Abstract
Septins are filament-forming GTP-binding proteins involved in many essential cellular events related to cytoskeletal dynamics and maintenance. Septins can self-assemble into heterocomplexes, which polymerize into highly organized, cell membrane-interacting filaments. The number of septin genes varies among organisms, and although their structure and function have been thoroughly studied in opisthokonts (including animals and fungi), no structural studies have been reported for other organisms. This makes the single septin from Chlamydomonas (CrSEPT) a particularly attractive model for investigating whether functional homopolymeric septin filaments also exist. CrSEPT was detected at the base of the flagella in Chlamydomonas, suggesting that CrSEPT is involved in the formation of a membrane-diffusion barrier. Using transmission electron microscopy, we observed that recombinant CrSEPT forms long filaments with dimensions comparable with those of the canonical structure described for opisthokonts. The GTP-binding domain of CrSEPT purified as a nucleotide-free monomer that hydrolyzes GTP and readily binds its analog guanosine 5′-3-O-(thio)triphosphate. We also found that upon nucleotide binding, CrSEPT formed dimers that were stabilized by an interface involving the ligand (G-interface). Across this interface, one monomer supplied a catalytic arginine to the opposing subunit, greatly accelerating the rate of GTP hydrolysis. This is the first report of an arginine finger observed in a septin and suggests that CrSEPT may act as its own GTP-activating protein. The finger is conserved in all algal septin sequences, suggesting a possible correlation between the ability to form homopolymeric filaments and the accelerated rate of hydrolysis that it provides.
Keywords: algae, arginine finger, Chlamydomonas, crystal structure, GTPase, filament, septin
Introduction
Septins are GTP-binding proteins involved in cytokinesis (1), membrane remodeling (2), and many other cellular events related to the cytoskeleton (3, 4). They were first described in yeast (1), but subsequent studies with other eukaryotic organisms have revealed that septin genes are also ubiquitous in animals (5). Multiple septin genes are present in all opisthokont eukaryotes (6), and septin genes and proteins have also been found in protozoa and some algae (7–9). This suggests that these proteins were present in a very early eukaryotic ancestor (9). However, the number of septin genes varies greatly from one species to another, there being only one in green algae, such as Chlamydomonas reinhardtii, but as many as 13 in humans.
Septins belong to the P-loop GTPase family and contain the classical G1, G3, and G4 conserved motifs (10) within a central GTP-binding domain (termed the G-domain), which is flanked by variable N and C termini. However, the biological significance of GTP binding (and hydrolysis) is still incompletely understood. The existence of a conserved motif located at the C terminus of the GTP-binding domain, called the septin unique element (SUE)3 (7, 11), seems to be a signature for septins, distinguishing them from other small GTPase. Within the C-terminal domain, a coiled-coil motif, frequently responsible for septin-septin interactions, is commonly present. However, this is not always the case, indicating that the coiled-coil is not essential for all septin-septin interactions (9).
One of the most notable characteristics of septins is their ability to assemble into oligomeric complexes to form filaments that are components of a variety of different highly organized structures (12–14). Their ability to spontaneously assemble in this way is thought to be key to the biological roles they play. Whereas the occurrence of heterofilaments (requiring the coexistence of distinct septin monomers) is well established, there are also a limited number of reports concerning the formation of homofilaments (consisting of a single septin) in vitro (15, 16), raising questions about their possible existence in vivo. To date their physiological relevance, if any, is uncertain, given that only heterofilaments have thus far been described in cells.
In this regard, it is interesting to note that several algal species possess only a single septin gene. Therefore, studies addressing the question of their assembly into possible homofilamentous assemblies, similar to those composed of multiple subunits, could be very informative. Thus far, only one report addresses this issue using a non-flagellate green alga (17). Their results show this unique septin to be localized within a ringlike structure in vivo and to be involved in cytokinesis. Furthermore, recombinantly expressed protein was capable of forming filamentous structures in vitro, implying that filaments composed of a single septin may play important physiological roles in some species (17).
The Chlamydomonas reinhardtii genome also codes for a single septin, and in a recent phylogenetic analysis (8), this was included in a branch close to the protists Tetrahymena and Paramecium, but forming a separate clade. Analysis of its similarity with other algal septins revealed sequence identities of 80, 55, and 33% with Volvox carteri, Monoraphidium neglectum, and Nannochloris bacillaris, respectively, all of which possess only a single septin gene. On the other hand, the search revealed 36% identity with Chlorella variabilis, a green algae that differs in having two different septin genes.
The only domain to be conserved when comparing C. reinhardtii with opisthokont septins is the central GTP-binding domain, which shares around 30% sequence identity with counterparts from Homo sapiens, Schistosoma mansoni, and Caenorhabditis elegans. Despite this low level of identity, it presents all of the traits expected for septins: three characteristic motifs within the GTP-binding domain (G1 (GXXGXGKST), G3 (DXXG), and G4 (XKXD)) and the SUE.
In the present study, we use the single septin from C. reinhardtii (CrSEPT), a single-cell model organism, to address the question of homofilament formation. We present the crystal structure of the GTP-binding domain of CrSEPT bound to GTPγS together with its characterization in terms of GTP-binding and hydrolysis. Based on the structural and mutagenesis data, we describe a catalytic arginine finger in CrSEPT and demonstrate that it forms long polymeric filaments in vitro. Additionally, CrSEPT was mainly immunolocated at the base of the flagella in Chlamydomonas cells.
Results
GTP binding and hydrolysis by CrSEPT
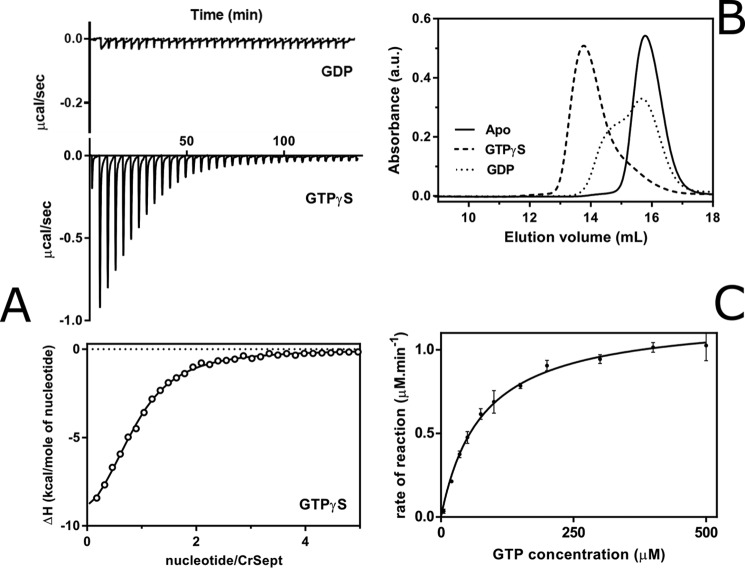
There has been substantial difficulty in obtaining recombinant full-length septins in stable form for subsequent biophysical characterization (18). To circumvent this difficulty, and taking into account the fact that the GTP-binding domain provides most of the structural elements necessary for the formation of oligomers and polymers (14, 19), the CrSEPT(86–393) construct was used here in all subsequent experiments. This construct, which lacks the variable N and C termini (hereafter called just CrSEPT) (Fig. 1A) was expressed in bacteria and purified to homogeneity. The nucleotide content of CrSEPT was analyzed by anion-exchange chromatography, where the recombinant protein was found to be free of bound nucleotide (data not shown). CrSEPT presents all of the elements expected for a GDP-GTP molecular switch (Fig. 1C), and therefore, binding and hydrolysis of guanine nucleotide are important biochemical properties that should be assessed. Nucleotide-binding affinities for CrSEPT were determined by isothermal titration calorimetry (ITC), showing an equilibrium dissociation constant (Kd) of 5.4 μm for GTPγS (Fig. 2A). GDP binding was not observable (Fig. 2A), presumably due to its low affinity, which prevented the estimation of the Kd under the experimental conditions used. The oligomeric state of CrSEPT was evaluated by SEC, yielding an elution profile with a single peak corresponding to the molecular mass of a monomer or, after preincubation with GTPγS, a dimer (Fig. 2B). Prior incubation with GDP resulted in a mixture of monomers and dimers (Fig. 2B). These results are very similar to those observed for human SEPT3 (20). In the GTP hydrolysis assay, CrSEPT showed a GTPase activity with a kcat of 2.41 min−1 (Fig. 2C). This value is about 40 times higher than that reported for human SEPT9 (kcat = 0.064 min−1) (21) and about 2 orders of magnitude higher than that of human SEPT2 (16), the Cdc3-Cdc12 complex in yeast (22) or the structurally related Ras GTPase (23).
