Abstract
Phototransduction is initiated when the absorption of light converts the 11-cis-retinal chromophore to its all-trans configuration in both rod and cone vertebrate photoreceptors. To sustain vision, 11-cis-retinal is continuously regenerated from its all-trans conformation through a series of enzymatic steps comprising the “visual or retinoid” cycle. Abnormalities in this cycle can compromise vision because of the diminished supply of 11-cis-retinal and the accumulation of toxic, constitutively active opsin. As shown previously for rod cells, attenuation of constitutively active opsin can be achieved with the unbleachable analogue, 11-cis-6-membered ring (11-cis-6mr)-retinal, which has therapeutic effects against certain degenerative retinal diseases. However, to discern the molecular mechanisms responsible for this action, pigment regeneration with this locked retinal analogue requires delineation also in cone cells. Here, we compared the regenerative properties of rod and green cone opsins with 11-cis-6mr-retinal and demonstrated that this retinal analogue could regenerate rod pigment but not green cone pigment. Based on structural modeling suggesting that Pro-205 in green cone opsin could prevent entry and binding of 11-cis-6mr-retinal, we initially mutated this residue to Ile, the corresponding residue in rhodopsin. However, this substitution did not enable green cone opsin to regenerate with 11-cis-6mr-retinal. Interestingly, deletion of 16 N-terminal amino acids in green cone opsin partially restored the binding of 11-cis-6mr-retinal. These results and our structural modeling indicate that a more complex binding pathway determines the regeneration of mammalian green cone opsin with chromophore analogues such as 11-cis-6mr-retinal.
Keywords: G protein-coupled receptor (GPCR), membrane protein, photoreceptor, retinoid, rhodopsin
Introduction
The visual signaling cascade starts in photoreceptor cells with the absorption of a photon by a visual pigment, which leads to electrical responses ultimately deciphered in the brain. In most vertebrates, there are two types of photoreceptor cells, rods and cones. They mediate vision under dim light or bright light through the action of their visual pigments Rho or cone opsins, respectively (1, 2). Rho and cone opsins belong to the same family of G protein-coupled receptors (GPCRs)3 and use 11-cis-retinal as the chromophore covalently attached to a Lys residue side chain in the apoprotein via a Schiff base. Although a protonated Schiff base bond is a common feature of all vertebrate visual pigments, their sensitivity across the light spectrum differs, which is attributed to specific interactions between the chromophore and neighboring amino acids in the binding pocket (3). Thus, changes in the amino acid composition of the retinal-binding pocket are critical for fine spectral tuning and result in different absorption maxima as follows: 420 nm for blue, 530 nm for green, and 560 nm for red human cone opsin, whereas Rho maximally absorbs at 498 nm (4, 5). As an exception, mouse and bird UV light-sensitive pigments possess deprotonated Schiff base in the dark state. Physiological and biochemical analyses have revealed differences in the molecular properties between Rho and cone opsins. Formation of the active Meta II state in response to light occurs faster in cones than rods (6). However, the shorter lifetime of this Meta II intermediate is related to the lower photosensitivity of cones (2, 7). In contrast, cone cells dark adapt faster, which is related to the rapid regeneration of their pigments (2). Furthermore, as compared with Rho, the ability of the retinal-binding pocket of cone pigments to accommodate geometric isomers, such as 9-cis-retinal, is more dynamic (8, 9).
Photon absorption triggers cis-chromophore isomerization to the all-trans-form, which is then released from the retinal-binding pocket after changes in the conformation of the opsin (10). Rod and cone photoreceptor cell outer segments and the retinal pigment epithelium then regulate a visual cycle to ensure a constant supply of newly regenerated 11-cis-retinal (11). Dysfunction of visual cycle enzymes can reduce the rate of chromophore regeneration, leading to a buildup of toxic all-trans-retinal by-products and an increase in free, constitutively active opsin (12–15), resulting in impaired visual function. Thus, mutations in the genes encoding retinal pigment epithelium 65 (RPE65) and lecithin:retinol acyltransferase (LRAT), key components of the visual cycle, are the most common cause of early onset blindness associated with Leber congenital amaurosis (LCA) (16–18). One of the therapies proposed to treat LCA is pigment restoration by pharmacological supplementation with analogues of retinal. In particular, treatment with 9-cis-retinoids has preserved retinal health in mouse models of LCA (19–23). Silencing of constitutively active opsin with locked retinals also has been suggested as a potential treatment for other retinal degenerative diseases (24, 25). However, the binding of locked retinals with cone opsins has not been extensively studied. Such information is needed to improve the design of retinal analogues as potential therapeutic agents.
Locked retinal analogues contain a ring between C10 and C13 of the chromophore to prevent light-stimulated isomerization around the C11=C12 double bond and eventual dissociation of the chromophore (26). Supplementation with a locked retinal could block free, constitutively active opsin and prevent light-stimulated chromophore release, thereby eliminating the accumulation of toxic retinoid by-products.
Specifically, 11-cis-6mr-retinal has been proposed as an effective candidate for such therapy. By using this retinal, toxic photoproducts could be curtailed without eliminating phototransduction in its entirety, because Rho regenerated with 11-cis-6mr-retinal exhibits residual light-dependent activity in vitro and in vivo (24). Transition of Rho regenerated with this locked retinal to the active Meta II-like state is achieved most likely due to an isomerization around another C13=C14 double bond (25, 27). Properties of Rho regenerated with 11-cis-6mr-retinal have been examined previously (24, 25); however, no equivalent studies have been performed for cone opsins. Therefore, we investigated the molecular mechanisms of green cone opsin regeneration with the locked chromophore analogue 11-cis-6mr-retinal as compared with Rho. We found that 11-cis-6mr-retinal was unable to regenerate green cone opsin, and we discerned that the N-terminal sequence of the opsin contributes to this property.
Results
Regeneration of rod and green cone opsin with 11-cis-6mr-retinal
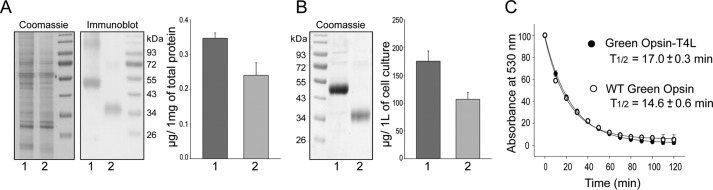
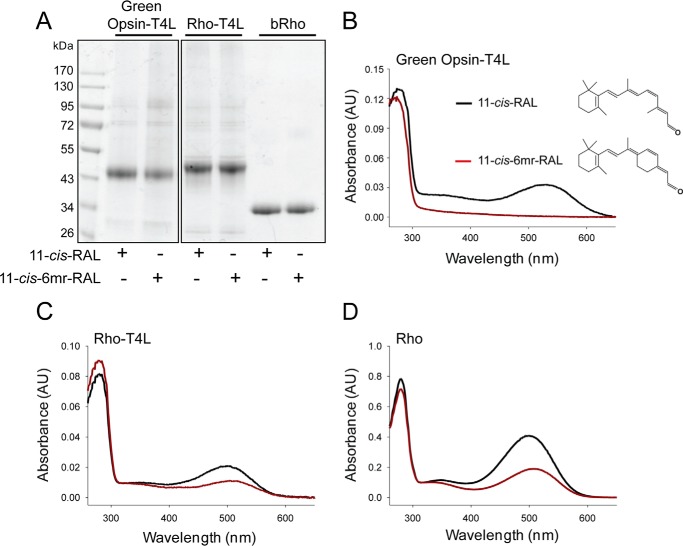
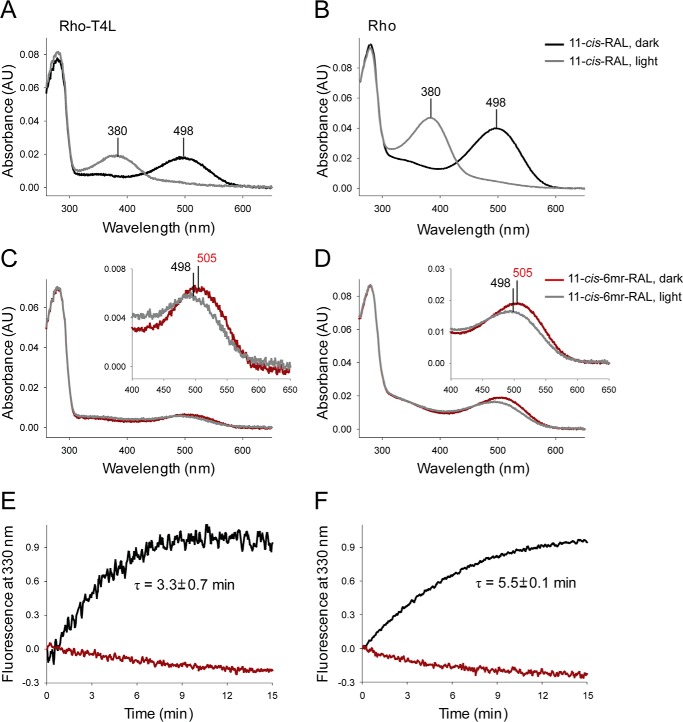
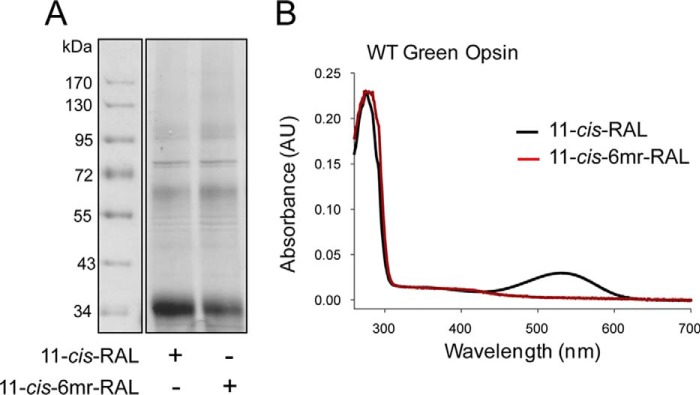
These studies compared the ability of the retinal-binding pocket in rod and green cone pigments to accommodate the 11-cis-6mr-retinal analogue. The properties of cone opsins often are difficult to study because they are less abundant in vivo and less stable in vitro when compared with Rho. To alleviate the problem of instability and to increase protein expression, we introduced a sequence from T4-lysozyme into the third intracellular loop of the green cone opsin sequence, a method that has proven successful in increasing the expression yield and stabilizing the structure of other GPCRs (28). Our green cone opsin-T4L construct was then expressed with greater efficiency (Fig. 1, A and B) and displayed enhanced thermal stability (Fig. 1C). Interestingly, its regenerative properties with native 11-cis-retinal chromophore were not affected (Fig. 2, A and B, black spectrum). However, efforts to regenerate green cone opsin-T4L with 11-cis-6mr-retinal were unsuccessful (Fig. 2B, red spectrum). To better understand this finding, we prepared a rod opsin-T4L construct to determine whether addition of the T4L sequence would prevent pigment regeneration with a locked retinal. Rod opsin-T4L was expressed in insect cells, and isolated membranes containing the receptor were demonstrated to bind both 