Abstract
Introduction
p14ARF stabilises nuclear p53, with a variable expression of p14ARF mRNA in breast cancers. In vitro, nuclear p14ARF binds Hdm2 to block Hdm2-dependent nucleocytoplasmic shuttling of p53, which is required before cytoplasmic degradation of p53. p14ARF is negatively regulated by p53 and through p53-independent pathways. No studies have yet examined levels of p14ARF protein expression in breast cancer and their relationship to Hdm2/p53 immunoreactivity or subcellular localisation. Previously, immunohistochemical expression of cytoplasmic p14ARF, p53 and Hdm2 has been described. HER-2 (c-erbB2/neu) predicts prognosis and interacts with the p14ARF/Hdm2 pathway to inactivate p14ARF and to influence Hdm2 activity and localisation. This study examined p14ARF and p53/Hdm2 expression and subcellular localisation by using immunohistochemistry in a series of invasive ductal breast cancers (IDCs) with concomitant ductal carcinoma in situ (DCIS), to evaluate whether findings in vitro were related to clinicopathological parameters such as HER-2 and their effect on patient outcome.
Methods
The 4C6 anti-p14ARF monoclonal antibody and Dako Envision Plus system were used to evaluate p14ARF expression in 103 patients; p53/Hdm2 staining was performed.
Results
p14ARF was evaluable in 96 patients, with nuclear p14ARF expression (modified Quick-score ≥ 3) in 79% (n = 76) of IDCs and in associated DCIS in 74 patients. Cytoplasmic p14ARF was detectable in 23 breast cancers. Nuclear and cytoplasmic p14ARF showed no correlation with p53 subcellular immunoreactivity. Increasing levels of cytoplasmic p14ARF were associated with nuclear and cytoplasmic Hdm2 expression (P < 0.001). Subcellular ARF expression was not associated with clinicopathological parameters, and although not an independent prognosticator, these preliminary findings suggest that cytoplasmic p14ARF might be associated with a better overall survival (P = 0.09; log rank). The association between HER-2 positivity and nuclear p14ARF (P = 0.038), as well as nuclear Hdm2 (P = 0.019), reflects the in vitro findings of HER-2 interaction with the ARF/Hdm2 pathway. Cytoplasmic p53 and Hdm2 expression might have biological implications, through an association of cytoplasmic p53 with increased tumour proliferation (P = 0.005), and an improved overall survival (P = 0.002, log rank) in cytoplasmic Hdm2-expressing tumours, that independently predict favourable overall survival (P = 0.02) and disease-free survival (P = 0.03).
Conclusions
Nuclear p14ARF expression is similar in IDCs and DCIS and is associated with Hdm2 immunoreactivity. Nuclear p14ARF and Hdm2 might be regulated by HER-2. Clearly, our findings in vivo suggest a complexity of p14ARF/Hdm2 and p53 pathways in which consideration of cytoplasmic p14ARF and Hdm2 might have tumorigenic implications.
Keywords: ARF protein, ductal carcinoma in situ, Hdm2, HER-2, immunohistochemistry
Introduction
The development and growth of breast cancers result from the inactivation of p53 or retinoblastoma (pRb) tumour suppressor proteins that regulate cell cycle control. Such regulatory pathways trigger cell cycle arrest or apoptosis in response to intracellular challenges such as DNA damage, hypoxia and oncogene activation, with preservation of genomic stability [1]. pRb maintains checkpoint integrity through binding and blocking E2F transcription factors, a process reinforced by the G1 cyclin-dependent kinase inhibitor p16INK4a [2]. Similarly, p53 accumulates to transcriptionally activate the cyclin-dependent kinase inhibitor p21WAFI/CIPI, as well as its own negative regulator Hdm2 (human Mdm2; murine double minute 2), which terminates the p53 response. Nuclear retention of p53 underlines its tumour suppressor response and suggests an additional pathway for disabling p53 over and above its mutation in more than 50% of human cancers [3-5]. In an autoregulatory feedback loop, Hdm2 maintains low levels of p53 in normal non-stressed cells and inhibits nuclear p53 through multiple and diverse mechanisms. Hdm2 binds p53 to inhibit its transactivation function and shuttles p53 from the nucleus to the cytoplasm to facilitate its degradation [6-8]. Hdm2 is also an E3 ubiquitin ligase that targets p53 for the ubiquitin-dependent 26S proteosome in the cytoplasm [9].
The INK4a/ARF gene locus on chromosome 9p21 encodes p16INK4a and p14ARF, both of which act in tumour surveillance and link the pRb and p53 pathways [2,10-14]. p14ARF is encoded from an alternative, but partly overlapping, reading frame together with p16INK4a [15], such that ARF has a separate first exon (1β) that splices into common exons 2 and 3, shared with p16INK4a. These proteins independently target two cell cycle control pathways, with p16INK4a inhibiting cyclin D1/cyclin-dependent kinases within the pRb pathway and p14ARF inhibiting the oncoprotein Hdm2 within the p53 pathway [10-14]. ARF expression and activation occur through mitogenic signals such as Myc, E1A, E2F1, Ras and v-Abl, stabilising p53, followed by cell cycle arrest or apoptosis [16,17]. Consequently, ARF connects the pRb and p53 pathways, with excessive proliferative signalling via pRb activating arrest mechanisms by p53. This suggests that loss of ARF function is a major contributor to carcinogenesis in humans [17]. p14ARF is a true tumour suppressor protein; tumours develop spontaneously in ARF-null mice [14,18]. Promoter hypermethylation of the INK4a and ARF genes is a major mechanism of their inactivation, followed by hemizygous deletions [2,10,11,19]. Breast cancers rarely demonstrate homozygous deletions of either gene, with no mutations of ARF [2,19-21].
ARF expression in vitro and in vivo is associated with a G1 or G2 cell cycle arrest or apoptosis after inappropriate mitogenic stimuli or DNA damage [10-13,22,23]. Although these effects in large part relate to activation of the p53 pathway, recent reports suggest that p14ARF inhibits growth independently of p53 [12,17,23,24]. Low or undetectable p14ARF expression in normal tissues and its function independently of p53 in highly proliferating, homeostatic tissues, in comparison with tumours, suggests that p14ARF function has yet to be completely established [17,24]. Subcellular localisation of p14ARF is preferentially nucleolar or nuclear, where it binds Hdm2 to inhibit the latter's activities towards p53 [15,25-27]. ARF-bound Hdm2 blocks Hdm2-dependent nucleocytoplasmic shuttling of p53, to produce nuclear retention and activation of p53 [13,27]. Further, ARF averts degradation of p53 by inhibiting Hdm2-dependent ubiquitination of p53 in vivo [13,28,29]. p14ARF acts upstream of p53 and is subject to its negative feedback regulation, suggesting that p53 mutations or its inactivation by HDM2 amplification are often accompanied by overexpression of ARF [10-14,30]. p53-positive tumours are also likely to have sustained epistatic mutations such as HDM2 amplification or ARF loss [11]. Inactivation of ARF might occur through the overexpression of several ARF repressors, including Twist and Tbx-2 in breast cancers and Bmi-1 in other tumours [31,32].
Nuclear import and export is a feature of both p53 and Hdm2 with implications for their functional regulation, such that cytoplasmic p53 is associated with tumours with a poor prognosis [3-6,8]. Just as Hdm2 regulates the nuclear export of p53, in vivo evidence suggests that ARF influences the subcellular localisation of Hdm2 [21]. Cytoplasmic ARF occurs through binding to Pex19p with evidence of weak to moderate cytoplasmic staining in human tumours, whereas two studies disregarded cytoplasmic p14ARF expression [33-37]. In a single in vitro study, mouse p19ARF binds a cytoplasmic protein Pex19p (cloned from mouse testis) in normal cells (NIH 3T3) [33]. Consequently, the subcellular localisation of these proteins and the relationship between their levels of expression are likely to be important in evaluating breast cancers and their development from pre-invasive ductal carcinoma in situ (DCIS) to invasive disease.
Previous studies have examined p14ARF mRNA expression in breast cancers, with evidence suggesting altered expression and an association with p53 [38,39]. The aim of this study was to evaluate the levels of protein expression for p14ARF in relation to Hdm2 and p53 using immunohistochemistry in DCIS and invasive ductal breast cancers (IDCs), including the study of their subcellular localisation. So far, no studies have examined p14ARF/Hdm2 and p53 in relation to clinicopathological parameters and prognosis in breast cancer. We have shown nuclear p14ARF expression in 79% of IDCs and in associated DCIS in 74 patients. Cytoplasmic p14ARF was detectable in 23 breast cancers. Levels of expression and subcellular localisation for p14ARF, Hdm2 and p53 were similar in IDCs and DCIS. ARF expression showed no correlation with p53 immunoreactivity and was associated with Hdm2 nuclear and cytoplasmic expression. HER-2-positive breast cancers were associated with nuclear p14ARF and nuclear Hdm2 immunoreactivity. Our preliminary findings suggest that cytoplasmic p14ARF and cytoplasmic Hdm2-expressing breast cancers might be associated with a better outcome.