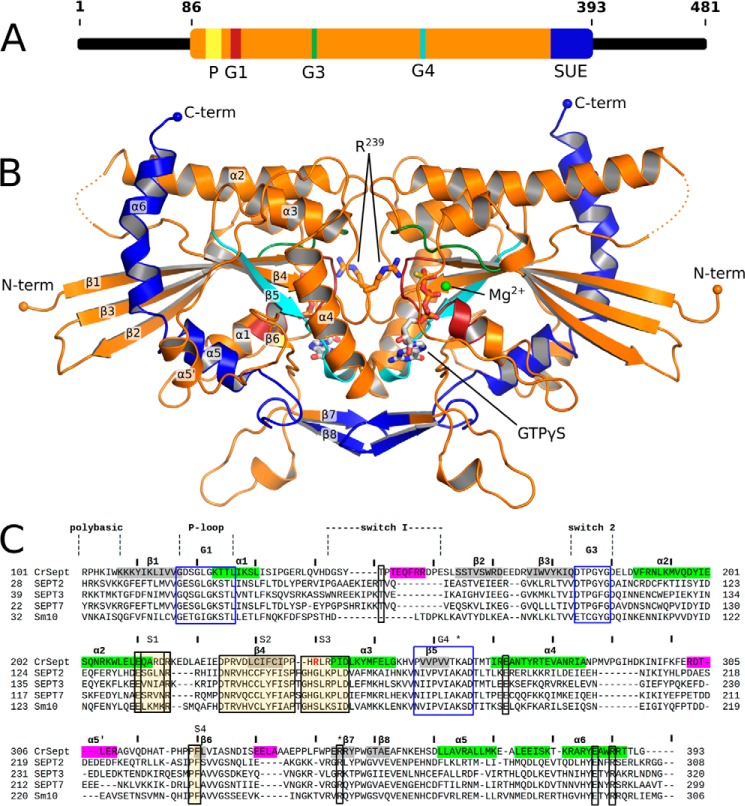
Figure 1.
The structure of dimeric CrSEPT. A, schematic representation of the domain organization of CrSEPT, with the residue positions indicated. CrSEPT contains a polybasic region (P, yellow); the GTPase domain (orange), containing three of the five GTPase conserved motifs (G1 (red), G3 (green), and G4 (cyan) and a SUE (dark blue). B, schematic representation of the GTP-bound structure of CrSEPT. The conserved motifs are shown according to the color code established in A. C, structure-based sequence alignment of CrSEPT (PDB entry 5IRR) and other septin structures available (SEPT2 (2QNR), SEPT3 (3SOP), SEPT7 (3TW4), and SmSEPT10 (4KVA)). The elements of secondary structure are shown in the CrSEPT sequence in gray (β-strands) and green (α-helices). Important structural regions common to small GTPases are shown in transparent boxes, whereas septin-specific motifs (S1–S4) are shown within shaded boxes. In magenta are shown structural regions, which adopt an unusual conformation or are not normally observed when compared with other septins. Single residues that are highly conserved are marked in transparent boxes. Numbers indicate amino acid positions according to CrSEPT. The residue corresponding to the arginine finger is marked in red. *, basic residues that pack over the guanine base.
Figure 2.
Biochemical characterization of CrSEPT. A, GTPγS binding affinity for CrSEPT was determined using ITC. Top, raw data (for GDP and GTPγS); bottom, best fit to the data for GTPγS. The binding isotherm has been fitted to a single-site binding model, yielding a Kd of 5.4 ± 0.3 μm (n = 0.84). GDP binding was not detectable (data not shown). B, influence of the nucleotide on the oligomeric state of CrSEPT. Aliquots of CrSEPT (10 μm), either nucleotide-free (solid line, monomer), complexed to GTPγS (dashed line, dimer), or complexed to GDP (dotted line) were analyzed by SEC on a Superdex 200 10/300 GL column and monitored at 280 nm. C, GTP hydrolysis by CrSEPT was assayed by measuring Pi release as a function of time. Initial rates were obtained from the slopes of phosphate-accumulation curves and fitted to a Michaelis-Menten model. Experiments were performed in triplicate, and the mean and S.D. values are reported. We obtained a kcat value of 2.41 ± 0.02 min−1. GraphPad Prism version 6.0 was used for all data fitting.
Overall structure of CrSEPT GTP-binding domain
To understand the molecular basis of the high GTP hydrolysis rate presented by CrSEPT, we solved the crystal structure of its GTP-binding domain (Fig. 1B) in complex with GTPγS. The structure of CrSEPT was solved to a resolution of 2.0 Å by single anomalous dispersion phasing, using the selenium signal of crystals grown from selenomethionine (SeMet)-derivatized protein. A summary of data collection and refinement is given in Table 1. The GTP binding pockets of the two protomers in the asymmetric unit are packed face-to-face, forming the G interface commonly found in septin filaments (Fig. 1B). The structure is unusual in that the switch I region of both monomers shows continuous electron density (Fig. 1D). The overall fold of the GTP-binding domain of CrSEPT is similar to that of septins described previously (Fig. 1B), as observed by a mean root mean square deviation of 1.8 Å between all main-chain atoms when superimposed on S. mansoni SEPT10 (SmSEPT10) or human SEPT2, SEPT7, or SEPT3. Nevertheless, in several respects, the GTP-binding domain of CrSEPT is the most unusual to have been reported to date.
Table 1.
Data collection, processing, and refinement statistics for CrSEPT
Values in parentheses correspond to the highest resolution shell. ML, maximum likelihood; RMSD, root mean square deviation.
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 177.58, 39.41, 130.90 |
| β (degrees) | 118.50 |
| Detector | PILATUS 2M |
| X-ray source | DLS I04-1 |
| Wavelength (Å) | 0.9200 |
| Resolution range (Å) | 43.63 to 2.04 (2.09 to 2.04) |
| Multiplicity | 4.1 (3.9) |
| Rmeas (%) | 9.4 (83.2) |
| CC (1/2) | 0.997 (0.615) |
| Completeness(%) | 97.1 (95.9) |
| Total reflections | 207,077 (14,179) |
| Unique reflections | 49,936 (3646) |
| I /σ(I) | 11.4 (2.0) |
| Refinement parameters | |
| Reflections used for refinement | 49,930 |
| R (%) | 19.46 |
| Rfree(%) | 23.22 |
| No. of protein atoms | 4532 |
| No. of ligand atoms | 66 |
| No. of water atoms | 338 |
| B (Å2) | 32.48 |
| Coordinate error (ML-based) (Å) | 0.24 |
| Phase error (degrees) | 22.55 |
| Ramachandran plot | |
| Favored (%) | 98.01 |
| Allowed (%) | 1.99 |
| Outliers (%) | 0.0 |
| All-atom Clashscore | 2.74 |
| RMSD from ideal geometry | |
| Bond lengths (Å) | 0.003 |
| bond angles (degrees) | 0.639 |
| PDB code | 5IRR |
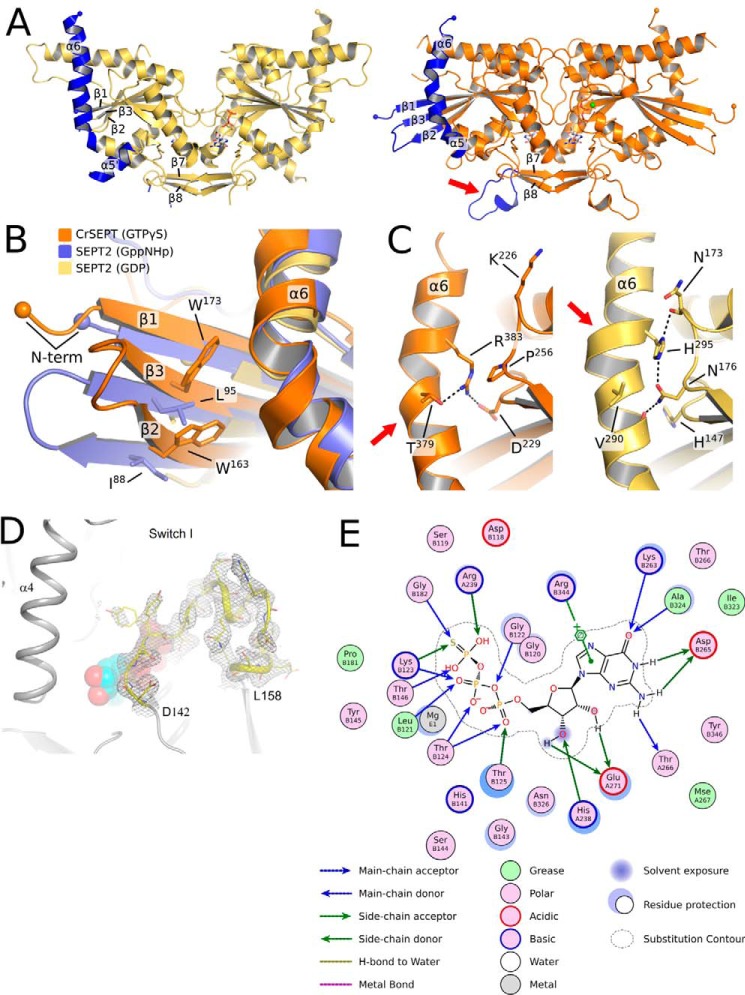
The first three strands of the central β-sheet are longer than those found in the majority of other septin structures (Fig. 3B), enabling two tryptophan residues from strands 2 and 3 (Trp-163 and Trp-173, respectively) to make contact with helix α6 (Fig. 3B). The structures of SmSEPT10 and human SEPT2 also present elongated strands, but only when bound to either GTP or one of its analogues, as is the case for CrSEPT. In SmSEPT10, the β3 strand is known to undergo slippage as a function of the nature of nucleotide (GDP or GTP) present, and the hydrogen-bonding pattern observed here for CrSEPT is entirely consistent with the GTP-bound state (24).
