Abstract
In response to limited nutrients, fungal cells exit the primary growth phase, enter the stationary phase, and cease proliferation. Although fundamental to microbial physiology in many environments, the regulation of this transition is poorly understood but likely involves many transcriptional regulators. These may include the sirtuins, which deacetylate acetyllysine residues of histones and epigenetically regulate global transcription. Therefore, we investigated the role of a nuclear sirtuin, sirtuin E (SirE), from the ascomycete fungus Aspergillus nidulans. An A. nidulans strain with a disrupted sirE gene (SirEΔ) accumulated more acetylated histone H3 during the stationary growth phase when sirE was expressed at increased levels in the wild type. SirEΔ exhibited decreased mycelial autolysis, conidiophore development, sterigmatocystin biosynthesis, and production of extracellular hydrolases. Moreover, the transcription of the genes involved in these processes was also decreased, indicating that SirE is a histone deacetylase that up-regulates these activities in the stationary growth phase. Transcriptome analyses indicated that SirE repressed primary carbon and nitrogen metabolism and cell-wall synthesis. Chromatin immunoprecipitation demonstrated that SirE deacetylates acetylated Lys-9 residues in histone H3 at the gene promoters of α-1,3-glucan synthase (agsB), glycolytic phosphofructokinase (pfkA), and glyceraldehyde 3-phosphate (gpdA), indicating that SirE represses the expression of these primary metabolic genes. In summary, these results indicate that SirE facilitates the metabolic transition from the primary growth phase to the stationary phase. Because the observed gene expression profiles in stationary phase matched those resulting from carbon starvation, SirE appears to control this metabolic transition via a mechanism associated with the starvation response.
Keywords: Aspergillus, glucose metabolism, secondary metabolism, sirtuin, sporulation, autolysis
Introduction
Histone deacetylase (HDAC)2 deacetylates distinct Lys residues of histones, leading to heterochromatin formation that silences gene expression. Silent information regulator 2 protein (sirtuin) is a Class III HDAC that requires NAD+ for deacetylation (1, 2), and it is involved in various cellular processes such as metabolism, energy conservation, development, stress responses, and genome integrity (3). Sirtuins are further categorized into Classes I–IV according to their amino acid sequences (4). Class I sirtuins comprise subgroups Ia, Ib, and Ic. Class Ia contains mammalian SIRT1 as well as Saccharomyces cerevisiae Sir2p and Hst1p that deacetylate acetylated histones H3 and H4 (1, 5). SIRT1 is the best characterized, and it also deacetylates non-histone substrates (6). Both mammalian SIRT2 and SIRT3 have been categorized into Class Ib. SIRT2 deacetylates histone H4 as well as cytosolic α-tubulin. S. cerevisiae Hst2p is a cytosolic isoform of this class that deacetylates histone H4 in vitro (2). Class Ic sirtuin is distributed among known fungi, and it involves S. cerevisiae nuclear Hst3p and mitochondrial Hst4p (4). Class II and III sirtuins have other functions besides protein deacetylation. For example, the Class II sirtuin SIRT4 transfers ADP-ribose to glutamate dehydrogenase, and the Class III mitochondrial SIRT5 catalyzes desuccinylation and demalonylation in addition to deacetylation (7, 8).
The genomes of filamentous ascomycete fungi predict genes for Class I–III sirtuins whereas evolutionally related S. cerevisiae produces only the Class I sirtuins Sir2p, Hst1p, Hst2p, Hst3p, and Hst4p (4). Aspergillus nidulans SirA is a Class Ia sirtuin that deacetylates the acetylated lysine residue 16 on histone H4 (H4K16ac) and regulates the expression of genes that synthesize the secondary metabolites penicillin and sterigmatocystin (9), whereas its Magnaporthe oryzae counterpart deacetylates non-histone substrates to control plant pathogenicity (10). The A. nidulans HstA is a Class II sirtuin that has presumed histone deacetylase activity and represses the expression of genes involved in the production of penicillin and norsolorinic acid (11). Aspergillus oryzae AoHstD is a Class Ic sirtuin that is associated with the production of conidia and secondary metabolites (12) and stress tolerance (13). The functions of filamentous fungal sirtuins are thus versatile, although the roles that they play in regulating primary metabolism are limited. The Class Ic sirtuin of A. nidulans (AN1226; SirE) is presumed (9), and its function remains unknown.
Fungal cells that exhaust nutrients exit the primary or exponential growth phase, enter a stationary phase, and cease proliferation. Entry into the stationary phase accompanies the production of secondary metabolites, some of which serve as pharmaceuticals, whereas others are life-threatening mycotoxins, and the development of sexual spores and conidia. The fungal mechanism of transcriptional regulation involves specific transcription factors for producing secondary metabolites and conidia (14, 15). Autolysis is another stationary-phase event during which the fungus self-digests vegetative cells. Such self-destruction accompanies the production of extracellular β-1,3-glucanase EngB, chitinase B (ChiB), and N-acetyl-d-glucosaminidase NagA, which degrade cell wall glucan and chitin to result in cell lysis (16–18). These stationary-phase events are presumably associated with the fungal response to nutrient limitation. Although this mechanism is not fully understood, the transcription repressor CreA of A. nidulans responds to exogenous glucose and regulates gene expression of the secondary metabolite and conidial production (19–21). Regardless of extensive research into the expression of stationary-phase genes, how glucose/nutrient limitation regulates gene expression of the primary-phase mechanisms is not fully understood. Furthermore, little is known about the transcription regulation that determines the transition between the primary growth and stationary phases, especially that mediated by sirtuins.
The present study clarified the role of the Class Ic sirtuin SirE of A. nidulans that deacetylates acetylated Lys residues 9, 18, and 56 of histone H3 (H3K9ac, H3K18ac, and H3K56ac) during the stationary growth phase. Transcriptomic, genetic, and physiological analyses revealed that SirE up-regulates the gene expression of molecules involved in autolysis, conidial development, and biosynthesis of sterigmatocystin during the stationary growth phase of cultured A. nidulans. The transcriptome indicated that SirE represses gene expression in primary metabolic processes such as glycolysis, the tricarboxylic acid (TCA) cycle, and nitrate assimilation. Along with these mechanisms, SirE repressed the expression of the glycolytic gpdA and pfkA genes and of agsB that encodes α-1,3-glucan synthase for normal cell wall synthesis by decreasing levels of the histone H3 acetylation on the gene promoters. Our findings suggest that SirE is the missing coordinator of the global primary and secondary metabolic mechanisms that function during the transition from the exponential growth phase to the stationary phase.
Results
Fungal sirtuin E is an NAD+-dependent histone deacetylase
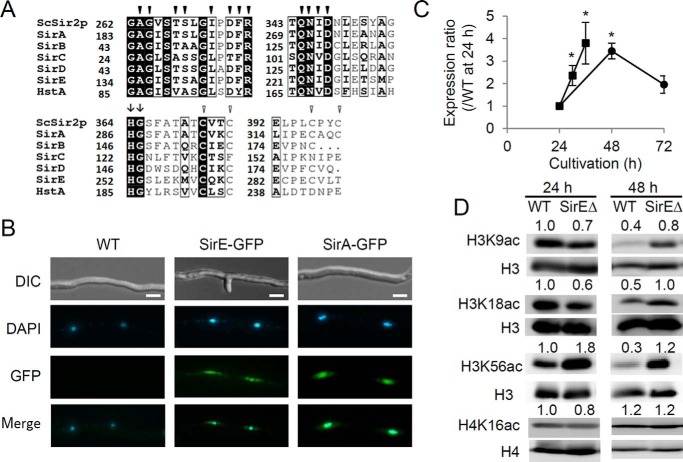
Phylogenetic analysis identified four predicted A. nidulans proteins (gene identities (IDs) ANID_07461, ANID_01782, ANID_11873, and ANID_01226) that belong to the same clade as SirA and HstA, both of which are Class III HDACs of this fungus (9, 11). A conserved sirtuin homology domain of ∼275 residues comprising NAD+-binding sites and a zinc finger motif (22, 23) characterized these proteins (Fig. 1A and supplemental Fig. S1). His-Gly residues that are found in all eukaryotic and prokaryotic sirtuins (24) were located at the middle of these predicted proteins. These results indicated that these are fungal sirtuin proteins, and they were designated SirB, SirC, SirD, and SirE, respectively. The amino acid sequences of all sirtuins except SirB contained motifs for nuclear localization signals (NLSs) (supplemental Fig. S1B). The pat4-, pat7-, and bipartite-type NLS motifs were predicted in the amino acid sequence of SirA, whereas the pat4-and pat7-type NLS motifs were found in those of SirC and SirD, respectively. SirE encoded two pat4- and six pat7-type NLS motifs. These findings predicted the nuclear localization of these sirtuins. We generated A. nidulans strains producing fusion proteins between green fluorescent protein (GFP) and SirE under the control of the constitutive gpdA gene promoter, and microscopy indicated that fluorescence emitted by the SirE-GFP protein co-localized with nuclei that were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Fig. 1B). This finding indicated that SirE functions in nuclei. A similar investigation of SirA-GFP generated essentially the same results, consistent with its function as a nuclear HDAC (9). This study focused on SirE in the Class Ic sirtuin family.