11-cis-retinal and 11-cis-6mr-retinal to form a pigment with properties similar to opsin isolated from bovine retinas (Rho) (Fig. 2, C and D). Rho-T4L and Rho exhibited similar spectroscopic properties. Pigments regenerated with 11-cis-retinal showed a maximum absorbance at 498 nm and similar sensitivity to light. The absorbance intensity of Rho regenerated with 11-cis-6mr-retinal was 50–60% compared with that of Rho regenerated with 11-cis-retinal for both constructs. Illumination with bright light through a 480–520-nm bandpass filter resulted in formation of the Meta II active state with an absorption maximum at 380 nm (Fig. 3, A and B). Upon regeneration with 11-cis-6mr-retinal, both Rho-T4L and Rho displayed a peak of absorption with a maximum at 505 nm, which transitioned to ∼498 nm after light exposure (Fig. 3, C and D). The observed nominal decline in absorbance at 498 nm could be due to a smaller extinction coefficient of the formed photoproduct, as observed previously (29, 30). A Meta II decay assay detected the release of chromophore from pigments regenerated with 11-cis-retinal upon illumination, which indicated the formation of a functional Schiff base between the chromophore and opsin despite the insertion of the T4L-lysozyme sequence (Fig. 3, E and F, black traces). However, no chromophore release was detected when pigments were regenerated with 11-cis-6mr-retinal (Fig. 3, E and F, red traces), confirming a much higher resistance of the Schiff base to hydrolysis in the locked retinal-regenerated rod opsin. These results indicate that addition of T4L to the rod opsin sequence had no negative effects on pigment regeneration either with 11-cis-retinal or the locked 11-cis-6mr-retinal. Therefore, to understand why regeneration did not occur in green cone opsin-T4L with 11-cis-6mr-retinal, we analyzed wild-type (WT) green cone opsin. Membranes isolated from insect cells containing WT green cone opsin were regenerated with either native 11-cis-retinal or 11-cis-6mr-retinal and purified by 1D4 immunoaffinity chromatography (Fig. 4, A and B). UV-visible spectra of the purified proteins indicated that WT green cone opsin regenerated successfully with 11-cis-retinal but not with 11-cis-6mr-retinal (Fig. 4B, black and red traces, respectively).
Figure 1.
Comparison of membrane expression levels and thermal stability at 37 °C between green cone opsin-T4L and WT green cone opsin. A, representative immunoblots indicating expression levels of green cone opsin-T4L (lane 1) and WT green cone opsin (lane 2) in membranes isolated from insect cells and detected with the anti-Rho C-terminal 1D4 tag antibody (middle panel). Five μg of total protein was loaded to each well. Coomassie Blue-stained SDS-polyacrylamide gel loading control is shown in the left panel. Band intensities were quantified from three independent experiments and normalized to 1 mg of total protein extract (right panel). Purified Rho with known concentration was used to derive the standard curve. B, representative Coomassie Blue-stained SDS-polyacrylamide gel comparing levels of green cone opsin-T4L and WT green cone opsin after their immunoaffinity purification from the same volume of cell culture (left panel). Band intensities were quantified from three independent experiments and normalized to micrograms of purified protein from 1 liter of cell culture (right panel). Purified Rho with known concentration was used to derive the standard curve. Quantification of these results was obtained with ImageJ software. Band intensities of both the monomer and the dimer were used for the analysis. C, half-lives of WT and T4L-conjugated green cone opsins after heating at 37 °C for 2 h.
Figure 2.
Binding of 11-cis-retinal chromophore and its locked 11-cis-6mr-retinal analogue to green cone opsin and rod opsin. A, SDS-polyacrylamide gel of regenerated and purified pigments: green cone opsin-T4L, Rho-T4L, and bovine Rho (Rho). Pigment regeneration was carried out in membranes isolated from insect cells, which then were purified by 1D4 immunoaffinity chromatography. Two μg of protein were loaded on each lane of the SDS-polyacrylamide gel, and the gel was stained with Coomassie Blue. Proteins were deglycosylated with PNGase F before loading onto the gel. B, structures of 11-cis-retinal and 11-cis-6mr-retinal (right) and UV-visible absorption spectra of reconstituted green cone opsin-T4L (left). C, UV-visible absorption spectra of reconstituted Rho-T4L. D, UV-visible absorption spectra of reconstituted Rho. Spectra of pigments regenerated with 11-cis-retinal (11-cis-RAL) are shown in black, and spectra of pigments regenerated with 11-cis-6mr-retinal (11-cis-6mr-RAL) are featured in red.
Figure 3.
Spectra of reconstituted Rho-T4L compared with those of bovine Rho. A and B, UV-visible absorption spectra of Rho-T4L and Rho regenerated with 11-cis-retinal chromophore in the dark (black spectra) and after light illumination (gray spectra). C and D, UV-visible absorption spectra of Rho-T4L and Rho regenerated with 11-cis-6mr-retinal in the dark (red spectra) and after light illumination (gray spectra). E and F, chromophore release from purified Rho-T4L and bovine Rho reconstituted with 11-cis-retinal and 11-cis-6mr-retinal. Changes in the intrinsic Trp fluorescence were measured after a 15-s illumination through a 420–520-nm bandpass filter. E, chromophore release from Rho-T4L. F, chromophore release from Rho. Black spectra indicate samples regenerated with 11-cis-retinal, and red spectra indicate those regenerated with 11-cis-6mr-retinal.
Figure 4.
Binding of 11-cis-retinal chromophore and its locked 11-cis-6mr-retinal analogue to WT green cone opsin. A, SDS-polyacrylamide gel of regenerated and purified WT green cone opsin. Pigment regeneration was carried out in membranes isolated from insect cells, which then were purified by 1D4 immunoaffinity chromatography. Two μg of protein were loaded on each lane of the SDS-polyacrylamide gel, and the gel was stained with Coomassie Blue. Proteins were deglycosylated with PNGase F before loading onto the gel. B, UV-visible absorption spectra of WT green cone opsin reconstituted with either 11-cis-retinal (black spectrum) or 11-cis-6mr-retinal (red spectrum).
Specificity of the chromophore-binding pockets in rod opsin and green cone opsin
Because residues in the binding pocket could be causing the inability of green cone opsin to bind 11-cis-6mr-retinal, we compared the binding pockets of rod opsin with the binding pocket in the model of green cone opsin. Rosetta was used to calculate pairwise interaction energies between 11-cis-retinal and surrounding residues for rod opsin and green cone opsin. Next, 11-cis-retinal was replaced in the binding pocket by 11-cis-6mr-retinal for rod opsin and green cone opsin, and pairwise interaction energies between 11-cis-6mr-retinal and surrounding residues were calculated by Rosetta. This analysis revealed residues in the binding pocket that show a difference in interaction energy of 11-cis-retinal versus 11-cis-6mr-retinal (Δligand) for each protein. Comparing Δligand between corresponding residues of rod opsin and green cone opsin indicated differentially affected residues that are candidates for the ligand binding specificity of green cone opsin (Table 1).
Table 1.
Differential energetic interactions of binding pocket residues with chromophore.
| Rhodopsin, residue |
GCPa, residueb |
GCPΔligand − rhodopsinΔligandc | ||
|---|---|---|---|---|
| Number | Type | Number | Type | |
| 86 | Met | 102 | Glu | 0.46 |
| 91 | Phe | 107 | Ser | 2.236 |
| 118 | Thr | 134 | Ser | 1.411 |
| 122 | Glu | 138 | Ile | 0.616 |
| 189 | Ile | 205 | Pro | 0.292 |
| 207 | Met | 223 | Leu | 0.471 |
| 208 | Phe | 224 | Met | 0.417 |
| 212 | Phe | 228 | Cys | 0.281 |
| 298 | Ser | 314 | Ala | 0.724 |
| 299 | Ala | 315 | Thr | 0.445 |
| 300 | Val | 316 | Ile | 1.043 |
a GCP means WT green cone opsin.
b This corresponds to the equivalent sequence-aligned residue in rhodopsin.
c Δligand = E11cis-6mr-retinal − E11cis-retinal, where E is the energy of interaction between the subscripted chromophore and the listed residue. Energy values are given in Rosetta energy units.