Figure 3.
Comparison between the structures of SEPT2 and CrSEPT. A, schematic representation of SEPT2 (left, PDB entry 2QNR) and CrSEPT (right). In both structures, β1, β2, β3, α5′, α6, and the loop before the β hairpin (β7 and β8) are highlighted (in blue). The latter is indicated with an arrow. These are the most notable structural differences when compared with SEPT2, particularly the protrusion of the three β-strands. B, superimposition of CrSEPT, SEPT2 bound to Gpp(NH)p (PDB entry 3FTQ), and SEPT2 bound to GDP (PDB entry 2QNR). Side chains of CrSEPT Trp-163 and Trp-173 and SEPT2 Ile-88 and Leu-95 are shown. C, side-by-side close view of α6 region from CrSEPT (left) and GDP-bound SEPT2 (right). Black dashed lines highlight the interactions between the conserved His-195 and Arg-383 and neighbor residues, SEPT2 and CrSEPT, respectively. D, continuous electron density (gray mesh) is observed in the composite omit map for the switch I region. Residues at the beginning and end of the region are labeled explicitly, and the GTPγS is shown as solid spheres. E, schematic representation of the GTP-binding site generated with Coot using a method described by Clark and Labute (61). All residues within 4.5 Å of the ligand are represented, and the size of the halo around the residues represents the extent to which the solvent-accessible surface area was affected by ligand binding. Blue and green arrows indicate hydrogen bonds from main-chain and side-chain atoms, respectively. A π-anion interaction involving Arg-344 is explicitly represented.
Other major differences include the α5′ and α6 helices. The former is greatly reduced in length and orientated differently from that typically observed. The latter is normally straight, being only slightly distorted at its center due to an α-aneurism where one turn of a π-helix leads to local widening (25). In CrSEPT, the aneurism has been lost, and the helix as a whole is significantly curved, bringing its C-terminal portion (residues 378–392) back toward the core of the GTP-binding domain. The curvature is caused by an unwinding at its center, which leads to the loss or weakening of the hydrogen bonds involving the carbonyl groups of residues 375–379. This is partially compensated by the side chain of Arg-383, which points inwards and forms a salt bridge with Asp-229 (Fig. 3C). Finally, the region comprising residues 329–343, which precedes the SUE, includes an α-helical turn that projects into the solvent, forming a protuberance, whereas equivalent residues in other structures form a small β-sheet (Fig. 3A).
The GTP-binding pocket and the arginine finger
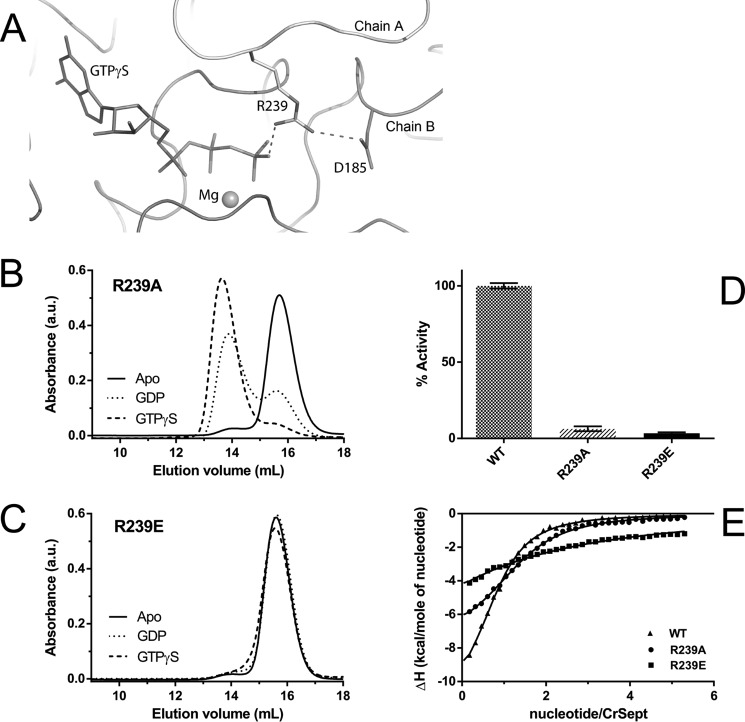
Switches I and II are completely ordered in CrSEPT. Switch I is one of the most variable regions among all septins both in terms of structure and sequence, and in this respect, CrSEPT is no exception (Fig. 1C). Switch II is more highly conserved than switch I, but in CrSEPT, it adopts an unusual conformation soon after Gly-182, enabling Asp-185 to form a salt bridge with Arg-239 from the opposite subunit (Fig. 4A). The latter forms the arginine finger, which will be discussed below. The switch II regions from the two monomers do not form main-chain hydrogen bonds across the G-interface, as has been observed in SEPT3 and SEPT7 (20, 26, 27).
Figure 4.
GTPase Activity of CrSEPT employs a catalytic arginine finger. A, hydrogen-bond interaction between Arg-239 (chain B) and the γ-phosphate of GTPγS within the CrSEPT nucleotide-binding pocket of chain A. The Arg-239 also interacts via a hydrogen bond with Asp-185 (from the switch II region). The magnesium is shown as a sphere. B and C, influence of the nucleotide on the oligomeric state of the R239A and R239E mutants. The mutation to Ala (R239A) did not influence GTP-dependent dimerization, whereas substitution by Glu (R239E) completely prevented dimerization. D, both mutants were unable to hydrolyze GTP. E, GTPγS binding affinities for CrSEPT mutants were determined using ITC, as in Fig. 2A. The following Kd values were obtained from the fits: R239A, 7.6 ± 0.4 μm; R239E, 103 ± 4 μm; WT, 5.4 ± 0.3 μm. Experiments were performed in triplicate, and the mean and S. D. values are reported.