Figure 1.
Identification of fungal SirE. A, partial amino acid sequences of predicted A. nidulans sirtuins. Filled and unfilled arrowheads, respectively, indicate conserved NAD+-binding site and zinc finger residues. Arrows, highly conserved His-Gly. Numbers of amino acid residues are shown on the left. B, fluorescence microscopy of A. nidulans producing GFP and SirE or SirA fusion proteins. Nuclei are stained with DAPI. Scale bars, 2 μm. C, PCR-quantified sirE transcript. Strain A26 was cultured in liquid (●) and solid agar (■) GMM. Values are relative to actA transcript at 24 h, which was taken as 1. Error bars represent S.E. D, Western blots of A. nidulans nuclear extracts. A. nidulans A26 (WT) and SirEΔ were cultured for 24 and 48 h, and their nuclear extracts (20 μg) were separated by SDS-PAGE. Left, antibodies that detected signals. Relative signal intensity is shown above the panels. Data are means ± S.E. of three experiments. *, p < 0.05 versus 24 h.
Sirtuin E deacetylates acetylated histone H3 at the stationary growth phase
Time-dependent changes in sirE transcripts were investigated in fungal cells cultured in liquid medium. Quantitative PCR indicated that the cells accumulated 3.3-fold more sirE transcripts during the stationary (48 h) than the exponential (24 h) growth phase (Fig. 1C). Western blotting indicated that the fungus accumulated only 40, 50, and 30% H3K9ac, H3K18ac, and H3K56ac, respectively, during the stationary growth phase compared with the exponential growth phase (Fig. 1D). Signals for H4K16ac did not decrease during the stationary growth phase. These findings were consistent with the notion that sirE encodes an HDAC that deacetylates H3K9ac, H3K18ac, and H3K56ac during the stationary growth phase.
We replaced the chromosomal sirE region of the A26 strain with a fused DNA fragment comprising the 5′- and 3′-regions of the sirE and the argB (transformation marker) genes using homologous recombination, and Southern blot analysis confirmed that chromosomes of one strain lacked intact sirE (supplemental Fig. S2). That strain, namely SirEΔ, was cultured in liquid minimal medium to the stationary growth phase, and then the acetylation of nuclear histones was analyzed. Western blots of SirEΔ nuclear extracts exhibited signals for H3K9ac, H3K18ac, and H3K56ac of histone H3 that were 2–4-fold more intense than those of the WT strain (Fig. 1D). Acetylation levels of H4 Lys-16 (Fig. 1D) did not differ between the strains, indicating that sirE deacetylates H3K9ac, H3K18ac, and H3K56ac. The preference for H3K9ac and H3K18ac was similar to that of human SIRT6 and SIRT7, and the preference for H3K56ac was similar to that of human SIRT6 and yeast Hst3p and Hst4p, respectively (25–27), both of which belong to the same clade as SirE in the phylogenetic tree (supplemental Fig. S1A) (12). When analyzing exponentially growing cells, a lack of sirE did not increase signals for H3K9ac and H3K18ac but did increase signal for H3K56ac by 1.8-fold (Fig. 1D). These results indicated that SirE deacetylates H3K9ac and H3K18ac during the stationary growth phase and H3K56ac during both the stationary and the exponential growth phases.
SirE up-regulates fungal autolysis
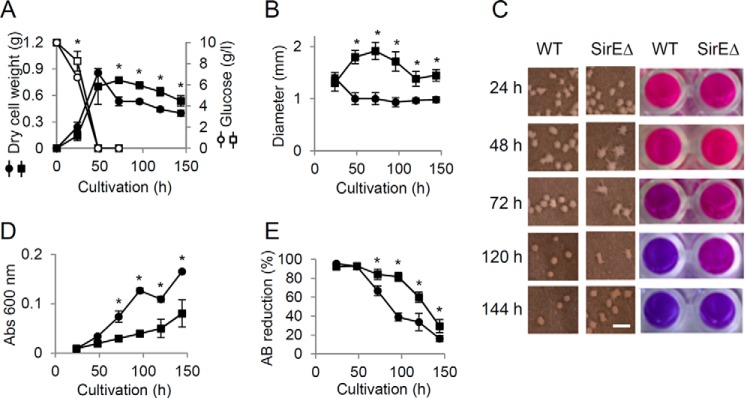
The weight of WT cells cultured in liquid medium gradually decreased after entry into the stationary growth phase, whereas that of SirEΔ decreased to a lesser extent (Fig. 2A). Larger mycelial pellets were generated by SirEΔ than by WT, and these remained larger for up to 144 h (Fig. 2, B and C). The culture medium of WT became more turbid than that of SirEΔ over the 144-h incubation (Fig. 2D). This indicates that WT dispersed more intracellular ingredients due to cell lysis. Viable cells were quantified by measuring the cellular reducing activity of alamarBlue (AB) (28). Exponentially growing WT and SirEΔ cells reduced AB to red pigments (Fig. 2C), indicating that the strains were viable. After 96-h culture, the WT and SirEΔ reduced 61 and 18% of the AB, and finally (∼144 h), >70% of the cells of both the strains lost viability (Fig. 2E). These results indicated that SirE prevents prolonged cell viability during the stationary growth phase. The decreased dry cell mass, disorganized pellet morphology, and reduced numbers of viable cells are characteristic of autolyzed mycelia (29). The activities of autolytic chitinase, β-1,3-endoglucanase and N-acetyl-β-d-glucosaminidase were increased during the stationary growth phase but to a lesser extent by SirEΔ than by the WT (Fig. 3, A–C). Fewer transcripts of their encoding chiB, engA, and nagA genes were accumulated during the stationary growth phase by SirEΔ than the WT (Fig. 3D). These results indicated that SirE is required for the normal expression of autolytic genes and hence for the autolysis of stationary phase cells.
Figure 2.
SirE up-regulates fungal autolysis during stationary growth phase. A. nidulans A26 (WT) (● and ○) and SirEΔ (■ and □) were cultured in liquid GMM at 30 °C. A, time-dependent changes in cell mass (● and ■) and residual glucose (○ and □). B, diameter of pelleted WT and SirEΔ mycelia. C, morphology of mycelial pellets (left) and AB assay of viable cells (right). The scale bar indicates 1 mm. D, time-dependent increase in optical density of culture supernatant due to autolytic release of cell contents into culture medium. E, reduced AB determined by absorbance (Abs) at 570 and 600 nm. Data are means ± S.E. of three experiments. Error bars represent S.E. *, p < 0.05 versus WT.
Figure 3.
