., Muller D., Moënne-Loccoz Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 4:6261. 10.1038/srep06261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Catara V., Sutra L., Morineau A., Achouak W., Christen R., Gardan L. (2002). Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1749–1758. 10.1099/00207713-52-5-1749 [DOI] [PubMed] [Google Scholar]
- Cook R. J., Thomashow L. S., Weller D. M., Fujimoto D., Mazzola M., Bangera G., et al. (1995). Molecular mechanisms of defense by rhizobacteria against root disease. Proc. Natl. Acad. Sci. U.S.A. 92, 4197–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillerot O., Prigent-Combaret C., Caballero-Mellado J., Moënne-Loccoz Y. (2009). Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 48, 505–512. 10.1111/j.1472-765X.2009.02566.x [DOI] [PubMed] [Google Scholar]
- Duffy B. K., Défago G. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnan S., Morrissey J. P., O'Gara F., Boyd E. F. (2004). Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J. Clin. Microbiol. 42, 5783–5792. 10.1128/JCM.42.12.5783-5792.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frapolli M., Défago G., Moënne-Loccoz Y. (2007). Multilocus sequence analysis of biocontrol fluorescent Pseudomonas spp. producing the antifungal compound 2,4-diacetylphloroglucinol. Environ. Microbiol. 9, 1939–1955. 10.1111/j.1462-2920.2007.01310.x [DOI] [PubMed] [Google Scholar]
- Frapolli M., Défago G., Moënne-Loccoz Y. (2010). Denaturing gradient gel electrophoretic analysis of dominant 2,4-diacetylphloroglucinol biosynthetic phlD alleles in fluorescent Pseudomonas from soils suppressive or conducive to black root rot of tobacco. Soil Biol. Biochem. 42, 649–656. 10.1016/j.soilbio.2010.01.005 [DOI] [Google Scholar]
- Frapolli M., Pothier J. F., Défago G., Moënne-Loccoz Y. (2012). Evolutionary history of synthesis pathway genes for phloroglucinol and cyanide antimicrobials in plant-associated fluorescent pseudomonads. Mol. Phylogenet. Evol. 63, 877–890. 10.1016/j.ympev.2012.02.030 [DOI] [PubMed] [Google Scholar]
- Garrido-Sanz D., Arrebola E., Martínez-Granero F., García-Méndez S., Muriel C., Blanco-Romero E., et al. (2017). Classification of isolates from the Pseudomonas fluorescens complex into phylogenomic groups based in group-specific markers. Front. Microbiol. 8:413. 10.3389/fmicb.2017.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Sanz D., Meier-Kolthoff J. P., Göker M., Martín M., Rivilla R., Redondo-Nieto M. (2016). Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 11:e0150183. 10.1371/journal.pone.0150183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila M., Peña A., Mulet M., Lalucat J., García-Valdés E. (2015). Phylogenomics and systematics in Pseudomonas. Evol. Genomic Microbiol. 6:214 10.3389/fmicb.2015.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont P. A. D., Vancanneyt M., Lefèvre M., Vandemeulebroecke K., Vauterin L., Brosch R., et al. (1996). Ability of Biolog and Biotype-100 systems to reveal the taxonomic diversity of the pseudomonads. Syst. Appl. Microbiol. 19, 510–527. 10.1016/S0723-2020(96)80024-5 [DOI] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hasegawa M., Kishino H., Yano T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. [DOI] [PubMed] [Google Scholar]
- Hayashi A., Saitou H., Mori T., Matano I., Sugisaki H., Maruyama K. (2012). Molecular and catalytic properties of monoacetylphloroglucinol acetyltransferase from Pseudomonas sp. YGJ3. Biosci. Biotechnol. Biochem. 76, 559–566. 10.1271/bbb.110860 [DOI] [PubMed] [Google Scholar]
- Hilario E., Buckley T. R., Young J. M. (2004). Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek 86, 51–64. 10.1023/B:ANTO.0000024910.57117.16 [DOI] [PubMed] [Google Scholar]
- Iavicoli A., Boutet E., Buchala A., Métraux J.-P. (2003). Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 16, 851–858. 10.1094/MPMI.2003.16.10.851 [DOI] [PubMed] [Google Scholar]
- Kichenaradja P., Siguier P., Pérochon J., Chandler M. (2010). ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 38, D62–D68. 10.1093/nar/gkp947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidarsa T. A., Goebel N. C., Zabriskie T. M., Loper J. E. (2011). Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 81, 395–414. 10.1111/j.1365-2958.2011.07697.x [DOI] [PubMed] [Google Scholar]
- Kupferschmied P., Maurhofer M., Keel C. (2013). Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front. Plant Sci. 4:287. 10.3389/fpls.2013.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Brinkman F. S. (2009). Islandviewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665. 10.1093/bioinformatics/btp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R. (2010). Mesquite: a Modular System for Evolutionary Analysis. 2011; Version 2.75. Available online at: http://mesquiteproject.org.
