Abstract
The Streptococcus pyogenes phospholipase A2 (SlaA) gene is highly conserved in the M3 serotype of group A S. pyogenes, which often involves hypervirulent clones. However, the role of SlaA in S. pyogenes pathogenesis is unclear. Herein, we report that SlaA induces the expression of intercellular adhesion molecule 1 (ICAM1) and vascular cell adhesion molecule 1 (VCAM1) via the arachidonic acid signaling cascade. Notably, recombinant SlaA induced ICAM1 and VCAM1 expression in human umbilical vein endothelial cells (HUVECs), resulting in enhanced adhesion of human monocytic leukemia (THP-1) cells. However, C134A, a variant enzyme with no enzymatic activity, did not induce such events. In addition, culture supernatants from S. pyogenes SSI-1 enhanced the adhesion of THP-1 cells to HUVECs, but culture supernatants from the ΔslaA isogenic mutant strain had limited effects. Aspirin, a cyclooxygenase 2 inhibitor, prevented the adhesion of THP-1 cells to HUVECs and did not induce ICAM1 and VCAM1 expression in HUVECs treated with SlaA. However, zileuton, a 5-lipoxygenase inhibitor, did not exhibit such effects. Furthermore, pre-administration of aspirin in mice intravenously injected with SlaA attenuated the transcriptional abundance of ICAM1 and VCAM1 in the aorta. These results suggested that SlaA from S. pyogenes stimulates the expression of adhesion molecules in vascular endothelial cells. Thus, SlaA contributes to the inflammation of vascular endothelial cells upon S. pyogenes infection.
Keywords: S. pyogenes, phospholipase A2, SlaA, HUVEC, ICAM1, VCAM1
Introduction
Streptococcus pyogenes is a gram-positive bacterium that can cause superficial infections, such as pharyngitis and pyoderma; invasive infections, such as necrotizing fasciitis and streptococcal toxic shock syndrome; and post-infectious diseases, such as rheumatic fever (Cunningham, 2000). S. pyogenes produces many extracellular molecules that contribute to host–pathogen interactions (Musser and Krause, 1998; Cunningham, 2000; Fischetti, 2000; Kurosawa et al., 2016) and most of the extracellular molecules possess enzymatic activities against host tissues and the immune system (Terao et al., 2006, 2008; Honda-Ogawa et al., 2013). Epidemiological studies have shown that M3 serotype strains are the second most common cause of invasive infections in the United States, Canada, Western Europe, Japan, and Israel (Murakami et al., 2002; Terao et al., 2002; Li et al., 2003; Moses et al., 2003; Muller et al., 2003; Nakagawa et al., 2003; Schmitz et al., 2003). Surveillance studies have also revealed that M3 strains cause a higher rate of severe invasive diseases such as necrotizing fasciitis and death than other M-type strains (O'Brien et al., 2002; Sharkawy et al., 2002).
S. pyogenes is a diverse species with strain-specific virulence genes derived from multiple prophages (Banks et al., 2002; Beres and Musser, 2007). Genome sequencing analysis of S. pyogenes M1–M6, M12, M18, M28, and M49 serotype strains indicated that these strains have 4–8 prophages or prophage-like elements (Nozawa et al., 2011). We identified that M3 serotype strain SSI-1 possesses the phospholipase A2 (PLA2) gene, designated as slaA, on the prophage genome. Beres et al. (2004) reported that the enhanced capacity for invasive infection incurred by M3 correlated with the acquisition of prophage genomes encoding the slaA gene. SlaA has a conserved region of amino acid residues found in several secreted PLA2 enzymes, such as those found in snake venom (Pearson et al., 1993).
Sitkiewicz et al. (2007) reported that SlaA enhanced the binding of S. pyogenes to human epithelial cells, such as immortalized pharyngeal cells and human tracheobronchial epithelial cells. However, the pathogenesis due to SlaA in host cells remains unclear. Yokote et al. (1993) reported that endogenous PLA2 from human endothelial cells induces the expression of intercellular adhesion molecule 1 (ICAM1) and vascular cell adhesion molecule 1 (VCAM1). In addition, Zhu et al. (1999) reported that cytosolic PLA2 in human eosinophils stimulated the expression of β1- and β2-integrin, resulting in binding to ICAM1 and VCAM1. In this study, we hypothesized that SlaA induces the expression of adhesion molecules. To confirm this hypothesis, we examined the adhesion of human monocytic leukemia cell line (THP-1) cells to human umbilical vein endothelial cells (HUVECs) and the association of this with the expression of adhesion molecules, such as ICAM1, VCAM1, E-selectin, and P-selectin, on HUVECs treated with SlaA.
