Abstract
Muscle and bone are closely associated in both anatomy and function, but the mechanisms that coordinate their synergistic action remain poorly defined. Myostatin, a myokine secreted by muscles, has been shown to inhibit muscle growth, and the disruption of the myostatin gene has been reported to cause muscle hypertrophy and increase bone mass. Extracellular vesicle-exosomes that carry microRNA (miRNA), mRNA, and proteins are known to perform an important role in cell-cell communication. We hypothesized that myostatin may play a crucial role in muscle-bone interactions and may promote direct effects on osteocytes and on osteocyte-derived exosomal miRNAs, thereby indirectly influencing the function of other bone cells. We report herein that myostatin promotes expression of several bone regulators such as sclerostin (SOST), DKK1, and RANKL in cultured osteocytic (Ocy454) cells, concomitant with the suppression of miR-218 in both parent Ocy454 cells and derived exosomes. Exosomes produced by Ocy454 cells that had been pretreated with myostatin could be taken up by osteoblastic MC3T3 cells, resulting in a marked reduction of Runx2, a key regulator of osteoblastic differentiation, and in decreased osteoblastic differentiation via the down-regulation of the Wnt signaling pathway. Importantly, the inhibitory effect of myostatin-modified osteocytic exosomes on osteoblast differentiation is completely reversed by expression of exogenous miR-218, through a mechanism involving miR-218-mediated inhibition of SOST. Together, our findings indicate that myostatin directly influences osteocyte function and thereby inhibits osteoblastic differentiation, at least in part, through the suppression of osteocyte-derived exosomal miR-218, suggesting a novel mechanism in muscle-bone communication.
Keywords: bone, exosome (vesicle), microRNA (miRNA), myostatin, osteoblast, osteocyte, skeletal muscle metabolism, Wnt signaling
Introduction
Muscle and bone form a functional unit that undergoes changes of mass in a coordinated, usually symmetrical, manner (1, 2). Mechanical loading of bone by muscle is one of the larger sources of load experienced by the skeleton; muscle mass is closely correlated with bone mass, suggesting an essential role of muscle activity in maintaining a balance between bone formation and bone resorption (3–5). Trained athletes, especially those who engage in weight-dependent activities, often have greater muscle and bone mass and strength than individuals who are less physically active (6). It is reported that in the United States among older adults with an average age of 70 years, the prevalence of sarcopenia is 36.5% (7). Sarcopenia with decline in skeletal muscle mass and function has been linked to senile osteoporosis (8). Particularly, extreme immobilization, such as that after spinal cord injury, leads to marked bone loss and muscle paralysis (9–13). However, the mechanisms by which muscle interacts with bone to determine mass and function remain poorly defined.
Comprising ∼95% of total bone cells, osteocytes are deeply embedded in cave-like lacunae in the bone matrix (14–16). Osteocytes have many dendritic processes extending through canaliculi to form a highly connected communication network between themselves and also between osteoblasts and osteoclasts at the bone surface (17). Osteocytes are widely believed to be a principal mechanosensor of bone that is able to respond to mechanical stimuli, transducing load signals into anabolic chemical signals to recruit osteoblasts and osteoclasts (15, 17, 18). Osteocytes are the major source of sclerostin and receptor activator of nuclear factor κB ligand (RANKL),2 two of the major signals in bone metabolism (19–21). As such, osteocytes have been considered an orchestrator of bone remodeling and mineral metabolism (15, 22). Recent work has revealed that muscle may positively or negatively influence bone growth by secreting osteogenic myokines, such as insulin-like growth factor 1 (IGF1), fibroblast growth factor 2 (FGF2), irisin, and myostatin (23–26). However, it remains unknown as to whether these myokines exert a direct effect on osteocytes and, if so, the mechanisms of action.
Myostatin, a hormone secreted by muscles and a member of the transforming growth factor β superfamily of growth factors, has been shown to inhibit muscle differentiation and growth (27–29). Myostatin activates signaling upon binding to the activin receptor IIB (ActRIIB), ultimately resulting in Smad2/3 phosphorylation followed by translocation to the nucleus to modulate transcription of numerous genes (30). Myostatin can increase during sarcopenia, cancer, infection, traumatic musculoskeletal injury, and bed rest, whereas resistance training decreases myostatin levels (2, 31–33). The absence of the myostatin gene in knock-out mice models not only increases muscle mass but also tends to result in an increase in bone density and strength (28). As such, the study of myostatin provides a unique opportunity to elucidate the mechanisms by which muscle regulates bone cell function and, hence, bone substance. Inhibition of myostatin by a soluble activin IIB receptor leads to increased bone formation in mice (34). It is possible that factors released from muscle, such as myostatin or other myokines, may directly or indirectly modulate secretory products of osteocytes that may then alter the function of osteoblasts and osteoclasts.
Exosomes are extracellular organelles (∼30–100 nm in size) that are released by cells by exocytosis and engulfed by cells through phagocytosis. Exosomes play an important role in intercellular communication and have pleiotropic effects on physiological functions of neighboring cells (35–38). Exosomes carry proteins, mRNA, and microRNA (miRNA) (37). miRNA is a single-stranded non-protein-coding RNA that binds to the target messenger RNA and functions primarily as a gene repressor. Thus, exosomes are considered to have regulatory functions through its component miRNA (37, 38). Emerging evidence indicates that miRNAs play important roles in regulating osteoclast and osteoblast function (39). During osteoblast differentiation, the miR-218 gene is induced and acts to promote the differentiation of undifferentiated bone marrow cells by stimulating the Wnt signaling pathway through the inactivation of Wnt inhibitors sclerostin (encoded by SOST gene), dickkopf WNT signaling pathway inhibitor 2(DKK2), and secreted frizzled-related protein 2 (sFRP2) (40). Osteoclast-derived exosomal miR-214-3p transfers to osteoblasts to inhibit bone formation (41). Recent observation suggested that osteocytes shed microvesicles containing not-yet-defined bone regulatory factors that may induce osteoblast differentiation.3 Thus, it is likely that myostatin may exert a role in muscle-bone communication by regulation of the release of exosomal miRNAs from osteocytes.
Wnt/β-catenin signaling pathway is a key determinant of bone mass by controlling bone formation and bone resorption (14, 43–46). Sclerostin and DKK1 act by binding to the Wnt co-receptors LRP5/6 to inhibit Wnt signaling, thereby contributing to bone loss (15). Osteoclastogenesis and osteoclast activity are regulated by RANKL and osteoprotegerin (OPG); specifically, RANKL activates osteoclast differentiation, whereas OPG reduces the activity of RANKL (46–48). We recently reported that electrically stimulated muscle contraction reverses elevations in bone resorption and increases Wnt signaling in bone-derived cells after spinal cord transection (49). Interestingly, injection of irisin profoundly stimulates cortical bone mass and strength in mice accompanied by dramatically increased osteoblastic bone formation and diminished osteoblast inhibitors, such as SOST (25). Moreover, the activation of Wnt/β-catenin signaling is widely believed to be a critical component of mechanotransduction that is orchestrated by the osteocyte (15). Of note, secreted factors by muscle cells have recently been shown to preserve osteocyte viability through the activation of β-catenin (50).
Hence, we hypothesized that (i) myostatin, and possibly other muscle-related factors, may play a essential role in muscle-bone interaction that is mediated through the action(s) of the osteocyte, (ii) myostatin has a direct effect on the osteocyte and osteocytic-derived exosomal miRNAs, thereby indirectly influencing the function of the osteoblast and the osteoclast, and (iii) the increased presence of myostatin alters osteocyte-derived exosomal miRNAs, whose change results in inhibition of osteoblast activity and/or promotion of osteoclast activity. To test these hypotheses, osteocytic cells, Ocy454, were challenged with myostatin and, several possible effects of myostatin on osteoblast and osteoclast biology were determined. Specifically, the studies performed included the expression of bone metabolism-related genes, protein, and miRNA in Ocy454 parent cells and their exosomes, differentiation of the osteoblast and osteoclast when co-cultured with osteocytic exosomes, and relationship of the osteocytic exosome-mediated effects to the Wnt signaling pathways. Our findings demonstrate a previously unidentified role of osteocyte-derived exosomal miR-218 in myostatin-mediated inhibition of osteoblast differentiation and thus reveal a novel mechanism of muscle-bone communication.