- Mazurier S., Corberand T., Lemanceau P., Raaijmakers J. M. (2009). Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 3, 977–991. 10.1038/ismej.2009.33 [DOI] [PubMed] [Google Scholar]
- Migula W. (1894). Über ein neues system der bakterien. Arb. Bakteriol. Inst. Karlsruhe 1, 235–238. [Google Scholar]
- Mirza M. S., Mehnaz S., Normand P., Prigent-Combaret C., Moënne-Loccoz Y., Bally R., et al. (2006). Molecular characterization and PCR detection of a nitrogen-fixing Pseudomonas strain promoting rice growth. Biol. Fertil. Soils 43, 163–170. 10.1007/s00374-006-0074-9 [DOI] [Google Scholar]
- Mithani A., Hein J., Preston G. M. (2010). Comparative analysis of metabolic networks provides insight into the evolution of plant pathogenic and non-pathogenic lifestyles in Pseudomonas. Mol. Biol. Evol. 28, 483–499. 10.1093/molbev/msq213 [DOI] [PubMed] [Google Scholar]
- Moynihan J. A., Morrissey J. P., Coppoolse E. R., Stiekema W. J., O'Gara F., Boyd E. F. (2009). Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Appl. Environ. Microbiol. 75, 2122–2131. 10.1128/AEM.02052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet M., Lalucat J., García-Valdés E. (2010). DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12, 1513–1530. 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- Munsch P., Geoffroy V. A., Alatossava T., Meyer J.-M. (2000). Application of siderotyping for characterization of Pseudomonas tolaasii and Pseudomonas reactans isolates associated with brown blotch disease of cultivated mushrooms. Appl. Environ. Microbiol. 66, 4834–4841. 10.1128/AEM.66.11.4834-4841.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien H. E., Thakur S., Guttman D. S. (2011). Evolution of plant pathogenesis in Pseudomonas syringae: a genomics perspective. Annu. Rev. Phytopathol. 49, 269–289. 10.1146/annurev-phyto-072910-095242 [DOI] [PubMed] [Google Scholar]
- Ortet P., Barakat M., Lalaouna D., Fochesato S., Barbe V., Vacherie B., et al. (2011). Complete genome sequence of a beneficial plant root-associated bacterium, Pseudomonas brassicacearum. J. Bacteriol. 193:3146. 10.1128/JB.00411-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A., Frapolli M., Fischer-Le Saux M., Gruffaz C., Meyer J.-M., Défago G., et al. (2011). Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34, 180–188. 10.1016/j.syapm.2010.10.005 [DOI] [PubMed] [Google Scholar]
- Ramette A., Moënne-Loccoz Y., Défago G. (2001). Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Mol. Plant Microbe Interact. 14, 639–652. 10.1094/MPMI.2001.14.5.639 [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 9126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.-A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbour Laboratory Press. [Google Scholar]
- Sanguin H., Kroneinsen L., Gazengel K., Kyselková M., Remenant B., Prigent-Combaret C., et al. (2008). Development of a 16S rRNA microarray approach for the monitoring of rhizosphere Pseudomonas populations associated with the decline of take-all disease of wheat. Soil Biol. Biochem. 40, 1028–1039. 10.1016/j.soilbio.2007.11.023 [DOI] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnider-Keel U., Seematter A., Maurhofer M., Blumer C., Duffy B., Gigot-Bonnefoy C., et al. (2000). Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182, 1215–1225. 10.1128/JB.182.5.1215-1225.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner K., Hagn A., Kyselková M., Moënne-Loccoz Y., Welzl G., Munch J. C., et al. (2010). Comparison of barley succession and take-all disease as environmental factors shaping the rhizobacterial community during take-all decline. Appl. Environ. Microbiol. 76, 4703–4712. 10.1128/AEM.00481-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trantas E. A., Licciardello G., Almeida N. F., Witek K., Strano C. P., Duxbury Z., et al. (2015). Comparative genomic analysis of multiple strains of two unusual plant pathogens: Pseudomonas corrugata and Pseudomonas mediterranea. Front. Microbiol. 6:811. 10.3389/fmicb.2015.00811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Moënne-Loccoz Y., Dubost A., Gonçalves-Martins M., Muller D., Prigent-Combaret C. (2016). Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front. Plant Sci. 7:1212. 10.3389/fpls.2016.01212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Ramette A., Punjasamarnwong P., Zala M., Natsch A., Moënne-Loccoz Y., et al. (2001). Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol. Ecol. 37, 105–116. 10.1111/j.1574-6941.2001.tb00858.x [DOI] [Google Scholar]
