Abstract
Biofilms are a collective mode of bacterial life in which a self-produced matrix confines cells in close proximity to each other. Biofilms confer many advantages, including protection from chemicals (including antibiotics), entrapment of useful extracellular enzymes and nutrients, as well as opportunities for efficient recycling of molecules from dead cells. Biofilm matrices are aqueous gel-like structures composed of polysaccharides, proteins, and DNA stabilized by intermolecular interactions that may include non-polar connections. Recently, polysaccharides extracted from biofilms produced by species of the Burkholderia cepacia complex were shown to possess clusters of rhamnose, a 6-deoxy sugar with non-polar characteristics. Molecular dynamics simulations are well suited to characterizing the structure and dynamics of polysaccharides, but only relatively few such studies exist of their interaction with non-polar molecules. Here we report an investigation into the hydrophobic properties of the exopolysaccharide produced by Burkholderia multivorans strain C1576. Fluorescence experiments with two hydrophobic fluorescent probes established that this polysaccharide complexes hydrophobic species, and NMR experiments confirmed these interactions. Molecular simulations to model the hydrodynamics of the polysaccharide and the interaction with guest species revealed a very flexible, amphiphilic carbohydrate chain that has frequent dynamic interactions with apolar molecules; both hexane and a long-chain fatty acid belonging to the quorum-sensing system of B. multivorans were tested. A possible role of the non-polar domains of the exopolysaccharide in facilitating the diffusion of aliphatic species toward specific targets within the biofilm aqueous matrix is proposed.
Keywords: biofilm, carbohydrate function, molecular modeling, NMR, polysaccharide, quorum sensing, Burkholderia multivorans
Introduction
Biofilms are a collective mode of bacterial life in which a self-produced matrix confines cells in close proximity to each other. Biofilms are the most common lifestyle of bacteria, and they are also implicated in persistent infections that are difficult to eradicate (1).
Biofilms confer many advantages on bacteria (2), including protection, the slow diffusion of useful extracellular enzymes and nutrients through the biofilm matrix, as well as opportunities for easy recycling of molecules from dead cells. Further, the limited dispersion of low-molecular-mass signaling (quorum-sensing) molecules throughout the biofilm results in easier communication between cells (3).
Biofilm matrices have a gel-like structure composed of a macromolecular network in an aqueous environment. The matrix network consists mainly of biopolymers (proteins, polysaccharides, and DNA) in variable amounts depending on the bacterial species. Exopolysaccharides (Epols)2 are the most abundant component of the biofilm matrix and are thought to be primarily responsible for the stability of the matrix (4). However, the chemical structure of biofilm Epols is only known for a few bacteria, and their precise role remains unclear. The biofilm macromolecules are presumed to interact via non-covalent intermolecular forces (hydrogen bonding as well as van der Waals and ionic interactions) to form the three-dimensional architecture of the biofilm matrix, but the details of the specific interactions between matrix components remain to be determined (5). The biofilm must ensure that hydrophilic species, important for bacterial survival and adaptation to specific environments, are not dispersed in water, thus solubilizing the biofilm. This can be done by strong polar interactions, as it happens in gels of alginate, the exopolysaccharide produced by Pseudomonas aeruginosa (6). However, non-polar or hydrophobic interactions also have the potential to stabilize the biofilm structure and could effectively retain non-polar species.
Our investigation into the structure of Epols produced by members of the Burkholderia cepacia complex revealed that these bacteria are able to produce a variety of Epols when cultured on agar Petri dishes in the presence of different culture media (7). Further, biofilm growth conditions may alter the composition of the Epols. For the Burkholderia multivorans reference strain C1576, yeast extract mannitol medium promoted the biosynthesis of the Epol cepacian (8), whereas, on Mueller-Hinton medium, the bacterium produced a rhamno-mannan containing α-d-rhamnose and α-d-mannose residues in equimolar ratios (9), with 50% O-methyl substitution on C-3 of the 2-linked rhamnose residues (Fig. 1a). The nature of this substitution, random or regularly occurring, has not been defined.
Figure 1.
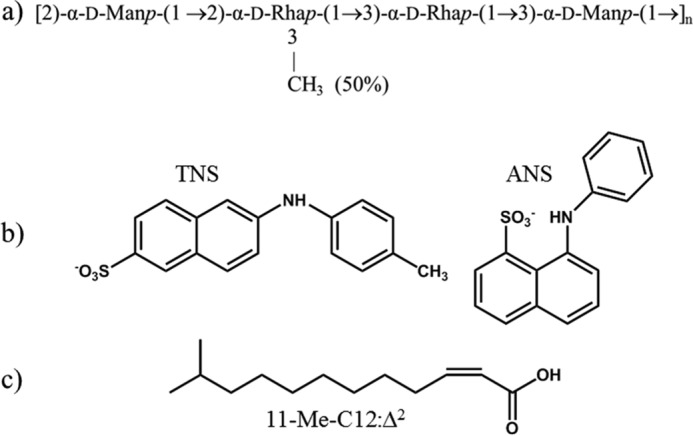
a, repeating unit of the exopolysaccharide produced by B. multivorans C1576. b, structures of TNS and ANS. c, structure of the signaling 11-Me-C12:Δ2 molecule.
