Abstract
Human myosin VIIa (MYO7A) is an actin-linked motor protein associated with human Usher syndrome (USH) type 1B, which causes human congenital hearing and visual loss. Although it has been thought that the role of human myosin VIIa is critical for USH1 protein tethering with actin and transportation along actin bundles in inner-ear hair cells, myosin VIIa's motor function remains unclear. Here, we studied the motor function of the tail-truncated human myosin VIIa dimer (HM7AΔTail/LZ) at the single-molecule level. We found that the HM7AΔTail/LZ moves processively on single actin filaments with a step size of 35 nm. Dwell-time distribution analysis indicated an average waiting time of 3.4 s, yielding ∼0.3 s−1 for the mechanical turnover rate; hence, the velocity of HM7AΔTail/LZ was extremely slow, at 11 nm·s−1. We also examined HM7AΔTail/LZ movement on various actin structures in demembranated cells. HM7AΔTail/LZ showed unidirectional movement on actin structures at cell edges, such as lamellipodia and filopodia. However, HM7AΔTail/LZ frequently missed steps on actin tracks and exhibited bidirectional movement at stress fibers, which was not observed with tail-truncated myosin Va. These results suggest that the movement of the human myosin VIIa motor protein is more efficient on lamellipodial and filopodial actin tracks than on stress fibers, which are composed of actin filaments with different polarity, and that the actin structures influence the characteristics of cargo transportation by human myosin VIIa. In conclusion, myosin VIIa movement appears to be suitable for translocating USH1 proteins on stereocilia actin bundles in inner-ear hair cells.
Keywords: cell permeabilization, molecular motor, myosin, processivity, single-molecule biophysics
Introduction
Myosins are actin-based molecular motor proteins that play important roles in diverse cell functions such as force production, cell motility, morphosis, cytokinesis, vesicle, or macromolecule transportation and so on (1). Myosin constitutes a superfamily with >35 subtypes, and it is thought that each myosin superfamily member has a specific physiological function in diverse cell motility processes (2). All myosins contain a conserved motor domain that binds actin and converts chemical energy of ATP to mechanical energy. The following neck domain binds calmodulin (CaM)2 or CaM-like light chains through IQ motifs3 and constitutes a lever arm when myosin moves on actin filaments. Tail domains are often critical for regulation of the motile activity, for selecting proper binding proteins, and for targeting of the motor to the specific intracellular location. Many myosins have a coiled-coil domain after the neck region to facilitate its dimer formation. Several myosins, e.g. myosin VI, myosin VIIa, myosin X, and MyoM (3), have a stable, single α-helix (SAH) within the neck domain, which is considered to work as flexible lever arm (3, 4). Although myosin superfamily members share this domain structure, it is thought that each myosin has its unique motility activity that is closely related to its physiological function. For instance, some myosin family members, such as myosin V, are suitable for cargo transportation, whereas others are appropriate for a large force production. Therefore, it is important to uncover the specific nature of specific myosin motor to understand its physiological function.
Myosin VIIa is composed of a motor domain, a neck domain with five IQ motifs, an SAH plus short coiled-coil domain, and a tail domain that contains two tandem MyTH4/FERM domains inserted by an SH3 domain (Fig. 1) (5). In humans, myosin VIIa is essential for normal hearing and vision, and the mutations often cause severe sensory defects, USH type 1B (6), and mild non-syndromic hearing loss, DFNB2; DFNA11 (7–9). Human myosin VIIa is expressed in a wide variety of cell types (10), but its function is best studied in auditory systems. In stereocilia in inner-ear hair cells, myosin VIIa is present at pericuticular necklace, actin meshwork in cuticular plate, and tip-links (11, 12). Although myosin VIIa broadly exists along stereocilia and is localized to the ankle links (13, 14), a recent study has revealed that myosin VIIa is particularly localized at upper tip-link density with the scaffold proteins, sans and harmonin B (12). From its tip-link localization, it is considered that myosin VIIa plays a crucial role in mechanoelectrical transduction in stereocilia of hair cells. Myosin VIIa is also present throughout retinal pigment epithelium cells and photoreceptor cells in eye and is especially abundant around apical actin-rich structures (16–18). Based on the mutational studies, it is considered that myosin VIIa is associated with the melanosome distribution and phagosome transfer in retinal pigment epithelium cells and opsin transport in photoreceptor cells (19–21).
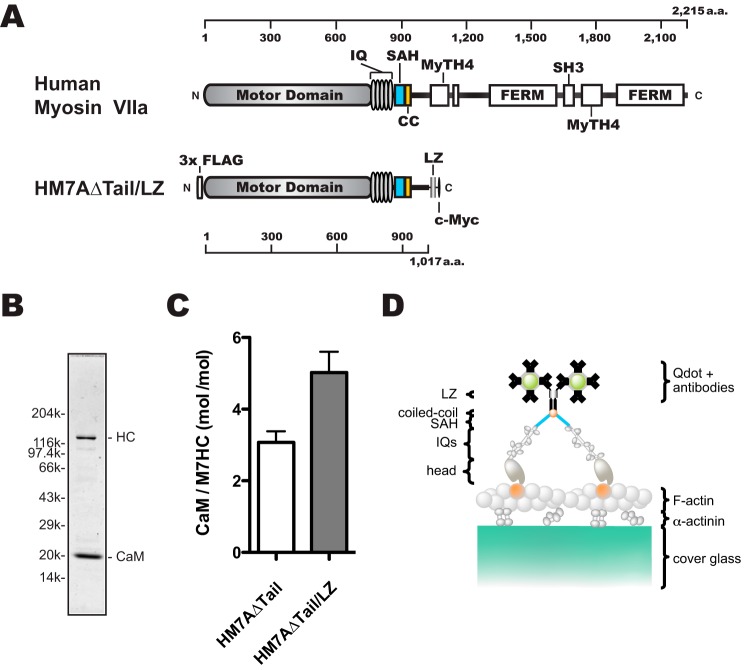
Figure 1.
Schematic diagrams of human myosin VIIa dimer construct (HM7AΔTail/LZ) used in this study. A, schematic diagram of human myosin VIIa construct. Upper, human myosin VIIa (1–2215 amino acids). Lower, forced dimer of human myosin VIIa (without tail domain) construct. The construct consisted of 3× FLAG, motor domain, 5× IQ motifs, an SAH domain, and LZ and c-Myc domains. The amino acid (a.a.) numbers of human myosin VIIa are shown at the top and bottom in A. B, SDS-PAGE of the purified HM7AΔTail/LZ. The myosin VIIa heavy chain was co-expressed with calmodulin and purified with anti-FLAG antibody agarose. Note that purified HM7AΔTail/LZ contains exogenous calmodulin that is added through all of the purification steps. Right: HC, HM7AΔTail/LZ heavy chain. Left, molecular mass markers described under “Experimental Procedures.” C, stoichiometry of CaM bound to HM7AΔTail and HM7AΔTail/LZ. Molar ratios of calmodulin: HM7AΔTail and HM7ADTail/LZ were estimated as described under “Experimental Procedures.” The means ± S.E. were 3.1 ± 0.3 (n = 4) and 5.0 ± 0.6 (n = 6), respectively. The p value of unpaired t test was 0.03 between HM7AΔTail and HM7AΔTail/LZ. D, schematic diagram of TIRF motility assay. HM7AΔTail/LZ labeled with Qdot525 is shown on an actin filament attached to α-actinin-coated cover glass. It is anticipated that HM7AΔTail/LZ dimerizes at the LZ motif with two Qdots at the maximum.
At least 12 proteins can bind to the tail domain of myosin VIIa (10), and the binding is associated with transportation of cargo and proper localization of myosin VIIa in cells. It has been reported that the tail domain of Drosophila myosin VIIa is also responsible for the regulation of the ATPase activity at the motor domain (22) and that the tail inhibition of ATPase activity is attributed to the folding of the tail domain back to the head/neck domain. The tail-dependent regulation mechanism is similar to those of myosin Va (23–27) and myosin X (28). The mechanoenzymatic inhibition is thought to be released by the binding of the Ca2+ ion or a cargo protein, MyRIP (22, 29, 30), directly involved in the motor domain-based motility of myosin VIIa.
