Abstract
Conifers II (cupressophytes), comprising about 400 tree species in five families, are the most diverse group of living gymnosperms. Their plastid genomes (plastomes) are highly variable in size and organization, but such variation has never been systematically studied. In this study, we assessed the potential mechanisms underlying the evolution of cupressophyte plastomes. We analyzed the plastomes of 24 representative genera in all of the five cupressophyte families, focusing on their variation in size, noncoding DNA content, and nucleotide substitution rates. Using a tree-based method, we further inferred the ancestral plastomic organizations of internal nodes and evaluated the inversions across the evolutionary history of cupressophytes. Our data showed that variation in plastome size is statistically associated with the dynamics of noncoding DNA content, which results in different degrees of plastomic compactness among the cupressophyte families. The degrees of plastomic inversions also vary among the families, with the number of inversions per genus ranging from 0 in Araucariaceae to 1.27 in Cupressaceae. In addition, we demonstrated that synonymous substitution rates are significantly correlated with plastome size as well as degree of inversions. These data suggest that in cupressophytes, mutation rates play a critical role in driving the evolution of plastomic size while plastomic inversions evolve in a neutral manner.
Keywords: conifer, cupressophyte, plastome reduction, compaction, inversion, mutation rate
Introduction
In land plants, plastid genomes (plastomes) typically contain 90–100 genes and range from 120 to 160 kb (Green 2011). In contrast to mitochondrial genomes, with overwhelming amount of noncoding DNA (Christensen 2014), plastomes are characterized by dense genes, estimated at about 0.73–1.01 genes per kb (Jansen and Ruhlman 2012). Therefore, expansion or reduction of plastomes was thought mainly to be associated with a gain or loss of genes.
Pelargonium x hortorum has the largest known plastome (217,942 bp in size; Chumley et al. 2006) whose inverted repeat (IR) contains 39 duplicated genes and is approximately 3-fold longer than those of other angiosperms (typically 20–30 kb) (Wicke et al. 2011). The smallest plastome sequenced to date is within the endo-parasitic species of Pilostyles, containing only five functional genes (Bellot and Renner 2015). Notably, loss of many photosynthetic genes from the plastomes of parasitic plants clearly indicates selection for removal of nonessential genes (Krause 2008). Other suggested benefits of selectively reduced plastomes include an increase in replication rates (McCoy et al. 2008) and streamlining available resources (Wu et al. 2009). Unfortunately, the viewpoint that selection favors reduction suggests a one-way ticket of evolution, which cannot explain expanded plastomes within a group of closely related plants.
Lynch and Conery (2003) proposed that genome size is positively associated with degree of random genetic drift but inversely with mutation rates. Subsequently, mutation rates rather than random genetic drift were considered the major driving force in organelle genomic evolution (Lynch et al. 2006). The finding that bi- and uni-parentally inherited plastomes do not significantly differ in size or compactness supports the low effect of random genetic drift on plastomic evolution (Crosby and Smith 2012). However, the inverse relationship between genome size and mutation rates was recently challenged by the co-existence of enlarged genome and accelerated synonymous substitution rates in the mitochondrial genomes of Silene (Sloan et al. 2012). Although a handful of exceptions have been noted, the effect of random genetic drift and mutation rates gives an alternative explanation for organelle genomic evolution (Smith 2016).
Similarly, suggested mechanisms responsible for plastomic rearrangements are diverse. Some studies have shown that rearrangement breakpoints are associated with short repeats, regardless of whether the plastomes contain IRs (Haberle et al. 2008; Guisinger et al. 2011; Weng et al. 2014) or lack IRs (Wu, Lin, et al. 2011; Guo et al. 2014; Li et al. 2016). Lack of efficient DNA repair proteins was also proposed as a potential mechanism linking the highly rearranged plastomes of Geraniaceae (Guisinger et al. 2011; Weng et al. 2014). On the basis of certain conserved gene clusters in most plastomes, plastomic rearrangements were hypothesized to be selectively constrained (Green 2011; Wicke et al. 2011). However, disruptions of conserved gene clusters were recently reported in two cupressophyte genera, Taxus (Hsu et al. 2014) and Sciadopitys (Hsu et al. 2016). Therefore, whether there is selective constraint on plastomic rearrangements of cupressophytes is still an open question.
Cupressophytes, a group of conifers other than Pinaceae (or conifers I), were designated as conifers II (Ran et al. 2010). They comprise two orders—Cupressales (including Cupressaceae, Sciadopityaceae, and Taxaceae) and Araucariales (containing Araucariaceae and Podocarpaceae)—and have diverged for more than 251 Myr (Leslie et al. 2012). The plastomes of both cupressophytes and Pinaceae have lost IRs, but they differ in several aspects. All 11 plastid NDH genes are retained in cupressophytes, but these genes are absent or pseudogenized in Pinaceae (Braukmann et al. 2009). As compared to Pinaceae, Cryptomeria japonica, a cupressophyte species, shows 2.1–4.4% increased gene content (Lin et al. 2010). Whether this finding indicates that cupressophyte plastomes have undergone compaction requires comparative inspection. Among Pinaceae genera, variation in the plastomic organization is limited to inversions of two specific long fragments (Wu, Lin, et al. 2011). In contrast, plastomic inversions are diverse in cupressophytes. For example, two Cupressaceous genera, Calocedrus and Cryptomeria, are distinguished from each other by five long plastomic inversions (Wu and Chaw 2014).