This exopolysaccharide, hereafter referred to as EpolC1576, attracted our attention because of the presence of non-polar chemical moieties: 6-deoxy sugars (rhamnoses) have a lower polarity because of replacement of the conventional C6 hydroxyl methyl with a methyl group. In addition, partial-O-methyl substitution on the Rha C-3 position produces a polysaccharide chain with a relative lack of hydroxyl groups, which is thus potentially less hydrophilic than is the norm for saccharides. Although the importance of hydrophobic interactions for carbohydrate-protein binding has been recognized for some time (10), carbohydrates, with their multitude of pendant hydroxyls, are usually considered to be hydrophilic, with only a few cases reported of saccharides forming hydrophobic complexes. The best known system is that of the cyclodextrin ring, where the all-equatorial arrangement of the glucose hydroxyl groups, together with the α-(1→4) glycosidic linkages, creates a carbohydrate annular ribbon with hydrophilic -OH groups protruding from the ribbon edges and non-polar -CH groups populating the ribbon plane. This geometry results in the well known ability of cyclodextrins to complex aromatic and aliphatic molecules into their cavity (11). Among the numerous investigations of the interaction of cyclodextrins with hydrophobic species, one of the authors (R. R.) investigated their inclusion properties with two aromatic probes (12) that are usually used to identify hydrophobic sites in proteins (13, 14): 2-(p-toluidinyl)-naphthalene-6-sulfonate (TNS) and 1-anilinonaphthalene-8-sulfonate (ANS). The structures of these probes are shown in Fig. 1b.
Recently, two investigations focused on the carbohydrate-lipid interactions in rhamnose-containing polysaccharides, native or synthetic O-methylated (15, 16). However, apart from these examples, the possible interaction of oligo- and polysaccharides with non-polar molecules is still a poorly investigated issue. We speculate that the presence of hydrophobic domains in biofilm Epols may promote association between macromolecules, either polysaccharide-polysaccharide or protein-polysaccharide, thus increasing the biofilm stability in aqueous environments. In addition, hydrophobic regions may favor interactions with non-polar low-molecular-mass species with specific biological activity, thus conferring to Epols, as well as to their producing organisms, the ability to modulate the activity of these components in an aqueous environment. Bacteria have evolved a cell-cell communication mechanism, known as quorum sensing, to coordinate the expression of genes useful for bacterial life, including biofilm formation and disruption (17). Interestingly, quorum-sensing signaling molecules composed of fatty acid chains have been described for B. multivorans (18), the species investigated in this study. Such signaling species are also important factors in the regulation of virulence and biofilm formation in a wide range of bacterial pathogens (19). As complexation of the polymer hydrophobic domains with signaling molecules has the potential to modulate their activity according to the stage of the bacterial life cycle, the possible interaction of such amphiphilic signaling molecules with the EpolC1576 saccharide is of great interest.
The gelling structure of the biofilm matrices, and their structural complexity, prohibits a full experimental structural definition of the biofilm matrix scaffolds. However, molecular simulation methods provide a basis for both interpreting sparse experimental data and for independently predicting conformational and dynamic properties of carbohydrates. In particular, molecular dynamics (MD) simulations are especially well suited to the characterization of the structure and dynamics of glycans and glycoconjugates (10). Indeed, MD simulations are increasingly used for conformational prediction of bacterial polysaccharide structures (20, 21). MD has been used to probe the interaction modes of oligosaccharides with molecules such as proteins (22, 23), but, to date, there have been few MD studies targeted at other molecular classes, such as lipids or hydrocarbons.
Here we report an investigation into the hydrophobic properties of EpolC1576. Our goal was first to confirm that EpolC1576 does complex hydrophobic species. We established this by demonstrating interaction of EpolC1576 with two hydrophobic fluorescent probes (the N-arylaminonapthalenesulfonate dyes ANS and TNS) using both fluorescence and NMR spectroscopy. Next we employed molecular simulations to model the hydrodynamics of EpolC1576 and the mode of hydrophobic interaction with guest species, especially the role of the EpolC1576 methyl-substituted rhamnose residues in creating non-polar domains suitable to establish and maintain hydrophobic interactions. As the Epol has no clear binding site, the interaction of a flexible Epol chain with a guest molecule is not a typical molecular docking problem; the location and the mode of interaction are unknown. We therefore used MD simulations to investigate the interaction of EpolC1576 with two hydrophobic species in water: simple aliphatic chains (non-polar hexane) and a more complex fatty acid signaling species found in B. multivorans cultures: cis-11-methyl-2-dodecenoic acid (11-Me-C12:Δ2, Fig. 1c) which has a hydrophobic aliphatic chain and a polar head group (18, 24). These studies enable us to make predictions of the conformation, dynamics, and modes of interaction of EpolC1576 with such hydrophobic species.