In the present study we attempted to clarify the unique function of myosin VIIa at the molecular level by analyzing the motor properties of human myosin VIIa. One of the best approaches to characterize the motor properties of myosin is single-molecule analysis. However, little is known about the motor properties of mammalian myosin VIIa, although a part of single-molecule characterization has been done in the Drosophila counterpart (31). In this study we characterized the properties of human myosin VIIa at a single-molecule level both on single actin filaments and actin structures in demembranated cells. The analyzed data reveal that human myosin VIIa is a very slow processive motor with a large 35-nm step size on single actin filaments, which is suitable for transporting proteins. Moreover, our results indicate that the movement of myosin VIIa motor is more efficient on parallel bundled-actin in filopodia rather than stress fibers, where actin polarity is bidirectional. It is thought that this motor property is suitable for myosin VIIa to effectively move on actin bundles in stereocilia of inner-ear hair cells as a USH1 protein transporter as well as tensor of tip-link.
Results
Expression and purification of human myosin VIIa dimer
We produced a tail-truncated human myosin VIIa dimer (HM7AΔTail/LZ) construct to study the motor properties (Fig. 1A). A leucine zipper (LZ) was introduced at 102 amino acid residues downstream of the predicted short coiled-coil to produce a stable dimer. Based upon the secondary structure prediction (SIMPA96, NPS@) (32), this region is composed of a number of random coils and is expected to be flexible. With the 88-amino acid sequence of myosin VIIa with a flexible nature, we anticipate that the innate coiled-coil property is not much influenced by the C-terminal LZ in the construct. N-terminal 3× FLAG was introduced in aid of purification by anti-FLAG antibody affinity chromatography. C-terminal c-Myc tag was added to conjugate quantum dots (Qdots) through anti-c-Myc antibodies. This construct was expressed in Sf9 cells using baculovirus expression system. To express the myosin VIIa construct with IQ domains, we co-expressed CaM with myosin VIIa heavy chain. The isolated HM7AΔTail/LZ heavy chain containing IQ domain was co-purified with CaM light chain (Fig. 1B). The apparent molecular mass of each construct determined from the mobility on SDS-PAGE was 130 kDa for HM7AΔTail/LZ heavy chain and 20 kDa for CaM light chains, which are consistent with the calculated molecular masses of these constructs (Fig. 1, A and B). We previously reported that human myosin VIIa could bind non-muscle regulatory light chain (RLC). In the present study we used CaM as the light chain because human myosin VIIa prefers CaM to RLC (30). Note that 5 μm CaM was exogenously added to the buffer throughout myosin VIIa preparation to obtain HM7AΔTail/LZ showing good motility. To estimate the number of calmodulin per heavy chain, myosin VIIa was co-precipitated with F-actin (supplemental Fig. 1). Consistent with the previous study, myosin VIIa without LZ showed substoichiometric CaM binding (30) (Fig. 1C). Interestingly, the five IQ motifs of myosin VIIa with LZ used in the present study were fully occupied with CaM, which suggests that the dimer formation of myosin VIIa may affect the association of light chains with the neck domain of human myosin VIIa.
Human myosin VIIa dimer moves processively on single actin filaments
We mixed HM7AΔTail/LZ with Qdot525 (molar ratio = 1:20) to examine the single-molecule movement (Fig. 1D). Under this condition, 4.7% and 0.12% of Qdots were calculated to bind one and two or more molecules, respectively, assuming a simple Poisson distribution. Therefore, >∼97% of the moving myosin VIIa is considered to be a single molecule. We prepared the flow cells in which actin filaments were immobilized, and the HM7AΔTail/LZ movement was observed under the total internal reflection fluorescence (TIRF) microscope. We found that HM7AΔTail/LZ moves successively on single actin filaments at a near-physiological ATP concentration (supplemental Movie 1). Fig. 2A shows a representative trace of the stepping of single HM7AΔTail/LZ molecule in the presence of 2 mm ATP. The histogram of observed step-size distribution of HM7AΔTail/LZ in the presence of 2 mm ATP was shown in Fig. 2B. The distribution of both plus- and minus-directed movement was symmetric, and the best fit to a single Gaussian equation yielded the mean step size of forward and backward steps of 34.7 ± 11.5 nm (mean ± S.D., n = 521) and −17.2 ± 16.5 nm (S.D., n = 21), respectively. This indicates that human myosin VIIa is a processive motor with large steps. The forward step size is slightly larger than that of Drosophila myosin VIIa (see Table 1) (31).
Figure 2.
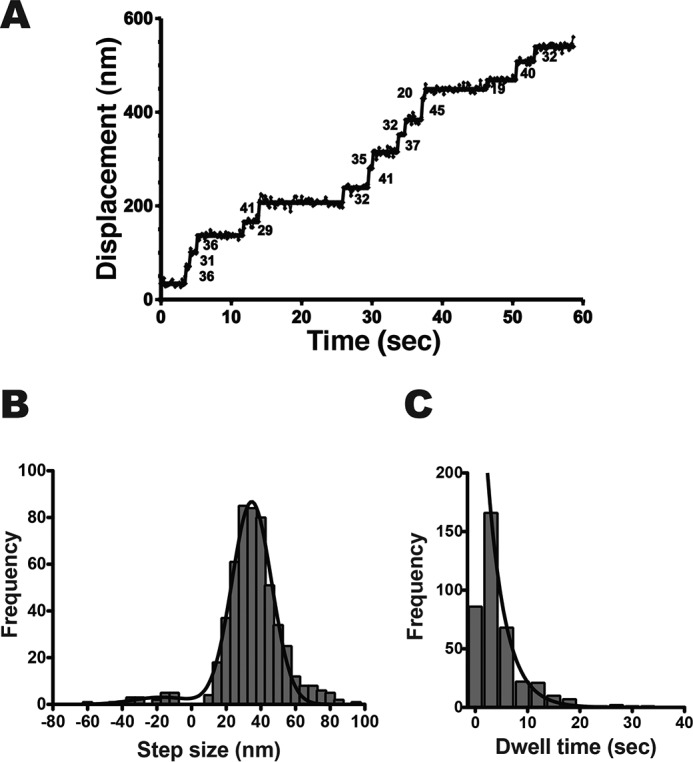
Processive movement of human myosin VIIa dimer on single actin filaments. A, a representative stepping trace of HM7AΔTail/LZ in the presence of 2 mm ATP. The fluorescence images were captured at 3.3 fps. The solid line represents the best fit to the trajectory. The numbers in the panel are displacement in nm of each step. B, step-size distribution of HM7AΔTail/LZ. The mean step-size of forward and backward steps are 37.5 ± 13.9 nm (mean ± S.D., n = 521) and −23.7 ± 13.4 nm (S.D., n = 21), respectively. The black solid line shows the best fit to the Gaussian equation with parameters of 34.7 ± 11.5 nm (mean ± S.D.) and −17.2 ± 16.5 nm (S.D.) for forward steps and backward steps, respectively. C, dwell-time distribution of HM7AΔTail/LZ. The solid line shows the best fit to a single exponential equation, ke−kt, where t and k represent time and rate constant (the first bin was excluded from the fitting). The average waiting time (τ = 1/k) is 3.4 ± 0.2 s (S.E., n = 387).
Table 1.