Materials and methods
Reagents and animals
Aspirin and zileuton were purchased from Merck. Aspirin was solubilized in phosphate-buffered saline (PBS) containing 0.5% ethanol (in vitro) or 0.5% CMC (in vivo). All other drugs were of analytical grade. Male, 10–12-week-old BALB/c mice (Nihon CLEA, Japan) were used in this study. Mice were maintained under standard conditions in accordance with our institutional guidelines. All animal experiments were approved by the Institutional Animal Care and Use Committee of Niigata University.
Bacterial strains
S. pyogenes clinical strains were generously provided by Dr. Kawabata of Osaka University (Table 1) (Murakami et al., 2002; Terao et al., 2002). All strains were grown in Todd Hewitt broth (Becton Dickinson, MD, USA) supplemented with 0.2% yeast extract (THY broth) at 37°C.
Table 1.
Expression of SlaA in various S. pyogenes strains.
| Strain name | M serotype | Reference | slaA (gene) | SlaA (protein) |
|---|---|---|---|---|
| NIH11 | 1 | Invasive | − | − |
| NIH17 | 1 | Invasive | − | − |
| NIH22 | 1 | Invasive | − | − |
| NIH27 | 1 | Invasive | − | − |
| TW3354 | 1 | Non-invasive | − | − |
| SSI-1 | 3 | Invasive | + | + |
| SSI-35 | 3 | Invasive | + | + |
| NIH1 | 3 | Invasive | + | + |
| NIH16 | 3 | Invasive | + | + |
| NIH21 | 3 | Invasive | + | + |
| TW3358 | 3 | Non-invasive | + | + |
| NIH2 | 4 | Invasive | − | − |
| NIH6 | 4 | Invasive | − | − |
| TW3392 | 4 | Non-invasive | + | + |
| TW3398 | 4 | Non-invasive | + | + |
| TW3400 | 4 | Non-invasive | + | + |
| TW3341 | 11 | Non-invasive | − | − |
| TW3360 | 11 | Non-invasive | − | − |
| TW3337 | 12 | Non-invasive | − | − |
| TW3344 | 12 | Non-invasive | − | − |
| NIH35 | 28 | Invasive | − | − |
| TW3357 | 28 | Non-invasive | − | − |
| TW3374 | 75 | Non-invasive | − | − |
| TW3364 | 75 | Non-invasive | − | − |
| TW3365 | 75 | Non-invasive | − | − |
| TW3419 | 89 | Non-invasive | − | − |
| TW3551 | 89 | Non-invasive | − | − |
+, Detected; −, not detected.
Detection of slaA gene
Chromosomal DNA from S. pyogenes was purified with Bactozol (Molecular Research Center, Inc., Cincinnati, OH, USA). The forward primer (5′-GGGGGATCCGGGATAAATGATAAAATGGAA-3′) and reverse primer (5′-CCCGAATTCTTAACATCCTATAGAACCTAC-3′) were used to amplify the slaA gene of various S. pyogenes strains.
Integration mutagenesis
The PCR product of the internal portion of the slaA gene was amplified using forward (5′-GGGAAGCTTATGAAAAAAGTAATAAATACTATTCTATAAGCTGCT-3′) and reverse (5′-CCCGGATCCTTAACATCCTATAGAACCTACTGTCTCAAAATATAC-3′) primers and ligated into a thermosensitive suicide vector pSET4s (Takamatsu et al., 2001a,b). pSET4s was kindly provided by Dr. Takamatsu (The National Agriculture and Food Research Organization, Japan). The constructed plasmid was transformed into wild-type strain SST-1 by electroporation (2.5 kV, 25 μF, and 600 Ω), and the inactivated mutant strain was selected on spectinomycin-containing agar plates.