Results
Interaction of fluorescent dyes with EpolC1576
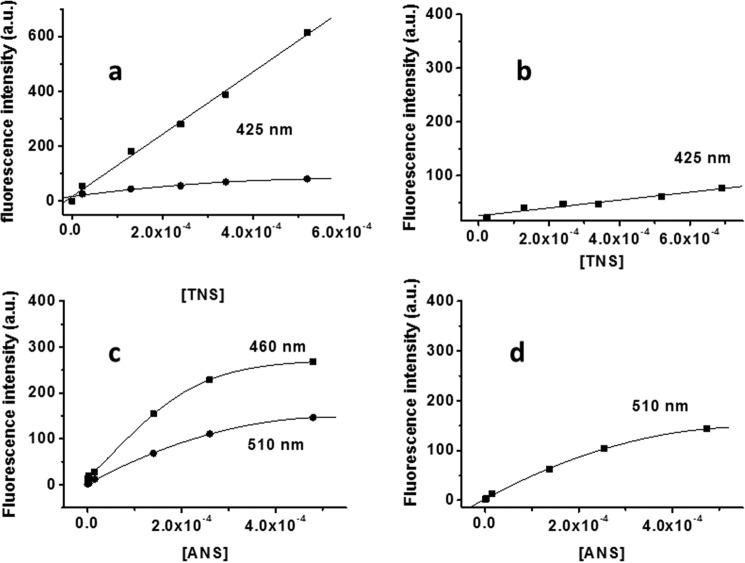
The fluorescence behavior of the aromatic TNS dye in the presence of EpolC1576 and dextran is shown in Fig. 2, a and b. As demonstrated for cyclodextrins, the complexation of TNS in a hydrophobic pocket produces an increase in fluorescence emission intensity because of the limited number of collisions with quencher solvent molecules. From spectroscopic data, it is clear that the fluorescence intensity of TNS alone in water and in the presence of dextran is practically identical. However, when corrected for the TNS self-absorption, the presence of EpolC1576 promotes a marked increase in fluorescence intensity (Fig. 2a). An almost identical behavior is observed for ANS in the presence of EpolC1576 and dextran (Fig. 2, c and d). Besides the increase in fluorescence intensity, the addition of ANS to EpolC1576 also produces a blue shift, from 510 to 450 nm, further indicating ANS complexation by the polysaccharide. In fact, as observed for tryptophan in proteins (25), fluorescent chromophores may change their emission maximum as a function of the polarity of the environment they experience. We speculate that the ANS-EpolC1576 interaction favors solution conformations of the polysaccharide that define a suitable binding site that decreases the solvent (water) interaction with the aromatic moiety. The slightly different behavior of the two dyes can be ascribed to their different structure (Fig. 1b).
Figure 2.
Fluorescence behavior of TNS and ANS in the presence of EpolC1576 (a and c) and dextran (b and d). ■, fluorescent dyes in the presence of polysaccharides; ●, fluorescent dyes in water. a.u., arbitrary units.
NMR investigation of the interaction between aromatic dyes and EpolC1576
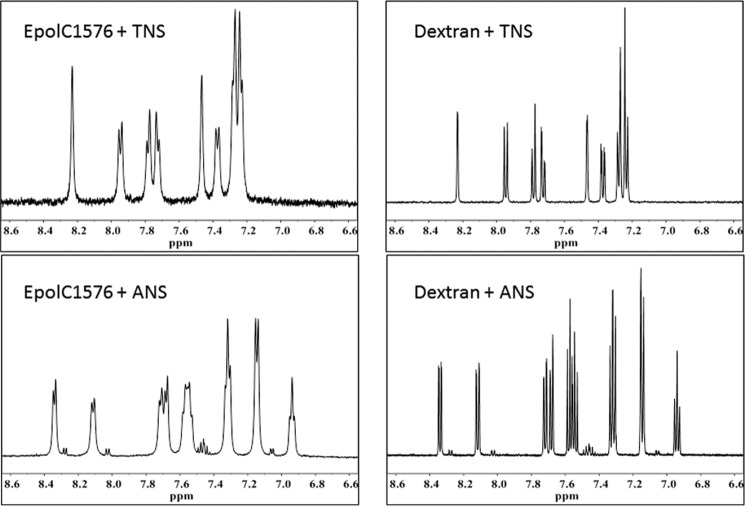
Independent evidence for TNS and ANS interaction with EpolC1576 was obtained by means of NMR spectroscopy, which indicates different dynamics for the two aromatic molecules in the presence of EpolC1576 and dextran, respectively. In Fig. 3, the aromatic portions of the ANS and TNS spectra obtained in the presence of EpolC1576 are compared with that in the presence of dextran. The spectra of both dyes in the presence of EpolC1576 show an increase in the line width of the aromatic resonance peaks (Fig. 3, left panels), which contrasts with the very sharp resonance lines obtained with dextran (Fig. 3, right panels). The increase in resonance line width indicates a decrease in molecular dynamics, which can be attributed to interactions with the non-polar domains of the EpolC1576 (26).
Figure 3.
The 1H NMR aromatic region of the spectra of TNS and ANS with EpolC1576 and dextran.
The data presented in Fig. 3 suggest that the presence of Rha and 3-O-methylated Rha residues in the polysaccharide-repeating unit produces domains able to interact with hydrophobic moieties. In fact, by comparison with data obtained in the presence of dextran, which exhibits a “conventional” saccharide chemistry, the interactions obtained with EpolC1576 can be interpreted as “assisted” by the non-polar groups of the polysaccharide chain. This feature may help in understanding the interactions required to set up and maintain the biofilm macromolecular matrix but also the possible molecular mechanisms involved in the biological activities of the matrix itself. In this context, the activity of signaling quorum-sensing molecules is particularly relevant for their implications in biofilm formation and bacterial pathogenicity. These species often exhibit a hydrophobic character, and interaction with the matrix polysaccharides may modulate their activity.
NMR investigation of the interaction between 11-Me-C12:Δ2 and EpolC1576
Having demonstrated the ability of EpolC1576 to interact with hydrophobic moieties, the possible interaction of 11-Me-C12:Δ2 with EpoLC1576 was investigated by NMR spectroscopy. Because of the very limited water solubility of the signaling molecule, a known amount of 11-Me-C12:Δ2 was solubilized with an aqueous solution of either EpolC1576 or dextran to measure the transfer of the signaling molecule in water fostered by polysaccharide interactions.