Parameters of single-molecule motility assay of myosin VIIa dimer
Experiments were done on single actin filaments in the presence of 2 mMATP (see “Experimental Procedures”). Mean ± S.E. is shown unless otherwise indicated. ND, not determined.
| Myosin Vlla | Step size | Dwell time, (τ) | Run length, (λ) | Velocity |
|---|---|---|---|---|
| nm | s | μm | nm s−1 | |
| Drosophila myosin VIIaa | ∼30 | ND | ND | 72 ± 20b |
| Human myosin VIIa (This study) | 34.7 ± 11.5b | 3.4 ± 0.2 | 0.71 ± 0.09 | 11.0 ± 0.6 |
| (n = 521) | (n = 387) | (n = 121) | (n = 86) |
a From Yang et al. (31).
b Mean ± S.D.
Dwell-time distribution was best fit to a single exponential curve (Fig. 2C), which gave the average waiting time (τ) of 3.4 ± 0.2 s (S.E., n = 387). The rate constant (0.29 ± 0.01 s−1) was similar to the Vmax value of ATPase activity in the previous study (30). This indicates that the stepping rate corresponds to the single turnover rate of ATP hydrolysis (0.32 ± 0.04 s−1) in saturating ATP concentration. This rate is 3–5 times slower than the Vmax value of Drosophila myosin VIIa (1.1–1.4 s−1) (31).
Fig. 3A shows a run length of HM7AΔTail/LZ in the presence of 2 mm ATP. The data were best fit to a single exponential equation to yield the average run length (λ) of 0.71 ± 0.09 μm (S.E., n = 121). This implies that HM7AΔTail/LZ walks ∼20 steps without dissociation on single actin filaments. The results indicate that the human myosin VIIa dimer moves processively on single actin filaments in physiological ATP concentration. Fig. 3B shows the histogram of the velocity of HM7AΔTail/LZ in 2 mm ATP. The average velocity was 11.0 ± 0.6 nm·s−1 (S.E., n = 86). The velocity is consistent with the calculated value, i.e. dwell time (0.29 s−1)/step size (35 nm) = 8.3 nm·s−1. This value was ∼8 times slower than that of Drosophila myosin VIIa, 72 nm·s−1 (31).
Figure 3.
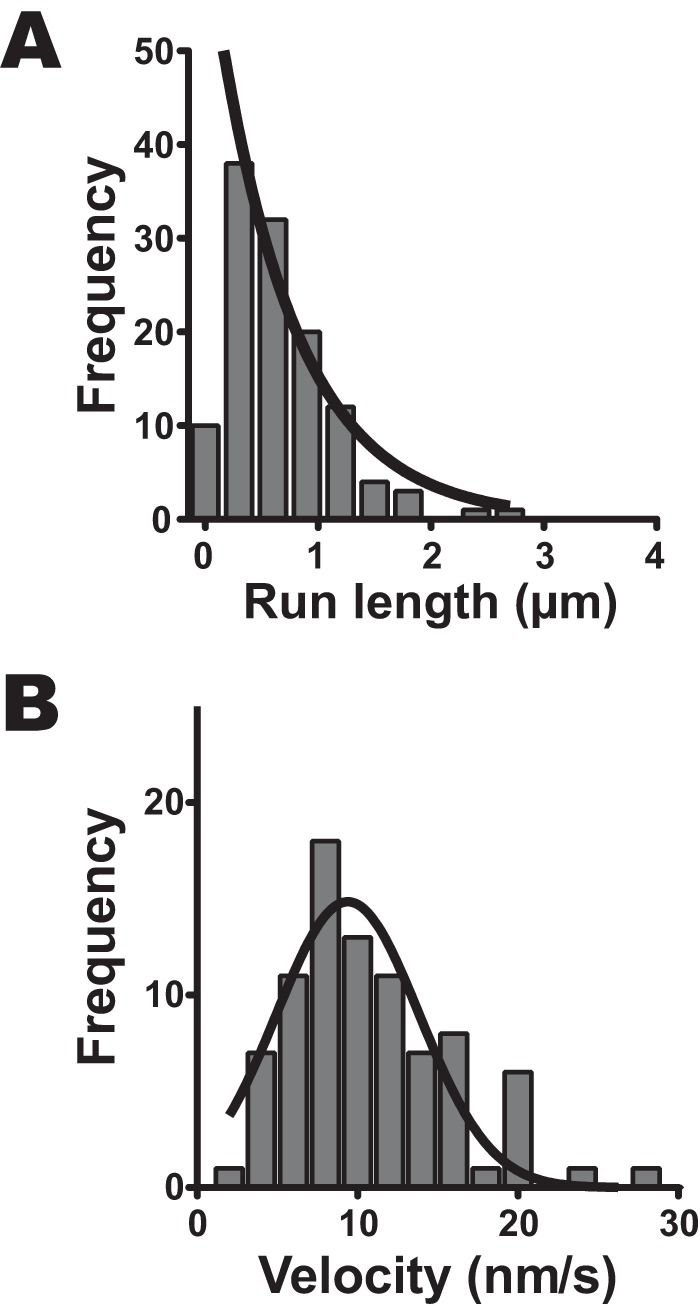
Run length and velocity of human myosin VIIa dimer on single actin filaments. A, run length of HM7AΔtail/LZ. The fluorescence images were captured at 2.0 fps. The solid line shows the best fit to a single exponential equation, R0e−r/λ, where R0 is the initial frequency extrapolated to zero run length, r is run length, and λ is average run length (the first bin is excluded from the fitting). The average run length was 0.71 ± 0.09 μm (mean ± S.E., n = 121). B, the velocity of HM7AΔTail/LZ. The average velocity of the histogram was 11.0 ± 0.6 nm·s−1 (S.E., n = 86). The best fit to Gaussian equation gives the mean velocity of 9.4 ± 0.5 nm·s−1 (S.E.).
Effective movement of human myosin VIIa dimer on filopodia
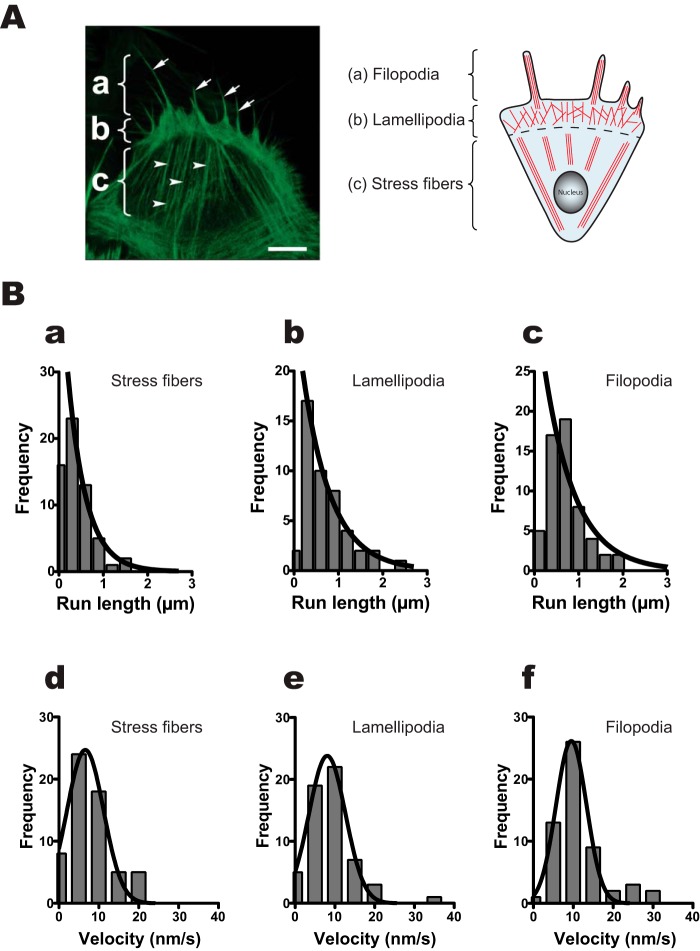
We asked the question of whether human myosin VIIa prefers specific actin structures for its movement in cells. To address this question, we used a demembranated cell system (33, 34). The right panel of Fig. 4A shows a schematic of typical actin structures of a moving cell (35). It is known that the moving cells make three different actin organizations (filopodia, lamellipodia, and stress fibers). Analogous to the right panel, we can observe those actin structures in mouse embryonic fibroblast (MEF)-3T3 cells. The MEF-3T3 cells were demembranated using Triton X-100 to incorporate purified HM7AΔTail/LZ attached on Qdot. Using this system, we examined the movement of HM7AΔTail/LZ on three different actin structures (Fig. 4B, supplemental Movies 2 and 3). The average run lengths on stress fibers, lamellipodia, and filopodia were 0.41 ± 0.03 μm (S.E., n = 60), 0.59 ± 0.06 μm (S.E., n = 57), and 0.69 ± 0.13 μm (S.E., n = 56), respectively (Fig. 4B, a–c; see also Table 2). The average run lengths on lamellipodia and filopodia are similar to that on single actin filaments (Fig. 3), whereas that on stress fibers was significantly shorter. This result suggests that the movement of HM7AΔTail/LZ is obstructed on stress fibers. On the other hand, the mean apparent velocities on stress fibers, lamellipodia, and filopodia were 6.6 ± 0.6 nm·s−1 (S.E., n = 60), 8.1 ± 0.2 nm·s−1 (S.E., n = 57), and 9.5 ± 0.4 nm·s−1 (S.E., n = 56), respectively (Fig. 4B, d–f, and Table 2). It should be noted that the displacement was measured as projected values to the movement axis. Therefore, the movement with variable stepping angles toward the overall axis of the movement yields slower apparent velocity.