Using plastomic data of six genera, we previously showed that cupressophyte plastomes are labile in their genome size and organization (Wu and Chaw 2014). As of May 2016, plastomes of 21 cupressophyte genera were available in GenBank (supplementary table S1, Supplementary Material online). To increase the diversity of samples, we determined four plastomes representing three and one key genera that have never been sequenced in the two largest cupressophyte families—Cupressaceae (149 species of trees and shrubs; Christenhusz and Byng 2016) and Podocarpaceae (187 species of trees and shrubs), respectively. Therefore, we could conduct the broadest plastomic analysis across 24 genera of the five cupressophytes families to date. Using other seed plant plastomes as references, we examined whether variation in the plastome size of cupressophytes is toward expansion or reduction. Furthermore, we used a tree-based method to infer the ancestral plastomic organization in the tree nodes of cupressophytes, then calculated the inversions between species. These analyses allowed us to estimate plastomic inversions across the evolutionary history of cupressophytes. Finally, we assessed potential mechanisms underlying the evolution of plastome size and inversion in cupressophytes.
Materials and Methods
Plastome Sequencing, Assembly, and Annotation
Total DNA was extracted from 2 g fresh leaves of Callitris rhomboidea (voucher Chaw 1505), Chamaecyparis formosensis (voucher Chaw 1506), Dacrycarpus imbricatus (voucher Chaw 1507), and Taxodium distichum (voucher Chaw 1508) using a CTAB protocol (Stewart and Via 1993) with 10 mg/L PVP-40K (Sigma). For each species, approximately 2 GB of 90-bp paired-end reads were sequenced on an Illumina GAII at Yourgene Bioscience (New Taipei City). After removal of adapters and quality trimming with a 0.05 error probability, reads were de novo assembled using CLC Genomics Workbench 5.5.1 (CLC Bio) with the options: word size = 31, bubble size = 50, minimum contig length = 1 kb, and mapping reads to contigs. Contigs with ≥ 30X sequencing depths were blast-searched against the plastome of Agathis dammara (NC023119) and those with <10−10E-values were considered plastomic contigs. Gaps between the plastomic contigs were filled with PCR amplicons obtained from specific primers locating on flanking regions of the contigs. The finished plastomes were imported into CLC Genomics Workbench to estimate average sequencing depths with the mapping parameters: mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, and similarity fraction = 0.8. Plastome annotation was performed in DOGMA (Wyman et al. 2004), and tRNA genes were predicted using tRNAscan-SE 1.21 (Schattner et al. 2005).
Exploration of Repeats
IRs were detected by using a BLASTN search against the examined plastome itself with the default settings and sequence identity cutoff = 90%. We discarded repeats < 200 bp.
Phylogenetic Analysis and Estimation of Nucleotide Substitution Rates
We extracted 80 common plastid protein-coding genes from Cycas and 24 sampled cupressophytes. Sequences of genes were aligned using ClustralW (codons) with the default settings implemented in Mega 6.06 (Tamura et al. 2013). We used RAxML v8.2.4 (Stamatakis 2014) to analyze a maximum likelihood (ML) tree based on concatenation of the 80 genes and a GTR + G + I model. Cycas was designated as an outgroup. Bootstrapping supports for the tree nodes were assessed from 1,000 nonparametric pseudo-replicates. This ML tree was the constraint topology for estimating synonymous (ds) and nonsynonymous (dn) substitution rates with the codeml program of PAML 4.8 (Yang 2007). The parameters were runmode = 0, seqtype = 1, CodonFreq = 2, estFreq = 0, and model = 1.
Correlation Analysis Using Phylogenetically Independent Contrasts Method
Species evolve in a hierarchical tree structure. Their traits are in general not phylogenetically independent. To avoid the effect of the phylogenetic nonindependence, we adopted the method proposed by Felsenstein (1985). This method computes differences or the so-called “contrasts” between examined traits of paired species rather than directly uses traits in correlation analyses. We used the R package “ape” to transform plastome size, noncoding content, and nucleotide substitution rates into contrasts that were independent of phylogenetic tree topology. The incorporated tree was the ML tree excluding the taxon Cycas taitungensis. We evaluated relatedness of the trasts using Spearman’s rank correlation test.