- Weller D. M., Mavrodi D. V., van Pelt J. A., Pieterse C. M. J., van Loon L. C., Bakker P. A. H. M. (2012). Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102, 403–412. 10.1094/PHYTO-08-11-0222 [DOI] [PubMed] [Google Scholar]
- Wolf Y. I., Koonin E. V. (2013). Genome reduction as the dominant mode of evolution. Bioessays 35, 829–837. 10.1002/bies.201300037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Kasai H., Arnold D. L., Jackson R. W., Vivian A., Harayama S. (2000). Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146, 2385–2394. 10.1099/00221287-146-10-2385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presence/absence of phlD gene in the 12 Pseudomonas type strains sought by PCR targeting phlD. MW, molecular weight; 1 and 6, negative control; 2, P. brassicacearum T; 3, P. kilonensis T; 4, P. thivervalensis T; 5, P. mediterranea T; 7, P. corrugata T; 8, P. protegens T; 9, 10, and 15, P. kilonensis F113 (positive control); 11, P. chlororaphis subsp. aurantiaca T; 12, P. chlororaphis subsp. aureofaciens T; 13, P. chlororaphis subsp. chlororaphis T; 14, P. tolaasii T; 16, P. marginalis T; 17, P. chlororaphis T.
Comparison of species tree of phl+ Pseudomonas strains and phlACBD tree for these bacteria. (A) Phylogenetic analysis based on concatenated housekeeping genes rpoB, gyrB, and rrs. When available, membership to the multilocus phylogenetic groups defined in Frapolli et al. (2007) is given in parenthesis. (B) Phylogenetic analysis of phl+ Pseudomonas based on nucleotide sequences for the operon phlACBD. In both panels, the Maximum Likelihood tree was inferred using PhyML model, and nodal robustness was assessed using 1,000 bootstrap replicates. Type strains are shown in bold and written in blue. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7).
DAPG production in Pseudomonas type strains. (A) HPLC chromatogram, UV, and mass spectra of DAPG standard. (B) HPLC detection of DAPG in Pseudomonas strain supernatants.
Phylogenetic analysis of the phl operon for the main Pseudomonas subgroups and betaproteobacterial species. When available, membership to the multilocus phylogenetic groups defined in Frapolli et al. (2007) is given in parenthesis. The Maximum Likelihood tree was inferred using PhyML model, and nodal robustness was assessed using 500 bootstrap replicates. The “P. corrugata” subgroup is highlighted in orange and the “P. protegens” subgroup in blue. Type strains are indicated in bold and red. Fluorescent Pseudomonas strains of uncertain taxonomic status are written as “sp.” and those misclassified were renamed based on rrs-rpoD-gyrB phylogeny and ANI data (See Tables S2–S7).
Pseudomonas strains and accession numbers for nucleotidic sequences used in this study.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. protegens species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. protegens species.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. chlororaphis species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. chlororaphis species.
Average nucleotide identity values (calculated using Blast algorithm) for the assignment of uncertain pseudomonads to the P. brassicacearum, P. kilonensis, and P. thivervalensis species.
Average nucleotide identity values (calculated using MUMmer algorithm) for the assignment of uncertain pseudomonads to the P. brassicacearum, P. kilonensis, and P. thivervalensis species.