Figure 4.
Movement of human myosin VIIa dimer on MEF-3T3 cells. A, left, typical actin structural image of MEF-3T3 cell. The MEF-3T3 cells were demembranated, fixed, and stained by Alexa Fluor 568-phalloidin as described under “Experimental Procedures.” a, filopodia; b, lamellipodia; c, stress fibers. Arrows and arrowheads indicate filopodia and stress fibers, respectively. The scale bar shows 10 μm. A, right, schematic diagram of actin structures in moving cells. a, filopodia; b, lamellipodia; c, stress fibers. B, run lengths and velocities of HM7AΔtail/LZ on stress fibers (a and d), lamellipodia (b and e), and filopodia (c and f). The fluorescence images were captured at 4 fps, and run lengths and the velocities were determined from the same traces. Solid lines in a–c are the best fit to an equation, R0e−r/λ. The average run lengths (λ) are 0.41 ± 0.03 μm (mean ± S.E., n = 60), 0.59 ± 0.06 μm (S.E., n = 57), and 0.69 ± 0.13 μm (S.E., n = 56) on stress fibers (a), lamellipodia (b), and filopodia (c), respectively. The bottom three panels show the velocities of HM7AΔTail/LZ on stress fibers (d), lamellipodia (e), and filopodia (f). The best fit to Gaussian equation gave the mean velocity of 6.6 ± 0.6 nm·s−1 (mean ± S.E., n = 60), 8.1 ± 0.2 nm·s−1 (S.E., n = 57), and 9.5 ± 0.4 nm·s−1 (S.E., n = 56) for stress fibers, lamellipodia, and filopodia, respectively.
Table 2.
Run lengths, velocities, and width of stepping angle for HM7AΔTail/LZ on different actin structures
Experiments were done on demembranated MEF-3T3 cells in the presence of 2 mMATP (see “Experimental Procedures”). Mean ± S.E. is shown.
| Actin structure | Run length (λ) | Velocity | S.D. (σ) for stepping anglea |
|---|---|---|---|
| μm | nm s−1 | Degree | |
| Stress fibers | 0.41 ± 0.03 | 6.6 ± 0.6 | 61.8 |
| (n = 60) | (n = 60) | (n = 274) | |
| Lamellipodia | 0.59 ± 0.06 | 8.1 ± 0.2 | 47.3 |
| (n = 57) | (n = 57) | (n = 263) | |
| Filopodia | 0.69 ± 0.13 | 9.5 ± 0.4 | 35.4 |
| (n = 56) | (n = 56) | (n = 270) |
a Residuals of each stepping angle to the moving direction are calculated, and the S.D. is shown.
Angular dependence of the movement of human myosin VIIa dimer on stress fibers, lamellipodia, and filopodia in MEF-3T3 cells
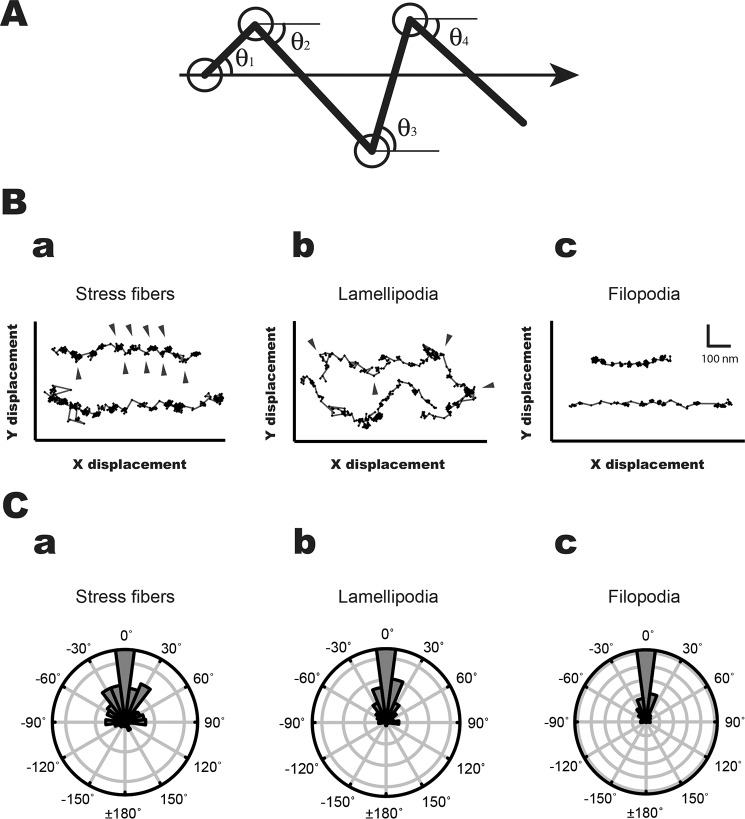
To analyze the angular dependence of stepping of HM7AΔTail/LZ on three actin structures in cells, we analyzed the HM7AΔTail/LZ movements using the fluorescence imaging with one-nanometer accuracy (FIONA) technique (61). To determine the individual steps precisely, we calibrated the length of the movement with a motor-driven stage on a TIRF microscope (supplemental Fig. S2; see also “Experimental Procedures”). Fig. 5B, a–c, shows the typical traces of HM7AΔTail/LZ movements on stress fibers, lamellipodia, and filopodia, respectively, in MEF-3T3 cells. The movement of HM7AΔTail/LZ was two-dimensional. Therefore, we analyzed the stepping orientation of HM7AΔTail/LZ to the moving direction (Fig. 5A). Although the HM7AΔTail/LZ steppings were mostly unidirectional on filopodia and lamellipodia, the stepping orientation of HM7AΔTail/LZ on stress fibers is widely distributed (Fig. 5, B and C). Note that the axis of stress fibers is taken as the X-direction. The HM7AΔTail/LZ movement showed notable Y-displacement (arrowheads in Fig. 5Ba). The result suggests that HM7AΔTail/LZ moves on different actin filaments within stress fibers. On the other hand, HM7AΔTail/LZ apparently moves straight in filopodia. In lamellipodia, HM7AΔTail/LZ seems to move various directions and showed apparent backward movement (Fig. 5Bb). The arrowheads in Fig. 5Bb show the lateral positions of the moving direction. In lamellipodia the mean angle of the lateral position was 84.6 ± 4.2° (S.E., n = 31), which was slightly larger than the angle of ARP2/3 junction, 70° (36, 37). On the other hand, the orientations of mouse myosin Va HMM (heavy meromyosin) on stress fibers, lamellipodia, and filopodia were similar to one another with low angle variation (supplemental Figs. S3 and S4). The result suggests that human myosin VIIa frequently changes the actin tracks, whereas myosin Va steppings are relatively on the same actin tracks, which is attributed to the difference in the stepping properties of myosin VIIa and myosin Va motors.