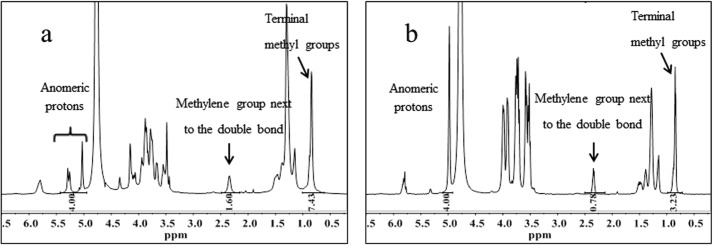
The integration of the polysaccharide H-1 resonance signals with respect to selected peaks of the signaling molecules (-CH2- at 2.3 ppm and -CH3 at 1.0 ppm) indicates that, indeed, the presence of EpolC1576 doubled the solubilization of 11-Me-C12:Δ2 in water (Fig. 4a) (-CH2-/H-1 = 0.40 and -CH3/H-1 = 1.86) with respect to that in the presence of dextran (-CH2-/H-1 = 0.20 and -CH3/H-1 = 0.81) (Fig. 4b). This very interesting result prompted us to investigate in more detail the possible mechanisms of interaction of the 11-Me-C12:Δ2 molecule with EpolC1576 by means of molecular modeling.
Figure 4.
a and b, 1H NMR spectra of signaling 11-Me-C12:Δ2 molecules in the presence of EpolC1576 (a) and dextran (b). Integration values of the H-1 anomeric proton signals and selected signals of the signaling species are indicated below the spectra.
Hydrodynamics of the EpolC1576 molecule
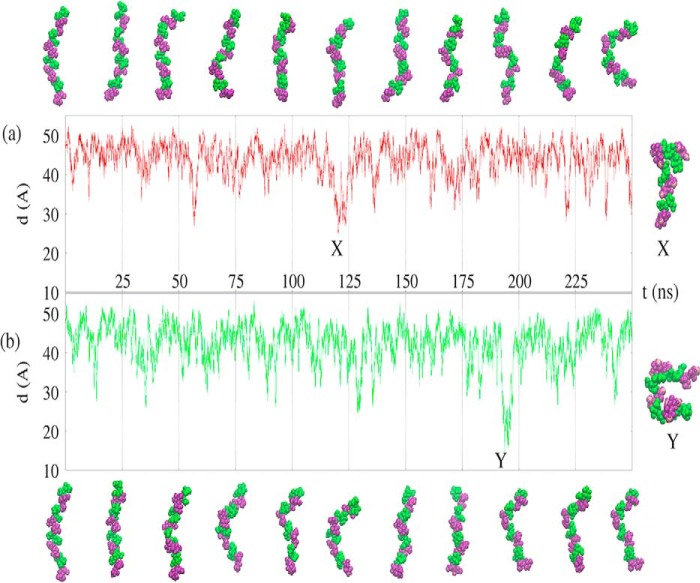
The simulations of the EpolC1576 tetramer (16 monosaccharides) in the presence of explicit water molecules and apolar guest molecules demonstrate that EpolC1576 behaves as a dynamic flexible random coil in aqueous solution (Fig. 5). In both aqueous simulations, EpolC1576 is primarily in a relatively elongated conformation, with regular and frequent conformational changes, as indicated by the end-to-end distance graphs for the 250-ns simulation with the hexane guest (Fig. 5a) and the 250-ns simulation with the signaling molecule (Fig. 5b). The EpolC1576 tetramer bends and straightens rapidly throughout both simulations, and the chain does not form a regular structure, as can be seen from the simulation snapshots in Fig. 5 for the hexane simulation (a) and the simulation with the signaling molecule (b). Occasionally, more compressed conformations stabilized by self-self interactions form, as shown by the examples X and Y in Fig. 5, a and b, respectively, but these more compact conformations do not persist for long periods of time in the aqueous medium. Therefore, EpolC1576 is a highly flexible carbohydrate chain that can perform rapid conformational adaptations to changing environments.
Figure 5.
EpolC1576 hydrodynamics. a and b, the end-to-end molecular distance in angstroms (d(A)) over the course of the simulation with three hexane molecules (a, red) and the simulation with the signaling molecule (b, green). The conformational snapshots of the EpolC1576 molecule are similar for the simulations with hexane (a) and signaling molecule (b). Mannose residues are colored green and rhamnose purple. Guest molecules, waters, and counterions are not shown. Snapshots of the most compressed conformations at X and Y are shown on the right.
Interaction with apolar guest molecules
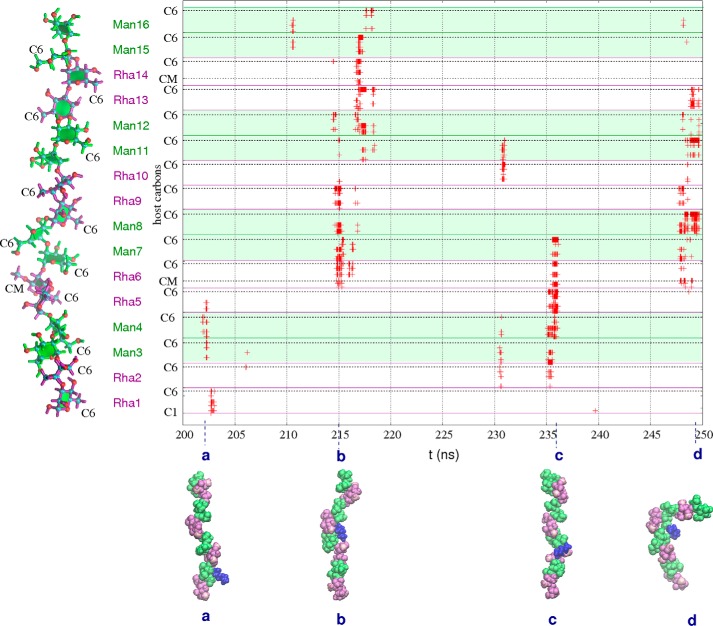
The aliphatic hexane chains interact frequently with EpolC1576 throughout the 250-ns simulation. The graph of close carbon-carbon contacts (<5 Å) for EpolC1576-hexane in the last 50 ns (Fig. 6) is representative of the entire simulation: close host-guest interactions occur often. In general, the EpolC1576-hexane host-guest interaction is very dynamic; although interactions typically involve two pairs of consecutive mannose/rhamnose residues, the site of the non-bonded contact changes constantly. Interactions typically involve at least one rhamnose methyl atom (either C6 or the 3-O-methyl substituent, as indicated on the y axis in Fig. 6). Interaction of the guest with multiple carbons in a single residue occurs frequently and indicates orientation of the guest hydrocarbon to face the plane of the pyranose ring, as for Rha6 in the interaction marked b in Fig. 6. Where interactions of the hexane guest occur simultaneously with four or more host residues, this indicates that the EpolC1576 strand has formed a pocket to encapsulate a hexane guest, as at 215 ns (Fig. 6b) and 235 ns (Fig. 6c). These pockets are associated primarily with the contiguous rhamnose residues with partial O-methyl substitutions and edged with hydrophobic plane of the pyranose ring in the mannose residues. The formation of a pocket results in a prolonged host-guest interaction that may persist for up to 2 ns.