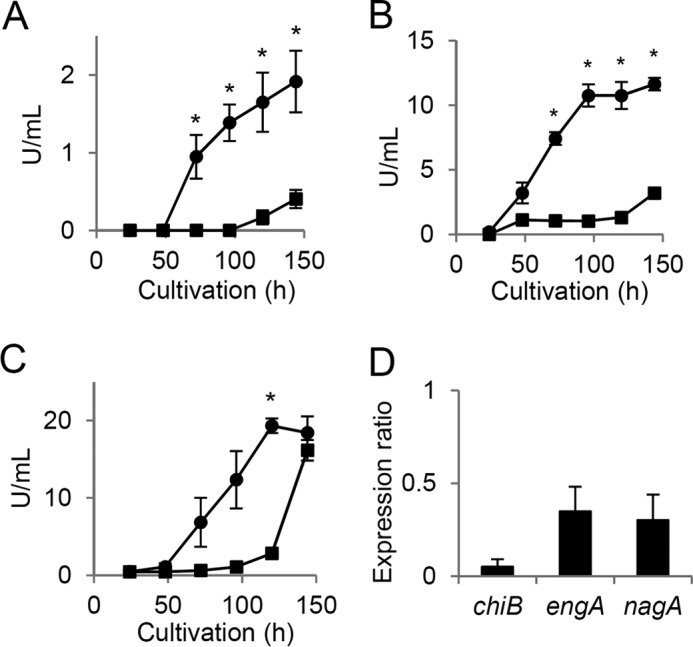
SirE up-regulates production of extracellular hydrolases. A. nidulans A26 (WT) (●) and SirEΔ (■) were cultured in liquid GMM at 30 °C, and then hydrolase activities in culture supernatants were determined. A, B, and C, chitinase, β-1,3-glucanase, and N-acetyl-d-glucosaminidase, respectively. Data are means ± S.E. of three experiments. *, p < 0.05 versus WT. D, amounts of gene transcripts in 48-h cultures of SirEΔ determine by real-time PCR. Values are relative to those of WT. Data are means ± S.E. of three experiments. Error bars represent S.E. p < 0.05.
SirE up-regulates conidial development
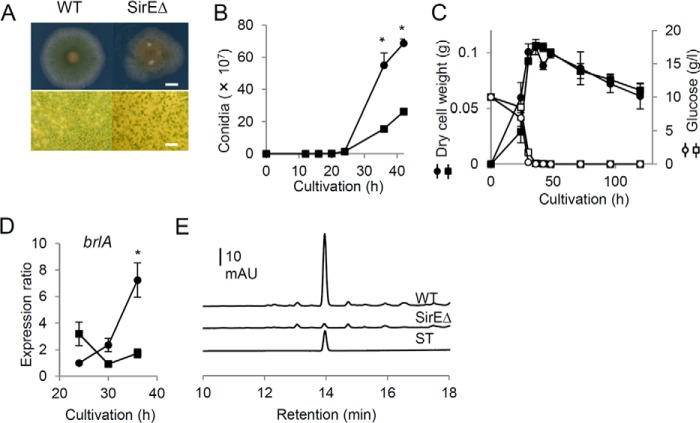
Macroscopic observation of fungal colonies on solid medium indicated that SirEΔ generated green colonies that exhibit less intense color than the WT, which microscopy showed was due to SirEΔ producing fewer conidiophores (Fig. 4A). Time-dependent changes in the numbers of conidia indicated that SirEΔ formed fewer conidia than the WT (Fig. 4B). The weight of SirEΔ and WT cells was the same (Fig. 4C), indicating that the difference was not due to a change in cell growth caused by a sirE deficiency. The expression of brlA, which is an essential transcription factor for initiating conidiophore development, was down-regulated in SirEΔ after 36 h in solid culture (Fig. 4D). Transcriptome analysis (see below) indicated that yA and wetA, the expression of which is induced by BrlA, were also down-regulated in SirEΔ (Table 1). These results showed that SirE induces conidiophore development during the stationary phase.
Figure 4.
Conidiophore development by WT and SirEΔ strains. A. nidulans A26 (WT) (● and ○) and SirEΔ (■ and □) were cultured on solid GMM at 30 °C. A, morphology of colonies after 72 h of culture. Scale bars, 10 mm (upper panel) and 10 μm (lower panel). B, time-dependent changes in numbers of conidia in colonies. C, cell mass (● and ■) and residual glucose (○ and □). D, transcript of brlA gene. Values are relative to actA transcript at 24 h, which was taken as 1. Data are means of three experiments. Error bars represent S.E. *, p < 0.05 versus WT. E, HPLC analysis of sterigmatocystin in mycelial extracts obtained from 48-h cultures. ST, commercial sterigmatocystin.
Table 1.
Genes for polysaccharide degradation and conidiophore development up-regulated by SirE during stationary phase
| Gene ID | Name | Putative products | SirEΔ/WTa | 24/48 hb | Carbon starvationc |
|---|---|---|---|---|---|
| log2R | log2R | log2R | |||
| Cellulose | |||||
| AN2828d | bglL | β-Glucosidase | −10.04 | −8.59 | 4.25 |
| AN4102 | bglA | β-Glucosidase | −2.45 | −0.98 | 4.54 |
| Hemicellulose | |||||
| AN8007d | abnC | α-1,5-l-Arabinosidase | −8.65 | −3.81 | 2.92 |
| AN8138d | aglC | α-Galactosidase | −5.42 | −3.83 | 1.67 |
| AN9380 | Chitin deacetylase | −1.48 | −0.08 | 8.14 | |
| Hemicellulose (pectin) | |||||
| AN2395d | β-Glucuronidase | −3.58 | −2.54 | 4.39 | |
| AN2463 | lacF | β-Galactosidase | −2.55 | −1.98 | −2.59 |
| Hemicellulose (mannan) | |||||
| AN2325d | Predicted mannanase | −6.59 | −3.32 | 5.17 | |
| AN1197d | Predicted mannanase | −5.32 | −1.07 | 2.94 | |
| AN0787d | mns1B | α-1,2-Mannosidase | −3.74 | −2.41 | 4.31 |
| Fungal cell wall degradation/remodeling proteins (autolytic enzymes) | |||||
| AN4825d | α-1,3-Glucosidase | −9.38 | −7.33 | 4.44 | |
| AN4871d | chiB | Chitinase class V | −7.92 | −4.15 | 3.47 |
| AN0245d | Predicted Glucanase | −6.11 | −1.80 | 5.62 | |
| AN1502d | nagA | N-Acetyl-β-glucosaminidase | −6.11 | −5.68 | 4.69 |
| AN9042d | agnC | α-1,3-Glucanase | −5.64 | −3.31 | 3.44 |
| AN0472d | engA | β-1,3-Glucosidase | −3.57 | −2.17 | 2.75 |
| AN0779d | β-1,3-Glucosidase | −2.34 | −1.21 | 1.34 | |
| Conidiophore development | |||||
| AN0973d | brlA | Transcription factor | −8.69 | −7.30 | 8.69 |
| AN6635 | yA | Conidial laccase | −3.69 | −0.22 | 3.69 |
| AN1937 | wetA | Regulatory protein of conidial development | −3.27 | −1.48 | 3.27 |
a Determined using DNA microarrays.
b Determined using mRNA sequencing.
c Determined using according to Szilágyi et al. (32).
d Overlap among genes up-regulated by SirE, stationary phase, and carbon starvation.
SirE up-regulates extracellular hydrolase and sterigmatocystin production
The total RNA of WT and SirEΔ cultured to the stationary growth phase was analyzed by high-throughput sequencing to identify genes that are regulated under the control of SirE. The total number of sequenced nucleotides covered 12-fold that of the A. nidulans genome, and 61–68% of the total reads were mapped to reference genes (supplemental Tables S2 and S3). Transcript profiles of WT and SirE were compared based on the numbers of fragments per kilobase in the exon region per million mapped reads. Among 9,000 genes (supplemental Table S4) comprising 82% of the total fungal genes, 701 and 748 were differentially expressed more or less than 2-fold, respectively (probability ≥ 0.8). Supplemental Table S5 shows 381 genes that were ≥8-fold down-regulated in SirEΔ. Gene ontology analysis indicated that a significant numbers of genes in the category of “biological process” were assigned to terms related to secondary metabolism (p < 1 × 10−6), which includes metabolic and biosynthetic processes of secondary metabolites, toxins, and sterigmatocystin (supplemental Table S6). This was consistent with the accumulation of sterigmatocystin in mycelia from the WT but not SirEΔ (Fig. 4E) and indicated that SirE up-regulates the biosynthesis of sterigmatocystin. Terms for conidiophore development included “asexual spore wall assembly” (p = 0.01) and “melanin biosynthetic process” (p = 6 × 10−4), which agree with the role of SirE in conidiophore development described above. The most abundant terms in the category of “molecular function” included “oxidoreductase activity,” “carboxylic ester hydrolase activity,” “heme binding,” and “tetrapyrrole binding” (p < 0.01 each).