Figure 5.
Stepping orientation of human myosin VIIa movements in MEF-3T3 cells. A, diagram of measurement of stepping orientation. The arrow shows the direction of the movement. The HM7AΔTail/LZ stepping angles (θ1, θ2, θ3, θ4, … θn) were defined by the stepping angle of fluorophore movement to the axis of the movement. B, the typical stepping traces of the movement in stress fibers (a), lamellipodia (b), and filopodia (c). The arrowheads indicate the lateral positions in stress fibers and lamellipodia. The HM7AΔTail/LZ frequently shows off-axis on stress fibers, whereas the direction of HM7AΔTail/LZ on filopodia is mostly straight. The movement of HM7AΔTail/LZ on lamellipodial actin was diverse. The scales of x and y axes are shown in c. C, polar plots for orientation of HM7AΔTail/LZ movement on stress fibers (a), lamellipodia (b), and filopodia (c). The stepping orientation of HM7AΔTail/LZ was measured and plotted as described under “Experimental Procedures.” The 0° means that the stepping orientation is parallel to the moving direction.
Discussion
In the present study the single-molecule motor characteristics of a tail-truncated human myosin VIIa dimer were investigated on single actin filaments and on actin structures in demembranated cells. We used the tail-truncated myosin VIIa construct because the tail domain inhibits the motor activity (22, 38), and the full-length myosin VIIa alone in cells does not translocate to the filopodial tips, suggesting that the motor activity and/or processive movement is inhibited.
It has been thought that the neck length of myosin is an important component for processive movement as it is related to the lever-arm length. It was previously reported that substoichiometric CaM light chains bind to the five IQ motifs of human myosin VIIa. We confirmed this substoichiometric binding of CaM with the tail-truncated myosin VIIa. Interestingly, we found that the tail-truncated myosin VIIa with LZ binds five CaM molecules. The result suggests that the dimer formation induces the full decoration of CaM light chains into the neck domain of myosin VIIa. The result implies that the dimer formation not only facilitates the processive movement with a hand-over-hand mechanism but also promotes the coupling between the ATP hydrolysis and the mechanical activity by changing the neck length and rigidity.
We found that the human myosin VIIa dimer was a slow processive motor (∼11 nm·s−1) with an ∼35-nm step size on single actin filaments. The average run length of human myosin VIIa on single actin filaments was ∼0.7 μm, which is shorter than that of a known value of typical processive motor, mammalian myosin Va (1–2 μm) (39). A similar run length and velocity of human myosin VIIa was also observed on filopodia in demembranated MEF-3T3 cells (∼0.7 μm and ∼10 nm·s−1, respectively). On the other hand, human myosin VIIa showed a shorter apparent run length (∼0.4 μm) and a slower velocity (∼7 nm·s−1) toward the long axis of stress fibers. This is primarily due to the fact that the movement in the stress fibers contains the component perpendicular to the long axis. Consistent with this view, the moving direction of human myosin VIIa is two-dimensional on stress fibers (Fig. 5Ba) and moves with wide-angle-stepping orientation (Fig. 5Ca). The movement of human myosin VIIa in lamellipodia was multiple directional, suggesting that human myosin VIIa frequently switches the actin track in the branched actin structure in lamellipodia. These results suggest that human myosin VIIa is flexible enough to be able to move on various actin structures.
In this study the processive movement of myosin Va tends to be straight on various actin structures (supplemental Fig. S3). This is consistent with the previous observation of the movement of myosin Va (34, 40, 41). However, the stepping orientation of human myosin VIIa is affected by cellular actin structures (Fig. 5B). Fig. 6 represents a model that explains the movement of human myosin VIIa. In this model three different actin structures (stress fibers, lamellipodia, and filopodia) are illustrated. Stress fiber is periodic actin-myosin II bundles, analogous to skeletal muscle sarcomeric structures (42) and is typically attached to focal adhesions. The filamentous structure of stress fiber is composed of 10–30 actin filaments (43) that are connected by actin bundling proteins such as α-actinin. Membrane protrusions called filopodia are observed during adhesion to the extracellular matrix, path-finding in neuronal growth-cone, and guidance during cell migration, etc. In filopodium, 10–30 actin filaments are tightly bundled by actin-bundling proteins such as fascin and fimbrin (44, 45) (Fig. 6). One of the most important differences between the two actin structures is the polarity of actin filaments. It is known that α-actinin cross-links both parallel and antiparallel actin filaments (46, 47); therefore, the overall polarity of actin filaments in stress fiber is not uniform but random. Because the stepping of myosin VIIa shows notable flexibility, it is anticipated that one of the two heads of myosin VIIa lands on adjacent actin filament with opposite polarity to the moving direction, which causes disruption of the movement (Fig. 6). Supporting this view, we observed myosin VIIa molecules stuck on stress fibers and disappeared from the TIRF field (supplemental Movie 2). This is consistent with a short run length of myosin VIIa on stress fibers (Fig. 4). On the other hand, the actin bundles in filopodia are unidirectional (44, 48), and the steppings of myosin VIIa were almost straight (Fig. 5), similar to that of myosin Va (supplemental Fig. S3). It should be noted that the run lengths or velocities of myosin VIIa on filopodia and single actin filaments were comparable, suggesting that human myosin VIIa motor does not have preference for single actin filaments or filopodial actin bundles.
Figure 6.
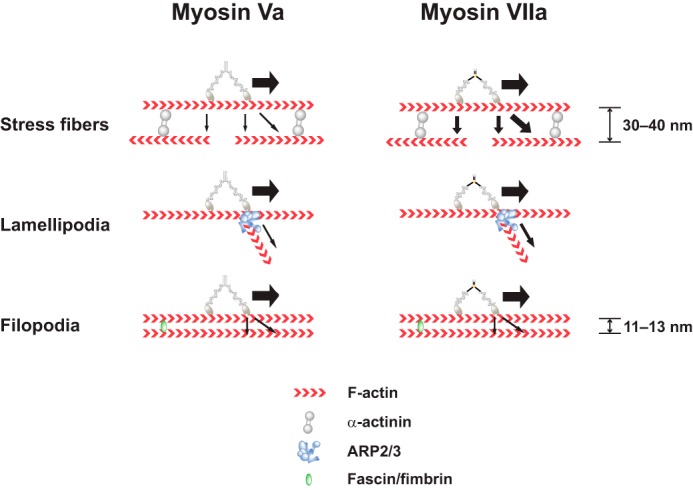
Model of myosin VIIa movement in cells. Shown is a model explaining the difference between myosin Va and myosin VIIa movement in three different actin structures (stress fibers, lamellipodia, and filopodia). The α-actinin, ARP2/3 complex, and fascin/fimbrin are the typical actin-associated structural proteins in stress fibers, lamellipodia, and filopodia, respectively. On stress fibers, myosin Va moves relatively straight along actin filaments, whereas myosin VIIa is frequently off the actin track. This may associate with actin with opposite polarity. On lamellipodia, myosin VIIa moves on branched actin in addition to the original actin track. On filopodia, both myosin Va and myosin VIIa move straight on parallel bundled actin. The space between actin bundles made by α-actinin is 30–40 nm (46), whereas the space by fascin is 11–13 nm (15, 60). The backward movement is not considered.