Results
Myostatin markedly increased the expression of SOST, DKK1, and RANKL in Ocy454 cells
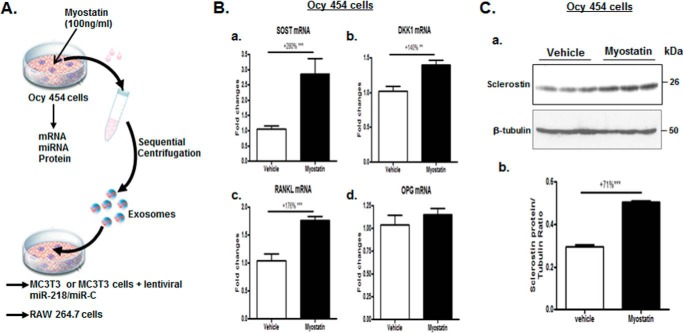
To address as to whether myostatin has a direct impact on osteocytes, Ocy454 cells were treated with 100 ng/ml recombinant myostatin protein or vehicle for 48 h, and total cellular RNA was isolated. Quantitative real-time PCR analysis was used to measure the levels of SOST, DKK1, RANKL, and OPG mRNA expression, and changes in expression of these genes in myostatin-treated Ocy454 cells were compared with those of vehicle-treated control cells. Dramatic increases were observed in mRNA expression of SOST, DKK1, and RANKL in response to the myostatin treatment by +280% (p < 0.001), +140% (p < 0.01), and +176% (p < 0.001), respectively (Fig. 1B). Myostain increased sclerostin protein levels by +71% (p < 0.001) (Fig. 1C). Thus, myostain positively modulates the expression of sclerostin at transcriptional and translational levels in Ocy454 cells.
Figure 1.
Effects of myostatin on SOST, DKK1, RANKL, and OPG mRNA and/or protein expression in Ocy454 cells. A, a schematic diagram illustrating experimental procedures employed. B and C, Ocy454 cells were differentiated for 12 days and then treated with 100 ng/ml myostatin or vehicle for 48 h. B, levels of SOST, DKK1, RANKL, and OPG mRNA were determined by real-time PCR. C, levels of SOST protein were determined by Western blotting. Images shown in C (a) are representative of Western blot analysis. Data shown are the mean values ± S.E. for three separate determinations. **, p < 0.01; ***, p < 0.001.
Myostatin inhibited expression of miR-218 in Ocy454 parent cells and released exosomes
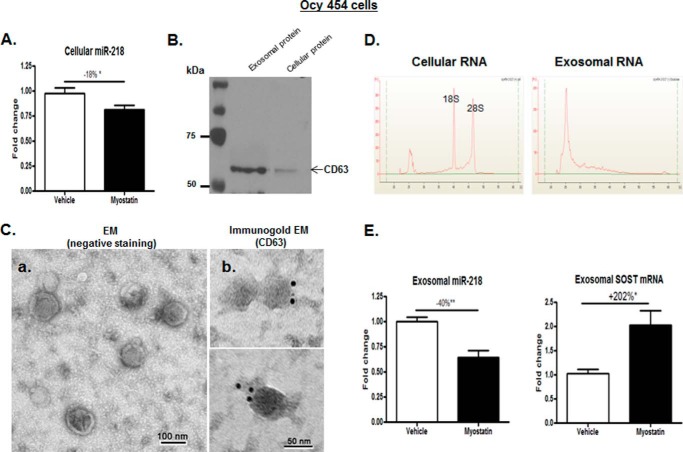
The simultaneous, coordinated up-regulation in osteocytes by myostatin of SOST and DKK1, Wnt inhibitors, suggests that a master regulator might be responsible. Because the miR-218 gene stimulates the Wnt pathway and promotes the differentiation of undifferentiated bone marrow cells by the inactivation of SOST and DKK2 during osteoblast differentiation (40), levels of miR-218 gene in Ocy454 cell that are altered in response to the myostatin were investigated. Interestingly, the expression of the miR-218 in myostatin-treated Ocy454 cells displayed a significant decrease by −18% (p < 0.05) compared with that in cells treated with vehicle (Fig. 2A). As such, myostatin appears to suppress the level of expression of the miR-218 gene, resulting in an increase in osteocyte SOST gene expression.
Figure 2.
Myostatin inhibited miR-218 expression in Ocy454 parent cells and their exosomes. As shown in Fig. 1A, Ocy454 cells were differentiated for 12 days and then treated with 100 ng/ml myostatin or vehicle for 48 h followed by extraction of total RNA from Ocy454 parent cells or the isolation of exosomes released form Cyc454 cells. A, levels of cellular miR-218 in Ocy454 parent cells were determined by real-time PCR. B, the Western blot showed that CD63 proteins are enriched in isolated exosome compared with that in cellular protein lysate. C, electron microscopy analysis of exosomes; a, exosomes were stained with uranyl acetate (EM (negative staining)); b, exosomes were labeled with 10-nm immunogold using antibody against exosomal membrane marker CD63 and stained with uranyl acetate (Immunogold EM (CD63)). Exosome morphology was then visualized using electron microscope Hitachi H7000. D, isolated cellular or exosomal RNA was analyzed using a Bioanalyzer. The results showed the absence of the ribosomal RNA peaks (18S and 28S rRNA) in exosomal RNA compared with that of cellular RNA. E, levels of miR-218 and SOST mRNA in exosomes produced by Ocy454 cells were determined by real-time PCR. Data shown are mean values ± S.E. for three separate determinations. *, p < 0.05 and **, p < 0.01.
Exosomes containing miRNAs are thought to be critical for cell-cell communication. Thus, studying the osteocyte-derived exosomal miRNA can indicate what cellular signals from the osteocytic parent cells send to the neighboring cells, such as osteoblasts and osteoclasts. Therefore, levels of osteocyte-derived exosomal miR-218 gene expression were examined. Ocy454 cells were treated with 100 ng/ml myostatin protein or vehicle for 48 h, as performed in the previous experiment, and exosomes released from cultured medium of the Ocy454 cells were then extracted by sequential centrifugation.
Serial assays were performed to confirm that the purified osteocyte-derived products were indeed exosomes. CD63 is a known protein marker of exosomes that is present on the surface of the exosome membrane (37, 51, 52). Western blot analysis revealed that levels of CD63 protein in exosomal fractions is 3–4-fold higher than those in cellular fraction (the parent cells) (Fig. 2B), suggesting an enrichment of CD63 protein expression in the isolated exosomes. Electron microscopy was used to examine morphology and structure of the exosomes obtained, which exhibited a typical morphology: round, roughly 50–100 nm in diameter, and cup-shaped (Fig. 2C). In parallel, the isolated exosomes were positively stained by immunogold using an antibody against CD63 (Fig. 2C). Additionally, cellular and exosomal RNA profiles were examined by the Agilent Bioanalyzer. The electropherograms of exosomal RNA had a distinct RNA profile compared with cellular RNA; the exosomal profile had significantly diminished 18S and 28S ribosomal RNA, indicating that purified products were indeed exosomes and not integral components of the parent cells (Fig. 2D).
Myostatin led to a reduction in levels of osteocyte-derived exosomal miR-218 in a time-dependent manner (supplemental Fig. 1). Consistent with the changes in the parent cells, exosomes obtained from Ocy454 cells after myostatin treatment for 48 h altered expression of miR-218 (−40%; p < 0.01) and SOST genes (+202% p < 0.05) compared with those from control cells treated with vehicle (Fig. 2E).
The myostatin-modified osteocytic exosomes inhibited osteoblastic differentiation in MC3T3 cells
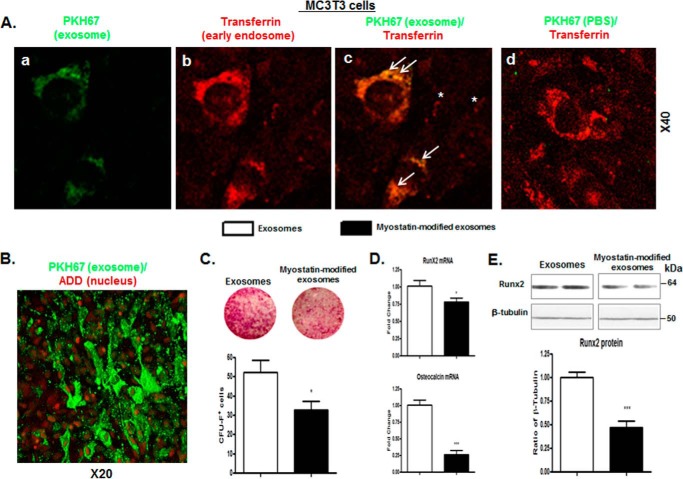
Whether osteocytic exosomes can directly influence differentiation of osteoblasts was investigated. The question was first addressed as to whether osteocyte-derived exosomes can be taken up by recipient cells-ostoblastic MC3T3 cells. After pretreatment with either myostatin or vehicle for 48 h, osteocyte-derived exosomes were isolated and labeled with PKH67 dye (green) and then added to cultures of MC3T3 cells for 30 min. In parallel, MC3T3 cells were preincubated with Texas Red-transferrin (red) to label early endosomes. Afterward, the MC3T3 cells were co-cultured with the PKH67-labeled exosomes or the PKH67-PBS control (containing no exosomes); the uptake was visualized by fluorescence microscopy. Confocal microscopy image showed evident uptake of the exosomes into early endosomes of MC3T3 cells as indicated by an increase of fluorescent intensity for PKH67 (Fig. 3, A, a–c) compared with MC3T3 cells co-cultured with the PKH67-PBS control (Fig. 3A, d). As early as 5 min after incubation, PKH67 was detected in early endosomes, suggesting a rapid uptake (supplemental Fig. 2). Internalization of exosomes is primarily an event in cytoplasm, as manifested by a distinct pattern of PKH67 staining in cytoplasm, which is in contrast to 7-aminoactinomycin D (7-ADD) staining in nuclei.