Detection of SlaA protein
S. pyogenes strains were incubated in THY broth at 37°C for 6 h to an OD610 of 0.3–0.4. The bacteria were collected, washed in PBS, and suspended in 1 ml of PBS. Bacteria (100 μl) were added to the HUVECs (2 ml, 6-well plate) and incubated at 37°C for 5 h. Culture medium was centrifuged at 3,000 × g for 15 min, and the culture supernatants were concentrated to 100 μl with Amicon Ultra Centrifugal Filters 10K (Merck Millipore, Billerica, MA, USA). The samples were mixed with 2% SDS-sample buffer containing 1% 2-mercaptoethanol, boiled for 3 min, then separated by SDS-PAGE using 12.5% gels, and transferred to polyvinylidene difluoride membranes (Merck Millipore). The membrane was blocked and incubated with an anti-SlaA antibody and HRP-conjugated secondary antibody. The membrane was treated with HRP substrates and analyzed using a chemiluminescence detector (Fujifilm, Tokyo, Japan). The anti-SlaA antibody was produced in rabbits intramuscularly administered SlaA protein (Eurofin, Tokyo, Japan).
Construction of recombinant SlaA
A recombinant (r) SlaA expression plasmid was constructed using a pGEX-6P-1 vector (GE Healthcare, Uppsala, Sweden). The forward primer (5′-GGGGGATCCGGGATAAATGATAAAATGGAA-3′) and reverse primer (5′-CCCGAATTCTTAACATCCTATAGAACCTAC-3′) were used to amplify the slaA gene of S. pyogenes strain SSI-1 by PCR. The resultant PCR fragment was cloned into a pGEX-6P-1 vector. The pGEX-6P-1 vector containing the slaA gene was transformed into Escherichia coli strain DH5α (TaKaRa, Shiga, Japan) by the heat shock method. The DH5α transformants were grown in Luria-Bertani broth (Nacalai Tesque, Kyoto, Japan) supplemented with 100 μg/ml ampicillin (Meiji Seika, Tokyo, Japan) to select for the pGEX-6P-1 vector. Then, the rSlaA protein was purified using glutathione-Sepharose 4B beads (GE Healthcare), and the GST tag was cleaved by PreScission Protease (GE Healthcare). The purified rSlaA protein was dialyzed against PBS. The amount of lipopolysaccharide (LPS) in 1 μg of purified rSlaA protein was determined to be less than 2 pg using the LPS detection kit (GenScript, Piscataway, NJ, USA).
Site-directed mutagenesis
The KOD-plus Mutagenesis kit (TOYOBO, Osaka, Japan) was used with the following primers to the modified plasmid (pGEX6P1-SlaA): C134A, 5′-GCCCAAAACCACGATAGTTGCTATAAGTGG-3′ and 5′-ACCTTGATCCAAAACATCTACTACTGGCAA-3′. The purified C134A were assayed for phospholipase A2 activity with a phospholipase A2 assay kit (Cayman Chemical, Ann Arbor, MI, USA). In line with a previous report (Nagiec et al., 2004), the enzymatic activity of C134A was not detected.
Phospholipase A2 activity
Phospholipase A2 activity of the recombinant protein (rSlaA and C134A) and culture supernatant of SSI-1 and ΔslaA isogenic mutant strain was measured by Phospholipase A2 colorimetric assay kit (Cayman, Michigan, USA).
Human monocytic cells
Human THP-1 monocytes were purchased from RIKEN (Tsukuba, Japan). These cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine. The culture was maintained at 37°C in 5% CO2. The cells were stained with 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Thermo Fisher Scientific, Eugene, OR, USA) at 37°C for 60 min, and then washed.
HUVECs
HUVECs were purchased from Lonza Inc. They were cultured to confluence in EGM-2 medium at 37°C in 5% CO2. HUVECs were used within 24 h after reaching confluence, between passages 3 and 5.
Assay for monocyte adhesion to endothelial cells
After the stimulation of HUVECs with SlaA, C134A, or various culture supernatants from S. pyogenes for 6 h, the cells were washed with EGM-2. CFSE-labeled THP-1 cells (4 × 105 cells/ml) were then layered over the HUVEC monolayers and incubated for 24 h at 37°C in 5% CO2. The cells were washed with EGM-2 and fixed in 1% glutaraldehyde in PBS. The fluorescent intensity of CFSE-labeled THP-1 cells was analyzed by fluorescent microscopy (BIOREVO BZ-9000; Keyence, Osaka, Japan) and the associated analysis software package (BZ-H2A).
Cytotoxicity assay
HUVECs (confluent) and THP-1 cells (4 × 105 cells/ml) were incubated with SlaA and C134A at 37°C for 24 h. Viable cells were determined using AlamarBlue cell-viability reagent (Bio-Rad, Kidlington, UK), in accordance with the manufacturer's instructions.