The guanine moiety of GTPγS forms hydrogen bonds with Asp-265 (from the G4 motif), Thr-266 and Ala-324 and two water molecules (anchored by the main chain of residues Gly-122, Lys-263, and Val-322) (Fig. 3E). Lys-263 from G4 stacks over the guanine base in conventional fashion. The water molecule that interacts with Asn-7 and Gly-122 (w6) is analogous to that observed previously in SmSEPT10. Only two water-mediated interactions between the protein and the ribose moiety were observed, but there is a large hydrogen-bond network involving the phosphate moieties together with 13 protein residues, four water molecules, and one Mg2+ ion. The latter is coordinated, as expected, by the β- and γ-phosphates, two water molecules (one held by Asp-179 from the G3 motif), Thr-146 from switch I, and Thr-124 from the P-loop. The main-chain interactions with the γ-phosphate are consistent with the universal switch mechanism (28), and Thr-146 is predicted to hold the catalytic water molecule in position for in-line attack during catalysis. However, this water is not observed in the present structure, suggesting that the conformation in the crystal is not fully primed for catalysis.
In general, the loop between strand β4 and helix α3 has a highly conserved sequence, which is characteristic for septin GTP-binding domains (5). In CrSEPT, this loop is two residues shorter and therefore assumes a different conformation. As a consequence, His-238, which would normally interact with the nucleotide phosphates, instead lies sandwiched between Glu-271 on one side and Lys-263 (from the opposite subunit) on the other. Glu-271 also forms a conserved salt bridge with Arg-344 across the G interface (Fig. 3E), as observed in other septins.
Most notable, however, within this otherwise highly conserved β4-α3 loop is Arg-239, which is unique to CrSEPT (Fig. 1C). Its side chain interacts directly with the γ-phosphate of the opposite subunit (Fig. 4A). This residue resembles the “arginine finger,” previously observed in small GTPases and their cognate GTPase-activating proteins (GAPs) (29, 30) and also in GIMAP7 (31), which has been implicated in accelerating GTP hydrolysis in both cases. In the complex between RhoGAP and RhoA/GDP/MgF3−, the arginine finger (Arg-85) from RhoGAP penetrates the RhoA active site and simultaneously interacts with one of the oxygens of the β-phosphate and one of the fluorides of MgF3−, straddling what would be the β- and γ-phosphates of the transition state (32). However, in CrSEPT, the density for Arg-239 clearly shows it interacting in trans but with only the γ-phosphate of the GTPγS. Its orientation is apparently stabilized by the presence of Asp-185 from the opposite subunit.
To characterize the relevance of Arg-239 for the catalytic mechanism of CrSEPT, mutagenesis studies were performed. We introduced a point mutation by replacing Arg-239 with alanine (R239A), which had little effect on the binding affinity for GTPγS, as determined by ITC measurements (Fig. 4E). On the other hand, it completely eliminated GTPase activity (Fig. 4D). The same mutant shows a behavior broadly similar to that of the wild-type protein with respect to dimerization (Fig. 4B). In fact, the mutant appears to show a greater tendency than the wild type to dimerize in the presence of GDP. This may be due to disfavoring the buried arginine at the interface of the wild-type enzyme in the absence of the γ-phosphate, which provides charge compensation. In contrast, the charge-reversal mutant (R239E) bound the nucleotide poorly, did not form a dimer in its presence, and also lacked enzymatic activity (Fig. 4). These results clearly show that Arg-239 is an important component of the catalytic machinery capable of accelerating the intrinsic hydrolytic activity of the GTPase. In this respect, it is noteworthy that CrSEPT presents a higher catalytic rate compared with most other septins (21), and this is dramatically reduced upon mutating Arg-239.
CrSEPT forms filaments in vitro
The majority of structures reported to date show the recurrent occurrence of two types of contact made by the GTP-binding domain within the crystal. These have been named the G and NC interfaces, and they lead to the formation of filaments within the crystal, which are similar to those seen for the SEPT2/6/7 heterocomplex. In the case of CrSEPT, the G interface is intact and represents the contact made between the two monomers of the asymmetric unit. However, no NC contact is observed, and the crystal packing, therefore, does not lead to the formation of the canonical filament. This is somewhat unexpected, because the septin unique element, which is characteristic of filament-forming septins and forms an important part of the NC interface, is present in CrSEPT (Fig. 1B). This apparent inconsistency guided experiments to directly observe whether CrSEPT is able to form filaments at all.
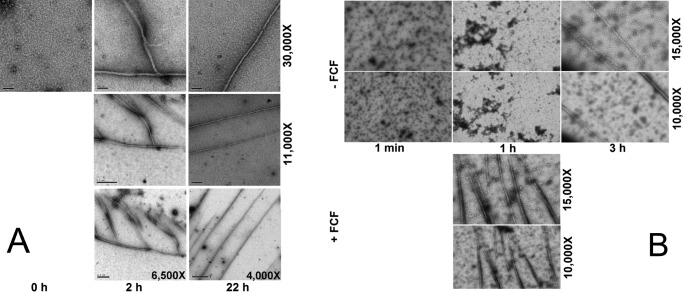
To address the capability of CrSEPT to form viable NC interactions, and therefore canonical septin filaments, without crystal packing constraints, we turned to transmission electron microscopy (TEM) analyses. Septins are known to polymerize under low-salt conditions, so we subjected purified CrSEPT to dialysis to evaluate whether it was able to form filaments in vitro, monitoring the process by TEM. Before dialysis, short rodlike structures were observed (Fig. 5A, panel 0 h), which probably correspond to dimers or to short oligomers forming during the initial stages of polymerization. These go on to form filaments with characteristic branches after 2 h of dialysis (Fig. 5A, panel 2 h). After further dialysis (22 h), the branches are no longer observed, and long homogeneous filaments remain (Fig. 5A, panel 22 h). These results show not only that the single septin from C. reinhardtii is able to homopolymerize, but also that this polymerization does not require the N- and C-terminal domains, because the filaments were observed with the CrSEPT construct, consisting of the GTP-binding domain alone. On the other hand, we did not observe bundles of filament or ring-like structures upon polymerization of CrSEPT. Because the N and C termini of yeast septins were shown to promote interfilament interactions (33), it is feasible that the full-size CrSEPT will form highly ordered structures observed in yeast septins (33) and S. mansoni septins (34).
Figure 5.
CrSEPT assembles into filaments in vitro. Shown are transmission electron micrographs of negatively stained septins at low salt concentration. A, samples produced by dialysis were taken after three different time intervals: 0, 2, and 22 h; B, sample produced by dilution. Top, samples were taken at three different times: within the first minute and after 1 and 3 h, respectively, in the presence of DMSO (control). Bottom, samples were taken only after the first hour, in the presence of 70 μm FCF. Nominal magnification for each panel is shown on the right.
Dynamics of septin filament assembly have been shown to be affected by forchlorfenuron (FCF; a compound known to affect septin filament assembly) in diverse organisms (34–36). Similar to the effect observed on the septin heterocomplex from the parasitic helminth S. mansoni (34), the presence of FCF accelerated the polymerization rate of CrSEPT under low-salt conditions, yielding long polymers after only 1 h (Fig. 5B). This result further supports the formation of septin-like CrSEPT homopolymeric filaments. Additionally, the ability of the R239E to form filaments under low-salt conditions was also monitored by TEM. We observed short oligomers for the mutant (data not shown), contrasting with long filaments for WT, suggesting that the lack of nucleotide binding indeed affects filament assembly in some way.
C. reinhardtii septin localizes mainly at the base of the flagella
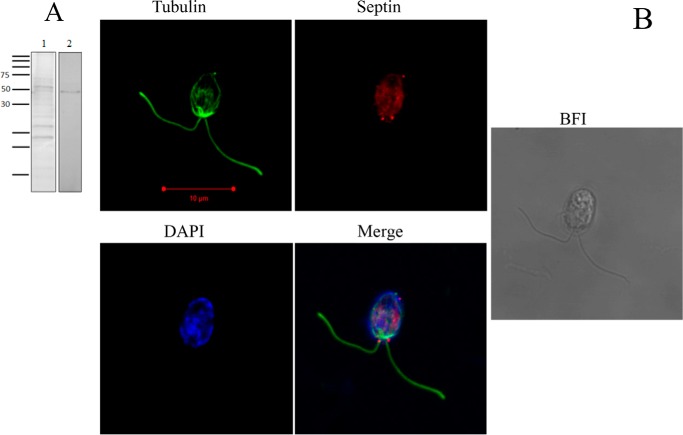
Once established that CrSEPT is able to polymerize in vitro, it was of interest to address the question of its subcellular localization with a view to shedding light on its possible functions. Full-length CrSEPT was used to produce an antibody, which was able to recognize the native protein in the crude extract of C. reinhardtii (Fig. 6A). Using this antibody, we then performed immunofluorescence experiments to localize the septin in C. reinhardtii cells. Fig. 6B shows a punctate distribution of septins in the cells, which is stronger at the base of the flagella (red). Control samples (without primary antibody) showed only an autofluorescent background due to chlorophyll (not shown).