Methods
Patients
The study included 103 patients aged 26–88 years (median 59 years) with IDC of the breast, in association with concomitant DCIS diagnosed between 1996 and 2000 at the Bristol Royal Infirmary, Bristol, UK (Table 1). Eligibility criteria were histological diagnosis of IDC and DCIS, no distant metastases, and unilateral tumour. Regional Ethics Committee approval was granted before commencement of the study. Axillary lymphadenopathy was evaluable in 90 patients: 37 (41%) of the patients were lymph node negative, and 53 (59%) were node positive (N1 [mobile ipsilateral] or N2 [fixed ipsilateral]). No axillary surgery was undertaken in the remaining 13 patients because of age-related co-morbidity.
Table 1.
Clinicopathological variables in the patients entered into this study
| Characteristic | No. of patients (%) |
| Total | 103 (100) |
| Age (years) | |
| ≤50 | 31 (30) |
| >50 | 72 (70) |
| Invasive tumours | |
| Lymph node status | |
| Negative | 37 (41) |
| Positive | 53 (59) |
| Not assessed | 13 |
| Invasive tumour grade | |
| Grade I | 16 (16) |
| Grade II | 51 (49) |
| Grade III | 36 (35) |
| Invasive tumour size (cm) | |
| ≤2 cm | 53 (52) |
| >2 cm | 48 (48) |
| Multifocal | 2 |
| Lymphovascular invasion | |
| Present | 50 (50) |
| Absent | 50 (50) |
| Not assessed | 3 |
| NPI | |
| GPG < 3.4 | 34 (39) |
| MPG 3.4–5.4 | 37 (42) |
| PPG > 5.4 | 17 (19) |
| Not possible to calculate | 15 |
| ER (quick-score) | |
| Positive (4–8) | 63 (64) |
| Negative (0–3) | 36 (36) |
| Not assessed | 4 |
| HER-2 IHC | |
| Negative (0/1+) | 87 (84) |
| Positive (2+/3+) | 16 (16) |
| Ki67 IHC | |
| Low proliferation, <10% | 51 (50) |
| High proliferation, ≥10% | 52 (50) |
| Ductal carcinoma in situ | |
| VNPC | |
| Grade I | 23 (24) |
| Grade II | 36 (37) |
| Grade III | 38 (39) |
| Not assessed | 6 |
| ER (quick-score) | |
| Positive (4–8) | 61 (64) |
| Negative (0–3) | 35 (36) |
| Not assessed | 7 |
| HER-2 IHC | |
| Negative (0/1+) | 77 (80) |
| Positive (2+/3+) | 19 (20) |
| Not assessed | 7 |
| Ki67 IHC | |
| Low proliferation, <10% | 61 (59) |
| High proliferation, ≥10% | 42 (41) |
ER, oestrogen receptor; GPG, good prognostic group; IHC, immunohistochemistry; MPG, moderate prognostic group; NPI, Nottingham Prognostic Index; PPG, poor prognostic group; VNPC, Van Nuys Pathologic classification.
Clinicopathological subgroups were analysed in accordance with the Nottingham Prognostic Index (NPI) and divided into good (GPG), moderate (MPG) and poor (PPG) prognostic groups as described, with a modification that included no assessment of the internal mammary lymph nodes [40]. Evaluation of the NPI was precluded in 15 patients because of non-evaluable regional lymphadenopathy and tumour size. Similarly, subgroups of DCIS were analysed in accordance with the Van Nuys Pathologic Classification (Table 1), which was not assessable in six patients [41]. The design of the study to include representative samples of synchronous IDC and DCIS precluded analysis by the Van Nuys Prognostic Index [41]. Adjuvant treatment groups comprised tamoxifen in 60 patients (27 GPG, 16 MPG, 4 PPG and 13 no NPI), and cyclophosphamide–methotrexate–5-fluorouracil-containing and anthracycline-containing regimes in 17 and 21 patients, respectively (4 GPG, 19 MPG, 13 PPG and 2 no NPI). Five patients received no adjuvant treatment. The median follow-up duration was 51 months (range 6–120 months). All were primary tumours with the exception of six local tumour recurrences, which were excluded from the analysis of patient outcome (see Table 4 and Fig. 3).
Table 4.
Relationships between clinicopathological variables, subcellular Hdm2 and patient outcome
| Variable | Univariate analysis | Multivariate analysis | ||
| 95% CI | P | 95% CI | P | |
| Overall survival | ||||
| Age | 0.99–1.04 | 0.4 | 0.98–2.90 | 0.009 |
| Lymph node status (+/-) | 2.79–153.9 | 0.004 | 2.65–161 | 0.003 |
| Invasive tumour grade (I, II, II) | 1.27–4.93 | 0.002 | 1.14–4.62 | 0.002 |
| Tumour size (cm) | 1.02–1.05 | <0.001 | 1.02–1.05 | <0.001 |
| NPI | 1.48–3.36 | <0.001 | 1.39–3.32 | <0.001 |
| ER quick-score | 0.66–0.92 | 0.002 | 0.68–0.98 | 0.03 |
| HER-2 score | 0.94–2.02 | 0.04 | 0.76–1.95 | 0.56 |
| Ki67 proliferative index | 0.99–1.00 | 0.4 | 0.99–1.00 | 0.65 |
| Lymphovascular invasion (+/-) | 1.46–9.38 | 0.006 | 0.89–6.69 | 0.08 |
| Hdm2 cytoplasm MQS | 0.66–0.95 | 0.01 | 0.67–0.97 | 0.02 |
| Hdm2 nucleus MQS | 0.68–1.01 | 0.07 | 0.69–1.05 | 0.13 |
| Disease-free survival | ||||
| Age | 0.97–1.02 | 0.9 | 0.99–1.06 | 0.06 |
| Lymph node status (+/-) | 2.29–25.3 | <0.001 | 2.06–25.6 | 0.002 |
| Invasive tumour grade (I, II, II) | 1.48–5.38 | 0.005 | 1.36–5.11 | 0.006 |
| Tumour size (cm) | 1.02–1.04 | <0.001 | 1.02–1.04 | <0.001 |
| NPI | 1.46–3.17 | <0.001 | 1.38–3.15 | <0.001 |
| ER quick-score | 0.66–0.89 | 0.001 | 0.68–0.95 | 0.01 |
| HER-2 score | 1.04–1.92 | 0.02 | 0.95–1.87 | 0.2 |
| Ki67 Proliferative index | 0.99–1.00 | 0.2 | 0.99–1.00 | 0.3 |
| Lymphovascular invasion (+/-) | 1.63–6.95 | 0.002 | 1.09–6.65 | 0.03 |
| Hdm2 cytoplasm MQS | 0.73–1.02 | 0.05 | 0.70–0.98 | 0.03 |
| Hdm2 nucleus MQS | 0.78–1.09 | 0.34 | 0.76–1.09 | 0.9 |
Confidence intervals (CI) and P values are given for the results of both the univariate and multivariate analyses. Data for the univariate analysis were evaluable in 97 patients (reflecting the exclusion of 6 local tumour recurrences as described in the Methods section) and included a multivariate analysis on 86 cases that excluded non-evaluable Nottingham Prognostic Index (NPI) (nodes, grade and size) in 11 patients. All clinicopathological variables and subcellular Hdm2 modified Quick-scores (MQSs) were analysed as a continuum, with lymph node status and lymphovascular invasion being assessed as present or absent. Univariate and multivariate analyses for cytoplasmic Hdm2 (and nuclear Hdm2) (overall survival) were evaluated on 77 and 70 patients, respectively (85 patients univariate [disease-free survival], and 77 patients multivariate, [disease-free survival]). The multivariate analysis is adjusted for NPI and treatment (tamoxifen/chemotherapy/none). Relationships that reached significance (P < 0.05) are highlighted in bold. ER, oestrogen receptor.
Figure 3.