In the category of “cellular component,” an abundant term was “extracellular region” (p = 6 × 10−8) (supplemental Table S6). This category contains (predicted) genes for polysaccharide degradation. The expression profiles of 100 genes for polysaccharide-utilizing enzymes listed by Saykhedkar et al. (30) were reanalyzed (supplemental Table S7). The results indicated that 20 of them were down-regulated ≥2-fold (Table 1) and that the genes for extracellular hydrolases were significantly down-regulated (p = 4 × 10−6) in SirEΔ. That the WT strain initiated the production of β-cellobiase, β-glucosidase, β-mannosidase, and β-galactosidase in 48-h cultures and continued to accumulate more, whereas those of SirEΔ generated less (supplemental Fig. S3), is consistent with the transcriptomes, which showed that SirEΔ accumulated fewer transcripts of the genes for β-glucosidase (bglL and bglA), α-1,2-mannosidase (mns1B), and β-galactosidase (lacF). SirEΔ produced less α-glucosidase and β-xylosidase activities than the WT (supplemental Fig. S3), whereas none of their encoding genes were altered, suggesting that WT expresses unannotated genes encoding these enzymes. Transcriptome analysis showed the cell wall-degrading hydrolases chiB, engA, and nagA (Table 1), which confirmed that SirE functions in up-regulating autolysis (Fig. 3).
Another transcriptome analysis found that the WT fungus produced more transcripts of genes for these polysaccharide hydrolases and autolytic enzyme genes during the stationary (48 h) than the exponential (24 h) growth phases (Table 1). These results indicated that SirE induces the expression of stationary-phase genes encoding extracellular hydrolases in addition to those for autolysis and conidiophore development.
Sirtuin E represses fungal primary metabolism during the stationary growth phase
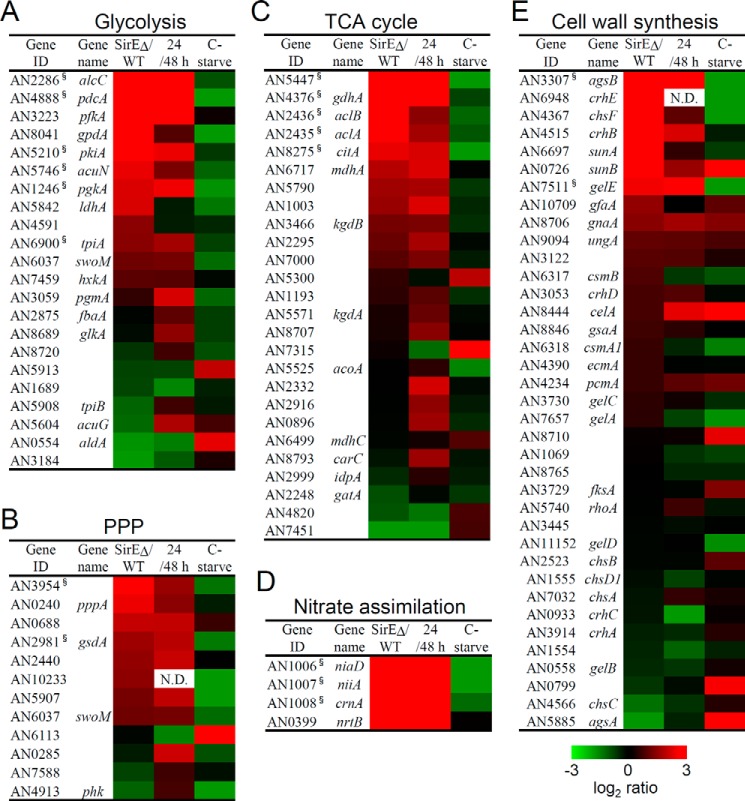
A transcriptomic comparison of WT and SirEΔ during the stationary growth phase revealed that SirEΔ up-regulated 300 genes ≥8-fold compared with the WT (supplemental Table S8). Their gene ontology (GO) was related to processes for organic acid metabolism and biosynthesis and for oxidoreductases with p < 0.01 (supplemental Table S6). These processes are related to fungal primary carbon metabolism; therefore, we selected the genes for the glycolytic and pentose phosphate pathways as well as the TCA cycle of this fungus. Fig. 5, A–C, show the expression of these genes as heat maps. Genes that were ≥2-fold up-regulated (supplemental Table S9) were significantly enriched in these gene sets (p < 0.01 each), indicating that these carbon metabolic processes were up-regulated in SirEΔ. We also found “nitrite transmembrane transporter activity” (p = 0.004) and “nitrite uptake transmembrane transporter activity” (p = 0.004) (supplemental Table S6). These terms were attached to genes for nitrate reductase (niaD), nitrite reductase (niiA), and nitrate transporters (nrtB, crnA, and nitA), which are used to assimilate nitrate as a nitrogen source (Fig. 5D). The transcriptome was also specifically enriched in the GO term “cell periphery” (p = 0.009) among the up-regulated genes of SirEΔ (supplemental Table S6). This term includes genes for synthesizing the cell wall, which is an essential mechanism of exponentially growing cells to shape normal mycelia. We found that SirE significantly down-regulated the 37 genes for cell-wall synthesis (Fig. 5E) described by de Groot et al. (31) (p = 0.008). Supplemental Table S9 lists those that were up-regulated in SirEΔ, among which genes for the major α-1,3-glucan synthase agsB, chitin synthase chsF, SUN family (for SIM1, UTH1, NCA3) proteins related to β-1,3-glucanases sunA and sunB, transglycosylase crhB, and β-1,3-transglycosylase gelE were up-regulated >4-fold.
Figure 5.
Transcriptome analysis of primary metabolism in A. nidulans. A. nidulans A26 (WT) and SirEΔ were cultured in liquid GMM at 30 °C. A–E, heat maps show ratios of fragments per kilobase in the exon region per million mapped reads between SirEΔ and WT at 48 h (stationary phase) determined by mRNA sequencing (SirEΔ/WT). Signal log ratios between exponential and stationary growth phase cultures of A. nidulans A26 were determined by DNA microarray analysis (24/48 h). Transcriptome data are from carbon-starved fungus (32) (C-starve). Genes for glycolysis (A), pentose phosphate pathway (PPP) (B), TCA cycle (C), and nitrate assimilation (D) were selected based on gene classification by KEGG. Genes for cell wall synthesis (E) were selected as described by de Groot et al. (31).
Growth phase-dependent changes in the transcriptome revealed by the DNA microarray analysis were compared with transcriptomic changes caused by sirE gene disruption (Fig. 5 and supplemental Table S9). The results indicated that significantly fewer transcripts of the genes described above for carbon and nitrogen utilization (which SirE down-regulated at stationary growth phase) were accumulated during the stationary than the exponential growth phase in the WT (p < 0.001). Most of the genes associated with the cell wall that were down-regulated by SirE were down-regulated during the stationary growth phase in the WT (Fig. 5). These findings indicated that A. nidulans SirE repressed these primary metabolic mechanisms at the level of transcription during the stationary growth phase.