Figure 3.
The myostatin-modified osteocytic exosomes were internalized into MC3T3 cells, inhibiting osteoblastic differentiation. As shown in Fig. 1A, Ocy454 cells were differentiated for 12 days and were treated with 100 ng/ml myostatin or vehicle for 48 h followed by the isolation of exosomes that are released form Cyc454 cells. MC3T3 cells were then co-cultured with the myostatin-modified osteocytic exosomes for 48 h. A and B, uptake of osteocytic exosomes by MC3T3 cells. 10 μg of the PKH67-labeled osteocytic exosomes or a PKH67-PBS control were added into per 75,000 MC3T3 cells and incubated at 37 °C for 30 min. The uptake of the fluorescently labeled exosomes by MC3T3 cells was detected with confocal fluorescence microscopy. PKH67 (green) was used to label the exosomes. Texas Red-transferrin (red, in A) or 7-AAD (red, in B) was used to detect the early endosomes and the nucleus (red) of the MC3T3 (red), respectively. PKH67 exosomes (green) were rapidly internalized into early endosomes labeled with Texas Red-transferring (yellow is indicative of co-localization of green and red). The arrows indicate co-localization; the asterisks indicate no co-localization. C–E, co-culturing myostatin-modified osteocytic exosomes with MC3T3 cells inhibits osteoblastic differentiation. C, representative images showing alkaline phosphatase staining (CFU-F) of cultures of MC3T3 cells and cell counts of alkaline phosphatase-positive cells. D, changes in mRNA levels of Runx2 and osteocalcin in MC3T3 cells were determined by real-time PCR. E, quantification of Runx2 protein content in cell lysate from MC3T3 cells assessed by immunoblot analysis (inset). Data are expressed as the mean ± S.E. for three separate determinations. *, p < 0.05; ***, p < 0.001 versus indicated group.
Using the same co-culture approach, the effect of myostatin-modified osteocytic exosomes on the differentiation of MC3T3 osteoblastic precursor cells in the presence of ascorbic acid and β-glycerophosphate was tested. The degree of osteoblastogenic differentiation of MC3T3 cells was determined by counting the number of cells that were positively stained for CFU-F; expression levels of pro-differentiation factors Runx2 and osteocalcin were also examined. A 3-day incubation of MC3T3 with exosomes isolated from osteocytes that were pretreated with myostatin (100 ng/ml) significantly reduced the number of CFU-F-positive-staining colonies (Fig. 3D, p < 0.05) and down-regulated Runx2 (p < 0.05) and osteocalcin (p < 0.01) mRNA expression (Fig. 3E) as well as Runx2 protein (Fig. 3F, p < 0.001) at day 10 compared with those of MC3T3 cells when incubated with control osteocytic exosomes.
Down-regulation of Wnt signaling contributed to the inhibition of osteoblastic differentiation by the myostatin-modified osteocytic exosomes
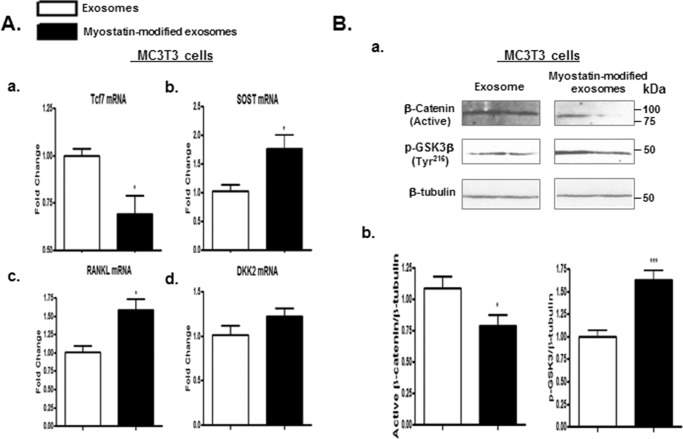
The molecular basis through which osteocytic exosomes mediate osteoblast differentiation was investigated. The effects of osteocytic exosomes on the Wnt/β-catenin signaling pathway, that is an important cascade controlling osteoblast activity, bone formation, and skeletal adaptation, were examined. Myostatin-modified osteocytic exosomes robustly reduced mRNA levels of activated Wnt signaling Tcf7 (Fig. 4A, a, p < 0.05) while significantly increasing mRNA expression of other Wnt-related genes SOST (p < 0.05) and RANKL (p < 0.05) (Fig. 4A, b and c) compared with those in control MC3T3 cell cultures. Levels of active β-catenin protein were examined using antibodies against active β-catenin (53), which is dephosphorylated at the Ser37 and Thr41 sites. Western blot studies revealed that levels of activated β-catenin protein were significantly reduced by myostatin-modified osteocytic exosomes (Fig. 4B, a and b; p < 0.05).
Figure 4.
Co-culturing MC3T3 cells with the myostatin-modified osteocyte-derived exosomes inactivated Wnt signaling. As shown in Fig. 1A, Ocy454 cells were differentiated for 12 days and were treated with 100 ng/ml myostatin or vehicle for 48 h followed by the isolation of exosomes released form Ocy454 cells. MC3T3 cells were then co-cultured with the myostatin-modified osteocytic exosomes for 48 h. A, changes in mRNA levels of Wnt signaling-related genes Tcf7, SOST, RANKL, and DKK2 in MC3T3 cells were determined by real-time PCR. B, Western blotting was performed on total proteins from MC3T3 cells. Images shown in (Ba) were representative Western blotting. Bb, blots in (Ba) were quantified by scanning densitometry and normalized relative to β-tubulin. Data are expressed as the mean ± S.E. for three separate determinations. *, p < 0.05; ***, p < 0.001 versus the indicated group.
Because intracellular β-catenin fate is determined by GSK3 enzyme-mediated proteasome ubiquitination (54), the effect of myostatin-modified osteocytic exosomes on GSK3 activity by measuring the protein levels of phospho-GSK3β at Tyr216, which is an activated phosphorylation site, was tested. An increase in phosphorylation of GSK-3 β at Tyr216 was observed in the MC3T3 cells treated with myostatin-modified osteocytic exosomes, indicating an activation of GSK3 activity (p < 0.001) that is associated with inactivation of β-catenin activity (Fig. 4B, a and b). Collectively, the cellular and molecular evidence indicates that myostatin-modified osteocytic exosomes down-regulate Wnt signaling and, as a result, inhibit osteoblastic differentiation and activity.
Expression of exogenous miR-218 reversed osteocytic exosome-mediated effects in osteoblasts
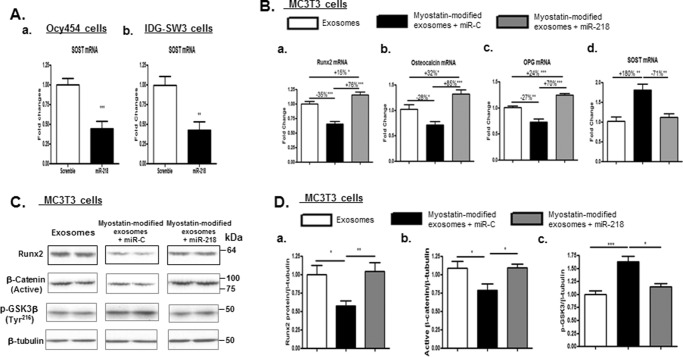
As previously demonstrated, the findings that myostatin inhibits expression of osteocyte-derived exosomal miR-218 (Fig. 2C) and that co-culturing myostatin-modified osteocytic exosomes with MC3TC cells suppresses differentiation of osteoblasts (Fig. 3) led us to postulate that osteocyte-derived exosomal miR-218 may play a key role in the regulation of osteoblast activity. To test this possibility, the effects of expression of exogenous miR-218 on osteocytic exosome-mediated osteoblast differentiation were evaluated in MC3T3 cells. Consistent with the inhibitory effects of miR-218 in SOST gene in MC3T3 cells (40), lentiviral expression of exogenous miR-218 dramatically suppressed mRNA expression of SOST by 2-fold in 2 osteocyte-like cell lines, Ocy454 cells and IDG-SW3 cells, confirming the efficacy of viral miR-218 on inhibiting SOST gene expression (Fig. 5A, a and b).