Figure 6.
CrSEPT immunodetection in Chlamydomonas. A, Western blot analysis was employed to examine the specificity of the anti-CrSEPT antibody to proteins extracted from Chlamydomonas cells. An extract of 10 ml of culture was prepared by using radioimmune precipitation buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) (61). Lane 1, total proteins extracted after resolution by 15% SDS-PAGE and stained with Coomassie Blue; lane 2, developed nitrocellulose membrane. B, septins are present mainly at the base of the flagella of C. reinhardtii. Shown is a confocal optical section of C. reinhardtii cells labeled with anti-CrSEPT. The immunolocalization of septin is shown as a bright red signal. This figure shows a punctate distribution of septins in the cells, stronger at the base of the flagella. A background fluorescent signal is present in the entire cell due to autofluorescence. Cells were also stained with anti-tubulin (green) and DAPI (blue). Panel 4 shows overlaid images from panels 1–3. Panel 5 shows the bright-field image (BFI) of Chlamydomonas cells.
Septins have been previously related to the formation of a diffusion barrier at the base of the primary cilium (37) and to ciliogenesis in Xenopus embryos (38). The localization of C. reinhardtii septin to the base of the flagella is consistent with a possible role in barrier formation, which has been described to involve a structure known as the “flagellar bracelet” (39, 40). Taken together, the in vitro and in vivo data point toward a probable role for CrSEPT in forming homopolymeric filaments that are organized at the base of the flagella to promote barrier formation.
When cells were incubated for 30 min in the presence of 75 μm FCF, they started to release their flagella, and this phenomenon was accelerated at higher concentration (after 15 min at 100 μm). Consequently, paralysis of the cells was observed, and after further incubation in the presence of FCF, they started to die. However, when cells, which had been exposed to FCF for up to 1 h at 75 μm, were subsequently transferred to fresh media without the compound, a partial recovery was observed after 24 h, with the growth of new flagella in most of the cells. The phenotype observed upon incubation was dependent upon FCF concentrations and time of exposure, as summarized in Table 2.
Table 2.
Effects of FCF treatment on C. reinhardtii
| FCF concentration | Incubation time |
||||||
|---|---|---|---|---|---|---|---|
| 15 min | 30 min | 1 h | 1.5 h | 2 h | 24 h | 30 min to 1 h removeda | |
| μm | |||||||
| 25 | NEb | NE | NE | NE | NE | ||
| 50 | NE | NE | ABc | AB | AB | AB | |
| 75 | Pd (∼50%) | P | P | P (starting to die) | Dead | Partial recovery | |
| 100 | P | P (starting to die) | Dead | ||||
a Phenotype observed after incubation with FCF for the indicated period followed by substitution with fresh medium not containing FCF and incubation for 24 h.
b NE, no effect.
c AB, abnormal behavior (cells less active).
d P, paralysis, observed as a result of flagellar release.
Discussion
Septin heterocomplexes typically present very low rates of intrinsic GTP hydrolysis. However, when expressed in isolation, some septins purify and are stable as nucleotide-free monomers, which tend to present much higher rates of catalysis (21). The relevance of such an observation is unclear, given that isolated septins are normally members of native heterofilaments. For example, in the case of human septins, these are based on core particles, which may be either heterohexamers or hetero-octamers (41). In yeast and C. elegans, the equivalent core particles, from which filaments are derived, are hetero-octamers and heterotetramers, respectively (11, 42), and a physiological function for a filament derived from a single septin has yet to be clearly identified. In this respect, the unique septin from C. reinhardtii represents an ideal model system to study homofilament formation and septin kinetics to be compared with data from individual septins from more complex systems.
CrSEPT readily polymerized under low-salt conditions, resulting in filaments with a diameter close to those reported for septin heterofilaments. This is suggestive of a similar overall architecture and would imply that CrSEPT probably fulfills its physiological role(s) in the form of filaments or complexes thereof rather than as a monomer, as is the case for Arf-like proteins (43). The localization of CrSEPT at the base of the flagella suggests that it could be acting as a membrane-diffusion barrier and, therefore, performing a role similar to that described for septins found in cilia and flagella in other organisms. The plasma membrane has been implicated in driving septin filament elongation (44) and thus could play a role at the base of the flagella in controlling polymerization for diffusion barrier formation. This could represent an original ancestral function for septins, which has been preserved throughout evolution.
The effect of FCF in causing flagella loss also has implications in the relevance of septins as a fundamental basal component of this structure. However, lacking more data, we cannot eliminate an alternative toxic effect of FCF, unrelated to septin action, as the main cause of flagella release, because this is a known cellular response to adverse conditions in Chlamydomonas (45). Nevertheless, the fact that septins were localized at the base of the flagella, which was the first structure to be disturbed by FCF treatment, is suggestive of septin involvement and should encourage further studies.
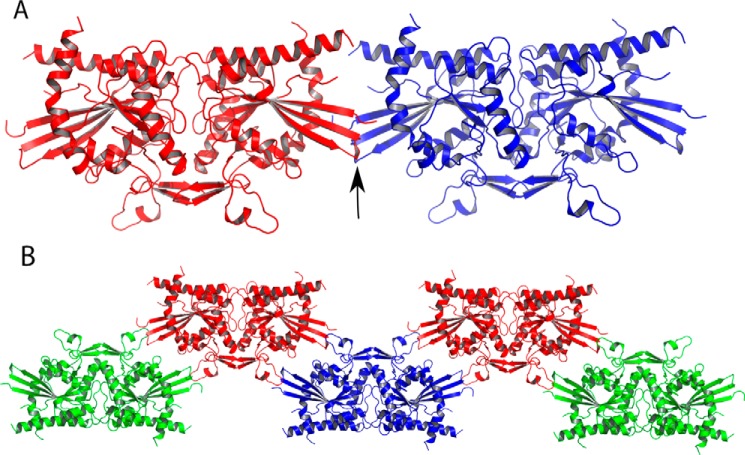
The formation of homopolymers by CrSEPT is consistent with the presence of the septin unique element at the C-terminal end of the GTP-binding domain. This region is a prominent feature in the formation of both the G and NC interfaces along a functional filament, most notably the latter (Fig. 1B). Also totally conserved in CrSEPT are the characteristic charged residues found within the C-terminal α-helix (α6) and at the end of helix α2 (residues Glu-211, Arg-216, Glu-385, and Arg-388). These normally form a characteristic network of salt bridges at the NC interface. Nevertheless, CrSEPT does not form filaments within the crystal, as have been frequently observed for other isolated septin GTP-binding domains. This is due to the absence of the NC contacts.
The absence of this interface is in part due to the protrusion of the first three strands of the main β-sheet, which would lead to steric hindrance with the neighboring subunit (Fig. 3A). This can be readily observed by superposing individual CrSEPT monomers across the NC interface of a homofilament composed of SEPT2 (Fig. 7A). However, only minor conformational adjustments to this region would be required to eliminate the bad contacts and generate a viable NC interface. Its absence in the structure reported here is therefore probably the result of crystal packing. Fig. 7B shows that distortions to the first three β-strands are not unexpected because they are directly involved in crystal contacts, involving residues 331–341 and 352–362 from a neighboring G-dimer. The absence of the NC contacts is entirely consistent with the fact that the charged residues, which form the conserved network of salt bridges, are largely disordered in CrSEPT because they presumably require these contacts for structural stability.
Figure 7.
NC canonical septin interfaces are not observed in CrSEPT crystals. A, superposing individual CrSEPT monomers across the NC interface of a homofilament composed of SEPT2. The arrow shows a minimal clash between the β-turns formed by strands β2 and β3 of each monomer. B, arrangement of CrSEPT dimers in crystal packing, showing how elements important for forming a canonical NC interface are unavailable because they are involved in packing contacts that stabilize the crystal.
Structural variation at the NC interface has been reported previously. In the case of the complex between human SEPT2 and the GTP analog, Gpp(NH)p, filaments are similarly not observed in the crystal due to the absence of the NC interface (19). Once again, this is almost certainly the consequence of crystal packing. Specifically, in the case of the SEPT2-Gpp(NH)p complex, the first three β-strands are observed to form a contiguous β-sheet with an identical region from a neighboring molecule.