Quantitative real-time RNA
Gene expression in HUVECs and the aorta was quantified using quantitative real-time PCR. Briefly, RNA was extracted from cell lysates using TRI Reagent (Molecular Research Center) and quantified by spectrometry at 260 and 280 nm. The RNA was reverse-transcribed using SuperScript VILO Master Mix (Thermo Fisher Scientific, Carlsbad, CA, USA), and quantitative real-time PCR with cDNA was performed with the StepOnePlus Real-time PCR system (Thermo Fisher Scientific), in accordance with the manufacturer's protocol. TaqMan probes, sense primers, and antisense primers for the expression of a housekeeping gene (GAPDH), ICAM1 (ICAM1), VCAM1 (VCAM1), E-selectin (SELE), or P-selectin (SELP) mRNA were purchased from Thermo Fisher Scientific.
Immunofluorescence analysis
rSlaA-treated HUVECs were fixed and permeabilized using a cell fixation and permeabilization kit (Thermo Fisher Scientific), in accordance with the manufacturer's instructions, followed by incubation of the cells in a blocking solution (Thermo Fisher Scientific) for 30 min. The samples were stained with PE anti-human ICAM1 antibody (Biolegend, San Diego, CA, USA) or PE anti-human VCAM1 antibody (Biolegend) in the blocking solution. After overnight incubation at 4°C in the dark, the cells were washed. The intensity of the fluorescence of each sample was analyzed by fluorescent microscopy (BioRevo model BZ-9000; Keyence) and the associated analysis software package (BZ-H2A).
In vivo analysis
Balb/c mice (five mice each) were injected i.v. with 30 μg/kg SlaA or PBS every 3 days for the indicated periods. Aspirin (100 mg/kg) and its vehicle (0.5% CMC) were administered i.p. 3 h prior to the injection of SlaA. The treated mice were sacrificed at several time points after this administration, after which the aorta was extirpated.
Statistical analysis
Data were analyzed using GraphPad Prism 6.05 (GraphPad Software, La Jolla, CA, USA). All results are expressed as the mean ± SEM. The group means were compared using one-way analysis of variance. P-values of 0.05 or less were considered statistically significant.
Results
S. pyogenes secretes SlaA into the culture supernatant
Table 1 shows the association between the status of the slaA gene and the clinical manifestations associated with S. pyogenes. In PCR analyses, the slaA gene was detected among M3 and M4 serotypes of S. pyogenes, but not in the M1, M11, M12, M28, M75, and M89 serotypes, under the experimental conditions applied. A previous study showed that S. pyogenes encoding the slaA gene significantly upregulated SlaA production when cultured on D562 human pharyngeal epithelial cells (Sitkiewicz et al., 2006). Therefore, we tested the release of the SlaA protein from 27 strains of S. pyogenes co-cultured with HUVECs. By western blot analyses, we found that the SlaA protein was only detected in culture supernatant from S. pyogenes encoding the slaA gene (Table 1).
Monocyte adhesion to endothelial cells
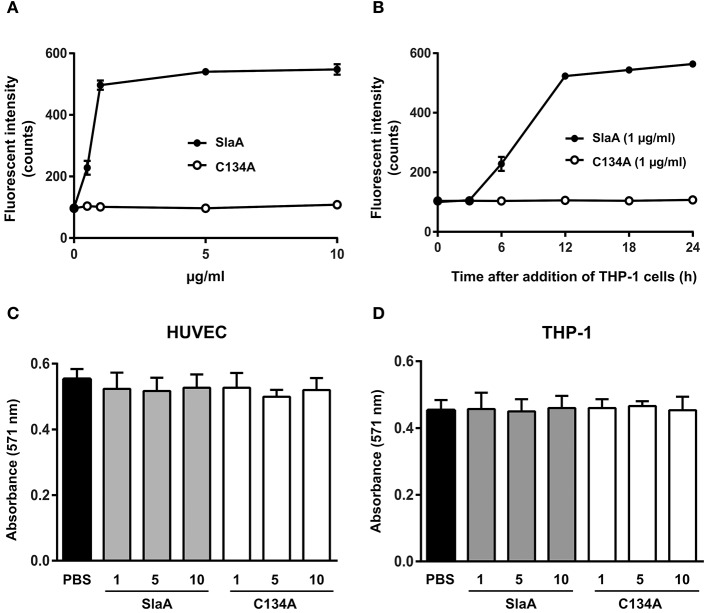
To investigate whether SlaA enhances the adhesion between HUVEC and THP-1 cells, we incubated SlaA-stimulated HUVEC with THP-1 cells. SlaA dose-dependently enhanced the adhesion of these two types of cells (Figure 1A). The interaction level peaked within 12 h and then remained at the same level (Figure 1B). The C134A variant, which does not have phospholipase A2 activity (Supplementary Figure 1), did not induce the adhesion of THP-1 cells to HUVECs (Figures 1A,B), showing that the enzymatic activity of SlaA is essential for this event.