Figure 5.
Expression of exogenous miR-218 reversed the inhibitory effects of myostatin-modified osteocyte-derived exosomes on osteoblastic differentiation. As shown in Fig. 1A, Ocy454 cells were differentiated for 12 days and were treated with 100 ng/ml myostatin or vehicle for 48 h followed by the isolation of exosomes released form Cyc454 cells. MC3T3 cells were then co-cultured with the myostatin-modified osteocytic exosomes in the presence of lentivirus-mediated exogenous expression of miR-218 or scramble control plasmid (miR-C) for 48 h. Total RNA or protein was isolated on day 3 and subjected to real-time PCR or Western blot analysis, respectively. A, efficacy of exogenous expression of miR-218 on inhibiting SOST mRNA expression in Ocy454 cells and IDG-SW3 cells. B, changes in mRNA levels of RunX2, osteocalcin, and OPG in MC3T3 cells were determined by real-time PCR. C, Western blotting was performed on total proteins from MC3T3 cells. Images shown were representative Western blottings. D, blots in C were quantified by scanning densitometry and normalized relative to β-tubulin. Data are expressed as the mean ± S.E. for three separate determinations. *, p < 0.05; **, p < 0.01, ***, p < 0.001 versus indicated group.
Consistent with our previous observation, myostatin-modified osteocytic exosomes demonstrated a similar inhibition of Runx2 (−35%, p < 0.001) and osteocalcin (−28%, p < 0.05) mRNA expression and up-regulation of SOST mRNA (+180%, p < 0.01), whereas viral expression of miR-218 markedly increased Runx2, osteocalcin, and OPG mRNA expression and decreased SOST mRNA as well as increasing levels of Runx2 protein by +76% (p < 0.001), +85% (p < 0.001), +70% (p < 0.001), −71% (p < 0.01), and +45% (p < 0.05) when compared with those of miR-control (Fig. 5B). The extent of these changes is all above or close to the levels of those of mRNA/protein observed in control cells, indicating a complete reversal of inhibitory effects on osteoblast differentiation that resulted from incubation with myostatin-modified osteocytic exosomes. Moreover, viral expression of miR-218 completely reversed the reduction of active β-catenin protein expression (p < 0.05) in MC3T3 cells induced by the treatment of myostatin-modified osteocytic exosomes (Fig. 5, C and D). The aforementioned effects were concurrent with inactivation of GSK3 activity, as reflected by decreased protein levels of phospho-GSK3β at Tyr216 (p < 0.05) (Fig. 5, C and D).
The myostatin-modified osteocytic exosomes had no direct effect on osteoclastic differentiation
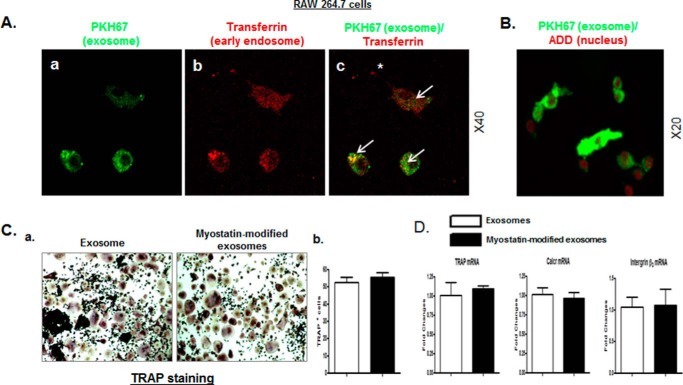
To test whether myostatin-modified osteocytic exosomes play a role in osteoclastic differentiation, the ability of osteoclasts to take up exosomes was investigated. Similar to the uptake of osteocytic exosomes by osteoblasts (Fig. 3, B and C), RAW 264.7 cells were able to rapidly take up the osteocytic exosomes into early endosomes at 5 and 30 min after co-culturing (supplemental Fig. 3 and Fig. 6A). The internalization of exosomes in RAW 264.7 cells occurred predominantly in cytoplasm (Fig. 6B).
Figure 6.
The myostatin-modified osteocyte-derived exosomes have no direct effect on osteoclastic differentiation. As shown in Fig. 1A, Ocy454 cells were differentiated for 12 days and were treated with 100 ng/ml myostatin or vehicle for 48 h followed by the isolation of exosomes released form Cyc454 cells. RAW 264.7 cells were then co-cultured with the myostatin-modified osteocytic exosomes for 48 h. A and B, uptake of osteocytic exosomes by RAW 264.7 cells was examined by using the approaches as described in Fig. 3, A and B. The arrows indicate co-localization; the asterisk indicates no co-localization. C and D, co-culture of myostatin-modified osteocytic exosomes with RAW 264.7 cells had no effect on osteoclastic differentiation. C, representative images showing TRAP staining of cultures of RAW 264.7 cells and cell counts of TRAP-positive cells. D, changes in mRNA levels of TRAP, calcitonin receptor (Calcr), and integrin β3 in RAW 264.7 cells were determined by real-time PCR. Data are expressed as the mean ± S.E. for three separate determinations.
After preculture with RANKL for 4 days, RAW 264.7 cells were further co-cultured with myostatin-modified osteocytic exosomes or osteocytic exosomes without any modification for two additional days. Unlike to the effect observed in the osteoblast, myostatin-modified osteocytic exosomes did not affect the osteoclastogenic differentiation as indicated by no change in the number of TRAP-positive colonies by TRAP staining (Fig. 6C). This was associated with no changes in the RANKL-induced stimulation of osteoclast markers of TRAP, calcitonin receptor (Calcr), and integrin β3 as determined by real-time PCR (Fig. 6D). Thus, in contrast to the negative biological impacts on osteoblasts, the myostatin-modified osteocytic exosomes do not appear to exert any direct effect on the differentiation of osteoclasts.
Discussion
Using the myokine myostatin as a probe, this study investigated how muscle-released products interact with bone cells to influence their function. One of our goals was to provide insight into the interaction of muscle and bone to coordinate cellular and molecular function, with resultant changes in bone mass. Here, we report for the first time that in response to myostatin, osteocytes react to produce greater amounts of SOST, RANKL, and DKK1, each of the products an important regulator in bone remodeling. Second, our study establishes myostatin as the first signaling product released by muscle that is able to directly target osteocyte-derived exosomes to regulate content of miR-218. Specifically, myostatin inhibits expression of miR-218 in the exosomes released from the osteocytes; the osteocyte-derived exosomes that contain reduced levels of miR-218 may then be transferred to osteoblasts to inhibit osteoblastic differentiation and, perhaps, bone formation. Furthermore, our data suggest that the inhibitory effect of myostatin-mediated suppression of osteocytic exosomal miR-218 on osteoblast function is associated with the down-regulation of the canonical Wnt signaling pathway. Of note, the suppressive effect of myostatin-modified osteocytic exosomes on osteoblast differentiation is completely counteracted by the expression of exogenous miR-218 through the down-regulation of SOST. These findings suggest a novel mechanism underlying muscle-bone communication to maintain homeostasis of the local musculoskeletal environment via osteocytes, at least in part, by the generation and release of miRNA inside of exosomes.
Our findings demonstrate that the treatment of osteocyte cells with myostatin leads to significant increases in the expression of SOST and DKK1, two Wnt signaling inhibitors, suggesting that myostatin may have an inhibitory impact on osteoblast activity and bone formation indirectly through products released by osteocytes. Whether there is a direct effect of myostatin on osteoblasts is unclear (28, 29). Literature regarding whether other muscle-secreted factors have direct effects on osteoblasts remains controversial. A recent study showed that direct incubation of preosteoblastic MC3T3 cells with conditioned medium from cultures of contracted muscle cells had no effect on proliferation, metabolic activity, or mineralization of osteoblasts,4 implying the absence of direct effect of muscle-produced products on osteoblasts in response to an exercise regimen. In contrast, irisin, a myokine generated by skeletal muscle during exercise, has been reported to directly enhance osteoblast differentiation, promote bone formation, and increase bone mass (25). Nonetheless, our evidence provides novel insight into how myostatin indirectly regulates bone formation via osteocytes. In addition, we have shown that myostatin significantly increases expression of RANKL in osteocytes, strongly suggesting that myostatin may indirectly promote osteoclast differentiation, activity, and viability, presumably associated with enhanced bone resorption.