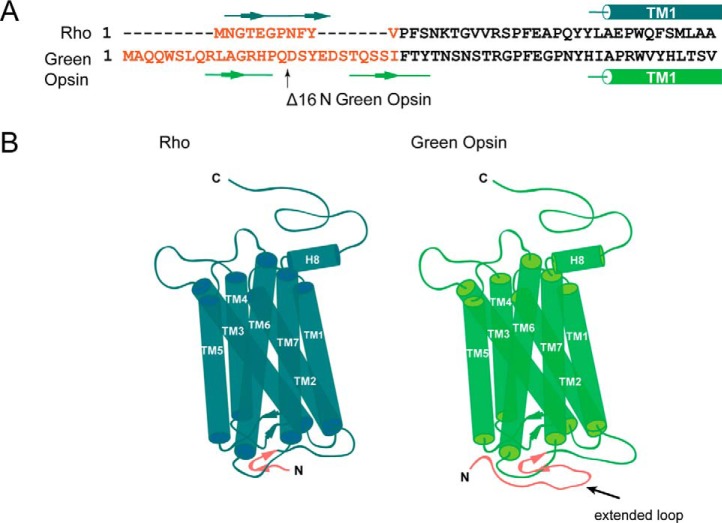
From this energetic analysis, Pro-205 in green cone opsin (Ile-189 in rod opsin) stands out within the context of previous functional data. Thus, mutation of Rho residue Ile-189 to a Pro residue, found at the equivalent position in vertebrate cone opsins, reduced the Meta II decay rate (31). When the Pro was mutated to Ile in chicken green opsin, the protein showed increased expression and thermal stability compared with the wild type (31). Conversion of this Pro residue also affected the Meta III decay rate (32). Moreover, removal of the 9-methyl group from 11-cis-retinal affected the rate of deactivation in salamander red cone pigments as well (33, 34), suggesting a steric sensitivity of cone pigments. Steric sensitivity in the binding pocket of rhodopsin also has been shown (35, 36). Therefore, we compared the binding pockets of rod opsin (Fig. 5A) and green cone opsin (Fig. 5B) between our constructs. Ile-189 in rod opsin was replaced with Pro-205 in green cone opsin. The position of this residue is directly adjacent to the polyene chain of the chromophore and opposite to a Trp residue on the other side of the binding pocket (Fig. 5B). We hypothesized that replacement of the Ile with the more rigid, ringed Pro would create a steric hindrance for the binding of 11-cis-6mr-retinal.
Figure 5.
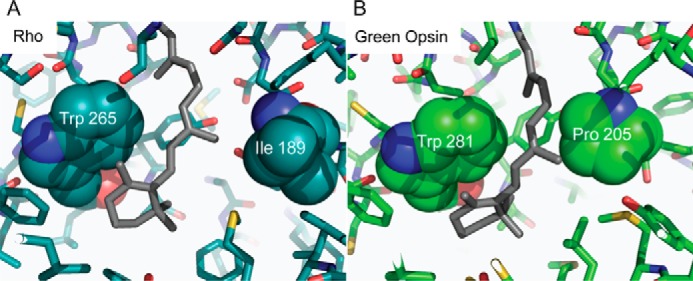
Comparison of the Rho and green cone pigment-binding pockets. A, in the Rho structure, 11-cis-retinal is flanked on either side by Trp-265 and Ile-189. B, predicted green cone pigment binding site shows Pro-205, which is less flexible than the corresponding Ile-189 residue in Rho, near the chromophore.
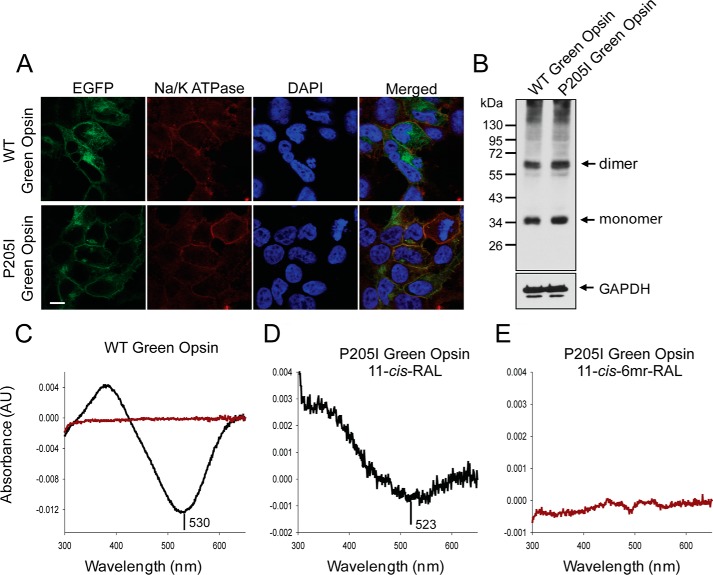
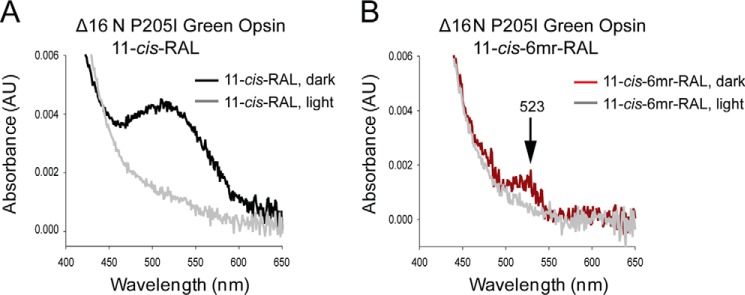
To verify experimentally whether the non-flexible bulk of Pro-205 prevents binding of 11-cis-6mr-retinal to green cone opsin, we mutated this residue to Ile, the corresponding amino acid in Rho. The P205I mutant was expressed either as a fusion protein with EGFP to validate its cell localization or in the pcDNA3.1(+) vector without the EGFP fusion for protein purification. The level of expression of the P205I mutant in transiently transfected HEK-293T cells was comparable with the expression of WT green cone opsin (Fig. 6B), and both proteins localized to the plasma membrane (Fig. 6A) and co-localized with the Na/K-ATPase used here as a plasma membrane localization marker. The P205I green cone opsin was regenerated during its biosynthesis in HEK-293T cells either with 11-cis-retinal or 11-cis-6mr-retinal, and 24 h later pigments were purified by 1D4 immunoaffinity chromatography. The UV-visible absorption spectra of the purified proteins revealed that similar to WT green cone opsin regeneration of the P205I green cone opsin mutant with 11-cis-6mr-retinal had not occurred (Fig. 6, C, red trace, and E). Moreover, this amino acid exchange resulted in much lower regeneration efficiency with 11-cis-retinal and thus a smaller absorption maximum peak as compared with the WT protein along with an ∼7-nm blue shift (Fig. 6, C and D).
Figure 6.
Expression, reconstitution, and purification of WT and P205I green cone opsin in HEK-293T cells. A, membrane localization of WT green cone opsin (top panel) and the P205I mutant (bottom panel) detected by of EGFP fluorescence in living cells (left panel). Immunolocalization of Na/K-ATPase used here as a plasma membrane localization marker (left-middle panel). Nuclei were stained with DAPI (middle-right panel). Merged image of all three panels shows co-localization of green opsin and Na/K ATPase (right panel). Scale bar, 7.5 μm. B, immunoblot indicating the expression levels of WT green cone opsin and the P205I mutant detected with the anti-Rho C-terminal 1D4 tag antibody. Proteins were deglycosylated with PNGase F before loading onto the gel. C, difference absorption spectrum of WT green cone opsin, regenerated with 11-cis-retinal or 11-cis-6mr-retinal, and purified by immunoaffinity chromatography (black and red trace, respectively). D and E, difference absorption spectra of the P205I mutant, regenerated with either 11-cis-retinal or 11-cis-6mr-retinal, and purified by immunoaffinity chromatography.