Identification of Locally Co-Linear Blocks
The progressiveMauve implemented in Mauve 2.4.0 (Darling et al. 2010) was used for multiple plastome alignment between Cycas and the 24 cupressophytes to identify locally co-linear blocks (LCBs). The plastome of Cycas was the reference because it likely retains the ancestral gene order for the seed plant plastome (Jansen and Ruhlman 2012). Before alignment, sequences of IRA were removed from the Cycas plastome because previous studies suggested loss of IRA from the plastomes of cupressophytes (Wu, Wang, et al. 2011; Wu and Chaw 2014). LCBs shared by all examined taxa were collected to generate a matrix of LCB order.
Reconstruction of Ancestral Plastomes and Calculation of Plastomic Inversions
Ancestral plastomes were inferred by using MLGO (Lin et al. 2013) with the ML tree and the LCB matrix mentioned above. We used GRIMM (Tesler 2002) to calculate the optimal number of inversions between plastomes.
Visualization of Plastome Maps
We used Circos 0.67 (Krzywinski et al. 2009) to draw plastome maps with their relative LCB orders. For the LCBs in inferred ancestral plastomes, their lengths were presumed to be the averages calculated from the corresponding LCBs between the two closest descendants. For example, the LCB lengths for the ancestral plastome C1 are the average lengths of the corresponding LCBs between Cupressus and Juniperus (fig. 4).
Fig. 4.—
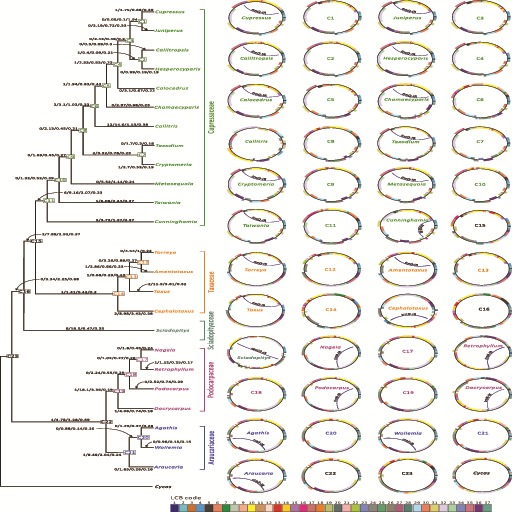
Estimates of inversion numbers, synonymous (ds), and nonsynonymous (dn) substitution rates in plastomes across the evolution of cupressophytes. The relationships among the 24 cupressophytes and their putative ancestors (nodes marked C1–C23) are indicated in the left tree modified from the ML tree in figure 1. Values on tree branches from left to right are inversion numbers, ds (×10−2), dn (×10−2), and dn/ds ratio. Ancestral and extant plastomes are visualized as circular maps at the right panel. Color boxes are locally co-linear blocks (LCBs) identified by comparisons of plastome sequences between the 24 cupressophytes and Cycas, with the latter’s IRA removed before the comparison. Within maps, specific IRs ≥200 bp are denoted and named by specific genes residing within them. Thin blue lines connecting two specific IRs show that homologous recombination might result.
Statistical Analysis
All statistical analyses involved use of R v3.2.0 (https://www.r-project.org/).
Results
Characteristics of the Four Newly Sequenced Plastomes
We elucidate the complete plastome sequences of three Cupressaceous species, Ca. rhomboidea (LC177555), Ch. formosensis (LC177668), and Ta. distichum (LC177556), and one Podocarpaceous species, Da. imbricatus (LC177621). Our assembly and read mapping confirmed that sequencing depths of the four plastome sequences were >100× (table 1). The four plastomes are circular molecules ranging from 121,117 to 133,811 bp, with the GC content from 34.71% to 37.23% (table 1; supplementary fig. S1, Supplementary Material online). They are not quadripartite. Among them, Callitris has the smallest set of plastid genes, only 115, whereas the remaining three have 121 plastid genes even though their plastome sizes differ from each other by 1.9 to 6.6 kb. Compared with the other two Cupressaceous species, Callitris has lost seven plastid genes (i.e., clpP, rps16, trnG-UCC, trnM-CAU, trnS-GGA, trnT-UGU, and trnV-UAC) but contains a duplicated rrn5 in the region between rrn16 and trnV-GAC.
Table 1.
Newly Sequenced Plastomes and Their Assembly Results
| Taxon | Size (bp) | Sequence depth (X) | GC content (%) |
|---|---|---|---|
| Callitris rhomboidea | 121,117 | 215.8 | 34.71 |
| Chamaecyparis formosana | 127,211 | 117.4 | 34.99 |
| Dacrycarpus imbricatus | 133,811 | 308.5 | 37.23 |
| Taxodium distichum | 131,954 | 244.1 | 35.26 |
The plastomes of Chamaecyparis, Dacrycarpus, and Taxodium have retained the conserved gene cluster rps2–atpI–atpH–atpF–atpA (abbreviated as rps2 gene cluster). However, in Callitris, the rps2 gene cluster splits into two sub-clusters (i.e., rps2–atpI and atpH–atpF–atpA) by a distance of 38.5 kb (supplementary fig. S2, Supplementary Material online). Among elucidated cupressophyte plastomes, disruption of the rps2 gene cluster was previously reported only in Sciadopitys, with two sub-clusters (i.e., rps2–atpI–atpH–atpF and atpA) separated by a distance of 82.5 kb (Hsu et al. 2016; also see supplementary fig. S2, Supplementary Material online). Clearly, the rps2 gene cluster has been differentially disrupted at least two times in the evolutionary history of cupressophytes.