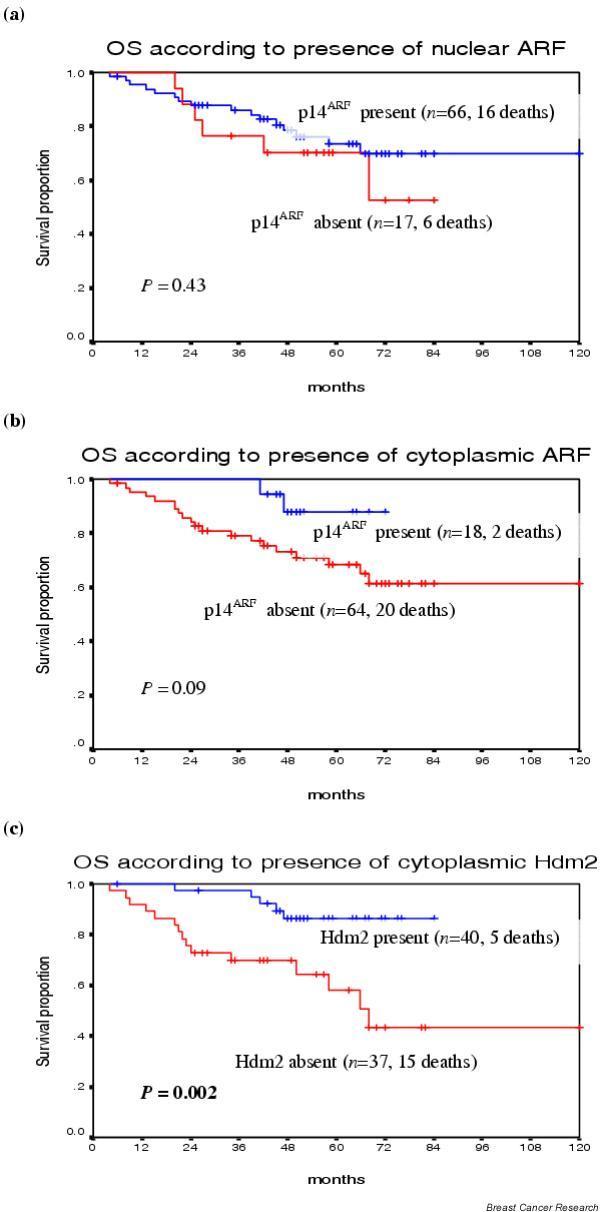
Relationships between p14ARF and Hdm2 localisation and overall survival. Overall survival curves according to nuclear p14ARF (a), cytoplasmic p14ARF (b) and cytoplasmic Hdm2 (c), with patient numbers and deaths in parentheses. The classification of 'present' or 'absent' p14ARF or Hdm2 expression was determined by an MQS of 3 or more to define positivity, compared with an MQS of <3 to define absent expression. Patient numbers reflect the exclusion of six local tumour recurrences from the analysis of patient outcome and include only recorded deaths in 23 patients. P values are given for the log ranks that reached significance.
Tumour samples were collected shortly after surgery and were fixed in buffered formalin for 24–48 hours at room temperature (20–22°C). Tumours were classified in accordance with the guidelines of the UK National Health Service Breast Screening Programme [42] and were graded by the modified Bloom's grading system described by Elston and Ellis [43]. Oestrogen receptor (ER) immunostaining was performed with a standard three-layered streptavidin–avidin–biotin horseradish peroxidase (HRP) method with a mouse anti-human ER primary antibody (M0747, 1:100 dilution; Dako, Ely, UK) and a biotinylated rabbit anti-mouse secondary antibody (E0354 1:350 dilution; Dako). Expression of ER was assessed with Quick-score (0–8) and classified as positive (4–8; more than 3) or negative (0–3; 3 or less) in five high-power fields [44].
Tumour proliferation was assessed with nuclear Ki67 immunostaining (polyclonal rabbit anti-human Ki67 antigen; A0047, 1:100 dilution; Dako). A goat anti-rabbit biotin-labelled polypeptide (E432, 1:400 dilution; Dako) was used as a secondary antibody. Tonsillar tissue was used as a positive control and primary antibody was replaced with Tris-buffered saline (TBS) as a negative control. Ki67 staining was evaluated as positive or negative, with low proliferation indicative of less than 10% of positive-staining cells, compared with high proliferation with at least 10% positivity [45]. HER-2 immunostaining was performed with the mouse monoclonal anti-HER-2 antibody (RTU-CB11; NovaCastra/Vector, Newcastle upon Tyne, UK) and the Envision Plus HRP system (K4006; Dako). HER-2 expression was scored according to the degree and proportion of membrane staining, with a score of 0 or 1+ defined as negative, and 2+ or 3+ as HER-2 positive [46]. Lymphovascular invasion was assessed as present or not, and was analysed together with ER, HER-2 and Ki67 in the Department of Pathology (by CS and CJC).
Immunohistochemistry
p14ARF immunoreactivity was evaluated with the mouse monoclonal antibody 4C6 (kindly provided by Dr Gordon Peters, Cancer Research UK, Lincoln's Inn Fields, London, UK) at a 1:200 dilution of 15 μg/ml. ARF immunostaining of IDC and DCIS was compared with formalin-fixed paraffin-embedded cell buttons, using H1299 non-small cell lung cancer cells as a positive control for p14ARF, and MCF-7 breast cancer cells as a negative control. Further negative controls replaced 4C6 with sero-matched IgG2a used on previously established p14ARF-expressing breast cancers. The 4C6 antibody with epitope recognition of carboxy-terminal p14ARF was chosen after testing of other mouse monoclonal antibodies including C-18 (Santa Cruz Biotechnology, California) and 14PO2 (LabVision/Neomarkers, Fremont, California). The specificity of 4C6 has been demonstrated previously [13,27].
Hdm2 expression was assessed with the mouse monoclonal antibody OP46 (Oncogene Research, CN Biosciences, Nottingham, UK) that detects C-terminal Hdm2 with specificity for the 90 kDa isoform [47]. OP46 was selected after testing with other antibodies including 2A10 and 3G5 (kindly given by AJ Levine, Rockefeller University, New York) both of which had technical or other limitations [48]. OP46 was used at a 1:80 dilution of 100 μg/ml and evaluated in the context of Hdm2 overexpressing A375 malignant melanoma cells, and negatively controlled with IgG2b, which was substituted for the primary antibody. Biotinylated rabbit anti-mouse IgG secondary antibody (E0354, 1:300 dilution of 1.26 mg/ml; Dako), together with the Strept-AB Complex/HRP (0377; Dako, Glostrup, Denmark), was used for p14ARF and Hdm2. Technical limitations precluded dual staining for ARF and Hdm2. p53 immunostaining was performed as described [49] by using the DO7 monoclonal antibody (NovaCastra/Vector). Breast carcinomas known to overexpress p53 with known TP53 gene mutations and protein accumulation were used as positive controls. Negative controls were obtained by omitting the primary antibodies.
Formalin-fixed paraffin sections of breast cancer tissue and cell pellets (controls) were mounted on 3-aminopropyl-triethoxysilane-coated (Sigma, Poole, Dorset, UK) glass slides and were baked for 30 min at 56–60°C before being dewaxed in Clearene (Surgipath Europe, Peterborough, UK). The tissue was rehydrated by sequential immersion in 100% and 50% ethanol to distilled water. Tissue sections were subjected to heat retrieval of antigens in citrate buffer (pH 6) in a pressure cooker for 3 min; after cooling they were incubated in 0.3% (v/v) hydrogen peroxide for 5 min. Subsequently, sections were washed in tapwater and TBS (pH 7.45). Before incubation with p14ARF and Hdm2 primary antibodies, sections were exposed to avidin and biotin blocking solutions (Vector Laboratories, Burlingame, California) for 15 min each. Further blocking was achieved through exposure to normal rabbit serum (diluted with TBS) for 30 min at room temperature. Primary antibody was applied and incubated at 4°C overnight (18 hours) for p14ARF and Hdm2, except for p53, which was incubated for 1 hour at room temperature. After washing with TBS, biotinylated rabbit anti-mouse secondary antibody was applied for 30 min at room temperature. The Strept-AB Complex/HRP was used for the detection of p14ARF and Hdm2, and involved an additional 30 min incubation with TBS/biotinylated streptavidin/HRP. Staining was revealed by development in the chromogen 3,3-diaminobenzidine tetrahydrochloride for 5–10 min before counterstaining with haematoxylin before mounting.