Regeneration of green cone opsin lacking 16 N-terminal amino acids
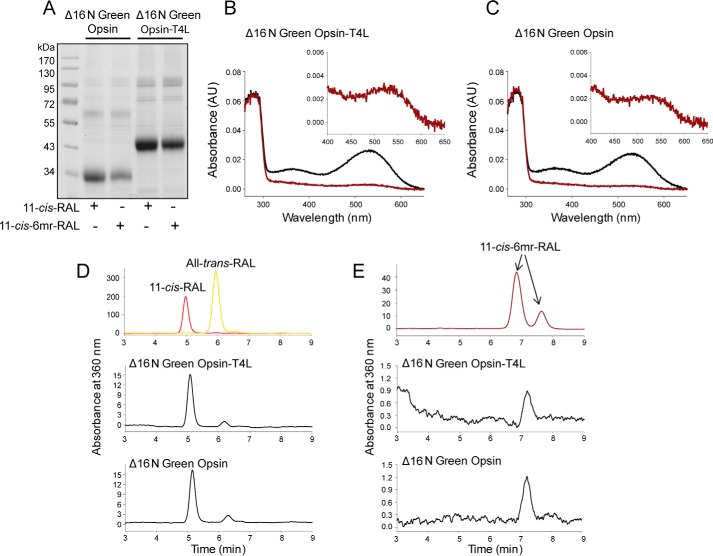
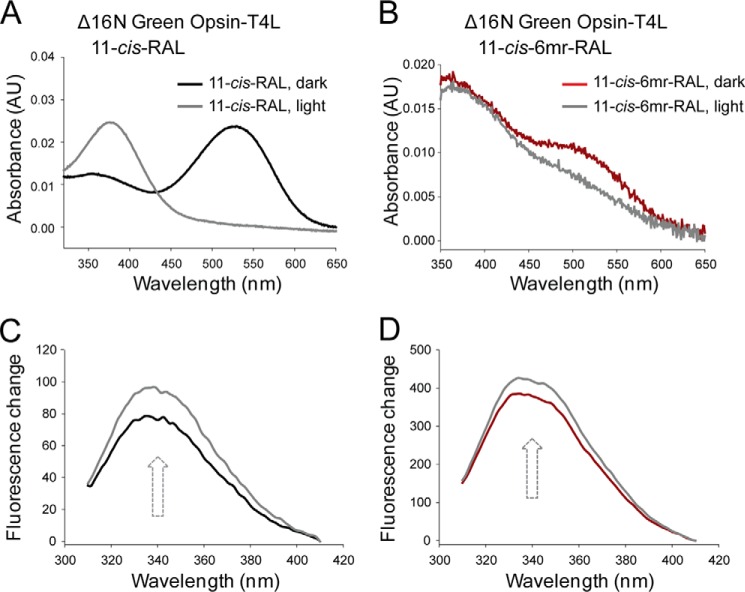
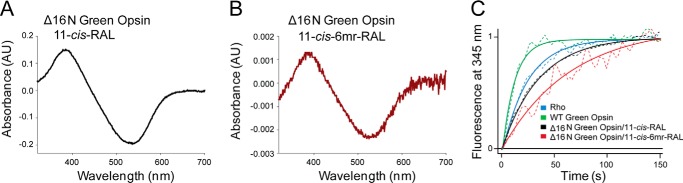
The longer N terminus of cone opsins includes an additional structural difference between rod and cone opsins (Fig. 7, A and B). To test whether this feature prevents regeneration with 11-cis-6mr-retinal, we deleted the 16 N-terminal amino acids in both the WT green cone opsin and the construct containing T4L to create Δ16N green opsin and Δ16N green opsin-T4L constructs, respectively. These constructs then were expressed in insect cells, and isolated membranes were regenerated with either 11-cis-retinal or 11-cis-6mr-retinal and purified by 1D4 immunoaffinity chromatography (Fig. 8A). Shortening the protein sequence had no deleterious effects on pigment regeneration with 11-cis-retinal (Fig. 8, B and C, black spectra). However, it did permit some regeneration to occur with 11-cis-6mr-retinal. UV-visible absorption measurements revealed a small but distinct peak in the 11-cis-6mr-retinal regenerated sample (Fig. 8, B and C, red spectra). Although regeneration was detected with 11-cis-6mr-retinal in both Δ16N green opsin and Δ16N green opsin-T4L, regeneration of Δ16N green opsin-T4L was more efficient (∼6%) than regeneration of Δ16N green opsin (∼2%), potentially due to the stabilizing effect of the T4L insertion. In the case of Rho, regeneration with 11-cis-6mr-retinal resulted in a shift of the absorption maximum toward longer wavelengths (maximum at 505 nm). However, whether such a shift occurs in Δ16N green cone opsin was difficult to ascertain, due to the small degree of regeneration. Nevertheless, incorporation of native chromophore and locked retinal into the Δ16N green opsin was confirmed by HPLC retinoid analysis (Fig. 8, D and E). 11-cis-6mr-Retinal elutes as four distinct peaks corresponding to 9,11-dicis, 11,13-dicis, 11-cis, and 9,11,13-tricis conformations under specific HPLC conditions (24, 25). However, with the HPLC conditions employed here, the 11-cis-6mr-retinal standard displayed two distinct peaks that eluted at 6.8 and 7.6 min (Fig. 8E, top panel), and only one small peak eluting at 7.1 min was found in samples extracted from both Δ16N green cone opsin-T4L and Δ16N green cone opsin (Fig. 8E, middle and bottom panels, respectively). The small shift in retinoid elution found in the experimental samples could be due to the difference in the amounts of standard retinoid and sample (∼40 times more standard was loaded onto the column). It could also suggest that only one isomer (most likely 11-cis) binds to the N-terminal truncated green cone opsin. Even though 11-cis-6mr-retinal could partially occupy the chromophore-binding pocket of Δ16N green cone opsin and cause the appearance of an absorption maximum peak at ∼530 nm, it failed to form a light-insensitive pigment (Fig. 9). Upon light exposure of Δ16N green opsin-T4L, the absorption peak at ∼530 nm decreased (Fig. 9B), and 11-cis-6mr-retinal eventually was released from the binding pocket, just as in the sample regenerated with 11-cis-retinal (Fig. 9A). Intrinsic fluorescence of Trp increased within 15 min after illumination due to chromophore release and un-quenching of Trp-281 in the binding pocket in both Δ16N green cone opsin-T4L samples regenerated with 11-cis-retinal and 11-cis-6mr-retinal (Fig. 9, C and D). This light sensitivity was confirmed in Δ16N green cone opsin regenerated with 11-cis-6mr-retinal and compared with Δ16N green cone opsin regenerated with 11-cis-retinal as shown by the difference spectra of light-illuminated and dark-adapted samples (Fig. 10, A and B). Interestingly, Δ16N green cone opsin regenerated with 11-cis-6mr-retinal resulted in functional pigment able to activate transducin similarly as the N-terminal truncated green opsin regenerated with 11-cis-retinal (Fig. 10C), but the initial activation rates were several times lower as compared with those for WT green cone opsin. Moreover, deletion of the same 16 N-terminal amino acids in the P205I green cone opsin mutant also restored partial regeneration with 11-cis-6mr-retinal resulting in formation of the light-sensitive bleachable pigment similar to that regenerated with native chromophore 11-cis-retinal (Fig. 11, A and B). Thus, the N terminus of green cone opsin clearly plays an important role in the chromophore binding specificity.
Figure 7.
Comparison of Rho sequence and structure with a green cone opsin homology model. A, sequence alignment of rod and green cone opsins were prepared in Clustal Omega; only N termini and beginning of TM1 in both Rho and green cone opsin are shown. The major difference between the N termini is shown in red. B, schematic structural model of Rho (created based on the Rho structure, PDB code 1U19) is shown on the left, and the schematic 3D homology model of green cone opsin (PDB code 1KPW) is on the right. The extended loop difference between the two N termini is shown in red.
Figure 8.
Binding of 11-cis-retinal and its locked 11-cis-6mr-retinal analogue to green cone opsin lacking its N-terminal 16 amino acids. A, SDS-polyacrylamide gel of regenerated and purified pigments, Δ16N green cone opsin-T4L and Δ16N green cone opsin. Pigment regeneration was carried out in membranes isolated from insect cells, which then were purified by 1D4 immunoaffinity chromatography. Two μg of protein were loaded on each lane of the SDS-polyacrylamide gel and stained with Coomassie Blue. Proteins were deglycosylated with PNGase F before loading onto the gel. B, UV-visible absorption spectra of reconstituted Δ16N green cone opsin-T4L. C, UV-visible absorption spectra of reconstituted Δ16N green cone opsin. Spectra of pigments regenerated with 11-cis-retinal are colored black, and those regenerated with 11-cis-6mr-retinal are colored red. D and E, HPLC analyses of retinoid oximes in Δ16N green cone opsin regenerated with 11-cis-retinal and 11-cis-6mr-retinal, respectively. Retinoids were identified based on their order of elution from an HPLC column compared with that of authentic standards. D, top panel, HPLC elution profile of 11-cis-retinal and all-trans-retinal. E, top panel, HPLC elution profile of 11-cis-6mr-retinal. D and E, middle panel, elution profile of retinoid extracted from Δ16N green cone opsin-T4L regenerated with 11-cis-retinal and 11-cis-6mr-retinal, respectively. D and E, bottom panel, retinoid elution profile extracted from Δ16N green cone opsin regenerated with 11-cis-retinal and 11-cis-6mr-retinal, respectively.
Figure 9.
Chromophore release from Δ16N green cone opsin-T4L and Δ16N green cone opsin regenerated with 11-cis-retinal or 11-cis-6mr-retinal. A and B, UV-visible spectra of Δ16N green cone opsin-T4L regenerated with either 11-cis-retinal (black spectrum) or 11-cis-6mr-retinal (red spectrum). Δ16N green cone opsin-T4L regenerated with either 11-cis-retinal or 11-cis-6mr-retinal was sensitive to light illumination (gray spectra). C and D, emission spectra of intrinsic Trp fluorescence were measured in the dark (black line for Δ16N green cone opsin-T4L regenerated with 11-cis-retinal and red line for Δ16N green cone opsin-T4L regenerated with 11-cis-6mr-retinal) and 15 min after a 15-s illumination through a 420–520-nm bandpass filter (gray line).
Figure 10.