Other variations at the NC interface have been observed in the crystal structures of SmSEPT10 and human SEPT3 (20, 24). In the former, β-strand slippage as a function of the nature of the bound nucleotide leads to strand projection into the interface, whereas in the latter, a dramatic rearrangement of the salt bridges causes a squeezing of the interface in which the centers of mass of the two subunits lie closer together. Taken together, all of these observations suggest that there is conformational plasticity among the components that make up the NC interface. Such plasticity is probably sufficient to allow for the minimal adjustments necessary for CrSEPT to eliminate the steric hindrance described above and generate a canonical NC interface. Nevertheless, the possibility of an alternative non-canonical filament cannot be entirely ruled out, particularly given the distant evolutionary relationship between algae and the remaining organisms for which structural information is available. Fig. 7B shows a possible non-canonical filament present within the crystal lattice. A similar zig-zag arrangement has been proposed for the septin-related GIMAP proteins (31, 46), although there is very little direct evidence to support it. On the contrary, based on the arguments laid out above, the most likely arrangement for the filaments observed by electron microscopy is that of an architecture broadly similar to that described previously. However, to definitively elucidate this issue, further studies regarding homofilament architecture are still required and encouraged.
Based on our structural data and mutagenesis experiments, it is justified to consider that Arg-239 acts as a catalytic arginine finger and is the principal reason that CrSEPT presents a higher catalytic rate compared with all other septins described to date. The kcat reported here, on the order of 2 min−1, is entirely consistent with that observed for Gα subunits of trimeric G proteins, which also possess a built-in arginine finger (47, 48). However, the catalytic water is absent, and the orientation of the arginine finger is different from that seen in complexes that imitate the transition state (32). It would necessarily undergo subtle rearrangements enabling the arginine finger to stabilize the negative charge, which develops on the transition state during catalysis, and the catalytic water to assume its correct position. Similar observations have been made for the complex between Cdc42Hs/GMPPNP and RhoGAP (49). Furthermore, the arginine finger may play a second role in favoring rapid catalysis by stabilizing the switch region via the salt bridge with Asp-185.
Arginine fingers are more commonly found in the auxiliary GAPs necessary for accelerating the hydrolysis of GTP via heterodimer formation. This would seem to imply that C. reinhardtii CrSEPT might act as its own GAP, and as far as we are aware this is the first direct evidence for such a mechanism applying to septins. This feature appears to have been lost in fungi and animal septins, which lack the corresponding arginine, possibly simultaneously with the appearance of multiple gene copies and consequently heterofilament assemblies. On the other hand, the Arg-239 is conserved in homologs from other algae species, such as V. carteri, N. bacillaris, M. neglectum, C. variabilis, Gonium pectorale, Coccomyxa subellipsoidea, and Klebsormidium flaccidum, suggesting it to be a common feature. The fact that most of algae genomes analyzed present a single septin gene copy suggests that a homofilament would be capable of performing the essential functions in these organisms. It would therefore appear that septin dynamics in this branch of evolution would be significantly different than those from other studied systems due to the predominance of homofilaments and high turnover of nucleotides. Therefore, further studies on algal septin filament formation and dynamics and comparison with the same aspects in other septin assemblies might provide important insights into the mechanism of such processes. Moreover, the ease with which GTP binding and GTPase activity can be measured in C. reinhardtii septin means that systematic studies using mutants could be readily employed to determine the influence of specific residues on both GTP hydrolysis and filament formation.
Experimental procedures
C. reinhardtii strain and culture conditions
C. reinhardtii cell wall-deficient CC-4351 (cw15 arg7-8 mt+) strain was maintained in Tris acetate phosphate (TAP), supplemented with 100 μg·ml−1 arginine and 2% agar (50) at 24 °C under continuous light. For RNA or protein isolation, cells were inoculated into liquid TAP medium, supplemented with 100 μg·ml−1 arginine, and allowed to grow under continuous light at 24 °C in an orbital shaker at 135 rpm until the culture reached mid-log growth.
Cloning procedures
Algae cells were harvested from 10 ml of culture by centrifugation and stored at −80 °C until RNA extraction. Total RNA was extracted as described previously (51), and the CrSEPT coding sequence was obtained by RT-PCR using cDNA as the template. cDNA was synthesized from total RNA via the gene-specific primer (accession number XM_001694125) by using a SuperScript II reverse transcriptase kit (Invitrogen Life Technologies). The full-length CrSEPT cDNA was subsequently amplified by PCR, introducing an in-frame 5′ BamHI site and a 3′ HindIII site. The PCR product was inserted into pGEM-T Easy vector (Promega) and confirmed by nucleotide sequencing (3130 Genetic Analyzer, Thermo Fisher Scientific). The R239A and R239E mutants of CrSEPT were generated using the QuickChange mutagenesis kit (Stratagene). All mutants were also confirmed by DNA sequencing.
Protein expression and purification
The full septin-encoding sequence (residues 1–481), named CrSEPT(1–481), and the truncated version comprising the GTP-binding domain (CrSEPT(86–393)) were subcloned into a modified pET28a expression plasmid, yielding N-terminally hexahistidine-tagged SUMO fusion proteins. The WT protein construct and mutants were overexpressed in Escherichia coli Rosetta (DE3) (Novagen), and the cells were grown in LB medium supplemented with 50 mg·liter−1 kanamycin at 37 °C. At a cell density corresponding to an absorbance of 0.8 at 600 nm, the temperature was reduced to 20 °C, and protein expression was induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside. After 16 h, cells were harvested by centrifugation and suspended in buffer A (50 mm Tris-HCl, pH 8, 500 mm NaCl, 10% glycerol, 5 mm mercaptoethanol). After cell lysis by sonication and removal of cell debris by centrifugation, clear lysates were loaded onto a nickel-nitrilotriacetic acid column (Qiagen) pre-equilibrated in buffer A. After resin-washing steps with buffer A, the protein was eluted with buffer A supplemented with 500 mm imidazole in one single step. Affinity tags were removed by protein incubation with the protease Ulp-1 at 4 °C for 2 h. The nickel-nitrilotriacetic acid affinity chromatography was performed for a second time, and the target protein was collected in the flow-through. Proteins were concentrated, and the buffer was exchanged, according to the subsequent experiment. Pure protein samples were stored at −80 °C. The mutants were purified likewise. To produce SeMet-labeled protein, M9 minimal medium supplemented with the antibiotics was inoculated with an overnight LB culture at a 1:100 dilution. Cells were grown to an A600 nm of 0.8 at 37 °C. Subsequently, an amino acid supplement (l-lysine, l-phenylalanine, l-threonine to a final concentration of 100 mg·liter−1 or l-isoleucine, l-leucine, l-valine, and l-SeMet to a final concentration of 50 mg·liter−1, respectively) was added to inhibit endogenous methionine biosynthesis and initiate SeMet incorporation. After 15 min, the bacterial culture was cooled to 20 °C, and protein expression was induced by the addition of 0.2 mm isopropyl 1-thio-β-d-galactopyranoside. Further growth, cell harvesting, and purification steps were conducted in the same way as for the native protein.
Determination of protein-bound nucleotide
Nucleotide determination was carried out according to the method described previously (52) with minor modifications, according to Ref. 20.
Hydrolytic rate parameters
GTP hydrolysis was measured by a continuous enzyme-coupled optical assay for the release of inorganic phosphate from GTP using the EnzChek phosphate assay kit (Thermo Fisher Scientific) according to the manufacturer's instructions. CrSEPT and mutants R239A and R239E (0.5 μm) were in 20 mm HEPES buffer, pH 7.5, 300 mm NaCl, 5% glycerol, 2 mm MgCl2.
ITC
ITC experiments were carried out at 25 °C in a VP-ITC calorimeter (Microcal) in phosphate buffer (20 mm phosphate, pH 7.5, 40 mm NaCl, 5% glycerol, 2 mm MgCl2), employing a CrSEPT concentration of 20 μm in the cell. Nucleotide concentrations in the syringe were 0.5 mm in the case of GTPγS and 2.3 mm for GDP (35 injections of 8 μl each for both titrations). In addition, the nucleotide binding to the mutants R239A and R239E was obtained using a similar protocol. The heat of dilution was subtracted from the binding curve, and binding isotherms were fitted and equilibrium dissociation constants calculated using the Microcal ORIGIN software.