Lamellipodium consists of actin filament network, and the actin filaments of lamellipodium often branches due to the actin branching protein, ARP2/3 (Fig. 6) (44, 48). The myosin VIIa movement is not always straight, and the trajectories were sometimes nonlinear (Fig. 5, B and C, and supplemental Movie 2). This movement agrees with the basic organization of lamellipodial actin meshwork. In lamellipodia, we often observed the myosin VIIa trajectory with a sharp change in direction (Fig. 5. Bb). On the other hand, the continuous straight movement toward filopodia was also observed in lamellipodia (supplemental Movie 3). It is thought that the former is due to the branching of actin filaments through ARP2/3 complex, and the latter is the movement on loosely organized inner cellular filopodial actin bundles in lamellipodia. These multiple movements of myosin VIIa suggest that human myosin VIIa is highly processive, and the neck region is flexible enough to move on different actin tracks. Alternatively, the actin-binding proteins present in lamellipodia might in part influence the paths taken by myosin VIIa and myosin Va.
The velocity of ∼11 nm·s−1 of human myosin VIIa is one of the slowest in myosin family (1). The value is 6–7-fold smaller than that of Drosophila myosin VIIa, which moves at 72 nm·s−1 (31) and is ∼100-fold smaller than mammalian myosin Va (39). Our finding suggests that human myosin VIIa is a suitable motor for slow cargo transportation on rigid actin bundles such as stereocilia. On the other hand, human myosin VIIa is also a suitable motor for maintaining and holding a tip-link complex composed of USH1 proteins at the desired location in the stereocilia in inner-ear cells with exerting force with low energy consumption. In this regard, myosin VIIa having a high duty ratio and an extremely slow cycling rate is an ideal motor to carry out these tasks. This view is consistent with the fact that Usher syndrome type 1B mutations (myosin VIIa mutations) are associated with altered ADP release rates during myosin VIIa ATPase cycle (49, 50). It is anticipated that the decrease or increase in the ADP release rate alters the duty ratio, which influences both processivity and stress maintenance capability of human myosin VIIa and leads to mislocalization of USH1 proteins at proper position in the stereocilia in inner-ear hair cells.
Experimental procedures
Materials
Restriction enzymes and modifying enzymes were purchased from New England Biolabs (Beverly, MA). Oligonucleotides were synthesized by Invitrogen (Thermo Fisher). Pfu Ultra II High-Fidelity DNA polymerase was purchased from Agilent. FluoSpheres sulfate (0.2 μm, 505/515), rhodamine-phalloidin, and streptavidin-conjugated Qdots were purchased from Invitrogen. Actin was prepared from rabbit skeletal muscle acetone powder according to Spudich and Watt (51). Recombinant human calmodulin (CALM2) was subcloned to pET vector (Novagen, EMD Millipore Corp., Merck KGaA) and expressed in an Escherichia coli strain, BL21(DE3) as described in Studier and Moffatt (52) and purified by phenyl-Sepharose column chromatography. The calmodulin cDNA was also transferred to pFastBac 1, generating baculovirus, and used for co-infection of Sf9 cells with myosin VIIa baculovirus to purify myosin VIIa as described below. The 3× FLAG peptides were synthesized by GenScipt Co.
Cloning of human myosin VIIa dimer cDNA
The cDNA fragments encoding human myosin VIIa (GenBankTM accession number NM000260) were obtained from human kidney cDNA library. The full-length myosin VIIa was subcloned into a modified pFastBacHT baculovirus transfer vector (Thermo Fisher) containing a 3× FLAG sequence at the 5′ end. HM7AΔTail cDNA was then made as described previously (38). This HM7AΔTail contains 1–1017 amino acids of human myosin VIIa. HM7AΔTail/LZ also contains this sequence and an LZ sequence (which corresponds to residues 250–281 of GCN4 protein) (53, 54) after a 1st linker sequence (amino acids SRACSLEEELLSK) and c-Myc (EQKLISEEDL) after a 2nd linker sequence (GGSGSTVPRARDPPVATMVSKG), and a stop codon was then created at the 3′ end of the construct by site-directed mutagenesis (55).
Expression and purification of human myosin VIIa dimer
To express the HM7AΔTail/LZ proteins, ∼2 × 109 Sf9 cells were coinfected with two viruses expressing HM7AΔTail/LZ proteins and calmodulin. The infected cells were cultured for 3 days at 28 °C. The cell pellets were suspended in buffer A (150 mm NaCl, 50 mm Tris-HCl, pH 8, 5 mm EGTA, 2 mm MgCl2, 1 mm ATP, 0.5 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 1 mm DTT, and 5 μm CaM) and gently homogenized using a sonicator. The cell suspension was centrifuged at 126,000 × g (Type 70 Ti rotor, Beckman) for 20 min. The supernatant was incubated with 20 unit/ml hexokinase and 20 mm glucose on ice for 20 min, and F-actin was added up to the final concentration of 0.1 mg/ml. After centrifugation at 164,000 × g for 1 h, the pellet was resuspended in buffer B containing 150 mm NaCl, 20 mm MOPS-KOH (pH 7.5), 10 mm ATP, 1 mm DTT, 1 mm EGTA, and 5 μm CaM. The suspension was centrifuged at 390,000 × g for 10 min with a Beckman TL-100 centrifuge (TLA-100.3 rotor). The supernatant was incubated with anti-FLAG antibody-conjugated agarose (Sigma) with gentle rotation for 1 h at 4 °C. After washing with buffer C (150 mm NaCl, 20 mm MOPS-KOH (pH 7.5), 1 mm EGTA, 1 mm DTT, and 5 μm CaM), the recombinant proteins were eluted with buffer C containing 0.1 mg/ml 3× FLAG peptide and 12.5% sucrose. An aliquot of the purified protein was flash-frozen in liquid nitrogen and stored at −80 °C. The protein was used within 1 day after thaw.
Determination of calmodulin content in human myosin VIIa dimer
About 30 μl of HM7AΔTail/LZ used in the present study was centrifuged at 100,000 × g for 10 min (TLA-100.3 rotor, Beckman) to remove aggregates. Phalloidin-stabilized F-actin was added to the supernatant at the final 0.05 mg/ml and centrifuged again to remove unbound CaM. The pellet was dissolved with SDS-PAGE sample buffer and loaded onto SDS-polyacrylamide gels. Purified turkey gizzard smooth muscle myosin and CaM were used for protein standards to eliminate the staining difference between heavy chain and calmodulin. The gels were stained with Coomassie Brilliant Blue G-250, and the gel images were scanned (Precision V750 pro, Epson) and analyzed (NIH ImageJ). The number of calmodulin per heavy chain was then calculated (Microsoft Excel) and plotted (GraphPad Prism). The concentration of CaM and smooth muscle myosin was determined by the percent extinction coefficients, E277 nm = 2.00 (56) and E280 nm = 5.66 (57), respectively.
Setup of TIRF microscope
The custom-made TIRF microscope system was built using IX83 microscope (Olympus, Co.) with a TIRF objective lens (UAPON100XOTIRF, 1.49 NA, Olympus), OPSL lasers (model Sapphire 488–50 CW CDRH and Sapphire 561-50 CW CDRH, Coherent, Inc.), and two ImagEM X2 (model C9100–23B, Hamamatsu Photonics K.K.). The dimensional calibration and the check of particle tracking precision were done using a ProScan III (Prior Scientific, Inc.). The position of FluoSpheres sulfate (0.2 μm, Invitrogen) fluorescence on a bottom dish was captured with TIRF microscope, and the stage of ProScan III was repeatedly moved to make artificial steps (10, 20, 40, 60, 80, and 100 nm) with 5-s intervals. The step sizes were then analyzed by FIONA as described below (supplemental Fig. S2). After calibration, the ProScan III stage was replaced to a manually controllable stage (B27-100C, Suruga Seiki Co. Ltd.) with a home-built stage pedestal. The difference of the calibrations between stages was corrected using S16 Stage Micrometer (PYSER-SGI, Ltd). The average number of video frames of stationary myosin VIIa-Qdot on single-actin filaments was 11.2 frames at 3.3 fps, which corresponds to the average waiting time of ∼3.4 s. The calculated S.E. value of the positions over 11.2 frames was 1.1 nm (σ = 3.75 nm), indicating that the tracking precision was ∼1 nm under the typical experimental conditions.