Immunostaining was assessed with a Zeiss Axioskop microscope with a 40 × Achrostig mat lens (×400 overall magnification) and a field diameter of 0.46 mm. In the neoplastic cell population for IDC and DCIS, the degree of staining intensity and the proportion of cells with p14ARF, Hdm2 and p53 immunoreactivity in the nucleus and cytoplasm were individually graded semiquantitatively to produce an intensity distribution score or H-score for each localisation, with invasive and pre-invasive components given separate scores [50]. Initial scoring was of 10 high-power fields; however, in view of the homogeneous staining this was reduced to 5 high-power fields. A modified Quick-score (MQS) was used to define a score of 0–3 for intensity (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) and 0–5 for distribution (0, no staining; 1, 1% or less; 2, 1–10%; 3, 11–33%; 4, 34–66%; 5, 67–100% of cells staining), giving an overall score of 0–8 [44]. Sections were scored independently by two pathologists (CS and CJC) in the Department of Pathology. Unless otherwise stated, scores were assessed as a continuum for the purposes of statistical correlation. For the purposes of describing p14ARF, Hdm2 and p53 immunoreactivity, tumour positivity was defined as a modified Quick-score of 3 or more (1+/2+) (see Table 2).
Table 2.
Immunohistochemical MQSs (0–8) for p14ARF, p53 and Hdm2 expression in the nucleus and cytoplasm of invasive ductal cancer and ductal carcinoma in situ
| Protein | Cancer type | Location | Tumour no. (%) | MQS 0–2 (0) | MQS 3–5 (1+) | MQS 6–8 (2+) |
| p14ARF | Invasive | Nucleus | 96 | 20 (21) | 31 (32) | 45 (47) |
| Cytoplasm | 95 | 72 (76) | 16 (17) | 7 (7) | ||
| DCIS | Nucleus | 91 | 17 (17) | 29 (32) | 45 (49) | |
| Cytoplasm | 91 | 70 (80) | 13 (14) | 8 (9) | ||
| p53 | Invasive | Nucleus | 93 | 62 (67) | 17 (18) | 14 (15) |
| Cytoplasm | 93 | 70 (75) | 18 (19) | 5 (5) | ||
| DCIS | Nucleus | 91 | 66 (73) | 12 (13) | 13 (14) | |
| Cytoplasm | 91 | 58 (62) | 27 (29) | 6 (6) | ||
| Hdm2 | Invasive | Nucleus | 92 | 49 (53) | 33 (36) | 10 (11) |
| Cytoplasm | 91 | 42 (46) | 33 (36) | 16 (18) | ||
| DCIS | Nucleus | 91 | 51 (56) | 25 (27) | 15 (16) | |
| Cytoplasm | 91 | 43 (47) | 27 (30) | 21 (23) |
Tumour numbers scored (MQS 0–8) by immunohistochemistry for p14ARF, p53 and Hdm2 expression in the nucleus and cytoplasm of invasive cancers and ductal carcinoma in situ (DCIS) are shown. Immunohistochemistry expression was assessed individually in the nucleus and the cytoplasm, and is defined as negative (0) (MQS 0–2), weak/moderate (1+) (MQS 3–5) and strong (2+) (MQS 6–8). Positivity for p14ARF, p53 and Hdm2 expression was defined as an MQS of 3 or more (1+/2+).
Statistical analysis
Data were analysed with the SPSS 10.0 for Windows (SPSS Incorporated, Chicago, Illinois) statistics software and summarised with descriptive statistics. The associations between p14ARF, p53 and Hdm2 and patient characteristics were assessed with the Spearman non-parametric test for continuous variables and the χ2 test and Fisher's exact test for categorical factors. The relationship between p14ARF localisation and the subcellular localisation of p53 and Hdm2 was analysed with the Spearman correlation coefficient (cc) for non-parametric data, with the p14ARF and p53/Hdm2 MQS used as continuous variables. Analyses of survival data were performed with the log-rank test and the Cox regression model, with survival curves computed with the Kaplan–Meier method. For p14ARF, p53 and Hdm2, univariate and multivariate analyses were performed and adjusted for the NPI score and treatment received (tamoxifen/chemotherapy/none). Because the NPI is based on nodal involvement, on tumour size and on grade, patients (n = 11) with non-evaluable lymphadenopathy and tumour size were excluded from the multivariate regression analyses (see Table 4).
Results
p14ARF levels of expression and subcellular localisation in IDC and DCIS
p14ARF immunoreactivity was evaluable in the nucleus and cytoplasm in 96 (93%) cases of IDC and in associated DCIS in 91 (88%) cases (Fig. 1 and Table 2). ARF expression (MQS ≥ 3) was detectable in 76 (79%) IDCs and was predominantly nuclear, with 23 breast cancers (24%) demonstrating cytoplasmic p14ARF. Nuclear ARF was homogeneous, with no distinct pattern of nucleolar staining or exclusive nucleolar p14ARF expression [30,34,35].
Figure 1.
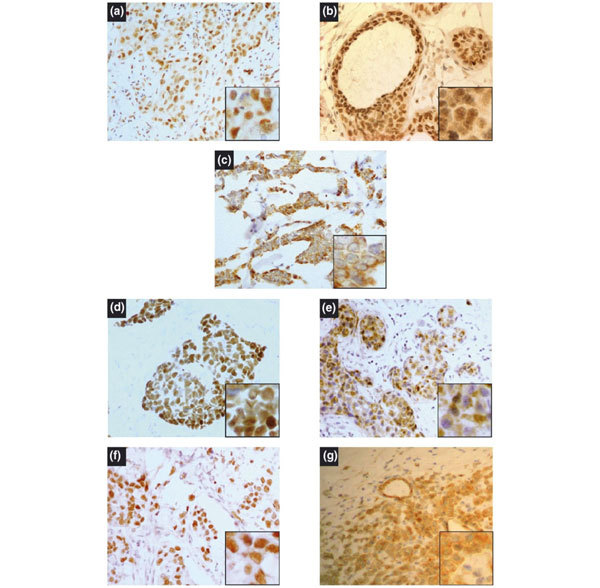
Examples of p14ARF, p53 and Hdm2 immunoreactivity in infiltrating ductal carcinoma of the breast. Immunostaining was performed as described in the Methods section, and nuclei were counterstained with haematoxylin. (a) A tumour showing nuclear p14ARF expression in invasive ductal cancer; (b) a similar nuclear p14ARF positivity in concomitant ductal carcinoma in situ (DCIS). (c) Cytoplasmic p14ARF expression in invasive ductal cancer is representative of similar expression in DCIS. Representative examples of expression of nuclear p53 (d), cytoplasmic p53 (e), and also nuclear Hdm2 (f) and cytoplasmic Hdm2 (g). High-power magnification (×400) (a-g). Insets show higher-power (×1000) views of the same fields.
Predominant nuclear ARF might have been 'masking' the accurate delineation of nucleolar p14ARF. Weak/moderate nuclear ARF expression (MQS 3–5) was evident in 31 (32%) cases, with half of these showing cytoplasmic ARF (n = 16). A loss of p14ARF nuclear immunostaining was observed in 20 (21%) invasive breast cancers, whereas the remaining 45 (47%) cases exhibited diffuse and strong nuclear staining (MQS 6–8; Table 2), with a similar intensity of cytoplasmic ARF in 7 cases only. A comparison showed that the levels of p14ARF expression in DCIS were similar to invasive disease (Table 2).
Relationships between p14ARF and p53 levels of expression and subcellular localisation in IDC and DCIS
p53 immunoreactivity and subcellular localisation were evaluable in 93 (90%) cases of IDC and in DCIS in 91 (88%) cases. Immunohistochemically, p53 was detectable in 31 (33%) IDCs (MQS ≥ 3), with similar levels of nuclear (31 cases) and cytoplasmic (23 cases) expression (25%), compared with p14ARF (Table 2). Weak to moderate (MQS 3–5) nuclear p53 expression was observed in 17 cases of IDC (18%), with similar cytoplasmic staining (18 cases). There was no nuclear or cytoplasmic p53 expression (MQS 0–2) in most IDCs (67% and 75%, respectively). Strong nuclear p53 expression (MQS 6–8) was evident in 15% of IDCs, with 5% of these expressing intense cytoplasmic staining. A comparison of the levels of p53 expression and subcellular localisation in DCIS were similar to those in IDC (Table 2).
Sixty-six percent (n = 46) of nuclear p14ARF-expressing breast cancers (1+/2+) were associated with absent p53 nuclear immunoreactivity, or the possibility of wild-type p53 (Table 3). By comparison, proportionately fewer or one-third of ARF-expressing tumours (n = 24) had nuclear p53 expression (1+/2+). These associations did not reach statistical significance. Nuclear p14ARF expression (1+/2+) showed no particular pattern of p53 subcellular immunoreactivity (data not shown).
Table 3.