Figure 6.
EpolC1576-hexane atomic contacts. Shown is a time series graph of the host-guest carbon-carbon contacts less than 5 Å for the last 50 ns of the simulation of EpolC1576 with three hexane guest molecules. Carbons are numbered consecutively along the y axis, from C1 in the Rha1 residue (bottom) to C6 in the Man16 residue (top). The specific locations of the C6 and CM (3-O-methyl substituent) atoms are indicated on the y axis. Interactions involving mannose residues are distinguished from rhamnose with background green shading. Representative snapshots of host-guest conformations are shown in a–d. Mannose residues are green, rhamnose purple, and O-methyl carbons pink.
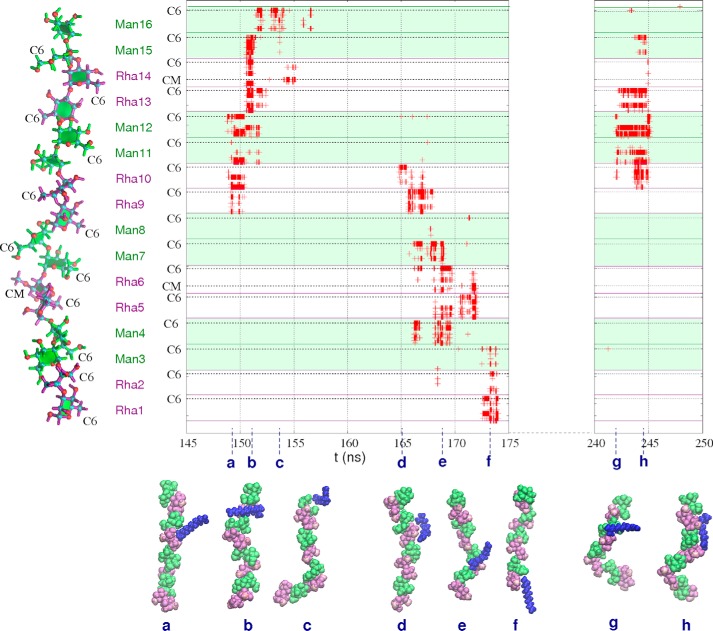
The aliphatic hexane chains interact more often with EpolC1576 than 11-Me-C12:Δ2, with 10% versus 8% of simulation time involving host-guests in close proximity. This is unsurprising, as there are more guest molecules in the hexane simulation, and they have faster diffusion than 11-Me-C12:Δ2. However, the increased length and/or amphiphilic nature of the signaling molecule serves to prolong the interaction in comparison with hexane, as can be seen from a comparison of the graphs of guest-host close carbon-carbon distances for EpolC1576/hexane (Fig. 6) and EpolC1576/11-Me-C12:Δ2 (Fig. 7). The EpolC1576/11-Me-C12:Δ2 simulation has three long periods with repeated close host-guest interactions: 148.6–155.5 ns (6.9 ns in duration), from 164.7–174.4 ns (9.7 ns), and from 241.9–245.6 ns (3.7 ns). Prolonged contact with the rhamnose methyl carbons (either C6 or the 3-O-methyl substituent, as indicated on the y axis in Fig. 7) are a clear feature of these extended interactions.
Figure 7.
EpolC1576-signaling molecule atomic contacts. Shown is a time series graph of carbon-carbon contacts less than 5 Å for the three long host-guest interactions in the simulation of EpolC1576 and the 11-Me-C12:Δ2 signaling guest molecule. The specific locations of the C6 and CM (3-O-methyl substituent) atoms are indicated on the y axis. Interactions involving mannose residues are distinguished from rhamnose with background green shading. Representative snapshots of host-guest conformations are shown in a–h. Mannose residues are green, rhamnose purple, and O-methyl carbons pink.
The first long interaction, from 148.6–155.5 ns, shows the EpolC1576 strand forming a pocket involving a pair of central recessed rhamnose residues to encapsulate the 11-Me-C12:Δ2 guest (Fig. 7b). Given the flexibility of the chain, EpolC1576 has the potential to form deeper hydrophobic pockets.