Single-molecule measurement using TIRF microscope
We made a tailless dimer construct with C-terminal c-Myc tag (HM7AΔTail/LZ) to attach Qdot fluorophores. Qdot 525-conjugated with goat F(ab′)2 anti-mouse IgG (Q11022MP, Invitrogen) was mixed with anti-c-Myc monoclonal antibodies (Clontech) at the Qdot/antibody ratio of 0.85:1, and HM7AΔTail/LZ protein was then labeled with the Qdot antibody at the ratio of 1:20 (myosin VIIa/Qdot). Flow chambers were prepared by using No. 1.5 glass coverslips and glass slides (Fisher). α-Actinin (A9776, Sigma) was used to immobilize F-actin, and the casein from bovine milk (07319–82, Nakarai Tesque, Japan) was used for glass-surface blocking. HM7AΔTail/LZ movement was then observed in a solution containing 25 mm KCl, 20 mm HEPES (pH 7.5), 5 mm MgCl2, 1 mm EGTA, 5 mm DTT, 12 μm CaM, 2 mm ATP, and O2 scavenger system containing glucose oxidase, catalase, and glucose. Experiments were performed at 22–23 °C.
Preparation of demembranated cells
MEF-3T3 cells were cultured on a fibronectin-coated glass bottom dish (35GC-1.5-14-C, MatTek, Co.) and treated with extraction buffer as described previously (33) with slight modifications. Briefly, the cells were treated with demembranation buffer (30 mm imidazole, pH 7.5, 70 mm KCl, 1 mm EGTA, 2 mm MgCl2, 0.5% Triton X-100, 4% polyethylene glycol (Mr 8000), and 250 nm Alexa Fluor 568 phalloidin (invitrogen)) for 5 min on ice and then washed twice with ice-cold PBS. The extracted cells were incubated on ice with blocking solution (2 mg/ml casein in PBS) before use.
Single-molecule data analysis
The movie data were captured using an open-source software (μManager, Dr. Vale's laboratory, UCSF) and analyzed by using in-house 2D Gaussian-fitting software (58). Step size, dwell time, velocity, and run length of HM7AΔTail/LZ were determined by using an in-house step-fitting software based on an algorithm described in Kerssemakers et al. (59). For image and data processing, we used ImageJ (National Institutes of Health) and Excel (Microsoft), respectively. For statistical calculations and the graph plotting, we alternatively used Prism (GraphPad Software, Inc.) and DeltaGraph (RockWare, Inc.). To determine stepping orientations of myosin, the movie data were analyzed by using 2D Gaussian-fitting software and depicted XY graph on Microsoft Excel software. The stepping trace was rotated as the angle of moving direction to be 0 degree, and the stepping orientation was then determined. The angle data were plotted with OriginPro software (OriginLab, Co.). The >100-nm or <−100-nm steps were judged as jumping and excluded from the data.
Confocal light microscopy
MEF-3T3 cells stained with Alexa Fluor 568 phalloidin were observed by using a laser-scanning confocal microscope, SP8 system (Leica Microsystems, Heidelberg, Germany).
SDS-PAGE and protein assays
SDS-PAGE was carried out using 4–20% gradient slab gel. Protein concentration was determined by densitometry of a Coomassie Brilliant Blue-stained polyacrylamide gel using smooth muscle myosin as a standard. Smooth muscle myosin heavy chain (204 kDa), β-galactosidase (116 kDa), phosphorylase b (97.4 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), smooth muscle myosin regulatory light chain (20 kDa), and α-lactalbumin (14.2 kDa) were used as the molecular mass standards.
Author contributions
O. S. performed the single-molecule experiments and analyses. S. K. and T. S. performed the cell biological experiments. Y. T. helped to set up the TIRF microscope. R. T. and T. M. developed step-size analysis software. T. M. W. developed the 2D Gaussian fitting software. T. S. and R. I. performed the molecular cloning. M. I. supervised the project. O. S. and M. I. conceived the study, designed the experiments, and wrote the manuscript with input from the other authors.
Supplementary Material
Acknowledgments
We thank Dr. Toshio Kitazawa for critical advice on physiological aspects and Munenori Ishibashi for a fruitful opinion on calmodulin contents in the myosin VIIa constructs.
This work was supported by National Institutes of Health Grant R01 DC006103 and by the University of Texas System TEXAS STARS PLUS award (to M. I.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Movies 1–3 and Figs. 1–4.
The consensus sequence: [I,L,V]QXXXRGXXX[R,K].
- CaM
- calmodulin
- HM7A
- human myosin VIIa
- ΔTail
- tail-deleted
- LZ
- leucine zipper
- USH
- Usher syndrome
- SAH
- stable single α-helix
- RLC
- regulatory light chain
- Qdot
- quantum dot
- TIRF
- total internal reflection fluorescence
- FIONA
- fluorescence imaging with one-nanometer accuracy
- MEF
- mouse embryonic fibroblasts
- fps
- frames/s.
References
- 1. Coluccio L. M. (2008) Myosins: a superfamily of molecular motors, Springer, Dordrecht, The Netherlands [Google Scholar]
- 2. Odronitz F., and Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knight P. J., Thirumurugan K., Xu Y., Wang F., Kalverda A. P., Stafford W. F. 3rd, Sellers J. R., and Peckham M. (2005) The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280, 34702–34708 [DOI] [PubMed] [Google Scholar]
- 4. Spink B. J., Sivaramakrishnan S., Lipfert J., Doniach S., and Spudich J. A. (2008) Long single α-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 15, 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weil D., Levy G., Sahly I., Levi-Acobas F., Blanchard S., El-Amraoui A., Crozet F., Philippe H., Abitbol M., and Petit C. (1996) Human myosin VIIA responsible for the Usher 1B syndrome: a predicted membrane-associated motor protein expressed in developing sensory epithelia. Proc. Natl. Acad. Sci. U.S.A. 93, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weil D., Blanchard S., Kaplan J., Guilford P., Gibson F., Walsh J., Mburu P., Varela A., Levilliers J., and Weston M. D. (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374, 60–61 [DOI] [PubMed] [Google Scholar]
- 7. Weil D., Küssel P., Blanchard S., Lévy G., Levi-Acobas F., Drira M., Ayadi H., and Petit C. (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat. Genet. 16, 191–193 [DOI] [PubMed] [Google Scholar]
- 8. Liu X. Z., Walsh J., Mburu P., Kendrick-Jones J., Cope M. J., Steel K. P., and Brown S. D. (1997) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat. Genet. 16, 188–190 [DOI] [PubMed] [Google Scholar]
- 9. Tamagawa Y., Ishikawa K., Ishikawa K., Ishida T., Kitamura K., Makino S., Tsuru T., and Ichimura K. (2002) Clinical presentation of DFNA11 (MYO7A). Adv. Otorhinolaryngol. 61, 79–84 [DOI] [PubMed] [Google Scholar]
- 10. El-Amraoui A. E., Bahloul A., and Petit C. (2008) Myosin VII in myosins: a superfamily of molecular motors, pp. 353–373, Springer, Dordrecht, The Netherlands [Google Scholar]
- 11. Hasson T., Gillespie P. G., Garcia J. A., MacDonald R. B., Zhao Y., Yee A. G., Mooseker M. S., and Corey D. P. (1997) Unconventional myosins in inner-ear sensory epithelia. J. Cell Biol. 137, 1287–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grati M., and Kachar B. (2011) Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. U.S.A. 108, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasson T., Walsh J., Cable J., Mooseker M. S., Brown S. D., and Steel K. P. (1997) Effects of shaker-1 mutations on myosin-VIIa protein and mRNA expression. Cell Motil. Cytoskeleton 37, 127–138 [DOI] [PubMed] [Google Scholar]
- 14. Morgan C. P., Krey J. F., Grati M., Zhao B., Fallen S., Kannan-Sundhari A., Liu X. Z., Choi D., Müller U., and Barr-Gillespie P. G. (2016) PDZD7-MYO7A complex identified in enriched stereocilia membranes. eLife 5, e18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bryan J., and Kane R. E. (1982) Actin gelation in sea urchin egg extracts. Methods Cell Biol. 25, 175–199 [DOI] [PubMed] [Google Scholar]