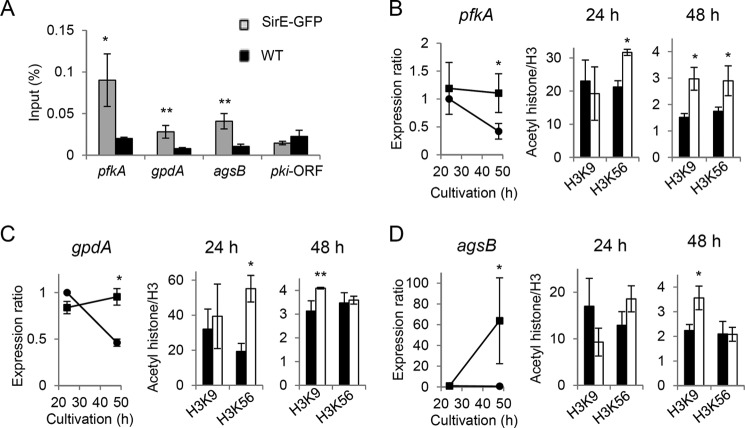
SirE deacetylates acetylated histone H3 to regulate primary metabolic genes
We selected pfkA and gpdA encoding 6-phosphofructokinase and glyceraldehyde-3-phosphate dehydrogenase, respectively, among the key glycolytic genes and agsB for cell-wall synthesis and investigated the SirE-dependent regulation of acetylhistones on the gene promoters. Chromatin immunoprecipitation (ChIP) using anti-GFP antibody and the SirE-GFP-producing strain (Fig. 1B) detected significant amounts of SirE-GFP binding to the promoters of the three genes but not to the coding region of pkiA (encoding pyruvate kinase; selected as the negative control; Fig. 6A). These findings indicated that SirE interacts with these primary metabolic genes promoters. Acetylation levels of histones on the gene promoters were measured by ChIP using anti-acetylhistone H3 antibodies. During the exponential growth phase (24 h), SirEΔ accumulated similarly high levels of H3K9ac on the pfkA promoter as the WT. Both strains accumulated normal amounts of pfkA transcripts, indicating that the acetylation level is sufficient for the gene promoter to be fully active. Upon entry into the stationary growth phase (48 h), both strains decreased the amount of H3K9ac on the gene promoter, whereas SirEΔ generated twice the amount of H3K9ac than the WT. These findings correlated with the amount of the pfkA transcript in SirEΔ being 2.6-fold higher than that in the WT (Fig. 6B) and are in agreement with the notion that SirE deacetylates H3K9ac to repress pfkA expression during the stationary growth phase. The H3K9ac levels on the gene promoters and transcripts of gpdA and agsB were also under the control of SirE (Fig. 6, C and D). These results indicated that SirE deacetylates H3K9ac to repress these primary metabolic genes during the stationary growth phase. By contrast, the levels of H3K56ac (Fig. 6) and of H3K18ac (supplemental Fig. S4) did not significantly correlate with the transcription of these genes, and the role of SirE in the acetylation of these residues remains obscure. Taken together, our results support the notion that SirE is a global repressor of primary metabolic processes in A. nidulans as it enters the stationary phase.
Figure 6.
Chromatin immunoprecipitation of primary metabolic genes. A, chromatin immunoprecipitation analyses using anti-GFP antibody determined SirE-GFP on the gene promoters of pfkA, gpdA, and agsB. A. nidulans producing SirE-GFP under gapA gene promoter (gray bars) and A26 (WT) (black bars) were cultured in GMM for 24 h. The coding region of pkiA (pkiA-ORF) served as a negative control. B–D, ChIP analyses measured acetylation levels on gene promoters of pfkA, gpdA, and agsB in A. nidulans A26 (WT) (black bars) and SirEΔ (white bars) cultured in GMM. Amounts of anti-H3K9ac and H3K56ac antibody-precipitated DNA are presented relative to that precipitated by anti-histone H3 antibody. The amount of DNA precipitated by negative controls without acetyllysine antibodies was <0.01. Left panels present levels of indicated gene transcripts. Data are means ± S.E. from three experiments. Error bars represent S.E. *, p < 0.05; **, p < 0.07.
Stationary phase regulation by SirE correlates with carbon starvation response
This study found that SirE targets and up-regulates the genes for autolysis, conidial development, and extracellular hydrolases and down-regulates those for primary carbon, nitrogen, and cell-wall metabolism. These are growth-phase dependent mechanisms among which some are reportedly regulated by carbon starvation. We compared the transcriptomes of the SirE-dependent genes with those of genes responding to carbon starvation (32). Venn diagrams showed that among 701 genes that were down-regulated by SirE, 490 and 311 were repressed during the stationary phase and by carbon starvation, respectively (Fig. 7A). This overlap included large populations of the primary metabolic genes described above and significant populations of the 215 genes involved in the glycolytic pathway, TCA cycle, pentose phosphate pathway, nitrate assimilation, and cell wall synthesis were shared by all three transcriptomes (Fig. 5 and supplemental Table S9). These results indicated that SirE-dependent repression during the stationary growth phase correlates with carbon starvation.
Figure 7.
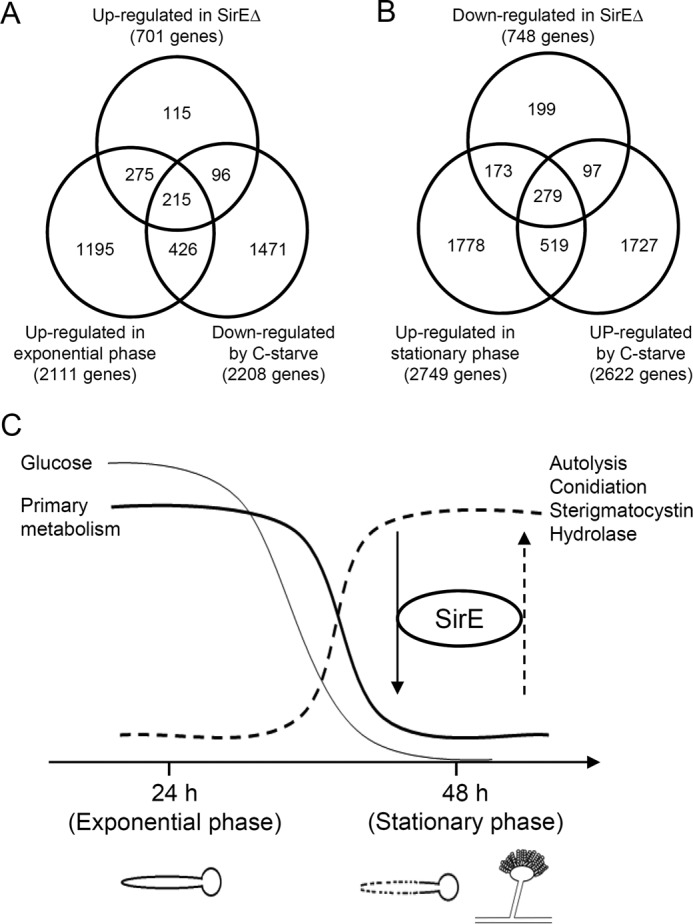
Correlation between sirE-dependent gene regulation and carbon starvation during growth phase. A and B, Venn diagrams show the numbers of genes identified by transcriptomes. Genes regulated by SirE (SirEΔ/WT), growth phase (24 h/48 h) (this study), and carbon starvation (C-starve) (42) were analyzed. A and B, up (≥1)- and down (≤−1)-regulated genes, respectively, in SirEΔ. C, model of transition from exponential to stationary growth phase driven by SirE.
Among the 748 genes up-regulated by SirE, 452 and 376 of them overlapped with those that were up-regulated during the stationary phase and by carbon starvation, respectively (Fig. 7B). Among these overlapping genes, 279 were common to the three transcriptomes and included the genes for fungal autolysis, conidiophore development, and carbon-assimilating hydrolases (Table 1). These findings imply that SirE-dependent transcription induction also correlates with the response to carbon starvation. The results of the comparison indicated that 30% of the up- or down-regulated genes were common between carbon starvation and the stationary phase (Fig. 7, A and B), which agrees with the general view that carbon starvation is a key environmental factor that causes the fungus to enter the stationary growth phase. Taken together, these results imply that SirE links gene regulation mechanisms during the stationary phase and during nutrient-poor conditions.