Extensive Variation in Plastome Size among Cupressophytes
We compared 24 cupressophyte species representing 24 different genera (supplementary table S1, Supplementary Material online). From concatenation of their 82 protein-coding genes, our phylogenetic analysis uncovered two subclades within cupressophytes. One is the Northern Hemisphere species, consisting of Cupressaceae (except Callitris, which is native to Australia), Taxaceae, and Sciadopityaceae, and the other the Southern Hemisphere species, including Araucariaceae and Podocarpaceae (fig. 1). The plastomes of Araucariaceae are the largest, ranging from 145.6 to 146.7 kb. In contrast, those of the other four families are <140 kb, with the Cupressaceae having the smallest, 129.2 ± 3.6 kb, on average.
Fig. 1.—
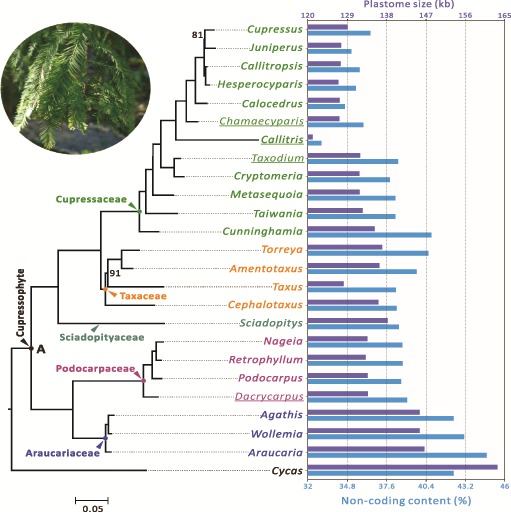
Variation in plastome size and noncoding content (%) among the sampled 24 cupressophyte genera. A maximum likelihood (ML) tree inferred from 80 plastid protein-coding genes is shown at the left with Cycas as the outgroup. Bootstrapping supports are shown when they are < 100%. For each genus, its plastome size and noncoding content are indicated by the colored bars in the right panel, with scales labeled on the top and bottom, respectively. Taxa sequenced in this study are underlined. Node A denotes the putative common ancestor of cupressophytes. Noncoding content (%) indicates the proportion of intergenic and intronic sequences.
In cupressophytes, larger plastomes tend to have higher proportions of intergenic and intronic sequences (hereafter designated noncoding content). Among the deciphered plastomes of cupressophytes (NCBI data up to May 2016), Araucaria is the largest and also has the highest noncoding content (44.6%), even higher than Cycas (42.2% in fig. 1). The plastome of Callitris is not only the smallest but also the most compact (33% noncoding content). These data prompted us to ask two questions. First, Have the plastomes of Araucariaceae undergone expansion? In other words, Has reduction occurred in non-Araucariaceae cupressophytes? Second, Are changes in noncoding content associated with variation in the plastome size across the cupressophyte families? To answer these questions, we included samples from five other major seed plant lineages for comparison.
Compaction Responsible for Reduction in Cupressophyte Plastomes
In figure 2, the plastome size and noncoding content were plotted for the 47 available plastomes of the five gymnosperm groups, with one species representing one genus. The angiosperms included a representative for each of the 59 sampled angiosperm orders. Except for gnetophytes, the typical IR-containing seed plants (i.e., ginkgo, cycads, and angiosperms) generally have larger plastomes than those without typical IRs (i.e., cupressophytes and Pinaceae). The noncoding contents of Pinaceae, ginkgo, cycads, and angiosperms range from 40% to 45% and do not significantly differ from one another (Wilcoxon 2-sided test, all P > 0.05 in supplementary fig. S3, Supplementary Material online). This finding suggests that (1) the absence or presence of typical IRs does not affect the estimated noncoding content, and (2) the noncoding content of seed plant ancestors is likely ≥ 40%. However, the noncoding content is significantly smaller in gnetophytes than in other groups (all P < 0.05 in supplementary fig. S3, Supplementary Material online), so among seed plants, gnetophytes contain the most compact plastomes.