Relationships between nuclear expression of p14ARF with nuclear p53 and Hdm2 in invasive breast cancers
| Nuclear p14ARF MQS | ||||
| 0 (n = 19) | 1+ (n = 28) | 2+ (n = 42) | ||
| Nuclear p53 MQS | 0 (n = 59) | 13 | 21 | 25 |
| 1+ (n = 17) | 2 | 5 | 10 | |
| 2+ (n = 13) | 4 | 2 | 7 | |
| Nuclear p14ARF MQS | ||||
| 0 (n = 16) | 1+ (n = 31) | 2+ (n = 41) | ||
| Nuclear Hdm2 MQS | 0 (n = 48) | 13 | 17 | 18 |
| 1+ (n = 31) | 2 | 10 | 19 | |
| 2+ (n = 9) | 1 | 4 | 4 | |
| Nuclear p53 MQS | ||||
| 0 (n = 58) | 1+ (n = 15) | 2+ (n = 11) | ||
| Nuclear Hdm2 MQS | 0 (n = 47) | 35 | 6 | 6 |
| 1+ (n = 34) | 23 | 7 | 4 | |
| 2+ (n = 3) | 0 | 2 | 1 | |
Tumour numbers scored (MQS 0–8) by immunohistochemistry for nuclear p14ARF are compared with those for nuclear p53; nuclear p14ARF with nuclear Hdm2; and nuclear p53 with nuclear Hdm2. Nuclear expression of p14ARF, Hdm2 and p53 is represented as negative (0) (MQS 0–2), weak/moderate (1+) (MQS 3–5) and strong (2+) (MQS 6–8). Tumour numbers reflect the combined assessment of protein pairs. Positivity for p14ARF, p53 and Hdm2 expression was defined as 1+/2+ (MQS ≥ 3).
Relationships between p14ARF and Hdm2 levels of expression and subcellular localisation in IDC and DCIS
Hdm2 expression and subcellular distribution were evaluable in 92 (89%) cases of IDC and in 91 cases of DCIS (Fig. 1 and Table 2). Nuclear Hdm2 was detectable (MQS ≥ 3) in 43 (47%) IDCs with a similar proportion (49 cases) of cytoplasmic staining (Table 2). A minority of IDCs strongly expressed nuclear and cytoplasmic Hdm2 (MQS 6–8) in 10 (11%) and 16 (18%) tumours, respectively. About half of invasive cancers were negative for nuclear (53%) and cytoplasmic (46%) Hdm2. A similar pattern for all levels of Hdm2 expression was seen in DCIS.
Proportional numbers of nuclear p14ARF-expressing breast cancers (1+/2+) either lacked nuclear Hdm2 expression (0) (49%, n = 35), as opposed to 51% (n = 37) expressing nuclear Hdm2 (1+/2+) (Table 3). Nuclear p14ARF showed no significant associations with nuclear Hdm2 expression (Fig. 2a). Surprisingly, the minority of cytoplasmic ARF-expressing breast cancers showed a significant association between increasing levels (MQS) of cytoplasmic p14ARF and nuclear Hdm2 expression (cc 0.475, P < 0.001) (Fig. 2c), as well as cytoplasmic Hdm2 expression (cc 0.461, P < 0.001) (Fig. 2d). Cytoplasmic ARF showed no other significant associations with p53. There were no associations between nuclear p53 and Hdm2 levels of expression or subcellular localisation (Table 3).
Figure 2.
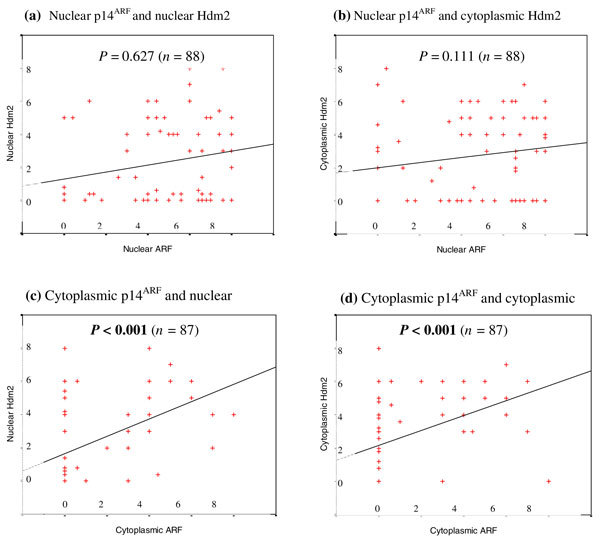
Relationships between subcellular localisation of p14ARF and Hdm2. Scatter plots according to nuclear p14ARF modified Quick-score (MQS) (a, b) and cytoplasmic p14ARF MQS (c, d). p14ARF and Hdm2 staining was analysed as described in the Methods section. (a, b) Distribution of nuclear p14ARF and nuclear Hdm2 (a), and also cytoplasmic Hdm2 (b), showed no significant correlations. (c, d) Distribution of cytoplasmic p14ARF and nuclear Hdm2 (cC), and also cytoplasmic Hdm2 (d), showed significant correlations (P < 0.001, respectively). P values in bold (P < 0.05) indicate significance.
Association of p14ARF, p53 and Hdm2 with clinicopathological factors
We investigated the relationships between nuclear and cytoplasmic levels of p14ARF expression and clinicopathological parameters (listed in Table 1), with ARF MQSs analysed as a continuum for the purposes of statistical analysis. There were no significant associations between either nuclear or cytoplasmic ARF with established prognostic indicators in invasive disease (lymph node involvement, large tumour size > 2 cm, increasing tumour histological grade, ER negativity, lymphovascular invasion, NPI and increased proliferation/Ki67), or in DCIS (Van Nuys Pathologic Classification, ER negativity and increased proliferation/Ki67). Nuclear ARF expression (defined as positive [MQS ≥ 3] or negative [MQS < 3]) showed a statistical relationship to HER-2 positivity (2+/3+) in IDCs (χ2 test, P = 0.038); however, this relates to a limited number of HER-2-expressing breast cancers (n = 14) in association with ARF. A further subset analysis of levels of p14ARF expression (negative [0–2], versus weak/moderate [3–5] versus strong [6–8]; Table 2) in relation to clinicopathological parameters (data not shown) showed no significant associations.
Nuclear and cytoplasmic levels of p53 expression (assessed as a MQS continuum) were analysed in relation to clinicopathological criteria as for p14ARF. Nuclear p53 levels were significantly associated with increased proliferation (Ki67) (cc 0.275, P = 0.008) in IDCs, with cytoplasmic p53 showing a similar association in DCIS (cc 0.295, P = 0.005). There was a possible significant trend in IDCs between nuclear p53 and the NPI (cc 0.210, P = 0.063). A subset analysis of p53 positivity (MQS ≥ 3) was evaluated in relation to clinicopathological parameters. This showed that nuclear p53 expression in invasive breast cancers (MQS ≥ 3 compared with MQS < 3) showed a significant correlation with loss of tumour differentiation (cc 0.243, P = 0.019).
We further analysed the relationship between nuclear and cytoplasmic Hdm2 expression and established parameters as described (Table 1), using the MQS as a continuum. In invasive tumours, increasing levels of nuclear and cytoplasmic Hdm2 expression were significantly associated with increased proliferation (Ki67) (cc 0.281, P = 0.007, and cc 0.224, P = 0.032, respectively). In DCIS nuclear Hdm2 was associated with HER-2 overexpression (2+/3+) (cc 0.249, P = 0.019), with no similar association with cytoplasmic Hdm2 (cc 0.181, P = 0.09). A further evaluation of Hdm2 positivity (MQS ≥ 3) to described parameters showed no other significant associations.
Relationship of clinicopathological factors to prognosis and the predictive potential of p14ARF in relation to p53 and Hdm2 expression
Overall survival (OS) and disease-free survival (DFS) were determined in 89 and 97 patients, respectively (primary tumours), with a median follow-up of 51 months (range 4–120 months). Disease relapses (local or distant recurrences) occurred in 30 women; of these, death was confirmed in 23 patients, with 8 suspected deaths in the absence of a recorded mortality date. Locoregional recurrence occurred at a median duration of 28.5 months (range 3–120 months) from diagnosis. Breast cancer-related mortality occurred at a median of 26 months (range 8–98 months) from presentation. The mean duration of OS and DFS were 93 months and 87 months, respectively. Four-year DFS and OS were 70% and 77%, respectively. The relationship of established clinicopathological features to OS and DFS were analysed with Cox's regression analysis (Table 4). Generally poor prognostic factors such as large tumour size, high tumour grade, lymph node metastases, ER negativity, HER-2 overexpression and NPI were significantly associated with decreased OS and DFS. High tumour proliferation (Ki67 on immunohistochemistry), although associated with a smaller percentage of patients remaining disease-free (DFS) and alive (OS) at 4 years, did not reach statistical significance on univariate or multivariate analysis.