Discussion
This study identified the novel fungal sirtuin isozyme SirE. This isozyme functionally correlated with the nutrient (carbon) starvation associated with the stationary growth phase and repressed the global primary metabolism (Fig. 7C). Yeast and mammalian sirtuins typically respond to nutrient limitation by maintaining metabolic homeostasis. For example, S. cerevisiae sirtuins repress the expression of genes for glycolysis and cell-wall biosynthesis like A. nidulans (33). This indicates that these organisms produce sirtuins with common functions and the capacity to adapt to nutrient-poor conditions. We also found that SirE decreased the global acetylation of chromosomal histone (Fig. 1D) and of the more specific pfkA, gpdA, and agsB gene promoters (Fig. 6, B–D) under nutrient-poor conditions. That the S. cerevisiae sirtuins regulate chromatin structure and silence the genes for central metabolic pathways (33) suggests that the primary regulation of metabolism driven by histone deacetylation among fungi is evolutionarily conserved. This present finding indicated that another function of sirtuin is the regulation of A. nidulans events during the stationary growth phase that mammals and yeasts lack, namely autolysis, conidial development, sterigmatocystin production, and extracellular hydrolase production. Although known A. nidulans sirtuins regulate the production of secondary metabolites and conidia (9, 11), none of them regulates primary metabolism. No specific fungal transcription factor for synthesizing secondary metabolites regulates primary metabolism. Thus, to date, a fungal transcription regulator that participates in both types of metabolism has not been identified. Our results indicated that SirE is the missing coordinator of global primary and stationary types of metabolism that function during the transition from primary- to stationary-phase gene expression.
Typical sirtuins deacetylate acetylated histones and silence gene expression, which cannot account for the ability of SirE to induce the mechanisms of gene expression during stationary phase metabolism. The general mechanism of the sirtuin induction of gene expression remains obscure, but we speculate that nuclear SirE down-regulates transcription repressors and indirectly derepresses the expression of stationary-phase genes. The close correlations between the genes induced by SirE and those induced by carbon starvation implied that the functions of such repressors are associated with carbon and energy metabolism in the fungus. The A. nidulans candidate for such repressors is CreA that represses the transcription of genes for utilizing alternative carbon sources to glucose (21) and for the extracellular hydrolases. In addition, although direct interaction with carbon starvation has not been established, transcription factors that repressed conidial formation and sterigmatocystin synthesis were identified (14, 15). SirE might down-regulate these transcription repressors at the level of transcription through the deacetylation of nucleosomal histones on their encoding genes. Although these speculations must await further investigation, this is unlikely for CreA because the findings of our transcriptome analyses suggested that the amount of creA transcripts was not increased in SirEΔ compared with the WT.
We consistently found that A. nidulans is faced with nutrient-poor conditions during the stationary phase that affect cytosolic NADH and NAD+ levels (9), and NAD+ is an indicator of cellular energy status and stimulates mammalian and yeast sirtuins. The A. nidulans Nudix hydrolase A, which is an intrinsic hydrolase of cellular NAD+, counteracts the effect to maintain constant cellular NAD+ levels during the stationary growth phase (9). This implies that the amount of NAD+ must remain constant for SirE to regulate gene expression during the stationary phase. Rather, the transcription activation of sirE (Fig. 1C) is likely to be key for SirE to regulate gene expression during the stationary growth phase. Although little is known about A. nidulans and even S. cerevisiae transcription factors that induce sirtuin gene expression, members of the Forkhead box O (FOXO) family of transcription factors modulate the amount of mammalian SIRT1 expression in response to nutrient-poor conditions (34). This family of proteins is distributed across phyla, and the A. nidulans genome encodes at least four proteins with the Forkhead domain motif (AN4521/fhpA, AN8858/mcnB, AN2845, and AN4985) that could be the targets of an investigation of gene expression during the stationary phase and the roles of fungal sirtuin.
Eukaryotes utilize several sirtuin isozymes to deacetylate different histone substrates, and our finding of A. nidulans SirE enabled comparisons of sirtuin substrates. Class Ic sirtuins are unique to fungi among which only S. cerevisiae Hst3p and Hst4p have been proven to deacetylate H3K56ac and are involved in DNA damage signaling (27). The present study found that A. nidulans SirE of the same Ic class deacetylates H3K56ac, indicating that the physiological substrates of the class Ic sirtuins are conserved between these fungi. Whether the deacetylating role of H3K9ac and H3K18ac is also shared between them will be clarified by a study of the effects of S. cerevisiae sirtuins on these residues. The Class Ia sirtuin of A. nidulans (SirA) uses H4 Lys-16 like mammalian SIRT1 and S. cerevisiae Sir2p (9) in agreement with the substrate specificity of canonical Class Ia sirtuins conserved beyond phyla. However, SirA does not use H3 Lys-9 as a substrate (9) unlike mammalian and yeast SIRT1 and Sir2p that use H3K9ac. Because both H4K16ac and H3K9ac are believed to be common modifications in eukaryotes and because A. nidulans regulates the acetylation of H3 Lys-9, SirE could compensate for the absence of H3 Lys-9 deacetylation activity in SirA.
Microorganisms integrate complicated metabolic mechanisms during both the exponential and stationary growth phases, and the transition between the phases imposes a challenge because cellular metabolic mechanisms must be redirected from primary to very different secondary types. Histone modification in general can regulate both global and specific gene sets and can direct the genome-wide transcription changes required during the growth-phase transition. SirE is recognized as being the first transcription regulator that controls the temporal expression of both primary and secondary types of metabolic mechanisms. The present findings provide the basis for future detailed understanding of the function of SirE and could contribute to the production of organic acids and novel pharmaceuticals via two metabolic mechanisms.
Experimental procedures
Strains, cultures, and media
Conidia from A. nidulans A26 (biA1), A89 (biA1, argB2) (Fungal Genetic Stock center, University of Missouri), and ABPUN (biA1, argB2; pyrG89; yA2; pyroA4) (35) were rotary shaken (20 rpm) at 30 °C in 500-ml flasks containing 200 ml of glucose minimal medium (GMM) containing 1% glucose as a carbon source (36) for the indicated periods. Solid medium containing 1.5% agar was inoculated with 1 × 106 conidia from these strains and incubated at 37 °C. Biotin (0.25 mg liter−1), arginine (0.2 mg liter−1), pyridoxine (0.1 mg liter−1), uracil (1.12 g liter−1), and uridine (1.2 g liter−1) were added to the culture medium for auxotrophic mutants.
Gene disruption of sirE
Three mutually primed DNA fragments of the 5′- and 3′-untranslated regions of the sirE genes and the argB gene were amplified by PCR using genomic DNA of A. nidulans A26 and Prime STAR HS DNA polymerase (Takara, Kyoto, Japan). The second PCR included these fragments and KOD Plus DNA polymerase (Toyobo, Tokyo, Japan), and a DNA cassette was generated for gene disruption. Supplemental Table S1 shows the primers used in this study. After purification using UltraClean 15 DNA purification kits (MO BIO Laboratories, Carlsbad, CA), the cassette was introduced into A. nidulans A89 as described (36), and total DNA in transformants was analyzed by Southern blotting. Gene disruptants were selected, and one strain was designated SirEΔ.
Fluorescence microscopy
Three mutually primed DNA fragments of the gpdA promoter, the sirE or sirA gene, and the egfp gene were amplified by PCR using the primers shown in supplemental Table S1 to construct plasmid pSirE-gfp and pSirA-gfp and produce SirE- or SirA-GFP fusion protein. The cassette was purified, digested with XbaI and SacII or NotI, and cloned into the same restriction sites of pBSpyrG (36) containing the pyrG gene. Plasmids pSirE-gfp and pSirA-gfp were introduced into meiotic progenies of ABPUN and either SirEΔ or the sirA gene disruptant (9). The designated strains were named SirE-GFP and SirA-GFP. Conidia of the WT and these strains were incubated on glass coverslips in GMM at 25 °C for 24 h, stained with DAPI (Wako), and then analyzed using an Axio Observer Z1 microscope (Carl Zeiss, Jena, Germany) with a 38HE filter set (excitation, BP470/40; emission, BP525/50) for GFP and a 49 filter set (excitation, G365; emission, BP445/50) for DAPI. ZEN 2.1 (Carl Zeiss) was used for image processing and analysis.
Quantitative PCR
Total RNA was extracted from A. nidulans strains cultured in liquid GMM for 24 and 48 h or on solid GMM for 24, 30, and 36 h using the RNeasy plant mini kit (Qiagen, Hilden, Germany). Single-strand cDNA was synthesized using PrimeScriptTM reverse transcriptase (Takara), and then quantitative real-time PCR proceeded as described (37). Supplemental Table S1 shows the gene-specific primers. The amounts of transcripts were normalized against those of actA encoding actin and are shown as relative values.