Our findings addressing the role of myostatin on osteoblast activity indirectly through osteocytes are consistent with a recent observation in which factors secreted from contracted muscle had indirect effects on osteoblasts through the stimulation of prostaglandin E2 production and reduction of sclerostin in osteocytes; however, it remains largely unknown what factors are exactly released from the contracted muscle.4 In this regard our study establishes myostatin as the first signaling product released by muscle that is able to directly target osteocytes and osteocyte-derived exosomal miRNA and, thereby, influence osteoblast function. Currently, it is unclear the manner in which myostatin is transported to the osteocyte that is deeply embedded in the bone matrix. Myostatin may be released into the general circulation to be presented to the dendritic processes of osteocyte protruding into vascular channels for cellular uptake (57),5 but certainly other mechanisms may exist to facilitate myostatin transport to the osteocyte. For example, the transport of myostatin may occur extracellularly through the lacunar-canalicular system within the bone matrix.
Among other mechanisms, expression of SOST has been shown to be controlled by miR-218, where miR-218 acts as an inhibitor of SOST and a potent activator of the Wnt signaling pathway, thus contributing to osteoblastogenesis (40). Excitingly, we found that in cultured osteocytes, the expression of miR-218 was significantly decreased by −18% upon challenge with myostatin. The myostatin-mediated down-regulation of miR-218 was associated with the increased levels of SOST mRNA and protein, suggesting that myostatin acts through miR-218 to influence SOST expression in osteocytes. Most strikingly, exosomal miR-218 shed from osteocytes is significantly down-regulated by myostatin by −40%, a change higher than that which occurs in the donor cells. Thus, myostain appears to manipulate miR-218 and potentially other miRNA gene profiles, not only within the osteocyte but also in the extracellular vesicles released. Future work is required to investigate how myostatin down-regulates miR-218 at transcriptional and/or post-transcriptional levels.
The release of osteocytic microvesicles was observed in a recent study using live-cell imaging in a mouse model expressing membrane-bound GFP specifically in osteocytes;5 however, the biological features of osteocyte-derived microvesicles, particularly exosomes, remained ill defined. In the present study, electron microscopy of the pellet obtained in the last centrifugation by a standard differential centrifugation revealed that Ocy454 cells secrete exosome-like microvesicles into the culture media. These vesicles are 50–100 nm in diameter, have a cup-shaped appearance, and are positively stained with immunogold-labeled CD63 antibody, consistent with previous descriptions of exosomes (36–38, 52, 58–60). Additional immune-blotting approaches confirmed that CD63 protein expression is enriched in the isolated exosomes. RNA profiles from cellular and exosomal fractions were compared using Agilent Bioanalyzer, indicating the absence of cytoplasmic contamination from donor cells and the presence of predominant amounts of small RNA (miRNA) species in the exosomes. These unique features are strongly confirmatory that the microvesicles released from Ocy454 cells in this study are indeed exosomes. To our knowledge our work is the first to systemically characterize osteocyte-derived exosomes at morphological, cellular, and molecular levels and, furthermore, to elucidate how miRNA expression (e.g. miR-218) of osteocytic-derived exosomes appears to be influenced by local environmental factors.
In this study the exosome-tracking experiments demonstrate that osteocyte-derived exosomes could be rapidly taken up by osteoblasts. When co-cultured with osteocyte-derived exosomes that were pretreated with myostatin, osteoblastic differentiation of MC3T3 cells was inhibited, including reductions in the number of CFU-F-positive cells and decreased levels of pro-differentiation factors Runx2 mRNA and protein as well as osteocalcin mRNA. These changes were accompanied by the inactivation of Wnt signaling activity, as evidenced by decreased Tcf7 and up-regulated SOST and RANKL as well as diminished levels of active β-catenin and increased phospho-GSK3β at Tyr216. It has been reported that in addition to targeting SOST, osteoblast-derived miR-218 also inhibits DKK2 and sFRP2 in cultured MC3T3 cells (40). It is unclear why the expression of DKK2 and sFRP2 were not altered in osteocytes by myostatin or in MC3T3 cells by myostatin-modified osteocytic exosomes (data not shown). It is possible that differences in other factors (e.g. other miRNAs, but not miR-218) targeting DKK2 and sFRP2 or other unknown effects of myostatin might have contributed to the distinct outcomes in two types of bone cells in their response to miR-218. Collectively, our data suggest that the osteocyte-derived exosomes containing reduced levels of miR-218 upon myostatin treatment could be transferred to osteoblasts to inhibit osteoblastic differentiation and perhaps bone formation, most likely through the down-regulation of canonical Wnt signaling pathway. Hence, this work represents the first study to characterize not only the uptake of osteocytic exosomes by osteoblast cells but also the consequence of such an event; that is, the modulation of osteoblast activity.
Considering the role of osteocyte-derived exosomal miR-218 in the regulation of osteoblastic differentiation, overexpression of this miRNA in osteoblasts is expected to exert beneficial effects on the differentiation of osteoblastic cells. Thus, the therapeutic effect of osteoblasts-targeted miR-218 activation by transducing lentiviral miR-218 was considered. Myostatin-modified osteocytic exosomes significantly down-regulated osteoblast activity in MC3T3 cells, and such an inhibitory effect was prevented by the forced expression of miR-218, through a mechanism involving miR-218-mediated inhibition of SOST. Down-regulation of osteoblast activity occurred in association with the resurgence of Wnt signaling activity, as reflected by up-regulated levels of active β-catenin and increased phospho- GSK3β at Tyr216. These findings further indicate that miR-218 plays a key role in myostatin-modified osteocytic exosomes in the control of osteoblastic differentiation through regulation of SOST. This mechanism suggests that miR-218 may be a therapeutic target and that the enhancement of miR-218 expression in osteocytes may promote osteoblast activity and, ultimately, increase bone formation.
Despite the profound inhibition of osteoblastic differentiation, the uptake of myostatin-modified osteocytic exosomes by osteoclasts has no direct effect on osteoclast activity. There was no change in osteoclastogenic differentiation as determined by TRAP staining and no changes in gene expressions of osteoclast markers TRAP, calcitonin receptor, and integrin β3. This effect is partially explained by the absence of change in RANKL mRNA expression in the osteocyte-derived exosomes in response to myostatin treatment (supplemental Fig. 4). These findings taken together with a recent observation that myostatin did not directly regulate expression of RANKL and M-CSF in mouse synovial fibroblasts and osteoblasts (61) suggests that the effect of myostatin on osteoclast differentiation, if any, should be attributable to either the paracrine action from other sources or the direct control over osteoclasts in an osteoblast or osteocytic exosome-independent mechanism. The former possibility was supported by our findings. First, myostatin significantly increased expression of RANKL mRNA in osteocytes and, presumably, increased soluble RANKL protein that was able to traffic to osteoclasts and indirectly promote osteoclast activity. Another possibility is that the uptake of the myostatin-modified osteocytic exosomes by osteoblasts greatly increased expression of RANKL (Fig. 4), thereby indirectly affecting osteoclast activity. We note that in the experiments presented here, osteocytes remain viable when challenged with 100 ng/ml myostatin, as assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (data not shown). Therefore, this study does not exclude the possibility that osteocytic exosomes might have a direct role on osteoclasts in other pathophysiological conditions. Consistent with this notion, osteocytic apoptotic bodies released during apoptosis have been shown to promote osteoclastogenesis, whereas factors present in the soluble fraction derived from both necrotic and healthy osteocytes were incapable of promoting osteoclastogenesis (62). In addition, myostatin has been recently reported to strongly accelerate RANKL-mediated osteoclast formation in in vitro cultured bone marrow-derived macrophages by a mechanism through Smad2-dependent regulation of nuclear factor of activated T-cells (NFATC1) (Fig. 7), suggesting a direct effect of myostatin on osteoclasts (61). Taken together, the findings from the present study define a direct action of myostatin on osteocytes to promote RANKL production as a novel mechanism of indirectly controlling osteoclast activity and bone resorption in an osteoblast or osteocytic exosomes-independent manner.
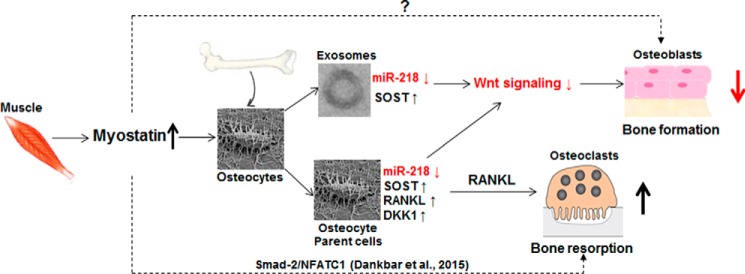
Figure 7.