Fig. 2.—
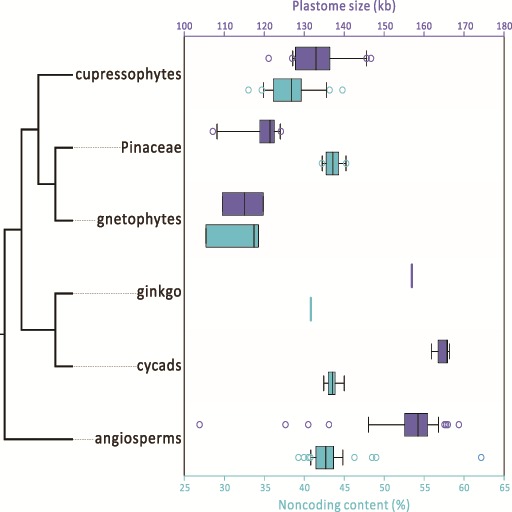
Boxplots for comparing plastome size (dark-blue) and noncoding content (light-blue) in the six seed plant groups. The tree topology (left) is based on the study of Wu, Wang, et al. (2011). Lines inside boxes are medians.
The noncoding content of cupressophytes is <40% except for Araucariaceae, Torrey of Taxaceae, and Cunninghamia of Cupressaceae (fig. 1). The noncoding content of Araucariaceae is 42.38–44.73% (fig. 1), which is not significantly different from that of Pinaceae, ginkgo, cycads, and angiosperms (Wilcoxon 2-sided test, all P > 0.05). Thus, the plastomes of Araucariaceae likely have retained the ancestral states in terms of noncoding content, whereas the remaining four cupressophyte families have undergone plastomic compaction, as indicated by their lower noncoding content (36.66–39.24%).
Accelerated Nucleotide Substitution Rates Coincide with Progressively Reduced Plastomes
Figure 1 clearly shows that the genera of Araucariaceae have larger plastomes and shorter branch lengths. To examine an association between plastome size and nucleotide substitution rate, we counted the branch length from the common ancestor (node A in fig. 1) to each of the 24 cupressophytes. The estimated rates varied from 0.122 to 0.348 substitutions per site, with Araucaria the slowest and Callitris the fastest. Furthermore, our independent contrasts analysis revealed that the substitution rates were inversely correlated with the noncoding content (Spearman’s rho = −0.661, P = 0.006; fig. 3a) and the plastome size (Spearman’s rho = −0.596, P = 0.0027; fig. 3b). These results strongly suggest that accelerated substitution rates together with decreased noncoding content have driven the plastomic reduction in cupressophytes.
Fig. 3.—
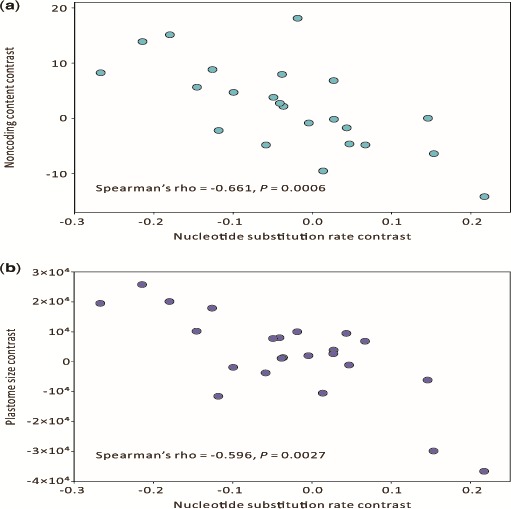
Relationships between nucleotide substitution rate and noncoding content (a) as well as plastome size (b) among the 24 sampled cupressophytes. The examined traits were transformed into independent contrasts, then these contrasts were evaluated using Spearman's rank correlation test.
Specific IR Systems in Generation of Isomeric Plastomes
In addition to the highly variable size, the plastomes of cupressophytes have been characterized by extensive inversions (Guo et al. 2014; Wu and Chaw, 2014; Hsu et al. 2014, 2016). However, these previous studies were based on comparing plastomes of a few species. To better understand variation in the plastomic organization across the five cupressophyte families, we constructed plastome maps of the 24 sampled cupressophytes and their putative ancestors on the basis of 37 locally collinear blocks (fig. 4). In the 24 living plastomes, we also highlight IRs that are ≥200 bp and common to each family or have been reported to be capable of inducing homologous recombination to generate isomeric plastomes.
Except for Callitris, all sampled genera of Cupressaceae and Taxaceae have a trnQ-IR (fig. 4), known to be associated with generation of isomeric plastomes in two cupressophyte genera, Cephalotaxus (Yi et al. 2013) and Juniperus (Guo et al. 2014). The presence of isomeric plastomes in Sciadopitys is associated with its unique rpoC2-IR (Hsu et al. 2016). Nevertheless, we found novel and family-specific IRs in Podocarpaceae (i.e., trnN-IR) and Araucariaceae (i.e., rrn5-IR), although their recombinant ability has not been confirmed. Collectively, every cupressophyte family has evolved its own specific short and novel IR systems for generating isomeric plastomes.