Chromophore release from Δ16N green cone opsin regenerated with 11-cis-retinal or 11-cis-6mr-retinal and it functional properties. A and B, UV-visible difference spectra of Δ16N green cone opsin regenerated with either 11-cis-retinal (black spectrum) or 11-cis-6mr-retinal (red spectrum). Δ16N green cone opsin regenerated with either 11-cis-retinal or 11-cis-6mr-retinal was sensitive to light illumination. C, activation of G protein by Δ16N green cone opsin regenerated with 11-cis-retinal or 11-cis-6mr-retinal. Shown is the intrinsic fluorescence increase of Gtα subunit upon interaction with photoactivated opsin pigment at 20 °C. The initial activation rates (k0) were calculated within the first 150 s from three independent experiments. The obtained k0 values were as follows: 0.042 ± 0.003 s−1 for Rho, 0.089 ± 0.002 s−1 for WT green opsin, and 0.029 ± 0.005 s−1 for Δ16N green cone opsin regenerated with 11-cis-retinal and 0.016 ± 0.002 s−1 for Δ16N green cone opsin regenerated with 11-cis-6mr-retinal.
Figure 11.
Spectral properties of Δ16N P205I green cone opsin mutant regenerated with 11-cis-retinal or 11-cis-6mr-retinal. A and B, UV-visible spectra of Δ16N P205I green cone opsin regenerated with either 11-cis-retinal (black spectrum) or 11-cis-6mr-retinal (red spectrum). Deletion of 16 N-terminal amino acids enabled partial regeneration of P205I mutant with 11-cis-6mr-retinal. Δ16N P205I green cone opsin regenerated with either 11-cis-retinal or 11-cis-6mr-retinal was sensitive to light illumination (gray spectra).
Our molecular modeling studies revealed that the N-terminal residues of green cone opsin interact with transmembrane helices five (TM5) and six (TM6) (Fig. 12A). These N-terminal residues are external to the binding site so only minimal structural differences were observed between WT and Δ16N green opsin in the binding site (Fig. 12B). 11-cis-6mr-Retinal is predicted to cause minor side-chain rearrangements in the binding pocket as compared with 11-cis-retinal (Fig. 12C).
Figure 12.
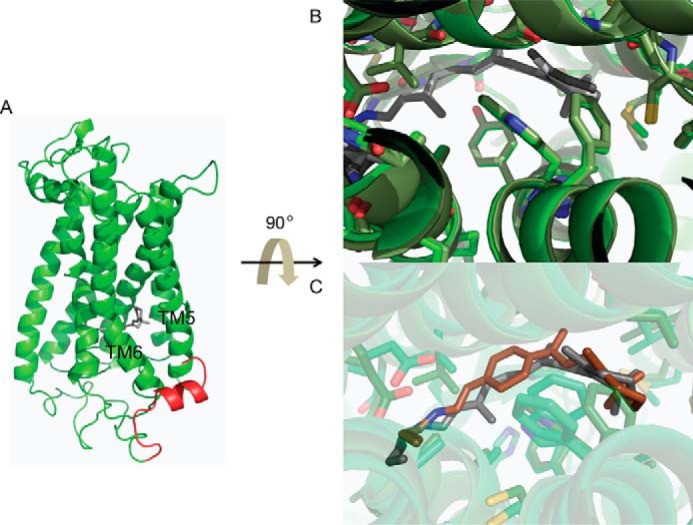
Model of green cone opsin and its binding pocket. A, overall structure of the green cone opsin. The N-terminal 16 amino acids are colored red. B, comparison of the WT (light green) and Δ16N (dark green) green cone opsin-binding sites. C, predicted binding site structure of Δ16N green cone opsin bound with either 11-cis-retinal (dark gray) or 11-cis-6mr-retinal (red).
Discussion
Rod opsin can accommodate retinal geometric isomers or analogues in addition to the native chromophore, 11-cis-retinal, in its binding pocket (26, 29, 37). One example of such an isomeric form is 9-cis-retinal, which binds to opsin both in vitro and in vivo, forming a functional pigment (38, 39). 9-cis-Retinal is more stable than 11-cis-retinal, and supplementation with this chromophore can benefit some retinal degenerative diseases (21, 23). An artificial Rho-like pigment also can be generated with locked retinals that contain a ring structure that prevents isomerization around the C11=C12 double bond (25, 26). This pigment is inactive or minimally active even in the presence of light, which again can be beneficial for the treatment of specific retinal disorders (24). Treatments with artificial chromophores affect the regeneration of both Rho and cone opsins. Although the regenerative properties of Rho with locked retinals were reported previously (26), similar studies for cone opsins were lacking. Therefore, pharmacological supplementation with these retinal analogues could not be implemented without further understanding of their effects on cone pigments.
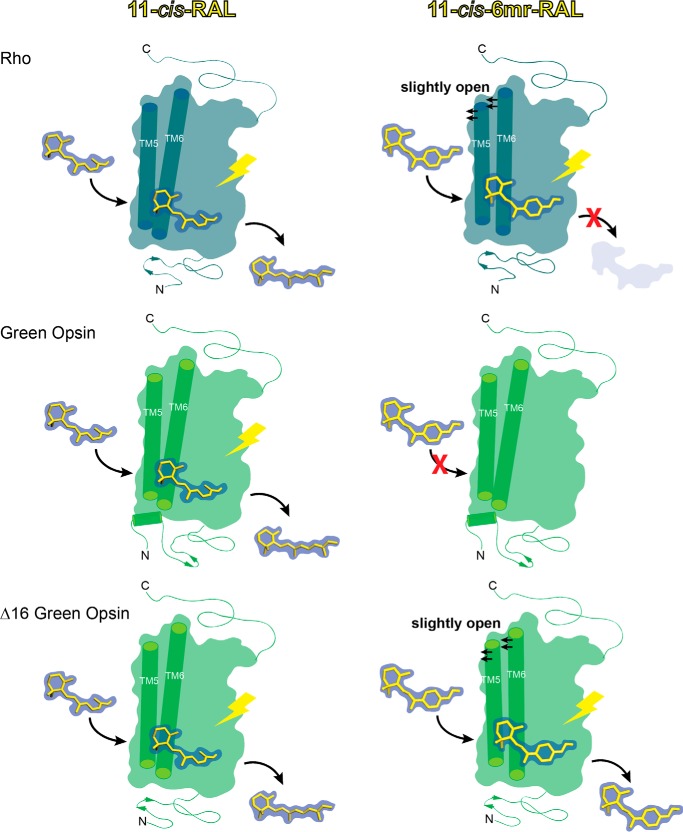
Here, we studied the regenerative properties of green cone opsin and compared them with those of rod opsin, as those two pigments are sensitive to light within a similar range of wavelengths (Fig. 13). The results demonstrate that green cone opsin cannot be regenerated with 11-cis-6mr-retinal. This could be due to an inability of the binding site to accommodate 11-cis-6mr-retinal. Based on structural comparisons between the binding site predicted for green cone opsin and the Rho-binding site from previous studies, we identified Pro-205 as a residue of notable difference in green cone opsin compared with the corresponding Ile-189 residue in Rho. However, the P205I mutation did not result in the binding of 11-cis-6mr-retinal and actually decreased the efficiency of regeneration with 11-cis-retinal. These effects could be due to a change in the stability of the binding pocket caused by the P205I mutation. As shown previously, mutation of Rho residue Ile-189 to a Pro residue, widely conserved in vertebrate cone opsins, increased the thermal stability of the chicken green cone pigment (31). Thus, such findings suggest that the normally lower stability of cone opsin compared with Rho (5) is of functional importance for efficient chromophore binding. Moreover, this residue substitution also altered the absorption spectrum causing a slight but significant blue shift of the λmax, similar to that of chicken green cone opsin P205I mutant (31). Changes in the environment around the chromophore and perturbation of its configuration or electrostatic state could explain the observed spectral shift. For example, human green cone opsin apoprotein also contains a Cl− ion-binding pocket near its chromophore-binding pocket. Such binding of Cl− noted in both green and red human cone opsins but not in blue cone opsin or rod opsin results in a red spectral shift (41). Therefore, the P205I mutant could destabilize the Cl−-binding site and alter its effect on the chromophore.
Figure 13.
Schematic representation of the green cone opsin regeneration process with 11-cis-6mr-retinal. 11-cis-Retinal binds to rod opsin via a protonated Schiff base in the retinal-binding pocket. After light illumination, 11-cis-retinal isomerizes to all-trans-retinal, which eventually dissociates from the binding pocket (top, left). Rod opsin can also bind 11-cis-6mr-retinal, but due to inhibition of its cis-trans isomerization around the C11=C12 double bond, its light-induced hydrolysis is inhibited (top, right). Similar to Rho, green cone opsin binds 11-cis-retinal and releases this chromophore after light illumination (middle, left). However, it does not bind 11-cis-6mr-retinal, possibly due to a tighter and more rigid binding pocket entry stabilized by its elongated N terminus (middle, right). Deletion of 16 N-terminal residues in green cone opsin not only allows binding of 11-cis-retinal (bottom, left) but also permits binding of the more bulky 11-cis-6mr-retinal (bottom, right).
Removing the N-terminal 16 amino acids from the green cone opsin allowed 11-cis-6mr-retinal to bind. Our model of the green cone pigment predicts that these N-terminal residues interact with TM5 and TM6. The proposed Rho dimer and oligomer structures indicate that the least sterically occluded area for retinal to enter the binding pocket is through the TM5-TM6 interface (42). In addition, molecular dynamics simulations also showed that TM5 and TM6 are the most probable points of retinal entry into both Rho (43, 44) and bacterio-opsin (45). Now, our modeling and experimental results suggest that the TM5-TM6 interface is of critical importance to the chromophore-binding pathway in green cone opsin. Removing the 16 N-terminal amino acids could result in increased flexibility of the TM5-TM6 interface due to loss of stabilizing contacts, and such increased flexibility then could allow the rigid 11-cis-6mr-retinal (compared with 11-cis-retinal) to reach the binding pocket.