A schema of the putative mechanisms by which myostatin may influence the function of bone cells. Myostatin influences osteoblastic differentiation through the osteocyte and its release of exosomal miR-218 in a mechanism that involves the activation of SOST and its inhibition of the Wnt signaling pathway. Additionally, myostatin promotes RANKL production in osteocytes and, in turn, has the potential to increase osteoclast differentiation, activity, and viability. We note that myostatin has been recently reported to directly accelerate RANKL-mediated osteoclast formation by a mechanism through transcription factor Smad2-dependent regulation of NFATC1 (nuclear factor of activated T-cells; Ref. 61). Whether there is a direct effect of myostatin on osteoblasts remains to be determined.
In summary, this study has defined novel mechanisms through which muscle exerts regulation of bone cell function (Fig. 7). Our findings provide the first in vitro evidence that osteocyte-derived exosomal miRNA works as a signal complex by which, in response to a message from muscle (e.g. myostatin), bone cells might respond and communicate to ensure that the mass and function of bone is regulated appropriately in concert with that of muscle. Future study will define whether the putative muscle-bone interaction that has been suggested in the present study may actually exist in the physiological environment. Our study supports the notion that inhibition of myostatin serves a dual benefit in the protection of muscle and bone against loss in osteopenia and sarcopenia (28). The regulation of the miR-218 gene in osteocytes emerges as an interesting research direction because miR-218 responds to a muscle-released hormone and, in turn, influences osteoblast activity by modulation of SOST and Wnt signaling, possibly among others signaling pathways. Thus, the manipulation of miR-218 in osteocytes can be considered as a future therapeutic strategy for the treatment of bone loss associated with the age-related sarcopenia, menopause, and immobilization (e.g. spinal cord injury, stroke, and other conditions associated with restricted weight-bearing activity). In a future study it will be relevant to determine whether other myokines, such as irisin and IL-6, have similar direct effects to that of myostatin on the osteocyte and its exosomes to influence the osteoblast and/or osteoclast. In addition, future work to characterize changes in osteocyte-derived exosomal miRNA profiles in response to muscle hormones will identify additional miRNA candidate genes in the regulation of function of the osteocyte with consequences to bone homeostasis.
Experimental procedures
Cell culture and treatment
Mouse osteocytic Ocy454 cells were maintained as described previously (63). To differentiate osteocytic phenotype, Ocy454 cells were seeded in a 10-cm culture plate at a density of 1 × 106 and cultured at 33 °C for 48 h. Cells were then cultured in 37 °C for an additional 10 days. At day 12, Ocy454 were treated with myostatin (100 ng/ml; R&D Systems, 788-G8-010) or PBS (vehicle) for 48 h followed by the isolation of exosome in conditioned medium, cellular total RNA, and proteins (as described in greater detail below). The dose of myostatin follows that utilized by previous studies (29, 64). IDG-SW3 cells were cultured as described previously (65, 66). For experimental treatment and co-culturing studies, FBS was depleted of exosomes by ultracentrifugation at 100,000 × g for 4 h at 4 °C followed by filtration through a 0.22-μm filter (Millipore) to sterilize.
For osteoblastogenesis, 2.5 × 104 MC3T3-E1 (subclone 4) cells, osteoblastic precursor cells were seeded and cultured in 12-well tissue culture plates as described in greater detail in the supplemental Materials and Methods At day 8, MC3T3 cells were co-cultured with myostatin-modified or control exosomes for 48 h followed by staining multicellular fibroblastoid colonies (CFU-F) using an alkaline phosphatase kit per manufacturer's recommendations (Sigma). For osteoclastogenesis, 2 × 103 RAW264.7 cells were seeded and cultured in 96-well tissue culture plates as described in greater detail in the supplemental Materials and Methods. At day 5, RAW264.7 cells were co-cultured with myostatin-modified or control exosomes for 48 h followed by TRAP staining using a kit (Sigma) per the manufacturer's instruction. A parallel set of osteoblasts or osteoclast cultures was used to extract total RNA or proteins, respectively.
For lentivirus packaging, 293TN cells were transfected with Lenti-miR218-2 (PMIRH218–2PA-1) or scramble control plasmid (miR-C, PMIRH000PA-1) plus the pPACKH1TM Packaging Plasmid mix (System Biosciences) using the Lipofectamine® LTX with Plus Reagent (Thermo Fisher Scientific). Supernatant was collected and concentrated 48 h post transfection. For viral transduction, targeted cells were infected with concentrated miR-218 or scramble lentiviral supernatant (multiplicity of infection 4) in the presence of 5 μg/ml Polybrene. Biological assays were performed 48 h post transduction. The methods are described in greater detail in the supplemental Materials and Methods.
Isolation of exosomes
A sequential centrifugation procedure was used to isolate exosomes as described previously (38, 59). Briefly, conditioned cell culture medium was prepurified by centrifugation at 2000 × g for 30 min at 4 °C to remove floating cells, and the supernatant was collected and filtered through a 0.2-μm syringe filter (Millipore) to remove cell debris. Exosomes were pelleted in a two-step ultracentrifugation using a Ti 50.2 rotor (Beckman) at 100,000 × g for 70 min at 4 °C with 1 PBS wash between 2 ultracentrifugations. The pelted exosomes were resuspended in PBS or in lysis buffer and stored at −80 °C for experimental analysis.
Western blot analysis
Proteins from cultured cells and purified exosomes were separated on a denatured SDS-polyacrylamide gel and were immunologically detected as described in greater detail in the supplemental Materials and Methods.
Electron microscopy
Exosomes from Ocy454 cells were fixed in 2.5% glutaraldehyde, loaded onto Formvar carbon-coated copper grids (Electron Microscopy Sciences, FCF-200-Cu), washed, and followed by negative staining with 2% uranyl acetate. The exosomes on grids were viewed using a Hitachi H7000 electron microscope at 75 kV. The electron microscope was equipped with an AMT Advantage HS digital camera (Danvers), and micrographs were recorded digitally as described previously (60). In a parallel study, the same procedures were performed except that before negative staining with uranyl acetate, the fixed exosomes in the copper grids were immunolabeled with anti-CD63 antibody followed by a 10-nm gold-labeled secondary antibody (Sigma). The methods are described in greater detail in supplemental Materials and Methods.
Visualization of exosome uptake
Isolated osteocytic exosomes were labeled with PKH67 Green Fluorescent Cell Linker Kit for General Cell Membrane Labeling (Sigma) according to the manufacturer's protocol (67). Before co-culture, MC3T3 cells or RAW264.7 cells were preincubated with 25 μg/ml Texas Red-transferrin (Life Technologies) for 30 min at 37 °C to label early endosomes (68). MC3T3 cells or RAW264.7 cells were then co-cultured with 10 μg of the PKH67-labeled exosomes or the PKH67-PBS control and incubated for 5 or 30 min at 37 °C. Uptake of exosomes by these cells was stopped by washing with 0.1% sodium azide PBS followed by fixation in 4% formaldehyde, mounting to Vectashield (Vector Laboratories), and visualizing with a fluorescence microscope (Zeiss Axioplan 2, Carl Zeiss). In a parallel study, instead of using Texas Red-transferrin to label early endosomes, MC3T3 or RAW264.7 cells were directly incubated with PKH67-labeled exosomes or the PKH67-PBS control. The binding of the exosomes to these cells was then visualized with a fluorescence microscope with 3% 7-ADD (BD Biosciences) to label nuclei. The methods are described in greater detail in the supplemental Materials and Methods.
RNA isolation and analysis of gene expression by quantitative PCR
Total RNA was isolated using the mirVana miRNA isolation kit (Invitrogen) according to the manufacturer's instructions. The quality of the RNA samples was assessed with an Agilent Bioanalyzer. For determination of mRNA levels,1 μg of total RNA was used to generate a cDNA library with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). For determination of miRNA levels, cDNA was synthesized from 1 μg of total RNA using the specific primers provided with the TaqMan miRNA assays (Life Technology) according to the protocol provided by the manufacturer. After dilution of cDNA libraries into water, levels of specific mRNAs or miRNAs were determined by Taqman Assay On Demand probesets (Applied Biosystems) or SYBR quantitative PCR (StepOnePlus, Life Technologies) using an ABI 7500 real time PCR machine. Primer sequences are available upon request. Changes in gene expression were calculated using the 2−ΔΔCt method using 18S RNA or U6 snRNA for normalization (69). The methods are described in greater detail in the supplemental Materials and Methods.