Western blotting
Nuclear fractions prepared from A. nidulans strains after culture in GMM for 24 and 48 h (38) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a stacking gel as described by Laemmli (39) and a 15% acrylamide separating gel, blotted onto nitrocellulose membranes, and incubated with anti-histone H3 (acetyl-Lys-9), anti-histone H3 (acetyl-Lys-18), anti-histone H3 (acetyl-Lys-56), anti-histone H3, anti-histone H4 antibodies ab10812, ab1191, ab76307, ab1791, and ab1015 (Abcam, Cambridge, UK) and anti-acetylhistone H4 (Lys-16) antibody 07-329 (Millipore, Billerica, MA) in TBS (25 mm Tris-HCl (pH 8.0), 140 mm NaCl, 3 mm KCl) containing 0.1% Tween 20 at a 1:1,000 dilution for 2 h. After three washes with TBS buffer, blots were incubated with a goat anti-rabbit IgG conjugated with horseradish peroxidase (Pierce) at a 1:20,000 dilution. Chemiluminescence was detected and quantified using the ECL detection system (GE Healthcare) and ImageQuant LAS4000 Mini (GE Healthcare).
Determination of mycelial mass, glucose, and autolysis
Mycelial dry weight was determined after liquid culture as described (17) and after growth on cellophane membranes covering solid GMM. Glucose in liquid culture filtrates or melted agar was determined using Glucose CII test kits (Wako). Culture supernatants were separated by centrifugation at 10,000 × g for 5 min, and then the optical density was spectroscopically determined in the supernatant by measuring absorbance at 600 nm. Fungal cell viability was determined using the AB cell viability reagent (Invitrogen). Fungal cultures (0.5 ml) and fresh GMM (1 ml) were mixed with the AB reagent (10% final concentration) in 24-well plates and incubated at 37 °C for 4 h in the dark, and then the absorbance of supernatants was analyzed at 570 and 600 nm. The ratio (%) of the reduction in AB was calculated as described (28).
Extracellular enzyme assays
Enzyme activity was measured in fungal culture supernatants obtained by centrifugation at 10,000 × g for 5 min. Chitinase activity was determined by liberating reducing sugars from colloidal chitin as described (40), and β-1,3-glucanase activity was assayed by modifying the published method (41) as follows. Culture supernatants (62.5 μl) and 10% laminarin (62.5 μl) were incubated at 37 °C for 16 h. Dinitrosalicylic acid reagent (375 μl) was added, the mixture was boiled for 5 min, and then precipitates were removed by centrifugation. The resulting solution was diluted, and absorption was determined at 535 nm. One unit was defined as the amount of chitinase and glucanase required to produce 1 μmol of N-acetylglucosamine and glucose/min, respectively. Other hydrolytic enzyme activities were measured in 25 mm sodium acetate (pH 6.0) at 37 °C. After enzyme reactions, the amounts of p-nitrophenol liberated from the 2 mm substrates p-nitrophenyl-α-d-glucopyranoside (α-glucosidase), p-nitrophenyl-β-d-glucopyranoside (β-glucosidase), N-acetyl-β-d-glucosaminide (N-acetyl-β-d-glucosaminidase), p-nitrophenyl-β-d-cellobioside (β-cellobiase), p-nitrophenyl-β-d-galactopyranoside (β-galactosidase), p-nitrophenyl-β-d-xyllopyranoside (β-xylosidase), and p-nitrophenyl-β-d-mannopyranoside (β-mannosidase) (Sigma) were quantified by measuring absorbance at 405 nm after adding equal amounts of 2 m sodium carbonate. A molar extinction coefficient of 18.5 mm−1 cm−1 was used for p-nitrophenylphenol. One unit was defined as the amount of enzyme required to produce 1 nmol of p-nitrophenol/min.
RNA sequencing
A. nidulans A26 and SirEΔ were cultured in 200 ml of GMM at 30 °C for 48 h, harvested by filtration, immediately frozen in liquid nitrogen, and powdered to prepare total RNA using RNeasy plant mini kits. Complementary DNA libraries were prepared from the total RNA, and nucleotides (∼1 Gb) were sequenced using a HiSeq4000 (Illumina, San Diego, CA) with a sequencing configuration for 50-bp single reads. Nucleotide sequences with Phred scores (Q) <20 and adapters were removed, and raw reads were filtered into clean reads and aligned to the reference A. nidulans FGSC A4 genomic DNA sequence (NCBI BioProject PRJNA13961) using Burrows-Wheeler Aligner (BWA) (42). Clean reads were mapped to the reference genes using Bowtie 2 (43). Gene expression was quantified using the RSEM software package (44). Significantly up-regulated or down-regulated genes (fold change ≥3) were detected using mean values of fragments per kilobase of exon per million mapped fragments and by calculating data from biological duplicates (probability ≥ 0.8). The RNA sequencing data are available under the NCBI BioProject (PRJNA337983) in GenBankTM. GO was analyzed using GO Term Finder published in AspGD (45). The genes were clustered to specific metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (46).
DNA microarray analysis
A. nidulans strain A26 was cultured in 200 ml of GMM at 30 °C for 24 and 48 h, and then total RNA was purified as described above. Transcriptomes were analyzed using a customized A. nidulans GeneChip as described (47). The signal log2 ratio between the data for the 24- and 48-h cultures was normalized by subtracting the difference between medians of data sets. Data have been published in the Gene Expression Omnibus (GEO) under accession number GSE85319.
Determination of sterigmatocystin
After culturing A. nidulans strains in GMM at 30 °C for 48 h, mycelia were collected by filtration and lyophilized. Ethyl acetate extracts were obtained from 10-mg samples overnight, dried in vacuo, and suspended in 200 μl of methanol for high-performance liquid chromatography (HPLC) separation using a 1200 Infinity Series instrument (Agilent Technologies, Palo Alto, CA) equipped with a 250 × 4.6-mm Purospher® Star RP-18 end-capped column with a particle size of 5 μm (Millipore). The solvent gradient comprising acetonitrile (solvent B) in 0.05% ammonium formate, water (pH 4.0; solvent A) was as follows: 40% B from 0 to 5 min, 40–80% B from 5 to 10 min, held at 80% B from 10 to 15 min, 80–100% B from 15 to 25 min, held at 100% B from 25 to 27 min, 100–60% B from 27 to 29 min, and held at 60% B from 29 to 35 min. The flow rate was 0.8 ml min−1, and absorption at 254 nm was monitored. The temperature of the column oven was 40 °C.
Chromatin immunoprecipitation
ChIP proceeded as described (48) by means of the antibodies (2 μg ml−1) that were used for Western blotting and 1 μg ml−1 anti-GFP antibody (catalogue number 3999-100, BioVision, Milpitas, CA). The QIAquick PCR purification kit (Qiagen) was used to purify DNA, which was then analyzed by quantitative real-time PCR as described above with gene-specific primers (supplemental Table S1). Relative amounts of DNA were calculated by dividing the amount of immunoprecipitated DNA by that of the input DNA.
Author contributions
N. T. supervised all laboratory experiments. E. I. designed experiments; acquired, analyzed, and interpreted all results; and wrote the manuscript. N. T. and M. S. designed, acquired, and interpreted all experiments and helped to prepare the manuscript. R. O. helped with fluorescence microscopy. K.-I. O. and S. M. supervised and helped write the manuscript. All authors read and approved the final submission.
Supplementary Material
Acknowledgment
We thank Norma Foster for critical reading of the manuscript.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grants-in-aid for Scientific Research 15H02487 and 24380041 (to N. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4 and Tables S1–S9.
- HDAC
- histone deacetylase
- sirtuin
- silent information regulator 2 protein
- AB
- alamarBlue
- NLS
- nuclear localization signal
- TCA
- tricarboxylic acid
- SirE
- sirtuin E
- H4K16ac
- acetylated lysine residue 16 on histone H4
- H3K9ac
- acetylated Lys residue 9 of histone H3
- H3K18ac
- acetylated Lys residue 18 of histone H3
- H3K56ac
- acetylated Lys residue 56 of histone H3
- GO
- gene ontology
- GMM
- glucose minimal medium
- ID
- identity.