CrSEPT oligomeric state dependence on the presence of nucleotide
Aliquots of CrSEPT(86–393) and the mutants (10 μm) in 20 mm HEPES buffer, pH 7.5, 40 mm NaCl, 5% glycerol, 2 mm MgCl2 were incubated with 100 μm GTPγS for 3 h at 20 °C and analyzed by SEC on a Superdex 200 10/300 GL column monitored at 280 nm.
Crystallization, data collection, and structure determination
Commercially available crystallization screens were initially used in hanging-drop vapor-diffusion experiments with CrSEPT at 2.8 mg·ml−1 (20 mm HEPES buffer, 300 mm NaCl, 5% glycerol, 2 mm MgCl2, pH 7.5). Crystals of protein preincubated with 0.5 mm GTPγS were obtained at 18 °C when equilibrated against a solution of 12.5% PEG 1000, 12.5% PEG 3350, 12.5% 2-methyl-2,4-pentanediol (MPD), 30 mm each of sodium nitrate, disodium hydrogen phosphate, ammonium sulfate, and 100 mm Bicine/Trizma (Tris base), pH 8.5. Before data collection, crystals were flash-frozen in liquid nitrogen. X-ray diffraction data were collected on beamline I04-1 of Diamond Light Source. The Xia2 software (53), a data-processing pipeline that uses XDS (54), CCP4 (55) and Aimless (56) processed the data. The structure of CrSEPT bound to GTPγS was solved by single anomalous dispersion using the anomalous signal of crystals grown with SeMet-labeled protein. Phasing was conducted with SHELX (57), and the initial model was automatically built using PHENIX (58). Alternating rounds of model rebuilding and refinement using COOT (59) and PHENIX, respectively, yielded the final model.
TEM of CrSEPT
The polymerization of CrSEPT was evaluated by using two different approaches to decrease the salt concentration. In the first method, the protein at 2.9 mg·ml−1 in crystallization buffer was dialyzed against 15 mm Tris-HCl, pH 8.0, containing 40 mm NaCl, 2 mm MgCl2, 0.5 mm GTP, 2 mm DTT for 2 and 22 h at 4 °C. In the second, a protein aliquot at 10 mg·ml−1 in 15 mm Tris-HCl, pH 8.0 buffer, containing 400 mm NaCl, 2 mm MgCl2, 5% glycerol was diluted to 1 mg·ml−1, to decrease the salt concentration to 40 mm, in the presence of 75 μm FCF or DMSO (control). In this case, aliquots were taken within the first minute and after 1 and 3 h. To visualize polymerization, aliquots (10 μl) were applied to glow-discharged carbon-coated grids and stained with 2% uranyl acetate. The grids were examined in a Tecnai-12 transmission electron microscope (FEI, Hillsboro, OR) under regular-dose conditions at an accelerating voltage of 80 keV.
Immunolocalization of CrSEPT in C. reinhardtii
A polyclonal antibody against the full-length recombinant CrSEPT(1–481) was raised in rabbits by Rhea Biotech (Campinas, SP, Brazil). Immunofluorescence was performed as described by Keller et al. (60). Primary antibodies used were rabbit anti-CrSEPT (1:20) and mouse anti-α-tubulin (Sigma; 1:20), and secondary antibodies were Alexa Fluor 555-conjugated goat anti-rabbit IgG (Life Technologies) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Life Technologies) at 1:200 dilution. Coverslips were washed and then mounted with Fluoromount G (Sigma-Aldrich). Confocal images were acquired with a Carl Zeiss LSM 780 microscope using a Plan-Apochromat 636/1.40 oil differential interference contrast objective.
Effects of FCF on C. reinhardtii
Aliquots of C. reinhardtii cultured for 7 days in TAP supplemented with arginine (100 μm·ml−1) were incubated with FCF at different concentrations (25–100 μm) and time intervals. The phenotypic effect was observed and recorded with the assistance of an optical microscope (Olympus BX53).
Author contributions
A. P. A. P. designed and performed most of the experiments under the supervision of A. P. U. A. and R. D.; H. C. and A. E. Z. acquired and analyzed confocal images; H. M. P. and J. B.-N. collected the diffraction data and performed the structure refinement; M. V. A. S. N. solved and analyzed the structure; F. M. F. designed the protein construct; V. E. G. and C. R. acquired and analyzed the transmission electron microscopy data; R. C. G. analyzed the structure and, together with A. P. U. A. (who conceived the project), finalized the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Joci N. A. Macedo, Juliana Cheleski, and Napoleão Valadares for helpful discussions and Isabel de Moraes for technical assistance.
This work was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) under Grants 2011/10152-7 (to A. P. U. A.), 2014/15546-1 (to R. C. G. and A. P. U. A.), and 2012/14223-9 (to H. M. P.) and Fellowships 12/21259-0 and 2013/20715-4 (to H. C. and A. E. Z., respectively). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5IRR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SUE
- septin unique element
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate
- GMPPNP
- guanosine 5′-[β,γ-imido]triphosphate
- CrSEPT
- septin from Chlamydomonas
- SeMet
- selenomethionine
- GAP
- GTPase-activating protein
- TEM
- transmission electron microscopy
- FCF
- forchlorfenuron
- TAP
- Tris acetate phosphate
- ITC
- isothermal titration calorimetry
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- SEC
- size-exclusion chromatography.
References
- 1. Hartwell L. H. (1971) Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69, 265–276 [DOI] [PubMed] [Google Scholar]
- 2. Ong K., Wloka C., Okada S., Svitkina T., and Bi E. (2014) Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat. Commun. 5, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., and Pringle J. R. (1996) The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119 [DOI] [PubMed] [Google Scholar]
- 4. Kinoshita M. (2006) Diversity of septin scaffolds. Curr. Opin. Cell Biol. 18, 54–60 [DOI] [PubMed] [Google Scholar]
- 5. Pan F., Malmberg R. L., and Momany M. (2007) Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindsey R., and Momany M. (2006) Septin localization across kingdoms: three themes with variations. Curr. Opin. Microbiol. 9, 559–565 [DOI] [PubMed] [Google Scholar]
- 7. Versele M., and Thorner J. (2005) Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15, 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wloga D., Strzyzewska-Jówko I., Gaertig J., and Jerka-Dziadosz M. (2008) Septins stabilize mitochondria in Tetrahymena thermophila. Eukaryot. Cell 7, 1373–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishihama R., Onishi M., and Pringle J. R. (2011) New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol. Chem. 392, 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bourne H. R., Sanders D. A., and McCormick F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 11. Versele M., Gullbrand B., Shulewitz M. J., Cid V. J., Bahmanyar S., Chen R. E., Barth P., Alber T., and Thorner J. (2004) Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 4568–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheffield P. J., Oliver C. J., Kremer B. E., Sheng S., Shao Z., and Macara I. G. (2003) Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J. Biol. Chem. 278, 3483–3488 [DOI] [PubMed] [Google Scholar]
- 13. Kinoshita M. (2003) The septins. Genome Biol. 4, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I. G., Weyand M., Stark H., and Wittinghofer A. (2007) Structural insight into filament formation by mammalian septins. Nature 449, 311–315 [DOI] [PubMed] [Google Scholar]
- 15. Mendoza M., Hyman A. A., and Glotzer M. (2002) GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 12, 1858–1863 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y.-W., Surka M. C., Reynaud D., Pace-Asciak C., and Trimble W. S. (2006) GTP binding and hydrolysis kinetics of human septin 2. FEBS J. 273, 3248–3260 [DOI] [PubMed] [Google Scholar]
- 17. Yamazaki T., Owari S., Ota S., Sumiya N., Yamamoto M., Watanabe K., Nagumo T., Miyamura S., and Kawano S. (2013) Localization and evolution of septins in algae. Plant J. 74, 605–614 [DOI] [PubMed] [Google Scholar]
- 18. Valadares N. F., and Garratt R. C. (2016) Septin crystallization for structural analysis. Methods Cell Biol. 136, 321–338 [DOI] [PubMed] [Google Scholar]