Statistics
Data is expressed as the mean ± S.E.; the number of independent samples (n) is provided in the legend of each figure. The statistical significance of differences among means was tested using one-way analysis of variance and a Newman-Keuls post hoc test to determine the significance of differences between individual pairs of means using a p value of 0.05 as the cutoff for significance. Statistical calculations were performed using Prism 4.0c (GraphPad Software, La Jolla, CA).
Author contributions
W. Q., Y. Q., Y. W., P. D. P., L. F. B., and W. A. B. were responsible for study design and data analysis and interpretation. Y. Q., Y. P., W. Z., C. C., H. K.-R., and J. P. conducted the bone biology study. The manuscript was written by Y. Q. and W. Q. and was revised and approved by all authors. W. Q. takes responsibility for the integrity of the data analysis.
Supplementary Material
This work was supported by the Veterans Health Administration, Rehabilitation Research, and Development Service (Grants 5I01RX001313, I01 RX002089, and 5I01RX000687 to W. Q.; B9212-C and B2020-C to W. A. B.). This work was also supported by NIH-UH3AR059655 to P. D P. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Materials and Methods and Figs. 1–4.
P. Veno, M. Prideaux, V. Dusevich, L. Bonewald, and S. Dallas, unpublished data.
H. Kondo, N. Zhao, M. Prideaux, Y. Kitase, J. Vallejo, S. Dallas, M. Brotto, and L. Bonewald, unpublished data.
P. Veno, M. Prideaux, V. Dusevich, L. Bonewald, S. Dallas, unpublished data.
- RANKL
- receptor activator of nuclear factor κB ligand
- miRNA
- microRNA
- sFRP2
- secreted frizzled-related protein 2
- OPG
- osteoprotegerin
- CFU-F
- colony forming unit-fibroblastic
- 7-ADD
- 7-aminoactinomycin D.
References
- 1. Schoenau E. (2005) From mechanostat theory to development of the “Functional Muscle-Bone-Unit.” J. Musculoskelet. Neuronal Interact. 5, 232–238 [PubMed] [Google Scholar]
- 2. Hamrick M. W. (2012) The skeletal muscle secretome: an emerging player in muscle-bone crosstalk. Bonekey Rep. 1, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloomfield S. A. (2010) Disuse osteopenia. Curr. Osteoporos. Rep. 8, 91–97 [DOI] [PubMed] [Google Scholar]
- 4. Karinkanta S., Piirtola M., Sievänen H., Uusi-Rasi K., and Kannus P. (2010) Physical therapy approaches to reduce fall and fracture risk among older adults. Nat. Rev. Endocrinol. 6, 396–407 [DOI] [PubMed] [Google Scholar]
- 5. Rittweger J., Beller G., Ehrig J., Jung C., Koch U., Ramolla J., Schmidt F., Newitt D., Majumdar S., Schiessl H., and Felsenberg D. (2000) Bone-muscle strength indices for the human lower leg. Bone 27, 319–326 [DOI] [PubMed] [Google Scholar]
- 6. Leigey D., Irrgang J., Francis K., Cohen P., and Wright V. (2009) Participation in high-impact sports predicts bone mineral density in senior Olympic athletes. Sports Health 1, 508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown J. C., Harhay M. O., and Harhay M. N. (2016) Sarcopenia and mortality among a population-based sample of community-dwelling older adults. J. Cachexia Sarcopenia Muscle 7, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clarke B. L., and Khosla S. (2010) Physiology of bone loss. Radiol. Clin. North Am. 48, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang S. D., Jiang L. S., and Dai L. Y. (2006) Mechanisms of osteoporosis in spinal cord injury. Clin. Endocrinol. (Oxf) 65, 555–565 [DOI] [PubMed] [Google Scholar]
- 10. Qin W., Bauman W. A., and Cardozo C. (2010) Bone and muscle loss after spinal cord injury: organ interactions. Ann. N.Y. Acad. Sci. 1211, 66–84 [DOI] [PubMed] [Google Scholar]
- 11. Qin W., Bauman W. A., and Cardozo C. P. (2010) Evolving concepts in neurogenic osteoporosis. Curr. Osteoporos. Rep. 8, 212–218 [DOI] [PubMed] [Google Scholar]
- 12. Bauman W. A., and Cardozo C. (2013) Immobilization Osteoporosis. In Osteoporosis Fourth Edition (Marcus R., Nelson D., and Rosen C. J., eds.) ch. 47, pp. 1139–1171, Academic Press, Orlanda, FL [Google Scholar]
- 13. Bauman W. A., Spungen A. M., Wang J., Pierson R. N. Jr., and Schwartz E. (1999) Continuous loss of bone during chronic immobilization: a monozygotic twin study. Osteoporos. Int. 10, 123–127 [DOI] [PubMed] [Google Scholar]
- 14. Bonewald L. F., and Johnson M. L. (2008) Osteocytes, mechanosensing and Wnt signaling. Bone 42, 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bonewald L. F. (2011) The amazing osteocyte. J. Bone Miner. Res. 26, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalajzic I., Matthews B. G., Torreggiani E., Harris M. A., Divieti Pajevic P., and Harris S. E. (2013) in vitro and in vivo approaches to study osteocyte biology. Bone 54, 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burra S., Nicolella D. P., Francis W. L., Freitas C. J., Mueschke N. J., Poole K., and Jiang J. X. (2010) Dendritic processes of osteocytes are mechanotransducers that induce the opening of hemichannels. Proc. Natl. Acad. Sci. U.S.A. 107, 13648–13653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batra N., Riquelme M. A., Burra S., Kar R., Gu S., and Jiang J. X. (2014) Direct regulation of osteocytic connexin 43 hemichannels through AKT kinase activated by mechanical stimulation. J. Biol. Chem. 289, 10582–10591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J. Q., Bonewald L. F., Kodama T., Wutz A., Wagner E. F., Penninger J. M., and Takayanagi H. (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17, 1231–1234 [DOI] [PubMed] [Google Scholar]
- 20. Xiong J., Onal M., Jilka R. L., Weinstein R. S., Manolagas S. C., and O'Brien C. A. (2011) Matrix-embedded cells control osteoclast formation. Nat. Med. 17, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wijenayaka A. R., Kogawa M., Lim H. P., Bonewald L. F., Findlay D. M., and Atkins G. J. (2011) Sclerostin stimulates osteocyte support of osteoclast activity by a RANKL-dependent pathway. PLoS ONE 6, e25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacobs C. R., Temiyasathit S., and Castillo A. B. (2010) Osteocyte mechanobiology and pericellular mechanics. Annu. Rev. Biomed. Eng. 12, 369–400 [DOI] [PubMed] [Google Scholar]
- 23. Hamrick M. W., McNeil P. L., and Patterson S. L. (2010) Role of muscle-derived growth factors in bone formation. J. Musculoskelet. Neuronal Interact. 10, 64–70 [PMC free article] [PubMed] [Google Scholar]
- 24. Colaianni G., Cuscito C., Mongelli T., Oranger A., Mori G., Brunetti G., Colucci S., Cinti S., and Grano M. (2014) Irisin enhances osteoblast differentiation in vitro. Int. J. Endocrinol. 2014, 902186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Colaianni G., Cuscito C., Mongelli T., Pignataro P., Buccoliero C., Liu P., Lu P., Sartini L., Di Comite M., Mori G., Di Benedetto A., Brunetti G., Yuen T., Sun L., Reseland J. E., et al. (2015) The myokine irisin increases cortical bone mass. Proc. Natl. Acad. Sci. U.S.A. 112, 12157–12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamrick M. W. (2011) A role for myokines in muscle-bone interactions. Exerc. Sport. Sci. Rev. 39, 43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McPherron A. C., Lawler A. M., and Lee S. J. (1997) Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 28. Elkasrawy M. N., and Hamrick M. W. (2010) Myostatin (GDF-8) as a key factor linking muscle mass and bone structure. J. Musculoskelet. Neuronal Interact. 10, 56–63 [PMC free article] [PubMed] [Google Scholar]
- 29. Bowser M., Herberg S., Arounleut P., Shi X., Fulzele S., Hill W. D., Isales C. M., and Hamrick M. W. (2013) Effects of the activin A-myostatin-follistatin system on aging bone and muscle progenitor cells. Exp. Gerontol. 48, 290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arounleut P., Bialek P., Liang L. F., Upadhyay S., Fulzele S., Johnson M., Elsalanty M., Isales C. M., and Hamrick M. W. (2013) A myostatin inhibitor (propeptide-Fc) increases muscle mass and muscle fiber size in aged mice but does not increase bone density or bone strength. Exp. Gerontol. 48, 898–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andrews N. A. (2013) The muscle-bone connection. IBMS boneKEy 10, 377, 10.1038/bonekey.2013.111 [DOI] [Google Scholar]
- 32. Kim J. S., Cross J. M., and Bamman M. M. (2005) Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am. J. Physiol. Endocrinol. Metab. 288, E1110–E1119 [DOI] [PubMed] [Google Scholar]
- 33. Buehring B., and Binkley N. (2013) Myostatin: the holy grail for muscle, bone, and fat? Curr. Osteoporos. Rep. 11, 407–414 [DOI] [PubMed] [Google Scholar]
- 34. Pearsall R. S., Canalis E., Cornwall-Brady M., Underwood K. W., Haigis B., Ucran J., Kumar R., Pobre E., Grinberg A., Werner E. D., Glatt V., Stadmeyer L., Smith D., Seehra J., and Bouxsein M. L. (2008) A soluble activin type IIA receptor induces bone formation and improves skeletal integrity. Proc. Natl. Acad. Sci. U.S.A. 105, 7082–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stoorvogel W., Kleijmeer M. J., Geuze H. J., and Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 36. van der Pol E., Böing A. N., Harrison P., Sturk A., and Nieuwland R. (2012) Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 64, 676–705 [DOI] [PubMed] [Google Scholar]
- 37. Mathivanan S., Ji H., and Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 38. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 39. Lian J. B., Stein G. S., van Wijnen A. J., Stein J. L., Hassan M. Q., Gaur T., and Zhang Y. (2012) MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 8, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hassan M. Q., Maeda Y., Taipaleenmaki H., Zhang W., Jafferji M., Gordon J. A., Li Z., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2012) miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 287, 42084–42092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li D., Liu J., Guo B., Liang C., Dang L., Lu C., He X., Cheung H. Y., Xu L., Lu C., He B., Liu B., Shaikh A. B., Li F., Wang L., et al. (2016) Osteoclast-derived exosomal miR-214–3p inhibits osteoblastic bone formation. Nat. Commun. 7, 10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deleted in proof.