Although we identified one residue of particular interest to test its effect on green cone opsin's ability to bind 11-cis-6mr-retinal, a systematic approach across multiple binding site residues could provide additional insights into the interaction of green cone opsin with 11-cis-6mr-retinal. Multiple and simultaneous interactions between residues could potentially be responsible for binding site specificity. Having identified the N-terminal 16 residues as a barrier to regeneration with 11-cis-6mr-retinal, such studies of the binding pocket should now be possible. However, removing the N-terminal 16 residues allowed only minimal chromophore regeneration, so deciphering the cause of the remaining difference in regeneration efficiency between Rho and Δ16N green cone opsin is an exciting future area of research that could allow the development of ligands targeting specific visual receptors. Moreover, we also need to understand why green cone opsin could be bleached when bound with 11-cis-6mr-retinal. This result and the limited regeneration indicate that binding is weak between green cone opsin and 11-cis-6mr-retinal. Now knowing that Δ16N green cone opsin allows 11-cis-6mr-retinal to reach the binding pocket will permit the design of improved chromophores that are incapable of being bleached.
Experimental procedures
Chemicals
n-Dodecyl β-d-maltoside (DDM) was obtained from Affymetrix Inc. (Maumee, OH). 11-cis-Retinal was a generous gift from Dr. Rosaline Crouch (Medical University of South Carolina, Charleston). 11-cis-6mr-retinal was provided by Novartis (Cambridge, MA). Mouse mAb anti-ATPase(Na+/K+)α5 originally obtained from The Developmental Studies Hybridoma Bank at University of Iowa (Iowa City) was a generous gift from Dr. Yoshikazu Imanishi.
Constructs
Human green cone opsin cDNA cloned into a pUC57 vector was synthesized by Genentech (San Francisco). To enable protein purification, the last 12 amino acids were replaced by the 1D4 tag (a TETSQVAPA amino acid sequence comprising the last 9 amino acids of rod opsin).
Constructs with T4L
The T4L sequence was inserted into human green cone opsin between amino acid positions 250 and 258, whereas amino acids 251–257 were deleted. For bovine opsin, amino acids 235–241 were replaced with the T4L sequence, which corresponds to the same position where the T4L sequence was inserted into green cone opsin.
Mutagenesis
The green cone opsin P205I mutant was constructed with the Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) according to the manufacturer's procedures.
For construction of human green cone opsin-EGFP WT and the P205I mutant, cDNA of human green cone opsin was amplified by PCR. EcoRI and SACII restriction sites were then introduced at the 5′- and 3′-ends with the following primers: for the cone opsin-EGFP construct, forward primer GTGGGGAATTCGCCATGAAGACCATCATCGCCCT and reverse primer TCTGGCCGCGGTGGCTGGAGCGACCTGA. The resulting amplified DNA was then cloned into the pEGFP-N3 vector (Clontech).
Additionally, WT green cone opsin and the P205I mutant were subcloned into a pcDNA3.1(+) vector (Invitrogen) following the manufacturer's protocol, and the resulting constructs were used for protein purification and UV-visible spectroscopy experiments. Moreover, constructs of WT green cone opsin, green cone opsin-T4L, and the P205I mutant lacking the sixteen N-terminal amino acids were also prepared. The sequence of each construct was confirmed by DNA sequencing.
Preparation of opsin membranes and pigment regeneration
Bovine ROS membranes were prepared from frozen retinas under dim red light (46). ROS membranes were suspended in 10 mm sodium phosphate, pH 7.0, and 50 mm hydroxylamine to a Rho concentration of 2 mg/ml, placed on ice, and illuminated with a 150-watt bulb for 30 min at a distance of 15 cm. Membranes then were centrifuged at 16,000 × g for 10 min, and the membranous pellet was washed four times with 10 mm sodium phosphate, pH 7.0, and 2% BSA followed by four washes with 10 mm sodium phosphate, pH 7.0, and two washes with 20 mm Bistris propane (BTP), 100 mm NaCl, pH 7.5. Rho and opsin concentrations were measured with a UV-visible spectrophotometer (Cary 50, Varian, Palo Alto, CA) and quantified using the absorption coefficients ϵ500 nm = 40,600 m−1 cm−1 and ϵ280 nm = 81,200 m−1 cm−1, respectively (47).
Regeneration of Rho with either 11-cis-retinal or 11-cis-6mr-retinal was attained by incubating opsin membranes with the respective retinal for 1 h at 4 °C. Regenerated pigments were purified by 1D4 immunoaffinity chromatography (48).
Expression of green cone opsin in insect cells and membrane isolation
All insect cell expression constructs were built in pFastBac HT vectors (Invitrogen) with their N-terminal His tags removed. Following the manufacturer's protocol, constructs were transformed into DH10 Bac to obtain a Bacmid for transfection with X-tremeGene 9 (Roche Diagnostics). The cell culture supernatant was collected 3–4 days after transfection with P1 virus. Then P2 and P3 viruses were collected similarly and stored. Large scale expression started with Sf9 cells at 3.5 × 106 cells/ml of culture that were infected with P3 stage virus at a 1:100 volume ratio in 3-liter flasks with shaking at 135 rpm in a 27.5 °C incubator for 2 days, and then cells were collected 48 h post-infection.
All subsequent purification steps were performed at 4 °C or on ice. To isolate membranes, cells were first homogenized with a Dounce homogenizer in a hypotonic buffer composed of 25 mm HEPES, pH 7.5, 10 mm MgCl2, 20 mm KCl, and EDTA-free complete protease inhibitor mixture (Roche Diagnostics GmbH, Mannheim, Germany). Then membranes were pelleted by centrifugation at 100,000 × g for 30 min. Membrane homogenization in the same hypotonic buffer and centrifugation were repeated twice and followed by three or four washes with a buffer composed of 25 mm HEPES, pH 7.5, 1.0 m NaCl, 10 mm MgCl2, 20 mm KCl, and EDTA-free complete protease inhibitor mixture. Washed membranes then were suspended in 50% (v/v) glycerol, flash-frozen with liquid nitrogen, and stored at −80 °C.
Expression of green cone opsins in HEK-293 cells and pigment reconstitution
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS (Hyclone, Logan, UT), 5 μg/ml plasmocin (InvivoGen, San Diego), and 1 unit/ml penicillin with 1 μg/ml streptomycin (Life Technologies, Inc.) at 37 °C under 5% CO2. Cells were transiently transfected with WT green cone opsin or P205I mutant constructs cloned into the pEGFP-N3 or pcDNA3.1(+) vectors (Clontech) with polyethyleneimine (49, 50). Twenty four h post-transfection, cone opsins were reconstituted with either 11-cis-retinal or 11-cis-6mr-retinal. Retinal was added to the cell culture from a DMSO stock solution to a final concentration of 10 μm, and then cells were incubated in the dark for 24 h at 37 °C with 5% CO2. To evaluate the membrane localization of WT green cone opsin and the P205I mutant, we used a TCS SP2 confocal microscope (Leica Microsystems Inc., Bannockburn, IL) to observe these proteins expressed as fusion proteins with EGFP (51).
Pigment purification by 1D4 immunoaffinity chromatography
Cells transiently transfected with WT green cone opsin or the P205I mutant were harvested from 15 10-cm plates, centrifuged at 800 × g, and the pellet was suspended in a buffer composed of 50 mm HEPES, 150 mm NaCl, 20 mm DDM, and protease inhibitor mixture, pH 7.0, and incubated for 1 h at 4 °C on a nutator. Alternatively, insect cell membranes with reconstituted cone opsins or regenerated bovine ROS membranes were solubilized with the above buffer. The lysate then was centrifuged at 100,000 × g for 1 h at 4 °C, and cone or rod pigments were purified from the supernatant by immunoaffinity chromatography with an anti-Rho C-terminal 1D4 antibody (52) immobilized on CNBr-activated agarose. Two-hundred to 400 μl of 6-mg 1D4/ml agarose beads were added to the supernatant and incubated for 1 h at 4 °C on the nutator. The resin was then transferred to a column and washed with 10 ml of buffer consisting of 50 mm HEPES, 150 mm NaCl, and 2 mm DDM, pH 7.0. Pigments were eluted with buffer consisting of 150 mm HEPES, 150 mm NaCl, and 2 mm DDM, pH 7.0, supplemented with 0.6 mg/ml of the TETSQVAPA peptide.
UV-visible spectroscopy of opsin pigments
UV-visible spectra were recorded in the dark from freshly purified cone or rod pigment samples. Spectra of the samples were then recorded after the samples were exposed to white light delivered from a Fiber-Light illuminator (150-watt lamp) (Dolan-Jenner, Boxborough, MA) at a distance of 10 cm for 3 min. To obtain a difference spectrum of purified cone pigments, the spectrum of the sample recorded in the dark was subtracted from the spectrum of the bleached sample.