Further, in the period 241.9–245.6 ns, the 11-Me-C12:Δ2 signaling molecule has a very dynamic interaction with EpolC1576. The shift of the interaction is clearly apparent in the graph in Fig. 7: the guest molecule “walks” down the carbohydrate chain over the course of 9 ns (see the snapshots in Fig. 7, d–f). In this manner, the 11-Me-C12:Δ2 signaling molecule could rapidly migrate across a biofilm matrix of saccharide chains. Further, the signaling molecule could form bridges between polysaccharide chains. This interaction also features the formation of a hydrophobic pocket at around 169 ns, shown in the snapshot in Fig. 7e.
The final long interaction from 241.9–245.6 ns shows the 11-carbon guest molecular interacting along its entire length with six consecutive residues of the saccharide chain (Fig. 7h). This provides further evidence for the range of possible EpolC1576/11-Me-C12:Δ2interactions with apolar molecules.
Discussion
Bacterial exopolysaccharides are generally considered biofilm components that mainly contribute to the setup and maintenance of matrix scaffold architecture (27–29), thus exhibiting a structural role. However, different bacteria biosynthesize different exopolysaccharides in biofilm. Therefore, their chemistry has to be preliminarily clarified to better understand the biological role of the matrix in the biofilm community (30).
We have demonstrated that the exopolysaccharide extracted from biofilms produced by B. multivorans strain C1576 has non-polar domains that enable this polymer to complex aromatic species. Spectroscopic NMR and fluorescence experiments showed interactions with aromatic molecules used previously to identify hydrophobic pockets in proteins and in cyclodextrins. Further, transfer experiments from organic phase to water carried out with the 11-Me-C12:Δ2 signaling molecule produced by B. multivorans indicated that this species can interact with the biofilm exopolysaccharide produced by B. multivorans.
We used molecular simulations to probe the mode of interaction between the EpolC1576 and apolar guest molecules modeling interactions with two hydrophobic species: both simple non-polar hexane molecules and the more complex 11-Me-C12:Δ2 signaling molecule, which has a hydrophobic chain and a polar headgroup. Our simulations indicate that EpolC1576 is a very flexible molecule with amphiphilic properties that can adapt its conformation to suit different environments. EpolC1576 forms a flexible random coil conformation in aqueous solution, and transient hydrophobic domains, or pockets, form dynamically in the chain that may accommodate non-polar guest molecules. The hydrophobic domains along the polysaccharide chain are associated primarily with the contiguous rhamnose residues with partial O-methyl substitutions and with the plane of the pyranose rings. There are two distinct modes of interaction between EpolC1576 and the guest molecules: short local interactions with the polymer that do not persist and longer interactions with a transient hydrophobic pocket or region in the dynamic EpolC1576 chain that capture the apolar guest for longer periods. Interactions with the polysaccharide demonstrated that the signaling molecule, which has very low solubility in water, could acquire mobility in the biofilm aqueous environment by moving from one non-polar domain on the polysaccharide chain to another. This mechanism, which is similar to a reverse-phase chromatographic process, may allow the active species to remain in the biofilm environment, ready to interact with a specific binding site by traveling along polysaccharide chains interconnected in the biofilm matrix network. In fact, a possible “sorptive” role of exopolysaccharides in biofilms is suggested in an article reporting a discussion held during the Biofilm 2007 conference (31). In addition, we further speculate that the EpolC1576 hydrophobic domains allow for complexation with antibiotics with aromatic or hydrophobic moieties, thus hindering their diffusion through the biofilm water channels to the bacterial cell surface or, at an extreme, sequestering them in the biofilm matrix.
Experimental procedures
Bacterial strain and bacterial growth
B. multivorans strain C1576 (LMG 16660) is a reference strain from the panel of B. cepacia complex strains (EP1), and it was purchased from the BCCMTM bacterium collection. Bacterial growth and polysaccharide purification were performed according to Protocol II described in Ref. 9.
Fluorescence spectroscopy experiments
The fluorescent probes ANS (Na+ salt) and TNS (K+ salt) were purchased from Sigma. EpolC1576 was dissolved in water at a concentration of octasaccharide repeating unit (4 Rha, 4 Man, 1 O-Me; molecular mass, 1246 Da) 2·10−4 m. A water solution of each fluorescent probes was added to the polysaccharide solution to have the proper final concentration in the range of 0–7·10−4 m. Fluorescence spectra were recorded on a PerkinElmer Life Sciences LS50B spectrofluorimeter thermostated at 25 °C and using 10-mm path length quartz cells. For ANS, the excitation wavelength was 365 nm, and emission was detected at 460 and 510 nm. For TNS, the excitation wavelength was 287 nm, and emission was detected at 425 nm. For comparison purposes, the same experiments were also carried out in the presence of dextran, as it has no non-polar domains along its backbone. The dextran concentration was established as for EpolC1576. Fluorescence intensity values were corrected for dye self-absorption according to Ref. 13.
NMR experiments
Proton NMR spectra were recorded on a 500-MHz Varian spectrometer operating at 25 °C. All samples were subsequently exchanged three times with 99.9% D2O by lyophilization and finally dissolved in 0.7 ml of 99.99% D2O. For the interactions experiments with aromatic dyes, the EpolC1576 concentration was 5.7·10−4 m, ANS was 1.5·10−3 m, and TNS was 2.3·10−3 m. For comparison purposes, NMR experiments were also carried out with dextran using the same polysaccharide and aromatic dye concentrations.