- 43. Holmen S. L., Zylstra C. R., Mukherjee A., Sigler R. E., Faugere M. C., Bouxsein M. L., Deng L., Clemens T. L., and Williams B. O. (2005) Essential role of β-catenin in postnatal bone acquisition. J. Biol. Chem. 280, 21162–21168 [DOI] [PubMed] [Google Scholar]
- 44. Glass D. A. 2nd, Bialek P., Ahn J. D., Starbuck M., Patel M. S., Clevers H., Taketo M. M., Long F., McMahon A. P., Lang R. A., and Karsenty G. (2005) Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell 8, 751–764 [DOI] [PubMed] [Google Scholar]
- 45. Kubota T., Michigami T., and Ozono K. (2009) Wnt signaling in bone metabolism. J. Bone Miner. Metab. 27, 265–271 [DOI] [PubMed] [Google Scholar]
- 46. Kramer I., Halleux C., Keller H., Pegurri M., Gooi J. H., Weber P. B., Feng J. Q., Bonewald L. F., and Kneissel M. (2010) Osteocyte Wnt/β-catenin signaling is required for normal bone homeostasis. Mol. Cell. Biol. 30, 3071–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takahashi N., Maeda K., Ishihara A., Uehara S., and Kobayashi Y. (2011) Regulatory mechanism of osteoclastogenesis by RANKL and Wnt signals. Front. Biosci. (Landmark Ed.) 16, 21–30 [DOI] [PubMed] [Google Scholar]
- 48. Zaidi M. (2007) Skeletal remodeling in health and disease. Nat. Med. 13, 791–801 [DOI] [PubMed] [Google Scholar]
- 49. Qin W., Sun L., Cao J., Peng Y., Collier L., Wu Y., Creasey G., Li J., Qin Y., Jarvis J., Bauman W. A., Zaidi M., and Cardozo C. (2013) The central nervous system (CNS)-independent anti-bone-resorptive activity of muscle contraction and the underlying molecular and cellular signatures. J. Biol. Chem. 288, 13511–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jähn K., Lara-Castillo N., Brotto L., Mo C. L., Johnson M. L., Brotto M., and Bonewald L. F. (2012) Skeletal muscle secreted factors prevent glucocorticoid-induced osteocyte apoptosis through activation of β-catenin. Eur. Cell Mater. 24, 197–209; discussion 209–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., and Fais S. (2009) High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 4, e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zeringer E., Li M., Barta T., Schageman J., Pedersen K. W., Neurauter A., Magdaleno S., Setterquist R., and Vlassov A. V. (2013) Methods for the extraction and RNA profiling of exosomes. World J. Methodol. 3, 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Noort M., van de Wetering M., and Clevers H. (2002) Identification of two novel regulated serines in the N terminus of β-catenin. Exp. Cell Res. 276, 264–272 [DOI] [PubMed] [Google Scholar]
- 54. Kockeritz L., Doble B., Patel S., and Woodgett J. R. (2006) Glycogen synthase kinase-3: an overview of an over-achieving protein kinase. Curr. Drug Targets 7, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 55. Deleted in proof.
- 56. Deleted in proof.
- 57. Kamioka H., Honjo T., and Takano-Yamamoto T. (2001) A three-dimensional distribution of osteocyte processes revealed by the combination of confocal laser scanning microscopy and differential interference contrast microscopy. Bone 28, 145–149 [DOI] [PubMed] [Google Scholar]
- 58. Lasser C., Eldh M., and Lotvall J. (2012) Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 59, e3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kogure T., and Patel T. (2013) Isolation of extracellular nanovesicle microRNA from liver cancer cells in culture. Methods Mol. Biol. 1024, 11–18 [DOI] [PubMed] [Google Scholar]
- 60. Mathias R. A., Lim J. W., Ji H., and Simpson R. J. (2009) Isolation of extracellular membranous vesicles for proteomic analysis. Methods Mol. Biol. 528, 227–242 [DOI] [PubMed] [Google Scholar]
- 61. Dankbar B., Fennen M., Brunert D., Hayer S., Frank S., Wehmeyer C., Beckmann D., Paruzel P., Bertrand J., Redlich K., Koers-Wunrau C., Stratis A., Korb-Pap A., and Pap T. (2015) Myostatin is a direct regulator of osteoclast differentiation and its inhibition reduces inflammatory joint destruction in mice. Nat. Med. 21, 1085–1090 [DOI] [PubMed] [Google Scholar]
- 62. Kogianni G., Mann V., and Noble B. S. (2008) Apoptotic bodies convey activity capable of initiating osteoclastogenesis and localized bone destruction. J. Bone Miner. Res. 23, 915–927 [DOI] [PubMed] [Google Scholar]
- 63. Spatz J. M., Wein M. N., Gooi J. H., Qu Y., Garr J. L., Liu S., Barry K. J., Uda Y., Lai F., Dedic C., Balcells-Camps M., Kronenberg H. M., Babij P., and Pajevic P. D. (2015) The Wnt inhibitor sclerostin is up-regulated by mechanical unloading in osteocytes in vitro. J. Biol. Chem. 290, 16744–16758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu J., Li Y., Shen W., Qiao C., Ambrosio F., Lavasani M., Nozaki M., Branca M. F., and Huard J. (2007) Relationships between transforming growth factor-β1, myostatin, and decorin: implications for skeletal muscle fibrosis. J. Biol. Chem. 282, 25852–25863 [DOI] [PubMed] [Google Scholar]
- 65. Ito N., Findlay D. M., Anderson P. H., Bonewald L. F., and Atkins G. J. (2013) Extracellular phosphate modulates the effect of 1alpha,25-dihydroxy vitamin D3 (1,25D) on osteocyte like cells. J. Steroid Biochem. Mol. Biol. 136, 183–186 [DOI] [PubMed] [Google Scholar]
- 66. Woo S. M., Rosser J., Dusevich V., Kalajzic I., and Bonewald L. F. (2011) Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J. Bone Miner. Res. 26, 2634–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lässer C., Alikhani V. S., Ekström K., Eldh M., Paredes P. T., Bossios A., Sjöstrand M., Gabrielsson S., Lötvall J., and Valadi H. (2011) Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morelli A. E., Larregina A. T., Shufesky W. J., Sullivan M. L., Stolz D. B., Papworth G. D., Zahorchak A. F., Logar A. J., Wang Z., Watkins S. C., Falo L. D. Jr., and Thomson A. W. (2004) Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 104, 3257–3266 [DOI] [PubMed] [Google Scholar]
- 69. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.