Chromophore release
Chromophore release was measured with an L55 fluorescence spectrometer (PerkinElmer Life Sciences) at 20 °C. All measurements were performed with 25 nm purified cone or rod pigment diluted in a buffer consisting of 10 mm BTP, 100 mm NaCl, and 1 mm DDM, pH 6.0. Samples were bleached by a Fiber-Light illuminator through a 420–520-nm bandpass filter for 15 s immediately before the fluorescence measurements. Bleaching was carried out at a distance of 10 cm. Spectrofluorometer slit settings were 5 nm at 295 nm for excitation, and 10 nm at 330 nm for emission collection. Changes in the intrinsic Trp fluorescence were recorded for 15–30 min. An increase of the intrinsic Trp fluorescence correlates with the decrease in the protonated Schiff base concentration (53). Alternatively, emission spectra were recorded between 300 and 450 nm after excitation at 295 nm.
HPLC analyses
Green cone opsin samples were denatured for 30 min at room temperature with 50% CH3OH in 40 mm NH2OH. The resulting retinal oximes were extracted with 600 μl of hexane, and their isomeric content was determined by normal phase HPLC with a Luna 10-μm PRE Silica (3) 100 Å, 250 × 4.6-mm column (Beckman, San Ramon, CA). Retinoids were eluted isocratically with 10% ethyl acetate in hexane at a flow rate of 2 ml/min. Their signals were detected by absorption at 360 nm (54, 55).
Transducin activation assay
The intrinsic fluorescence increase from Gtα was measured with an L55 luminescence spectrophotometer (PerkinElmer Life Sciences). Excitation and emission wavelengths were chosen at 300 and 345 nm, respectively. Gt was mixed with each pigment (Rho, WT green cone opsin or ΔΝ16 green cone opsin regenerated either with 11-cis-retinal or 11-cis-6mr-retinal) at the ratio of 10:1, with Gt at a concentration of 1000 nm, followed by the addition of 300 μm GTPγS to determine the GTPγS-induced complex dissociation and fluorescence changes. The samples were then illuminated for 1 min with a fiber light delivered through a 480–520-nm long pass wavelength filter. Gt activation rates were determined for the first 150 s of the Gt activation assay.
Modeling
We generated a model of the green cone opsin as follows. The 2.2 Å resolution crystal structure of Rho (PDB code 1U19) (56) was used as the template for the human green cone opsin protein sequence (UniProtKB, P04001). The bovine Rho sequence (UniProtKB- P02699) was aligned with the green cone opsin sequence with the EMBOSS Needle Pairwise Sequence Alignment tool (57). Given this sequence alignment, coordinates were initially assigned to the green cone opsin sequence with the MEDELLER server (58). Then the N-terminal 20 amino acids of the structure were rebuilt 60 times with the Rosetta protein structure prediction suite loop-building module (59), and the most energetically favorable model was finally selected.
Retinal was parameterized for use within Rosetta (60). Because the retinal ligand is bound to Lys-312 by a protonated Schiff base, the ligand-residue complex was treated as a non-canonical amino acid, allowing the structure to be seamlessly used within all components of the Rosetta framework. Coordinates for the retinal-lysine complex were extracted from the 2.2 Å resolution crystal structure of Rho (PDB code 1U19), and a Rosetta non-canonical amino acid parameter file was created. The fixed-backbone design protocol in Rosetta (61) was used to mutate Lys-312 to the retinal-lysine complex, which was introduced with the conformation present in the crystal structure. All other coordinates in the green cone opsin structure were kept constant. To remove any structural inconsistencies or clashes introduced into the green cone pigment model, the structure was relaxed with the Rosetta membrane fast relax protocol (40), and the most energetically favorable model was selected. This protocol is specialized to optimize membrane protein coordinates according to the Rosetta energy potentials that employ an implicit biological membrane. These modeling procedures were repeated to produce a model with 11-cis-6mr-retinal in the binding site.
Author contributions
B. J. and K. P. conceived and designed the experiments. B. J., K. K., N. S. A., W. S., J. Z., S. G., and D. S. conducted the experiments. M. M. synthesized 11-cis-6mr-retinal. K. K., N. S. A., W. S., D. S., K. P., and B. J. wrote the manuscript. B. J. and K. P. coordinated and oversaw the research project. All authors discussed the results and commented on the manuscript.
Acknowledgment
We thank Dr. Leslie T. Webster, Jr. for helpful comments on this manuscript.
This work was supported National Institutes of Health Grants EY025214 (to B. J.), EY009339 and EY027283 (to K. P.), EY024864 (to T. S. Kern), and EY025007 (to N. S. A.), Japan Society for the Promotion of Science Overseas Research Fellowship (to K. K.), and an Alcon Research Institute grant (to K. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GPCR
- G protein-coupled receptor
- LCA
- Leber congenital amaurosis
- DDM
- n-dodecyl β-d-maltoside
- BTP
- Bistris propane, 1,3-bis[tris(hydroxymethyl)methylamino]propane
- TM
- transmembrane
- PNGase F
- peptide:N-glycosidase F
- PDB
- Protein Data Bank
- ROS
- rod outer segment
- 11-cis-6mr
- 11-cis-6-membered ring
- T4L
- T4-lysozyme
- EGFP
- enhanced GFP.
References
- 1. Baylor D. A., Lamb T. D., and Yau K. W. (1979) Responses of retinal rods to single photons. J. Physiol. 288, 613–634 [PMC free article] [PubMed] [Google Scholar]
- 2. Wald G., Brown P. K., and Smith P. H. (1955) Iodopsin. J. Gen. Physiol. 38, 623–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fujimoto K., Hasegawa J., and Nakatsuji H. (2009) Color tuning mechanism of human red, green, and blue cone pigments: SAC-CI theoretical study. Bull. Chem. Soc. Jpn. 82, 1140–1148 [Google Scholar]
- 4. Merbs S. L., and Nathans J. (1992) Absorption spectra of human cone pigments. Nature 356, 433–435 [DOI] [PubMed] [Google Scholar]
- 5. Hofmann L., and Palczewski K. (2015) Advances in understanding the molecular basis of the first steps in color vision. Prog. Retin. Eye Res. 49, 46–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shichida Y., Okada T., Kandori H., Fukada Y., and Yoshizawa T. (1993) Nanosecond laser photolysis of iodopsin, a chicken red-sensitive cone visual pigment. Biochemistry 32, 10832–10838 [DOI] [PubMed] [Google Scholar]
- 7. Okada T., Matsuda T., Kandori H., Fukada Y., Yoshizawa T., and Shichida Y. (1994) Circular dichroism of metaiodopsin II and its binding to transducin: a comparative study between meta II intermediates of iodopsin and rhodopsin. Biochemistry 33, 4940–4946 [DOI] [PubMed] [Google Scholar]
- 8. Srinivasan S., Cordomí A., Ramon E., and Garriga P. (2016) Beyond spectral tuning: human cone visual pigments adopt different transient conformations for chromophore regeneration. Cell. Mol. Life Sci. 73, 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Srinivasan S., Ramon E., Cordomí A., and Garriga P. (2014) Binding specificity of retinal analogs to photoactivated visual pigments suggest mechanism for fine-tuning GPCR-ligand interactions. Chem. Biol. 21, 369–378 [DOI] [PubMed] [Google Scholar]
- 10. Palczewski K. (2006) G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Travis G. H., Golczak M., Moise A. R., and Palczewski K. (2007) Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol. 47, 469–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jäger S., Palczewski K., and Hofmann K. P. (1996) Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry 35, 2901–2908 [DOI] [PubMed] [Google Scholar]
- 13. Palczewski K., Jäger S., Buczyl̸ko J., Crouch R. K., Bredberg D. L., Hofmann K. P., Asson-Batres M. A., and Saari J. C. (1994) Rod outer segment retinol dehydrogenase: substrate specificity and role in phototransduction. Biochemistry 33, 13741–13750 [DOI] [PubMed] [Google Scholar]
- 14. Hofmann K. P., Pulvermüller A., Buczyl̸ko J., Van Hooser P., and Palczewski K. (1992) The role of arrestin and retinoids in the regeneration pathway of rhodopsin. J. Biol. Chem. 267, 15701–15706 [PubMed] [Google Scholar]
- 15. Fan J., Woodruff M. L., Cilluffo M. C., Crouch R. K., and Fain G. L. (2005) Opsin activation of transduction in the rods of dark-reared Rpe65 knockout mice. J. Physiol. 568, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson D. A., Gyürüs P., Fleischer L. L., Bingham E. L., McHenry C. L., Apfelstedt-Sylla E., Zrenner E., Lorenz B., Richards J. E., Jacobson S. G., Sieving P. A., and Gal A. (2000) Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Invest. Ophthalmol. Vis. Sci. 41, 4293–4299 [PubMed] [Google Scholar]
- 17. Sénéchal A., Humbert G., Surget M. O., Bazalgette C., Bazalgette C., Arnaud B., Arndt C., Laurent E., Brabet P., and Hamel C. P. (2006) Screening genes of the retinoid metabolism: novel LRAT mutation in leber congenital amaurosis. Am. J. Ophthalmol. 142, 702–704 [DOI] [PubMed] [Google Scholar]
- 18. Lorenz B., Gyürüs P., Preising M., Bremser D., Gu S., Andrassi M., Gerth C., and Gal A. (2000) Early-onset severe rod-cone dystrophy in young children with RPE65 mutations. Invest. Ophthalmol. Vis. Sci. 41, 2735–2742 [PubMed] [Google Scholar]