Degrees of Plastomic Inversions Are Associated with Substitution Rates
Comparison of the ancestral and extant plastomes allow for estimating inversions across the evolutionary course of cupressophytes (fig. 4). Degrees of plastomic inversions differed among the cupressophyte families. Excluding Sciadopityaceae, which contains only a single genus, the number of inversions per genus within families was 1.27 (= 28/22, total inversions divided by total number of genera, including ancestral and extant ones), 1 (= 6/6), 0.17 (= 1/6), and 0 (= 0/5) for Cupressaceae, Taxaceae, Podocarpaceae, and Araucariaceae, respectively. This estimated degree of plastomic inversions is apparently not biased by the total number of sampled genera within families. For example, Podocarpaceae and Taxaceae each have six genera (two ancestral and five extant ones; fig. 4), but their inversions per genus differ by 6-folds. Thus, these data suggest that the degrees of plastomic inversions are lineage-specific in cupressophytes.
We wondered about an association between inversions and substitution rates, because the latter also varied among the cupressophyte families (fig. 1). Therefore, we estimated and plotted synonymous (ds) and nonsynonymous (dn) substitution rates as well as their ratio (dn/ds) in figure 4. Theoretically, substitutions are selectively neutral at ds sites but constrained at dn sites. Hence, the dn/ds ratios should reflect the degree of selection pressure. In figure 4, estimating the correlation of inversions with substitution rates as well as dn/ds ratios is reasonable because they experienced the same time period evolutionarily on each of the branches between nodes. Among all tree branches, only that between the ancestors C1 and C3 showed a dn/ds ratio > 1 (dn/ds = 1.94; fig. 4). We detected a strong and significant correlation between ds and dn rates (Spearman’s rho = 0.95, P < 0.001 in supplementary fig. S4a, Supplementary Material online), which indicates a mechanism driving the evolution of both ds and dn rates. In addition, the number of inversions was significantly correlated with ds rates (Spearman’s rho = 0.566, P < 0.001 in supplementary fig. S4b, Supplementary Material online) and dn rates (rho = 0.576, P < 0.001 in supplementary fig. S4c, Supplementary Material online) but not dn/ds ratios (rho = 0.272, P = 0.068; data not shown).
Discussion
Mechanisms Underlying Plastome Compaction in Cupressophytes
Previously, Crosby and Smith (2012) discovered a strong linear relationship between plastome size and amount of noncoding sequences across photosynthetic organisms. In this study, we used “proportion” instead of “amount” for assessing the relationship. Moreover, we focused on variation among each of the six seed plant groups. We found strong positive correlations only within cupressophytes and gnetophytes (fig. 2), which suggests that the plastome sizes of these two lineages vary with the dynamics of their noncoding content. In contrast, the plastomes of Pinaceae do not show decreased noncoding content, although they are remarkably reduced (fig. 2). As compared to plastomes of their phototropic relatives, those of some parasitic plants are relatively reduced but not compact (Krause 2008; Wicke et al. 2013). Therefore, a reduced plastome does not necessarily evolve toward compaction.
In cupressophytes, mechanisms underlying plastome compaction are particularly perplexing. Loss of typical IRs does not directly lead to compaction of plastomes. For instance, the plastomes of Pinaceae have lost typical IRs but are not compact (fig. 2). In addition, our data show that smaller plastomes tended to have a greater degree of inversions. For example, the Cupressaceae have a mean plastomic size of 129.2 ± 3.6 kb with 1.27 times of inversions per genus, whereas the Araucariaceae have a mean plastomic size of 146 ± 0.5 kb but no inversion. However, we rule out the possibility that compaction results from inversions because in cupressophyte plastomes, inversions could lead to elongated rather than shortened nonsyntenic intergenic regions (Wu and Chaw 2014). In Silene species, their extremely expanded mitochondrial genomes were shown to be associated with numerous rearrangements (Sloan et al. 2012). Moreover, nonhomologous repairs were proposed to cause genomic expansion and rearrangements in organelles (Christensen 2014). Therefore, rearrangements appear to contribute to the expansion rather than reduction of organelle genomes.
Several hypotheses have been put forth to link plastomic compaction. For example, compaction might benefit the rapid replication of plastomes (McCoy et al. 2008), which, in turn, might cause incompatibility between plastid and nuclear genomes (Greiner et al. 2015). In addition, selection for nucleotides with less nitrogen content might have a competitive benefit for the plastomes of gnetophytes, which are compact with enriched AT content (Wu et al. 2009). Most plastomes are AT-rich (Smith, 2012). In the plastomes of cycads, substitutions appear to be AT-biased except in their typical IRs (Wu and Chaw 2015). In cupressophytes, loss of typical IRs is coupled with absence of conversion at the genes originally residing in the IRs (Zhu et al. 2016). This implies that AT-biased substitutions are plastome-wide in cupressophytes, and that accelerated substitution rates are expected to elevate AT content.