- 19. Sirajuddin M., Farkasovsky M., Zent E., and Wittinghofer A. (2009) GTP-induced conformational changes in septins and implications for function. Proc. Natl. Acad. Sci. 106, 16592–16597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macedo J. N., Valadares N. F., Marques I. A., Ferreira F. M., Damalio J. C., Pereira H. M., Garratt R. C., and Araujo A. P. (2013) The structure and properties of septin 3: a possible missing link in septin filament formation. Biochem. J. 450, 95–105 [DOI] [PubMed] [Google Scholar]
- 21. Zent E., and Wittinghofer A. (2014) Human septin isoforms and the GDP-GTP cycle. Biol. Chem. 395, 169–180 [DOI] [PubMed] [Google Scholar]
- 22. Farkasovsky M., Herter P., Voss B., and Wittinghofer A. (2005) Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol. Chem. 386, 643–656 [DOI] [PubMed] [Google Scholar]
- 23. Miao W., Eichelberger L., Baker L., and Marshall M. S. (1996) p120 Ras GTPase-activating protein interacts with Ras-GTP through specific conserved residues. J. Biol. Chem. 271, 15322–15329 [DOI] [PubMed] [Google Scholar]
- 24. Zeraik A. E., Pereira H. M., Santos Y. V., Brandão-Neto J., Spoerner M., Santos M. S., Colnago L. A., Garratt R. C., Araújo A. P., and DeMarco R. (2014) Crystal structure of a Schistosoma mansoni septin reveals the phenomenon of strand slippage in septins dependent on the nature of the bound nucleotide. J. Biol. Chem. 289, 7799–7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keefe L. J., Sondek J., Shortle D., and Lattman E. E. (1993) The α-aneurism: a structural motif revealed in an insertion mutant of Staphyloccocal nuclease. Proc. Natl. Acad. Sci. U.S.A. 90, 3275–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Serrão V. H., Alessandro F., Caldas V. E., Marcal R. L., Pereira H. D., Thiemann O. H., and Garratt R. C. (2011) Promiscuous interactions of human septins: the GTP binding domain of SEPT7 forms filaments within the crystal. FEBS Lett. 585, 3868–3873 [DOI] [PubMed] [Google Scholar]
- 27. Zent E., Vetter I., and Wittinghofer A. (2011) Structural and biochemical properties of Sept7, a unique septin required for filament formation. Biol. Chem. 392, 791–797 [DOI] [PubMed] [Google Scholar]
- 28. Vetter I. R., and Wittinghofer A. (2001) The guanine nucleotide binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 29. Bourne H. R. (1997) The arginine finger strikes again. Nature 389, 673–674 [DOI] [PubMed] [Google Scholar]
- 30. Zhang B., Zhang Y., Collins C. C., Johnson D. I., and Zheng Y. (1999) A built-in arginine finger triggers the self-stimulatory GTPase-activating activity of rho family GTPases. J. Biol. Chem. 274, 2609–2612 [DOI] [PubMed] [Google Scholar]
- 31. Schwefel D., Arasu B. S., Marino S. F., Lamprecht B., Köchert K., Rosenbaum E., Eichhorst J., Wiesner B., Behlke J., Rocks O., Mathas S., and Daumke O. (2013) Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure 21, 550–559 [DOI] [PubMed] [Google Scholar]
- 32. Graham D. L., Lowe P. N., Grime G. W., Marsh M., Rittinger K., Smerdon S. J., Gamblin S. J., and Eccleston J. F. (2002) MgF3− as a transition state analog of phosphoryl transfer. Chem. Biol. 9, 375–381 [DOI] [PubMed] [Google Scholar]
- 33. Bertin A., McMurray M. A., Grob P., Park S. S., Garcia G. 3rd, Patanwala I., Ng H. L., Alber T., Thorner J., and Nogales E. (2008) Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 8274–8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeraik A. E., Galkin V. E., Rinaldi G., Garratt R. C., Smout M. J., Loukas A., Mann V. H., Araujo A. P. U., DeMarco R., and Brindley P. J. (2014) Reversible paralysis of Schistosoma mansoni by forchlorfenuron, a phenylurea cytokinin that affects septins. Int. J. Parasitol. 44, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu Q., Nelson W. J., and Spiliotis E. T. (2008) Forchlorfenuron alters mammalian septin assembly, organization, and dynamics. J. Biol. Chem. 283, 29563–29571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeMay B. S., Meseroll R. A., Occhipinti P., and Gladfelter A. S. (2010) Cellular requirements for the small molecule forchlorfenuron to stabilize the septin cytoskeleton. Cytoskeleton 67, 383–399 [DOI] [PubMed] [Google Scholar]
- 37. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., and Nelson W. J. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim S. K., Shindo A., Park T. J., Oh E. C., Ghosh S., Gray R. S., Lewis R. A., Johnson C. A., Attie-Bittach T., Katsanis N., and Wallingford J. B. (2010) Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329, 1337–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiss R. L., Goodenough D. A., and Goodenough U. W. (1977) Membrane particle arrays associated with the basal body and with contractile vacuole secretion in Chlamydomonas. J. Cell Biol. 72, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caudron F., and Barral Y. (2009) Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16, 493–506 [DOI] [PubMed] [Google Scholar]
- 41. Sellin M. E., Sandblad L., Stenmark S., and Gullberg M. (2011) Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. John C. M., Hite R. K., Weirich C. S., Fitzgerald D. J., Jawhari H., Faty M., Schläpfer D., Kroschewski R., Winkler F. K., Walz T., Barral Y., and Steinmetz M. O. (2007) The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 26, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gillingham A. K., and Munro S. (2007) The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 44. Bridges A. A., Zhang H., Mehta S. B., Occhipinti P., Tani T., and Gladfelter A. S. (2014) Septin assemblies form by diffusion driven annealing on membranes. Proc. Natl. Acad. Sci. U.S.A. 111, 2146–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quarmby L. N. (2009) Deflagellation. In The Chlamydomonas Sourcebook: Cell Motility and Behavior, Vol. 3 (Witman G. B., ed) 2nd Ed., pp. 43–69, Academic Press, Inc., San Diego [Google Scholar]
- 46. Schwefel D., Fröhlich C., Eichhorst J., Wiesner B., Behlke J., Aravind L., and Daumke O. (2010) Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc. Natl. Acad. Sci. U.S.A. 107, 20299–20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Landis C. A., Masters S. B., Spada A., Pace A. M., Bourne H. R., and Vallar L. (1989) GTPase inhibiting mutations activate the α chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 340, 692–696 [DOI] [PubMed] [Google Scholar]
- 48. Graziano M. P., and Gilman A. G. (1989) Synthesis in Escherichia coli of GTPase deficient mutants of Gsα. J. Biol. Chem. 264, 15475–15482 [PubMed] [Google Scholar]
- 49. Rittinger K., Walker P. A., Eccleston J. F., Nurmahomed K., Owen D., Laue E., Gamblin S. J., and Smerdon S. J. (1997) Crystal structure of a small G protein in complex with the GTPase activating protein rhoGAP. Nature 388, 693–697 [DOI] [PubMed] [Google Scholar]
- 50. Hoober J. K. (1989) Chlamydomonas in the laboratory in The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use (Harris E. H., Stern D. B., and Witman G. B., eds) 2nd Ed., pp. 241–302, Academic Press, Inc., San Diego: [DOI] [PubMed] [Google Scholar]
- 51. Helliwell K. E., Wheeler G. L., Leptos K. C., Goldstein R. E., and Smith A. G. (2011) Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol. 28, 2921–2933 [DOI] [PubMed] [Google Scholar]
- 52. Seckler R., Wu G.-M., and Timasheff S. N. (1990) Interactions of tubulin with guanylyl-(P-y-methylene)diphosphonate: formation and assembly of a stoichiometric complex. J. Biol. Chem. 265, 7655–7661 [PubMed] [Google Scholar]
- 53. Winter G. (2010) Xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 [Google Scholar]
- 54. Kabsch W. (2010) XDS. Acta Crystallogr. D. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 57. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 58. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Keller L. C., Wemmer K. A., and Marshall W. F. (2010) Influence of centriole number on mitotic spindle length and symmetry. Cytoskeleton 67, 504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clark A. M., and Labute P. (2007) 2D depiction of protein-ligand complexes. J. Chem. Inf. Model. 47, 1933–1944 [DOI] [PubMed] [Google Scholar]