References
- 1. Imai S., Armstrong C. M., Kaeberlein M., and Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 2. Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., and Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Michan S., and Sinclair D. (2007) Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frye R. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 5. Li M., Petteys B. J., McClure J. M., Valsakumar V., Bekiranov S., Frank E. L., and Smith J. S. (2010) Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell. Biol. 30, 3329–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., and Gu W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 7. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., and Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 8. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., and Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimizu M., Masuo S., Fujita T., Doi Y., Kamimura Y., and Takaya N. (2012) Hydrolase controls cellular NAD, sirtuin, and secondary metabolites. Mol. Cell. Biol. 32, 3743–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernandez J., Marroquin-Guzman M., Nandakumar R., Shijo S., Cornwell K. M., Li G., and Wilson R. A. (2014) Plant defence suppression is mediated by a fungal sirtuin during rice infection by Magnaporthe oryzae. Mol. Microbiol. 94, 70–88 [DOI] [PubMed] [Google Scholar]
- 11. Shwab E. K., Bok J. W., Tribus M., Galehr J., Graessle S., and Keller N. P. (2007) Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6, 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawauchi M., Nishiura M., and Iwashita K. (2013) Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA. Eukaryot. Cell 12, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kawauchi M., and Iwashita K. (2014) Functional analysis of histone deacetylase and its role in stress response, drug resistance and solid-state cultivation in Aspergillus oryzae. J. Biosci. Bioeng. 118, 172–176 [DOI] [PubMed] [Google Scholar]
- 14. Yu J. H., Butchko R. A., Fernandes M., Keller N. P., Leonard T. J., and Adams T. H. (1996) Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29, 549–555 [DOI] [PubMed] [Google Scholar]
- 15. Adams T. H., Boylan M. T., and Timberlake W. E. (1988) brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362 [DOI] [PubMed] [Google Scholar]
- 16. Szilágyi M., Kwon N. J., Dorogi C., Pócsi I., Yu J. H., and Emri T. (2010) The extracellular β-1,3-endoglucanase EngA is involved in autolysis of Aspergillus nidulans. J. Appl. Microbiol. 109, 1498–1508 [DOI] [PubMed] [Google Scholar]
- 17. Yamazaki H., Yamazaki D., Takaya N., Takagi M., Ohta A., and Horiuchi H. (2007) A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr. Genet. 51, 89–98 [DOI] [PubMed] [Google Scholar]
- 18. Kim S., Matsuo I., Ajisaka K., Nakajima H., and Kitamoto K. (2002) Cloning and characterization of the nagA gene that encodes β-N-acetylglucosaminidase from Aspergillus nidulans and its expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66, 2168–2175 [DOI] [PubMed] [Google Scholar]
- 19. Shroff R. A., O'Connor S. M., Hynes M. J., Lockington R. A., and Kelly J. M. (1997) Null alleles of creA, the regulator of carbon catabolite repression in Aspergillus nidulans. Fungal Genet. Biol. 22, 28–38 [DOI] [PubMed] [Google Scholar]
- 20. Espeso E. A., and Peñalva M. A. (1992) Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol. Microbiol. 6, 1457–1465 [DOI] [PubMed] [Google Scholar]
- 21. Ruijter G. J., and Visser J. (1997) Carbon repression in aspergilli. FEMS Microbiol. Lett. 151, 103–114 [DOI] [PubMed] [Google Scholar]
- 22. Avalos J. L., Bever K. M., and Wolberger C. (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17, 855–868 [DOI] [PubMed] [Google Scholar]
- 23. Sherman J. M., Stone E. M., Freeman-Cook L. L., Brachmann C. B., Boeke J. D., and Pillus L. (1999) The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell 10, 3045–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frye R. A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 25. Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T. L., Barrett J. C., Chang H. Y., Bohr V. A., Ried T., Gozani O., and Chua K. F. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R. I., Paredes S., Young N. L., Chen K., Struhl K., Garcia B. A., Gozani O., Li W., and Chua K. F. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang B., Miller A., and Kirchmaier A. L. (2008) HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol. Biol. Cell 19, 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al-Nasiry S., Geusens N., Hanssens M., Luyten C., and Pijnenborg R. (2007) The use of Alamar Blue assay for quantitative analysis of viability, migration and invasion of choriocarcinoma cells. Hum. Reprod. 22, 1304–1309 [DOI] [PubMed] [Google Scholar]
- 29. Emri T., Molnár Z., Pusztahelyi T., and Pócsi I. (2004) Physiological and morphological changes in autolyzing Aspergillus nidulans cultures. Folia Microbiol. 49, 277–284 [DOI] [PubMed] [Google Scholar]
- 30. Saykhedkar S., Ray A., Ayoubi-Canaan P., Hartson S. D., Prade R., and Mort A. J. (2012) A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol. Biofuels 5, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Groot P. W., Brandt B. W., Horiuchi H., Ram A. F., de Koster C. G., and Klis F. M. (2009) Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 46, S72–S81 [DOI] [PubMed] [Google Scholar]
- 32. Szilágyi M., Miskei M., Karányi Z., Lenkey B., Pócsi I., and Emri T. (2013) Transcriptome changes initiated by carbon starvation in Aspergillus nidulans. Microbiology 159, 176–190 [DOI] [PubMed] [Google Scholar]
- 33. Li M., Valsakumar V., Poorey K., Bekiranov S., and Smith J. S. (2013) Genome-wide analysis of functional sirtuin chromatin targets in yeast. Genome Biol. 14, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nemoto S., Fergusson M. M., and Finkel T. (2004) Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 35. Motoyama T., Fujiwara M., Kojima N., Horiuchi H., Ohta A., and Takagi M. (1997) The Aspergillus nidulans genes chsA and chsD encode chitin synthases which have redundant functions in conidia formation. Mol. Gen. Genet. 253, 520–528 [DOI] [PubMed] [Google Scholar]
- 36. Takasaki K., Shoun H., Yamaguchi M., Takeo K., Nakamura A., Hoshino T., and Takaya N. (2004) Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. J. Biol. Chem. 279, 12414–12420 [DOI] [PubMed] [Google Scholar]
- 37. Shimizu M., Fujii T., Masuo S., Fujita K., and Takaya N. (2009) Proteomic analysis of Aspergillus nidulans cultured under hypoxic conditions. Proteomics 9, 7–19 [DOI] [PubMed] [Google Scholar]
- 38. Lee I., Oh J.-H., Keats Shwab E. K., Dagenais T. R., Andes D., and Keller N. P. (2009) HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 46, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Yanai K., Takaya N., Kojima N., Horiuchi H., Ohta A., and Takagi M. (1992) Purification of two chitinases from Rhizopus oligosporus and isolation and sequencing of the encoding genes. J. Bacteriol. 174, 7398–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saikia R., Singh B. P., Kumar R., and Arora D. K. (2005) Detection of pathogenesis-related proteins—chitinase and β-1,3-glucanase in induced chickpea. Curr. Sci. 89, 659–663 [Google Scholar]
- 42. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cerqueira G. C., Arnaud M.B., Inglis D.O., Skrzypek M.S., Binkley G., Simison M., Miyasato S. R., Binkley J., Orvis J., Shah P., Wymore F., Sherlock G., and Wortman J.R. (2014). The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 42, D705–D710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kanehisa M., Furumichi M., Tanabe M., Sato Y., and Morishima K. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Masuo S., Terabayashi Y., Shimizu M., Fujii T., Kitazume T., and Takaya N. (2010) Global gene expression analysis of Aspergillus nidulans reveals metabolic shift and transcription suppression under hypoxia. Mol. Genet. Genomics 284, 415–424 [DOI] [PubMed] [Google Scholar]
- 48. Bernreiter A., Ramon A., Fernández-Martínez J., Berger H., Araújo-Bazan L., Espeso E. A., Pachlinger R., Gallmetzer A., Anderl I., Scazzocchio C., and Strauss J. (2007) Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell. Biol. 27, 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.