- 19. Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., and Palczewski K. (2004) Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 279, 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maeda T., Maeda A., Matosky M., Okano K., Roos S., Tang J., and Palczewski K. (2009) Evaluation of potential therapies for a mouse model of human age-related macular degeneration caused by delayed all-trans-retinal clearance. Invest. Ophthalmol. Vis. Sci. 50, 4917–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maeda T., Maeda A., Casadesus G., Palczewski K., and Margaron P. (2009) Evaluation of 9-cis-retinyl acetate therapy in Rpe65−/− mice. Invest. Ophthalmol. Vis. Sci. 50, 4368–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maeda T., Cideciyan A. V., Maeda A., Golczak M., Aleman T. S., Jacobson S. G., and Palczewski K. (2009) Loss of cone photoreceptors caused by chromophore depletion is partially prevented by the artificial chromophore pro-drug, 9-cis-retinyl acetate. Hum. Mol. Genet. 18, 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeda T., Maeda A., Leahy P., Saperstein D. A., and Palczewski K. (2009) Effects of long-term administration of 9-cis-retinyl acetate on visual function in mice. Invest. Ophthalmol. Vis. Sci. 50, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuksa V., Bartl F., Maeda T., Jang G. F., Ritter E., Heck M., Van Hooser J. P., Liang Y., Filipek S., Gelb M. H., Hofmann K. P., and Palczewski K. (2002) Biochemical and physiological properties of rhodopsin regenerated with 11-cis-6-ring- and 7-ring-retinals. J. Biol. Chem. 277, 42315–42324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jang G. F., Kuksa V., Filipek S., Bartl F., Ritter E., Gelb M. H., Hofmann K. P., and Palczewski K. (2001) Mechanism of rhodopsin activation as examined with ring-constrained retinal analogs and the crystal structure of the ground state protein. J. Biol. Chem. 276, 26148–26153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhattacharya S., Ridge K. D., Knox B. E., and Khorana H. G. (1992) Light-stable rhodopsin. I. A rhodopsin analog reconstituted with a nonisomerizable 11-cis retinal derivative. J. Biol. Chem. 267, 6763–6769 [PubMed] [Google Scholar]
- 27. Shichida Y., Nakamura K., Yoshizawa T., Trehan A., Denny M., and Liu R. S. (1988) 9,13-dicis-rhodopsin and its one-photon-one-double-bond isomerization. Biochemistry 27, 6495–6499 [DOI] [PubMed] [Google Scholar]
- 28. Chun E., Thompson A. A., Liu W., Roth C. B., Griffith M. T., Katritch V., Kunken J., Xu F., Cherezov V., Hanson M. A., and Stevens R. C. (2012) Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Grip W. J., van Oostrum J., Bovee-Geurts P. H., van der Steen R., van Amsterdam L. J., Groesbeek M., and Lugtenburg J. (1990) 10,20-Methanorhodopsins: (7E,9E,13E)-10,20-methanorhodopsin and (7E,9Z,13Z)-10,20-methanorhodopsin. 11-cis-locked rhodopsin analog pigments with unusual thermal and photo-stability. Eur. J. Biochem. 191, 211–220 [DOI] [PubMed] [Google Scholar]
- 30. Gulati S., Jastrzebska B., Banerjee S., Placeres Á. L., Miszta P., Gao S., Gunderson K., Tochtrop G. P., Filipek S., Katayama K., Kiser P. D., Mogi M., Stewart P. L., and Palczewski K. (2017) Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc. Natl. Acad. Sci. U.S.A. 114, E2608–E2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuwayama S., Imai H., Hirano T., Terakita A., and Shichida Y. (2002) Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry 41, 15245–15252 [DOI] [PubMed] [Google Scholar]
- 32. Kuwayama S., Imai H., Morizumi T., and Shichida Y. (2005) Amino acid residues responsible for the meta-III decay rates in rod and cone visual pigments. Biochemistry 44, 2208–2215 [DOI] [PubMed] [Google Scholar]
- 33. Estevez M. E., Kolesnikov A. V., Ala-Laurila P., Crouch R. K., Govardovskii V. I., and Cornwall M. C. (2009) The 9-methyl group of retinal is essential for rapid Meta II decay and phototransduction quenching in red cones. J. Gen. Physiol. 134, 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Das J., Crouch R. K., Ma J. X., Oprian D. D., and Kono M. (2004) Role of the 9-methyl group of retinal in cone visual pigments. Biochemistry 43, 5532–5538 [DOI] [PubMed] [Google Scholar]
- 35. Ganter U. M., Schmid E. D., Perez-Sala D., Rando R. R., and Siebert F. (1989) Removal of the 9-methyl group of retinal inhibits signal transduction in the visual process. A Fourier transform infrared and biochemical investigation. Biochemistry 28, 5954–5962 [DOI] [PubMed] [Google Scholar]
- 36. Vogel R., Fan G. B., Sheves M., and Siebert F. (2000) The molecular origin of the inhibition of transducin activation in rhodopsin lacking the 9-methyl group of the retinal chromophore: a UV-Vis and FTIR spectroscopic study. Biochemistry 39, 8895–8908 [DOI] [PubMed] [Google Scholar]
- 37. Hu S., Franklin P. J., Wang J., Ruiz Silva B. E., Derguini F., Nakanishi K., and Chen A. H. (1994) Unbleachable rhodopsin with an 11-cis-locked eight-membered ring retinal: the visual transduction process. Biochemistry 33, 408–416 [DOI] [PubMed] [Google Scholar]
- 38. Spalink J. D., Reynolds A. H., Rentzepis P. M., Sperling W., and Applebury M. L. (1983) Bathorhodopsin intermediates from 11-cis-rhodopsin and 9-cis-rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 80, 1887–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Hooser J. P., Aleman T. S., He Y. G., Cideciyan A. V., Kuksa V., Pittler S. J., Stone E. M., Jacobson S. G., and Palczewski K. (2000) Rapid restoration of visual pigment and function with oral retinoid in a mouse model of childhood blindness. Proc. Natl. Acad. Sci. U.S.A. 97, 8623–8628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barth P., Schonbrun J., and Baker D. (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U.S.A. 104, 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z., Asenjo A. B., and Oprian D. D. (1993) Identification of the Cl(−)-binding site in the human red and green color vision pigments. Biochemistry 32, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 42. Fotiadis D., Jastrzebska B., Philippsen A., Müller D. J., Palczewski K., and Engel A. (2006) Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr. Opin. Struct. Biol. 16, 252–259 [DOI] [PubMed] [Google Scholar]
- 43. Wang T., and Duan Y. (2011) Retinal release from opsin in molecular dynamics simulations. J. Mol. Recognit. 24, 350–358 [DOI] [PubMed] [Google Scholar]
- 44. Wang T., and Duan Y. (2007) Chromophore channeling in the G-protein coupled receptor rhodopsin. J. Am. Chem. Soc. 129, 6970–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Isralewitz B., Izrailev S., and Schulten K. (1997) Binding pathway of retinal to bacterio-opsin: a prediction by molecular dynamics simulations. Biophys. J. 73, 2972–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Papermaster D. S. (1982) Preparation of retinal rod outer segments. Methods Enzymol. 81, 48–52 [DOI] [PubMed] [Google Scholar]
- 47. Wald G., and Brown P. K. (1953) The molecular excitation of rhodopsin. J. Gen. Physiol. 37, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salom D., Le Trong I., Pohl E., Ballesteros J. A., Stenkamp R. E., Palczewski K., and Lodowski D. T. (2006) Improvements in G protein-coupled receptor purification yield light stable rhodopsin crystals. J. Struct. Biol. 156, 497–504 [DOI] [PubMed] [Google Scholar]
- 49. Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., and Behr J. P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethyleneimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Y., and Tang H. (2015) High-throughput screening assays to identify small molecules preventing photoreceptor degeneration caused by the rhodopsin P23H mutation. Methods Mol. Biol. 1271, 369–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perkins B. D., Kainz P. M., O'Malley D. M., and Dowling J. E. (2002) Transgenic expression of a GFP-rhodopsin COOH-terminal fusion protein in zebrafish rod photoreceptors. Vis. Neurosci. 19, 257–264 [DOI] [PubMed] [Google Scholar]
- 52. MacKenzie D., Arendt A., Hargrave P., McDowell J. H., and Molday R. S. (1984) Localization of binding sites for carboxyl-terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry 23, 6544–6549 [DOI] [PubMed] [Google Scholar]
- 53. Farrens D. L., and Khorana H. G. (1995) Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 270, 5073–5076 [DOI] [PubMed] [Google Scholar]
- 54. Van Hooser J. P., Garwin G. G., and Saari J. C. (2000) Analysis of visual cycle in normal and transgenic mice. Methods Enzymol. 316, 565–575 [DOI] [PubMed] [Google Scholar]
- 55. Garwin G. G., and Saari J. C. (2000) High-performance liquid chromatography analysis of visual cycle retinoids. Methods Enzymol. 316, 313–324 [DOI] [PubMed] [Google Scholar]
- 56. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., and Buss V. (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 57. Rice P., Longden I., and Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 58. Kelm S., Shi J., and Deane C. M. (2010) MEDELLER: homology-based coordinate generation for membrane proteins. Bioinformatics 26, 2833–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee J., Lee D., Park H., Coutsias E. A., and Seok C. (2010) Protein loop modeling by using fragment assembly and analytical loop closure. Proteins 78, 3428–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaufmann K. W., Lemmon G. H., Deluca S. L., Sheehan J. H., and Meiler J. (2010) Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry 49, 2987–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., and Baker D. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 [DOI] [PubMed] [Google Scholar]