Excess noncoding DNAs might be mutational hazards, which should be eliminated from genomes with increased effective population size or accelerated mutation rates (Lynch et al. 2011). In a small effective population with severe genetic drift, elimination of deleterious mutations is less efficient, thereby leading to elevated dn/ds ratios (Ellegren 2008). Our comparison of the estimated dn/ds ratios did not reveal significant differences among the five families of cupressophytes, despite an exception between Araucariaceae and Cupressaceae (supplementary fig. S5, Supplementary Material online). These data imply that in cupressophytes, random genetic drift might have little effect on plastome compaction.
We showed a striking evolutionary trend in cupressophyte plastomes—the more compact the plastomes, the faster the substitution rates (fig. 3). This trend reinforces that variable substitution rates are associated with plastome-size variation in cupressophytes. Mutation rates are often estimated by measuring the ds rates, which generally are neutral (Nei et al. 2010). We demonstrated an inverse relationship between plastome size and ds rate after transforming them into independent contrasts (Spearman’s rho = −0.64, P = 0.001, supplementary fig. S6, Supplementary Material online). This finding agrees well with the notion that the evolution of organellar genomes is primarily driven by different mutation rates (Lynch et al. 2006). Furthermore, if mutation rates are crucial in shaping the genome complexity in cupressophytes, we would expect a relationship between nuclear and organelle genome sizes because the ds rates of nuclear and organelle genes are in general correlated (Drouin et al. 2008; Bromham et al. 2015). Indeed, supplementary table S2, Supplementary Material online indicates a positive correlation between the genome sizes of nuclei and plastids (Spearman’s rho = 0.46, P = 0.031) among the 22 sampled cupressophytes (C-values of Callitropsis and Hesperocyparis were unavailable in the Plant DNA C-values Database as of June 2016). Unfortunately, information on their mitochondrial genomes is wanting.
Why do the substitution rates of plastomes vary among cupressophytes? The Araucariaceae are all tall trees, whereas the Cupressaceae are trees or shrubs. Figure 3 also shows that the substitution rates of Araucariaceae are approximately 2- to 3-fold slower than those of the Cupressaceae. This finding echoes those of Lanfear et al. (2013) and Bromham et al. (2015) that rates of molecular evolution tend to be slower for taller than shorter plants.
Evolutionary Implications of Plastomic Inversions in Cupressophytes
Another remarkable feature of cupressophytes plastomes is their numerous inversions (fig. 4), hypothesized to result from loss of typical IRs (Wu and Chaw 2014). Based on a positive correlation of plastomic rearrangements with dn but not ds rates, Guisinger et al. (2011) and Weng et al. (2014) suggested that in the plastomes of Geraniaceae, numerous rearrangements might be ascribed to improper functioning of nuclear-encoded DNA repair proteins. In the plastomes of cupressophytes, a strongly positive correlation between dn and ds rates (supplementary fig. S4a, Supplementary Material online) can be best explained by the fact that both rates are dependent on mutation rates (Bromham et al. 2015; Wicke et al. 2016). This co-evolution of dn and ds rates interprets the positive relationship between dn rates and the degree of inversions because the latter also is associated with ds rates (supplementary fig. S4b, c, Supplementary Material online). A positive correlation between dn/ds ratios and rearrangements led to a conclusion of selection pressure on the extensively rearranged plastomes of Geraniaceae (Weng et al. 2014). However, in cupressophyte plastomes, the dn/ds ratios are not correlated with the degree of inversions. Therefore, we rule out that selection pressure affects plastomic inversions in cupressophytes.
Recent study of the IR-lacking clade of legumes showed that, like ds substitutions, plastomic rearrangements accumulated over time but leveled off because of the constraint on conserved gene clusters (Sveinsson and Cronk 2016). Such constraint might be weak in cupressophyte plastomes, because some conserved gene clusters have been disrupted in the plastomes of Taxus (Hsu et al. 2014), Sciadopitys (Hsu et al. 2016), and Callitris (supplementary fig. S2, Supplementary Material online). A positive correlation between the degree of inversions and ds rates (supplementary fig. S4b, Supplementary Material online) led us to propose that in cupressophyte plastomes, the plastomic inversions likely evolve in a neutral manner, in which variable mutation rates play a major driving force.
Blazier et al. (2016) proposed a competing relationship between typical IRs and short repeats because intra-molecular recombination was largely limited to the former, thus suppressing illegitimate recombination if the latter were not abundant. Nonetheless, the competing relationship of Blazier et al. is not held in the plastomes of cupressophytes because they have lost typical IRs. We discovered that cupressophyte families have evolved their own specific short IRs (fig. 4). Intriguingly, some of the short IRs, similar to typical IRs, can mediate homologous recombination, thereby resulting in the co-existence of major and minor isomeric plastomes (Yi et al. 2013; Guo et al. 2014; Hsu et al. 2016). However, isomeric plastomes mediated by typical IRs should be equally abundant inside plastids (Palmer 1983; Martin et al. 2013). In terms of recombinant ability, these specific short IRs of cupressophytes are apparently less efficient than typical IRs. They are also ineffective in suppressing illegitimate recombination, leading to generation of numerous inversions in the plastomes of cupressophytes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to the administrators of Taipei Botanical Garden and Dr Ya-Nan Wang for the gifts of fresh branchlets of Callitris rhomboidea, Chamaecyparis formosensis, Dacrycarpus imbricatus, and Taxodium distichum. We thank Ivan Hsu and Min-Shao Tsai for the help of DNA extraction and quality control. We also like to thanks the two anonymous reviewers for their critical reading and helpful comments. This work was supported by research grants from the Ministry of Science and Technology, Taiwan (MOST 103-2621-B-001-007-MY3), and from the Biodiversity Research Center of Academia Sinica to S.-M.C.