Interaction of EpolC1576 with cis-11-methyl-2-dodecenoic acid
The method used consisted of solubilizing an aliquot of cis-11-methyl-2-dodecenoic acid in a polysaccharide aqueous solution containing EpolC1576 or dextran. 0.4 μmol of EpolC1576 and 0.4 μmol of dextran (calculated for an octasaccharide repeating unit) were exchanged three times with 99.9% D2O by lyophilization, dissolved in 0.7 ml of 99.99% D2O, and added to two vials, each containing 9.4 μmol of cis-11-methyl-2-dodecenoic acid. The fatty acid was dissolved previously in CD3Cl, transferred to the two vials, and dried under a stream of N2. The two vials were gently shaken at room temperature for about 7 h, centrifuged at 14,500 × g for 10 min at room temperature to remove insoluble material, and transferred in NMR tubes for experiments. NMR spectra were processed using MestreNova software. Chemical shifts are expressed in parts per million and referred to the HOD signal, set as 4.768 ppm.
Molecular simulations
Molecular simulations using all-atom molecular dynamics were employed, modeling a tetramer (16 monosaccharides of EpolC1576, having one 3-O-methyl Rha every second repeating unit) with guest species in water. Two simulations were carried out: the first with three hexane molecules in explicit water and the second with the amphiphilic signaling species, also in explicit water and with counterions to neutralize the charge. All simulations were performed with the NAMD molecular dynamics program version 2.9 (employing NAMD CUDA extensions for calculation of long-range electrostatics and non-bonded forces on graphics processing units (32) and run on a 12 core, 4 TeslaM2090 GPU server with 64GB RAM per core and 6GB DDR5 per GPU). Water was simulated with the TIP3P model (33). EpolC1576 was modeled with the CHARMM36 additive force field for carbohydrates, (34, 35) with ad hoc extensions for the O-methyl groups, adapted from the parameters in existing residues. The hexane and 11-Me-C12:Δ2 molecules were modeled with the CGenFF parameters for the CHARMM general force field for small-molecule drug design (36), with the 11-Me-C12:Δ2 topology modeled on CGenFF propenoic acid and hexane. Initial configurations of EpolC1576 were built using our CarbBuilder software (37). The psfgen tool (http://www.ks.uiuc.edu/Research/vmd/plugins/psfgen/)3 was used to create “protein structure” (psf) files for modeling with a specified CHARMM force field and the NAMD molecular dynamics program. These initial oligosaccharide structures were optimized through 1000 steps of standard NAMD minimization in a vacuum and then solvated (using the solvate plugin to the Visual Molecular Dynamics (38) analysis package in a periodic cubic unit cell with a 80 × 80 × 80 Å cube. The simulations with the 11-Me-C12:Δ2 signaling molecule had one randomly distributed sodium ion to electrostatically neutralize the system. The two aqueous simulations ran for 250 ns and were preceded by a 30,000-step minimization phase, with a temperature control and equilibration regime involving 10 K temperature reassignments from 10 K, culminating in a maximum temperature of 300 K. Equations of motion were integrated using a Leap-Frog Verlet integrator with a step size of 1 fs and periodic boundary conditions. Simulations were performed under isothermal-isobaric (nPT) conditions at 300 K maintained using a Langevin piston barostat (39) and a Nosé-Hoover thermostat (40). Long-range electrostatic interactions were treated using particle mesh Ewald summation and particle mesh Ewald grid dimensions of 80. Non-bonded interactions were truncated with a switching function applied between 12.0 and 15.0 Å to groups with integer charge. The 1,4 interactions were not scaled, in accordance with the CHARMM force field recommendations.
Author contributions
M. K. performed and interpreted the molecular simulation experiments. P. C. performed the NMR and fluorescence experiments. M. D. contributed to the fluorescence experiments. M. K., P. C., and R. R. wrote the manuscript.
Acknowledgments
Computations were performed using facilities provided by the University of Cape Town ICTS High-Performance Computing team (http://hpc.uct.ac.za). (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.)
The South African Medical Research Council funding agency provided financial support for the modeling component of this work (SIR 415983). The University of Trieste is kindly acknowledged for financial support (FRA2013 and FRA2015). The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- Epol
- exopolysaccharide
- TNS
- 2-(p-toluidinyl)-naphthalene-6-sulfonate
- ANS
- 1-anilinonaphthalene-8-sulfonate
- MD
- molecular dynamics.