Univariate and multivariate analysis with the continuous MQS variables were used to investigate possible relationships between patient outcome data and levels of subcellular expression for p14ARF, Hdm2 and p53. When scores were measured as a continuum, neither nuclear nor cytoplasmic ARF expression was predictive of OS or DFS after univariate or multivariate analysis (data not shown). Similarly, a Kaplan–Meier analysis for OS and DFS of the nuclear p14ARF subgroups (negative [0–2], weak/moderate [3–5] and strong [6–8]) showed no significant relationship (data not shown). The small number of patients with strong cytoplasmic p14ARF precluded a similar analysis (see Table 2). Kaplan–Meier survival curves showed that the presence of cytoplasmic p14ARF (MQS ≥ 3) might be associated with a better OS compared with ARF negativity (MQS < 3) (P = 0.09, Fig. 3b), with a similar separation of DFS curves, although not statistically significant (data not shown). The presence or absence of nuclear p14ARF (MQS ≥ 3 compared with MQS < 3) was not significantly associated with patient outcome (Fig. 3a).
Subcellular levels of p53 expression were not shown to be independent prognosticators for OS and for DFS on univariate or multivariate analyses (data not shown). Kaplan–Meier analyses of nuclear p53 subgroups (negative [0–2], weak/moderate [3–5] and strong [6–8]) showed a significant association between strong nuclear p53 expression and a reduced DFS (P = 0.05; log rank), although patient numbers were small for this group (data not shown). Similarly, nuclear p53 expression (MQS ≥ 3 compared with MQS < 3) was associated with a reduced OS and DFS, although this was not statistically significant (data not shown). Nuclear Hdm2 expression was not an independent prognosticator for OS and for DFS on univariate or multivariate analyses. Cytoplasmic Hdm2 expression (analysed as a continuous MQS) was associated with improved OS (P = 0.01, univariate analysis) and DFS (P = 0.05, univariate) (Table 4). Cytoplasmic Hdm2 independently predicted for improved OS (P = 0.02, multivariate) and DFS (P = 0.03, multivariate) (Table 4). Kaplan–Meier survival curves showed that the presence of cytoplasmic Hdm2 (MQS ≥ 3 compared with MQS < 3) was associated with improved OS (P = 0.002) (Fig. 3c).
Discussion
p14ARF suppresses cancer cell growth, and several in vitro studies have reported its inhibition of Hdm2 to stabilise and activate p53, with a loss of ARF increasing p53 degradation. The relevance of these mechanisms in vivo requires further investigation, in view of ARF's proposed function as a tumour suppressor and the suggestion that p14ARF overexpression is a surrogate marker of dysregulation of the pRb and p53 pathways [30]. p14ARF mRNA expression in breast cancers underlines a variability with overexpression in 17–19% and underexpression in 24–26% [38,39]. There are few studies clarifying p14ARF protein levels in vivo, with low ARF expression in non-neoplastic epithelium [17,34,35]. The present study suggests an increased frequency of nuclear p14ARF overexpression in 47% of invasive and non-invasive breast cancers. Nuclear p14ARF overexpression has been shown by immunohistochemistry in 22% of B cell lymphomas to predict tumour aggression and outcome [30]. Possible abnormalities in the p53 pathways might be implicated [10-14,30,34]. p53-independent mechanisms might also contribute through oncogenic stimulation of ARF [16,17,30].
The lack of p14ARF expression in 21% of tumours is not dissimilar to other studies of ARF mRNA levels in breast cancers [38,39]. B cell lymphomas lacked nuclear p14ARF in 11% of cases, in association in large part with promoter hypermethylation, as reported for breast cancers [21,30,38]. Predominant nucleolar localisation of p14ARF is determined by amino-terminus and exon 2 carboxy-terminus, with evidence of this after immunofluorescence of a number of cancer cell lines [25,26] and exclusive nucleolar p14ARF in a subset of lymphomas and non-tumour tissue [25,26,30]. The 4C6 monoclonal antibody and other p14ARF monoclonal antibodies have verified the nuclear/nucleoplasmic localisation of ARF with intactness of its functional pathway, as well as its implications as a prognostic surrogate marker compared with nucleolar ARF [27,30].
This study shows nuclear p14ARF in the majority of breast cancers, with 24% cytoplasmic detection. Few other studies have attempted to analyse cytoplasmic p14ARF, although detectable on immunohistochemistry, with in vitro evidence of an ARF-binding cytoplasmic protein [33-37]. Cytoplasmic p14ARF has been observed in non-small cell lung cancers, in oral squamous carcinomas and in another study of lung and pancreatic tumours with the use of several monoclonal and polyclonal antibodies [34-37]. This study analysed cytoplasmic ARF in the context of positive and sero-matched negative controls as described, with no evidence of ARF expression in surrounding normal breast epithelial cells. Such observations might invoke an additional pathway of ARF regulation through changes in its subcellular localisation as for other tumour suppressors, such as p53 and p21WAF1/CIP1 [5,8,51,52]. Similar genomic aberrations might occur in DCIS and IDC, highlighting possible similarities in protein expression [53,54]. This study shows a consistent similarity in levels of protein expression and subcellular localisation for p14ARF, p53 and Hdm2 in DCIS and IDCs.
It is suggested that the upregulation of nuclear p14ARF expression is a consequence of cell cycle malfunction involving p53 and Hdm2 [30]. Increasing levels of nuclear ARF on immunohistochemistry are a measure of p53 inactivation by mutation, or by Hdm2 overexpression resulting in disruption of the p53–p14ARF negative feedback loop [30]. The implication is that p14ARF expression is associated with P53-deficient cell lines, suggesting a p53-mediated downregulation of ARF [10-14]. Although p53 immunostaining is not necessarily tightly correlated with TP53 gene function, 57% (17 cases) of p53-positive tumours in this study strongly expressed ARF, reflecting similar in vivo evidence showing concomitant p14ARF mRNA expression and p53 immunostaining in breast cancers, with a similar relationship between increasing levels of ARF protein and p53 mutations in B cell lymphomas (Table 3) [30,34,39]. Others have shown a lack of an inverse relationship between the two genes, suggesting that the ARF–p53 pathways are not strictly linear and that decreased ARF expression and TP53 mutations are not mutually exclusive [39,55]. ARF stabilises nuclear p53 and is postulated to congregate with Hdm2 and p53 in ternary complexes or 'nuclear bodies' that have yet to be demonstrated in vivo [10,25,27,30].
Double-fluorescent immunolabelling in B cell lymphomas revealed a partial co-localisation of nuclear p14ARF and p53, with no Hdm2 association, suggesting that the mechanisms underpinning ARF's role in inhibiting the nuclear export of p53 by Hdm2 inhibition remains to be defined [30]. Studies in vivo have not previously examined a possible association between ARF expression and p53 subcellular localisation in breast cancer. In large part, this interpretation depends on the accumulation of p53 that is likely to reflect mutant p53 as inferred from immunohistochemistry [30,34,49]. Proportionately more (55%) p53-expressing breast cancers showed exclusive nuclear p53 in association with nuclear ARF expression (data not shown).
Hdm2 expression is a feature of tumorigenesis, with overexpression attributed in part to gene amplifications in the minority of breast cancers [56-59]. Other contributing mechanisms include enhanced transcription and translation and an extended protein half-life. This study demonstrates a minority (11%) of Hdm2-overexpressing (MQS 6–8) invasive breast cancers and 16% of non-invasive breast cancers. Recent evidence shows that p14ARF overexpression is associated with increased levels of Hdm2 in cancer cells, with a similar finding in oral cancers and B cell lymphomas that indicate a direct association between p14ARF and Hdm2 expression determined by immunohistochemistry [27,30,35]. Similar findings are reflected in this series of breast cancers (Table 3).
An additional mechanism of Hdm2-mediated regulation by p14ARF involves intracellular compartmentation [21]. The subcellular localisation of Hdm2 involves its nucleolar localisation signal, as well as both nuclear import and export receptors [59]. In vitro immunofluorescent studies verify the ARF-mediated localisation of Hdm2 in the nucleolus and nucleoplasm [27]. Not uncommonly in colorectal cell lines and primary tumours, in vivo inactivation of ARF by methylation is associated with increasing cytoplasmic Hdm2 expression, emphasising the importance of p14ARF in nuclear Hdm2 localisation [21]. ARF-negative cancers, implying inactivation in this series, show no specific association with increased cytoplasmic Hdm2 expression (Fig. 2). There is a further suggestion in this series of breast cancers that nuclear ARF expression is associated with both nuclear and cytoplasmic Hdm2-expressing breast cancers, underlining the absence of a preferential nuclear localisation of Hdm2 (data not shown). Similarly, a study of colorectal cancers shows the lack of an exclusive relationship between ARF function and the subcellular localisation of Hdm2, in comparison with cell lines [21]. HER-2/neuneu might also regulate the subcellular localisation of Hdm2, with better prognostic HER-2-negative breast cancers associated with Hdm2 in both the nucleus and cytoplasm, in contrast to HER-2-mediated phosphorylation of Hdm2 to produce its nuclear localisation and the degradation of p53 [60]. Such findings could imply that the relationship between p14ARF and the subcellular localisation of Hdm2 is not as exclusive as that found in cell lines.