- 16. Hasson T., Heintzelman M. B., Santos-Sacchi J., Corey D. P., and Mooseker M. S. (1995) Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc. Natl. Acad. Sci. U.S.A. 92, 9815–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. el-Amraoui A., Sahly I., Picaud S., Sahel J., Abitbol M., and Petit C. (1996) Human Usher 1B/mouse shaker-1: the retinal phenotype discrepancy explained by the presence/absence of myosin VIIA in the photoreceptor cells. Hum. Mol. Genet. 5, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 18. Liu X., Vansant G., Udovichenko I. P., Wolfrum U., and Williams D. S. (1997) Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil. Cytoskeleton 37, 240–252 [DOI] [PubMed] [Google Scholar]
- 19. Liu X., Ondek B., and Williams D. S. (1998) Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat. Genet. 19, 117–118 [DOI] [PubMed] [Google Scholar]
- 20. Gibbs D., Kitamoto J., and Williams D. S. (2003) Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. U.S.A. 100, 6481–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X., Udovichenko I. P., Brown S. D., Steel K. P., and Williams D. S. (1999) Myosin VIIa participates in opsin transport through the photoreceptor cilium. J. Neurosci. 19, 6267–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Umeki N., Jung H. S., Watanabe S., Sakai T., Li X. D., Ikebe R., Craig R., and Ikebe M. (2009) The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc. Natl. Acad. Sci. U.S.A. 106, 8483–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X. D., Mabuchi K., Ikebe R., and Ikebe M. (2004) Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem. Biophys. Res. Commun. 315, 538–545 [DOI] [PubMed] [Google Scholar]
- 24. Li X. D., Jung H. S., Mabuchi K., Craig R., and Ikebe M. (2006) The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J. Biol. Chem. 281, 21789–21798 [DOI] [PubMed] [Google Scholar]
- 25. Li X. D., Jung H. S., Wang Q., Ikebe R., Craig R., and Ikebe M. (2008) The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc. Natl. Acad. Sci. U.S.A. 105, 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang F., Thirumurugan K., Stafford W. F., Hammer J. A 3rd, Knight P. J., and Sellers J. R. (2004) Regulated conformation of myosin V. J. Biol. Chem. 279, 2333–2336 [DOI] [PubMed] [Google Scholar]
- 27. Krementsov D. N., Krementsova E. B., and Trybus K. M. (2004) Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 164, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Umeki N., Jung H. S., Sakai T., Sato O., Ikebe R., and Ikebe M. (2011) Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol. 18, 783–788 [DOI] [PubMed] [Google Scholar]
- 29. El-Amraoui A., Schonn J. S., Küssel-Andermann P., Blanchard S., Desnos C., Henry J. P., Wolfrum U., Darchen F., and Petit C. (2002) MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep. 3, 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakai T., Jung H. S., Sato O., Yamada M. D., You D. J., Ikebe R., and Ikebe M. (2015) Structure and regulation of the movement of human myosin VIIA. J. Biol. Chem. 290, 17587–17598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y., Kovács M., Sakamoto T., Zhang F., Kiehart D. P., and Sellers J. R. (2006) Dimerized Drosophila myosin VIIa: a processive motor. Proc. Natl. Acad. Sci. U.S.A. 103, 5746–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Combet C., Blanchet C., Geourjon C., and Deléage G. (2000) NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 33. Sivaramakrishnan S., and Spudich J. A. (2009) Coupled myosin VI motors facilitate unidirectional movement on an F-actin network. J. Cell Biol. 187, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brawley C. M., and Rock R. S. (2009) Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 106, 9685–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parsons J. T., Horwitz A. R., and Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goley E. D., and Welch M. D. (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 37. Mullins R. D., Heuser J. A., and Pollard T. D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakai T., Umeki N., Ikebe R., and Ikebe M. (2011) Cargo binding activates myosin VIIA motor function in cells. Proc. Natl. Acad. Sci. U.S.A. 108, 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hodges A. R., Krementsova E. B., and Trybus K. M. (2007) Engineering the processive run length of Myosin V. J. Biol. Chem. 282, 27192–27197 [DOI] [PubMed] [Google Scholar]
- 40. Ricca B. L., and Rock R. S. (2010) The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys. J. 99, 1818–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ali M. Y., Previs S. B., Trybus K. M., Sweeney H. L., and Warshaw D. M. (2013) Myosin VI has a one track mind versus myosin Va when moving on actin bundles or at an intersection. Traffic 14, 70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tojkander S., Gateva G., and Lappalainen P. (2012) Actin stress fibers: assembly, dynamics, and biological roles. J. Cell Sci. 125, 1855–1864 [DOI] [PubMed] [Google Scholar]
- 43. Cramer L. P., Siebert M., and Mitchison T. J. (1997) Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136, 1287–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lewis A. K., and Bridgman P. C. (1992) Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119, 1219–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bretscher A., and Weber K. (1980) Villin is a major protein of the microvillus cytoskeleton which binds both G and F actin in a calcium-dependent manner. Cell 20, 839–847 [DOI] [PubMed] [Google Scholar]
- 46. Meyer R. K., and Aebi U. (1990) Bundling of actin filaments by α-actinin depends on its molecular length. J. Cell Biol. 110, 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taylor K. A., and Taylor D. W. (1994) Formation of two-dimensional complexes of F-actin and cross-linking proteins on lipid monolayers: demonstration of unipolar α-actinin-F-actin cross-linking. Biophys. J. 67, 1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mattila P. K., and Lappalainen P. (2008) Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe S., Ikebe R., and Ikebe M. (2006) Drosophila myosin VIIA is a high duty ratio motor with a unique kinetic mechanism. J. Biol. Chem. 281, 7151–7160 [DOI] [PubMed] [Google Scholar]
- 50. Watanabe S., Umeki N., Ikebe R., and Ikebe M. (2008) Impacts of Usher syndrome type IB mutations on human myosin VIIa motor function. Biochemistry 47, 9505–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spudich J. A., and Watt S. (1971) The regulation of rabbit skeletal muscle contraction: I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 52. Studier F. W., and Moffatt B. A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 53. Hinnebusch A. G. (1984) Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. U.S.A. 81, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O'Shea E. K., Klemm J. D., Kim P. S., and Alber T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 [DOI] [PubMed] [Google Scholar]
- 55. Weiner M. P., Costa G. L., Schoettlin W., Cline J., Mathur E., and Bauer J. C. (1994) Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151, 119–123 [DOI] [PubMed] [Google Scholar]
- 56. Klee C. B. (1977) Conformational transition accompanying the binding of Ca2+ to the protein activator of 3′,5′-cyclic adenosine monophosphate phosphodiesterase. Biochemistry 16, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 57. Olney J. J., Sellers J. R., and Cremo C. R. (1996) Structure and function of the 10 S conformation of smooth muscle myosin. J. Biol. Chem. 271, 20375–20384 [DOI] [PubMed] [Google Scholar]
- 58. Watanabe T. M., Iwane A. H., Tanaka H., Ikebe M., and Yanagida T. (2010) Mechanical characterization of one-headed myosin-V using optical tweezers. PLoS ONE 5, e12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., and Dogterom M. (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 60. Brown J. W., and McKnight C. J. (2010) Molecular model of the microvillar cytoskeleton and organization of the brush border. PLoS ONE 5, e9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003) Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.