Literature Cited
- Bellot S, Renner SS. 2015. The Plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol Evol. 8:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier, et al. 2016. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 117:1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann TW, Kuzmina M, Stefanović S. 2009. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet. 55:323–337. [DOI] [PubMed] [Google Scholar]
- Bromham L, Hua X, Lanfear R, Cowman PF. 2015. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am Nat. 185:507–524. [DOI] [PubMed] [Google Scholar]
- Chumley, et al. 2006. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23:2175–2190. [DOI] [PubMed] [Google Scholar]
- Christensen AC. 2014. Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biol Evol. 6:1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa 261:201–217. [Google Scholar]
- Crosby K, Smith DR. 2012. Does the mode of plastid inheritance influence plastid genome architecture? PLoS One 7:e46260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49:827–831. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2008. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 17:4586–4596. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125:1–15. [Google Scholar]
- Green BR. 2011. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66:34–44. [DOI] [PubMed] [Google Scholar]
- Greiner S, Sobanski J, Bock R. 2015. Why are most organelle genomes transmitted maternally? Bioessays 37:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28:583–600. [DOI] [PubMed] [Google Scholar]
- Guo, et al. 2014. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol. 6:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66:350–361. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Wu CS, Chaw SM. 2014. Ancient nuclear plastid DNA in the yew family (taxaceae). Genome Biol Evol. 6:2111–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Wu CS, Chaw SM. 2016. Birth of four chimeric plastid gene clusters in Japanese umbrella pine. Genome Biol Evol. 8:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. 2012. Plastid genome in seed plants In: Bock R, Knoop V, eds. Genomics of chloroplasts and mitochondria. Dordrecht, the Netherlands: Springer; p. 103–126. [Google Scholar]
- Krause K. 2008. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 54:111–121. [DOI] [PubMed] [Google Scholar]
- Krzywinski, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, et al. 2013. Taller plants have lower rates of molecular evolution. Nat Commun. 4:1879.. [DOI] [PubMed] [Google Scholar]
- Leslie, et al. 2012. Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci U S A. 109:16217–16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Huang JP, Wu CS, Hsu CY, Chaw SM. 2010. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol. 2:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Hu F, Tang J, Moret BM. 2013. Maximum likelihood phylogenetic reconstruction from high-resolution whole-genome data and a tree of 68 eukaryotes. Pac Symp Biocomput. 2013:285–296. [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. 2016. Evolution of short inverted repeat in cupressophytes, transfer of accD to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci Rep. 6:20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311:1727–1730. [DOI] [PubMed] [Google Scholar]
- Lynch M, Bobay LM, Catania F, Gout JF, Rho M. 2011. The repatterning of eukaryotic genomes by random genetic drift. Annu Rev Genomics Hum Genet. 12:347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Baurens FC, Cardi C, Aury JM, D'Hont A. 2013. The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS One 8:e67350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy SR, Kuehl JV, Boore JL, Raubeson LA. 2008. The complete plastid genome sequence of Welwitschia mirabilis: an unusually compact plastome with accelerated divergence rates. BMC Evol Biol. 8:130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. 2010. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 11:265–289. [DOI] [PubMed] [Google Scholar]
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature 301:92–93. [Google Scholar]
- Ran JH, Gao H, Wang XQ. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol. 54:136–149. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10:e1001241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2012. Updating our view of organelle genome nucleotide landscape. Front Genet. 3:175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2016. The mutational hazard hypothesis of organelle genome evolution: 10 years on. Mol Ecol. 25:3769–3775. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, Via LE. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750. [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveinsson C,, Cronk Q. 2016. Conserved gene clusters in the scrambled plastomes of IRLC legumes (Fabaceae: Trifolieae and Fabeae). bioRxiv 040188.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G. 2002. GRIMM: genome rearrangements web server. Bioinformatics 18:492–493. [DOI] [PubMed] [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31:645–659. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113:9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. 2009. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 52:115–124. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM. 2014. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol J. 12:344–353. [DOI] [PubMed] [Google Scholar]
- Wu CS, Chaw SM. 2015. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol Evol. 7:2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yi X, Gao L, Wang B, Su YJ, Wang T. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209:1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