Clinically, the implications of p14ARF levels of expression in breast cancer are unknown; two previous studies examined p14ARF mRNA expression in relation to prognostic parameters [38,39]. Variable p14ARF mRNA expression including both overexpression as well as decreased mRNA levels are reported to be significantly associated with poor prognostic criteria including p53 mutational status, peritumoural vessel invasion, lymph node metastases and negative progesterone receptors [38,39]. By comparison, nuclear p14ARF overexpression in B cell lymphomas with the use of immunohistochemistry predicts tumour aggression and reduced overall survival [30]. We find no clear associations between ARF levels and prognostic parameters or outcome, although the presence of cytoplasmic p14ARF as opposed to its absence suggests a better outcome (Fig. 3b). At present, a poor understanding of the implications of cytoplasmic ARF expression in tumours requires further study [35-37]. In vitro, HER-2/neuneu interacts with the p14ARF/Hdm2 pathway; some in vivo studies show that p14ARF mRNA overexpression in breast cancers is correlated with HER-2 negativity [38,39,61]. In vitro, HER-2 promotes Hdm2-mediated p53 degradation through the inactivation of ARF, and HER-2 further enhances mammary tumorigenesis in ARF heterozygous mice [60,61]. Our finding of an association between ARF and HER-2 expression is preliminary and suggests the importance of HER-2 in the ARF pathway.
Inactivation of p53 might occur through pathways other than mutation, involving p14ARF/Hdm2 and its degradation through subcellular localisation. Immunohistochemical p53 expression might detect up to 89% of TP53 point mutations in breast carcinoma specimens, although it does not always correlate with specific mutations in exons 5 to 9 [34,49]. Despite an established body of literature regarding the implications of p53 nuclear immunoreactivity, few or no in vivo studies have evaluated cytoplasmic p53. The cytoplasmic localisation of p53 is a prerequisite for its proteosomal degradation and has been implicated in patient prognosis [5,62]. A study of inflammatory breast cancers showed the presence of wild-type cytoplasmic p53 in 37% of cases [5]. We found a significant association of cytoplasmic p53 with increased tumour proliferation in DCIS, suggesting the value of its further investigation in future studies. Similarly, Hdm2 undergoes nucleocytoplasmic shuttling; highly proliferative, invasive breast cancers were associated with increasing levels of nuclear or cytoplasmic Hdm2 expression in this study. Immunohistochemical p53 and Hdm2 expression were also shown to be correlated with ki67 in B cell lymphomas [30]. A possible functional relevance of subcellular Hdm2 expression is suggested, although Ki67 was not an independent prognosticator in this study.
Our findings of an association between HER-2 and nuclear Hdm2 expression might support in vivo findings in which HER-2, through phosphoinositide 3-kinase/Akt-mediated phosphorylation, is preferentially associated with nuclear Hdm2 [60]. In the present study the association between the presence of cytoplasmic Hdm2 and improved outcome would initially seem counterintuitive, yet it is substantiated by recent evidence that HER-2-negative and Akt-negative cell lines are associated predominantly with cytoplasmic Hdm2 [60]. In vivo, the study of a small subset of breast cancers (21 tumours) confirmed these in vitro findings, suggesting an important relationship between HER-2 activation and the p14ARF/Hdm2 and p53 pathways [60].
Conclusion
Defining relevant pathways regulating p53 function are fundamental to an understanding of its tumour suppressor role and mechanisms of p53 inactivation, which might be independent of a p53 mutation. p14ARF and Hdm2 are key upstream and downstream regulators of p53, although in part they might be independent of p53. We have demonstrated a spectrum of tumour-specific p14ARF upregulation with predominant nuclear ARF expression in the absence of exclusive nucleolar staining in invasive and non-invasive breast cancers; we suggest that the presence of cytoplasmic p14ARF might be favourable in terms of breast cancer outcome. Predominant nuclear ARF levels showed no clear clinical implications about the degree of ARF expression versus its negativity. Clearly our in vivo findings do not completely mirror in vitro relationships between p14ARF/Hdm2 and p53 to suggest a greater complexity of these pathways in human solid tumours. Increasingly, evidence suggests the importance of evaluating the subcellular localisation of checkpoint proteins to include p53, Hdm2 and p14ARF [51,52]. Potentially, changes in the cellular distribution of these proteins might be driven by key tumorigenic processes to which HER-2 and other growth factor pathways might contribute.
Competing interests
None declared.
Abbreviations
cc = Spearman correlation coefficient; DCIS = ductal carcinoma in situ; DFS = disease-free survival; ER = oestrogen receptor; GPG = good prognostic group; HRP = horseradish peroxidase; IDC = invasive ductal cancer; MPG = moderate prognostic group; MQS = modified Quick-score; NPI = Nottingham Prognostic Index; OS = overall survival; PPG = poor prognostic group; pRb = retinoblastoma protein; TBS = Tris-buffered saline.
Acknowledgments
Acknowledgements
We thank Rosemary Greenwood (Statistician, Department of Research and Development, United Bristol Healthcare NHS Trust) for her contribution to statistical analyses, and Mr Paul Newcomb (Department of Medicine, University of Bristol) for his technical assistance. This work was funded by the University of Bristol Cancer Research Committee, the Joan Greenfield Fellowship and the United Bristol Healthcare NHS Trust Medical Research Committee. SBV was the recipient of a Ronald Raven Travelling Fellowship, and ZEW was a recipient of the King James IV Professorship, Royal College of Surgeons of Edinburgh, in recognition of this work.
Contributor Information
SB Vestey, Email: vesteyhaynes@hotmail.com.
C Sen, Email: Chandan.Sen@north-bristol.swest.nhs.uk.
CJ Calder, Email: caroline.calder@ubht.swest.nhs.uk.
CM Perks, Email: Claire.M.Perks@bristol.ac.uk.
M Pignatelli, Email: Massimo.Pignatelli@bristol.ac.uk.
ZE Winters, Email: zoe.winters@bristol.ac.uk.