References
- 1. Fux C. A., Costerton J. W., Stewart P. S., and Stoodley P. (2005) Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 [DOI] [PubMed] [Google Scholar]
- 2. Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., and Kjelleberg S. (2016) Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 [DOI] [PubMed] [Google Scholar]
- 3. Hawver L. A., Jung S. A., and Ng W.-L. (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Limoli D. H., Jones C. J., and Wozniak D. J. (June, 2015) Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr. 10.1128/microbiolspec.MB-0011-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flemming H.-C. (2016) EPS: Then and Now. Microorganisms 10.3390/microorganisms4040041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franklin M. J., Nivens D. E., Weadge J. T., and Howell P. L. (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuzzi B., Herasimenka Y., Silipo A., Lanzetta R., Liut G., Rizzo R., and Cescutti P. (2014) Versatility of the Burkholderia cepacia complex for the biosynthesis of exopolysaccharides: a comparative structural investigation. PLoS ONE 9, e94372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cescutti P., Bosco M., Picotti F., Impallomeni G., Leitão J. H., Richau J. A., and Sá-Correia I. (2000) Structural study of the exopolysaccharide produced by a clinical isolate of Burkholderia cepacia. Biochem. Biophys. Res. Commun. 273, 1088–1094 [DOI] [PubMed] [Google Scholar]
- 9. Dolfi S., Sveronis A., Silipo A., Rizzo R., and Cescutti P. (2015) A novel rhamno-mannan exopolysaccharide isolated from biofilms of Burkholderia multivorans C1576. Carbohydr. Res. 411, 42–48 [DOI] [PubMed] [Google Scholar]
- 10. Fadda E., and Woods R. J. (2010) Molecular simulations of carbohydrates and protein-carbohydrate interactions: motivation, issues and prospects. Drug. Discov. Today 15, 596–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Del Valle E. M. M. (2004) Cyclodextrins and their uses: a review. Process Biochem. 39, 1033–1046 [Google Scholar]
- 12. Crescenzi V., Gamini A., Palleschi A., and Rizzo R. (1986) Physico-chemical aspects of inclusion complex formation between cycloamyloses and aromatic guest molecules in water. Gazz. Chim. Ital. 116, 435–440 [Google Scholar]
- 13. McClure W. O., and Edelman G. M. (1967) Fluorescent probes for conformational states of proteins: II: the binding of 2-p-toluidinylnaphthalene-6-sulfonate to a-chymotrypsin. Biochemistry 6, 559–566 [DOI] [PubMed] [Google Scholar]
- 14. Edelman G. M., and McClure W. O. (1968) Fluorescent probes and the conformation of proteins. Accounts Chem. Res. 1, 65–70 [Google Scholar]
- 15. Cheon H.-S., Wang Y., Ma J., and Kishi Y. (2007) Complexation of fatty acids and fatty acid-CoAs with synthetic O-methylated polysaccharides. ChemBioChem 8, 353–359 [DOI] [PubMed] [Google Scholar]
- 16. Liu L., Bai Y., Sun N., Xia L., Lowary T. L., and Klassen J. S. (2012) Carbohydrate-lipid interactions: affinities of methylamannose polysaccharides for lipids in aqueous solutions. Chem. Eur. J. 18, 12059–12067 [DOI] [PubMed] [Google Scholar]
- 17. Coenye T. (2010) Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing. Future Microbiol. 5, 1087–1099 [DOI] [PubMed] [Google Scholar]
- 18. Deng Y., Wu J., Eberl L., and Zhang L.-H. (2010) Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl. Environ. Microb. 76, 4675–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He Y. W., and Zhang L. H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol. Rev. 32, 842–857 [DOI] [PubMed] [Google Scholar]
- 20. Galochkina T., Zlenko D., Nesterenko A., Kovalenko I., Strakhovskaya M., Averyanov A., and Rubin A. (2016) Conformational dynamics of the single lipopolysaccharide O-antigen in solution. ChemPhysChem 17, 2839–2853 [DOI] [PubMed] [Google Scholar]
- 21. Kuttel M. M., Jackson G. E., Mafata M., and Ravenscroft N. (2015) Capsular polysaccharide conformations in pneumococcal serotypes 19F and 19A. Carbohydr. Res. 406, 27–33 [DOI] [PubMed] [Google Scholar]
- 22. Bryce R. A., Hillier I. H., and Naismith J. H. (2001) Carbohydrate-protein recognition: molecular dynamics simulations and free energy analysis of oligosaccharide binding to concanavalin A. Biophys. J. 81, 1373–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colombo G., Meli M., Cañada J., Asensio J. L., and Jiménez-Barbero J. (2004) Toward the understanding of the structure and dynamics of protein-carbohydrate interactions: molecular dynamics studies of the complexes between hevein and oligosaccharidic ligands. Carbohydr. Res. 339, 985–994 [DOI] [PubMed] [Google Scholar]
- 24. Marques C. N., Davies D. G., and Sauer K. (2015) Control of biofilms with fatty acid signalling molecule cis-2-decenoic acid. Pharmaceuticals 8, 816–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vivian J. T., and Callis P. R. (2001) Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poveda A., and Jiménez-Barbero J. (1998) NMR studies of carbohydrate-protein interactions in solution. Chem. Soc. Rev. 27, 133–143 [Google Scholar]
- 27. Sutherland I. (2001) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9 [DOI] [PubMed] [Google Scholar]
- 28. Flemming H. C., and Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 29. Hobley L., Harkins C., MacPhee C. E., and Stanley-Wall N. R. (2015) Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39, 649–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Branda S. S., Vik S., Friedman L., and Kolter R. (2005) Biofilms: the matrix revisited. TRENDS Microbiol. 13, 20–26 [DOI] [PubMed] [Google Scholar]
- 31. Flemming H.-C., Neu T. R., and Wozniak D. J. (2007) The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 189, 7945–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stone J. E., Phillips J. C., Freddolino P. L., Hardy D. J., Trabuco L. G., and Schulten K. (2007) Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 28, 2618–2640 [DOI] [PubMed] [Google Scholar]
- 33. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (August, 1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 [Google Scholar]
- 34. Guvench O., Greene S. N., Kamath G., Brady J. W., Venable R. M., Pastor R. W., and Mackerell A. D. Jr. (2008) Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 29, 2543–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guvench O., Hatcher E. R., Venable R. M., Pastor R. W., Alexander J., and Mackerell A. D. Jr. (2009) CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 5, 2353–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., and Macerell A. D. Jr. (2010) CHARMM General Force Field (CGenFF): A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuttel M. M., Ståhle J., and Widmalm G. (2016) CarbBuilder: software for building molecular models of complex oligo- and polysaccharide structures. J. Comput. Chem. 37, 2098–2105 [DOI] [PubMed] [Google Scholar]
- 38. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 27, 28,, 33–38 [DOI] [PubMed] [Google Scholar]
- 39. Feller S.E., Zhang Y.H., Brooks B.R., Pastor R.W.. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 [Google Scholar]
- 40. Martyna G. J., Tobias D. J., and Klein M. L. (1994) Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 [Google Scholar]