References
- Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4a/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/S0959-437X(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Vande Woude GF. The ins and outs of p53. Nat Cell Biol. 2000;2:E178–E180. doi: 10.1038/35036427. [DOI] [PubMed] [Google Scholar]
- Laín S, Xirodimas D, Lane DP. Accumulating active p53 in the nucleus by inhibition of nuclear export: a novel strategy to promote the p53 tumor suppressor function. Exp Cell Res. 1999;253:315–324. doi: 10.1006/excr.1999.4672. [DOI] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci USA. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Pochampally R, Chen L, Traidej M, Wang Y, Chen J. Nuclear exclusion of p53 in a subset of tumors requires MDM2 function. Oncogene. 2000;19:232–240. doi: 10.1038/sj.onc.1203262. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/S0959-437X(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- Lloyd AC. p53 only ARF the story. Nat Cell Biol. 2000;2:E48–E50. doi: 10.1038/35004078. [DOI] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumour suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–100. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle D, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signalling via the ARF tumour suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci USA. 2003;100:15930–15935. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/S0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Silva J, Silva JM, Dominguez G, Garcia JM, Cantos B, Rodriguez R, Larrondo FJ, Provencio M, Espana P, Bonilla F. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- Hui R, Macmillan RD, Kenny FS, Musgrove EA, Blamey RW, Nicholson RI, Robertson JFR, Sutherland RL. INK4a gene expression and methylation in primary breast cancer: overexpression of p16INK4a messenger RNA is a marker of poor prognosis. Clin Cancer Res. 2000;6:2777–2787. [PubMed] [Google Scholar]
- Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, Watkins DN, Capella G, Peinado MA, Matias-Guiu X, Prat J, Baylin SB, Herman JG. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res. 2001;61:2816–2821. [PubMed] [Google Scholar]
- Khan SH, Moritsugu J, Wahl GM. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc Natl Acad Sci USA. 2000;97:3266–3271. doi: 10.1073/pnas.050560997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003;22:1822–1835. doi: 10.1038/sj.onc.1206303. [DOI] [PubMed] [Google Scholar]
- O'Leary KA, Mendrysa SM, Vaccaro A, Perry ME. Mdm2 regulates p53 independently of p19ARF in homeostatic tissues. Mol Cell Biol. 2004;24:186–191. doi: 10.1128/MCB.24.1.186-191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Darmanian AP, Mann GJ, Kefford RF. Two arginine rich domains in the p14ARF tumour suppressor mediate nucleolar localization. Oncogene. 2000;19:2978–2985. doi: 10.1038/sj.onc.1203629. [DOI] [PubMed] [Google Scholar]
- Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Laín S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Desterro JMP, Saville MK, Howard S, Sparks A, Hay RT, Lane DP. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Sanchez-Aguilera A, Sanchez-Beato M, Garcia JF, Prieto I, Pollan M, Piris MA. p14ARF nuclear overexpression in aggressive B-cell lymphomas is a sensor of malfunction of the common tumor suppressor pathways. Blood. 2002;99:1411–1418. doi: 10.1182/blood.V99.4.1411. [DOI] [PubMed] [Google Scholar]
- Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJL, Scheijen B, Voncken J-W, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via IK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara T, Kaul SC, Kato J-y, Reddel RR, Nomura H, Wadhwa R. Pex19p dampens the p19ARF-p53-p21WAF1 tumor suppressor pathway. J Biol Chem. 2001;276:18649–18652. doi: 10.1074/jbc.C100011200. [DOI] [PubMed] [Google Scholar]
- Vonlanthen S, Heighway J, Tschan MP, Borner MM, Altermatt HJ, Kappeler A, Tobler A, Fey MF, Thatcher N, Yarbrough WG, Betticher DC. Expression of p16INK4a/p16α and p19ARF/p16β is frequently altered in non-small cell lung cancer and correlates with p53 overexpression. Oncogene. 1998;17:2779–2785. doi: 10.1038/sj.onc.1202501. [DOI] [PubMed] [Google Scholar]
- Sano T, Hikino T, Xue Q, Saito T, Kashiwabara K, Oyama T, Nakajima T. Immunohistochemical inactivation of p14ARF concomitant with MDM2 overexpression inversely correlates with p53 overexpression in oral squamous cell carcinoma. Pathology Int. 2000;50:709–716. doi: 10.1046/j.1440-1827.2000.01109.x. [DOI] [PubMed] [Google Scholar]
- Geradts J, Wilentz RE, Roberts H. Immunohistochemical detection of the alternate INK4a-encoded tumor suppressor protein p14ARF in archival human cancers and cell lines using commercial antibodies: correlation with p16INK4a expression. Mod Pathol. 2001;14:1162–1168. doi: 10.1038/modpathol.3880452. [DOI] [PubMed] [Google Scholar]
- Vestey S, Winters ZE. Immunohistochemical detection of the alternate INK4A-encoded tumor suppressor protein p14ARF in archival human cancers and cell lines using commercial antibodies: correlation with p16INK4a expression. Mod Pathol. 2002;15:1106–1107. doi: 10.1097/01.MP.0000030090.45666.DB. (letter) [DOI] [PubMed] [Google Scholar]
- Silva J, Domínguez G, Silva JM, García JM, Gallego I, Corbacho C, Provencio M, Espana P, Bonilla F. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene. 2001;20:4586–4590. doi: 10.1038/sj.onc.1204617. [DOI] [PubMed] [Google Scholar]
- Domínguez G, Silva J, Silva JM, Garcia JM, Larrondo FJ, Vargas J, Sanfrutos L, Provencio M, Espana P, Bonilla F. Different expression of P14ARF defines two groups of breast carcinomas in terms of TP73 expression and TP53 mutational status. Genes Chromosomes Cancer. 2001;31:99–106. doi: 10.1002/gcc.1123.abs. [DOI] [PubMed] [Google Scholar]
- Galea MH, Blamey RW, Elston CW, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat. 1992;22:207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- Silverstein MJ, Poller DN, Waisman JR. Prognostic classification of breast ductal carcinoma in situ. Lancet. 1995;345:1154–1157. doi: 10.1016/S0140-6736(95)90982-6. [DOI] [PubMed] [Google Scholar]
- National Breast Screening Programme . Pathology Reporting in Breast Cancer Screening. 2. Sheffield, UK: NHSBSP Publications; 1995. [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer: the value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Leake R, Barnes D, Pinder S, Ellis I, Anderson L, Anderson T, Adamson R, Rhodes T, Miller K, Walker R. Immunohistochemical detection of steroid receptors in breast cancer: a working protocol. UK Receptor Group, UK NEQAS, The Scottish Breast Cancer Pathology Group, and The Receptor and Biomarker Study Group of the EORTC. J Clin Pathol. 2000;53:634–635. doi: 10.1136/jcp.53.8.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GP, Butt IS, Prasad R, Knox WF, Bundred NJ. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer. 2004;90:423–429. doi: 10.1038/sj.bjc.6601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes A, Jasani B, Anderson E, Dodson AR, Balaton AJ. Evaluation of HER-2/neu immunohistochemical assay sensitivity and scoring on formalin-fixed and paraffin-processed cell lines and breast tumours: a comparative study involving results from laboratories in 21 countries. Am J Clin Pathol. 2002;118:408–417. doi: 10.1309/97WN-W6UX-XJWT-02H2. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Girlando S, Dalla Palma P, Pellegrini S, Fina P, Doglioni C, Bevilacqua G, Barbareschi M. mdm2 gene alterations and mdm2 protein expression in breast carcinomas. J Pathol. 1995;175:31–38. doi: 10.1002/path.1711750106. [DOI] [PubMed] [Google Scholar]
- O'Neill M, Campbell SJ, Save V, Thompson AM, Hall PA. An immunochemical analysis of mdm2 expression in human breast cancer and the identification of a growth-regulated cross-reacting species p170. J Pathol. 1998;186:254–261. doi: 10.1002/(SICI)1096-9896(1998110)186:3<254::AID-PATH185>3.3.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Sjögren S, Inganas M, Norberg T, Lindgren A, Nordgren H, Holmberg L, Bergh J. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- Kinsel LB, Szabo E, Greene GL, Konrath J, Leight GS, McCarty KS., Jr Immunocytochemical analysis of estrogen receptors as a predictor of prognosis in breast cancer patients: comparison with quantitative biochemical methods. Cancer Res. 1989;49:1052–1056. [PubMed] [Google Scholar]
- Winters ZE, Hunt NC, Bradburn MJ, Royds JA, Turley H, Harris AL, Norbury CJ. Subcellular localisation of cyclin B, Cdc2 and p21WAF1/CIP1 in breast cancer: association with prognosis. Eur J Cancer. 2001;37:2405–2412. doi: 10.1016/S0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Winters ZE, Leek RD, Bradburn MJ, Norbury CJ, Harris AL. Cytoplasmic p21WAF1/CIP1 expression is correlated with HER-2/neu in breast cancer and is an independent predictor of prognosis. Breast Cancer Res. 2003;5:R242–R249. doi: 10.1186/bcr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuukasjarvi T, Tanner M, Pennanen S, Karhu R, Kallioniemi O-P, Isola J. Genetic changes in intraductal breast cancer detected by comparative genomic hybridization. Am J Pathol. 1997;150:1465–1471. [PMC free article] [PubMed] [Google Scholar]
- Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res. 1999;59:1433–1436. [PubMed] [Google Scholar]
- Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14ARF is independent of p16INK4a methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Girlando S, Palma PD, Pellegrini S, Fina P, Doglioni C, Bevilacqua G, Barbareschi M. Mdm2 gene alterations and mdm2 protein expression in breast carcinomas. J Pathol. 1995;175:31–38. doi: 10.1002/path.1711750106. [DOI] [PubMed] [Google Scholar]
- Bueso-Ramos CE, Manshouri T, Haidar MA, Yang Y, McCown P, Ordonez N, Glassman A, Sneige N, Albitar M. Abnormal expression of MDM-2 in breast carcinomas. Breast Cancer Res Treat. 1996;37:179–188. doi: 10.1007/BF01806499. [DOI] [PubMed] [Google Scholar]
- Lohrum MAE, Ashcroft M, Kubbutat MHG, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung M-C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- D'Amico M, Wu K, Di Vizio D, Reutens AT, Stahl M, Fu M, Albanese C, Russell RG, Muller WJ, White M, Negassa A, Lee HW, DePinho RA, Pestell RG. The role of Ink4a/Arf in ErbB2 mammary gland tumorigenesis. Cancer Res. 2003;63:3395–3402. [PubMed] [Google Scholar]
- Stenmark-Askmalm M, Stal O, Sullivan S, Ferraud L, Sun XF, Carstensen J, Nordenskjold B. Cellular accumulation of p53 protein: an independent prognostic factor in stage II breast cancer. Eur J Cancer. 1994;30A:175–180. doi: 10.1016/0959-8049(94)90082-5. [DOI] [PubMed] [Google Scholar]