Figure 1.
SlaA stimulated the adhesion of THP-1 cells to HUVECs. (A) HUVECs were incubated with various concentrations (A) or 1 μg/ml (B) of SlaA or C134A at 37°C for 6 h. The cells were incubated with CFSE-labeled THP-1 cells at 37°C for 24 h (A) or the indicated times (B). These cells were washed and the labeled THP-1 cells were viewed with a fluorescent microscope. The fluorescent intensity of CFSE-labeled THP-1 cells was quantified as described in Materials and Methods. (C,D) HUVECs (C) and THP-1 cells (D) were incubated with various concentrations of SlaA and C134A at 37°C for 24 h, and then stained with AlamarBlue. The results shown represent the mean ± SEM; n = 4.
To determine whether SlaA caused cytotoxicity, SlaA and C134A were incubated with HUVECs or THP-1 cells at 37°C for 24 h, and each type of treated cell was stained with AlamarBlue. SlaA and C134A did not have a detrimental effect on the growth, cell morphology, and membrane integrity (Figures 1C,D).
S. pyogenes strain SSI-1 culture supernatant enhances the interaction between HUVECs and THP-1 cells
We next confirmed the effect of S. pyogenes strain SSI-1 cultured supernatant on the adhesion of THP-1 cells to HUVECs. The phospholipase A2 enzymatic activity of culture supernatant of ΔslaA isogenic mutant was not detected (Supplementary Figure 1). S. pyogenes wild-type strain on a HUVEC monolayer expressed SlaA in culture supernatant, but ΔslaA isogenic mutant strain did not (Figure 2A). The culture supernatant of the wild-type strain enhanced the adhesion of THP-1 cells to HUVECs, but that of ΔslaA isogenic mutant had a limited effect under our experimental conditions (Figure 2B).
Figure 2.
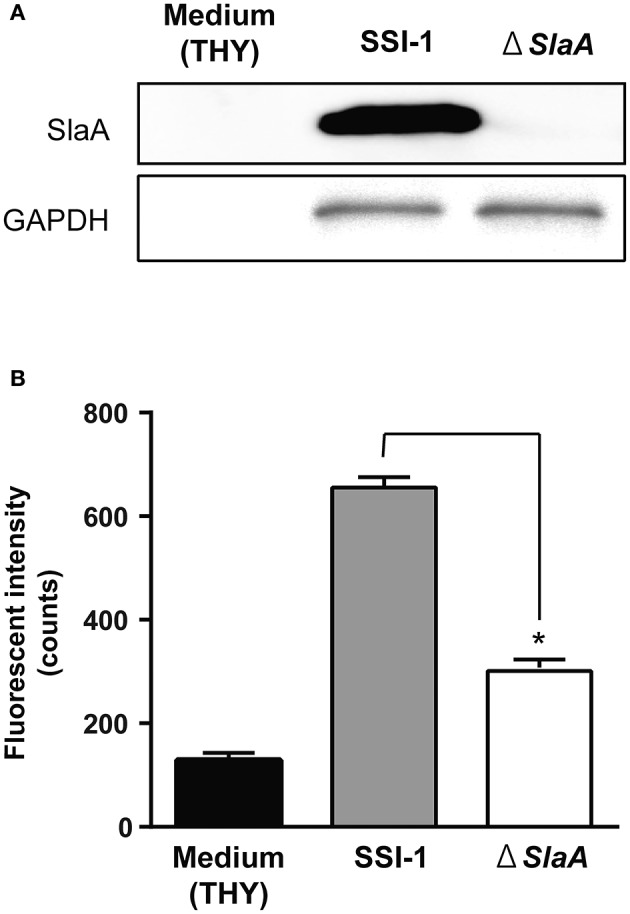
Streptococcous pyogenes culture supernatant induces the adhesion of THP-1 cells to HUVECs. (A) Protein expression of SlaA and GAPDH/Plr from S. pyogenes wild-type strain SSI-I and its ΔSlaA isogenic mutant strain was detected by Western blotting. (B) HUVECs were pre-incubated with THY broth, S. pyogenes SSI-I culture supernatant, or its ΔSlaA mutant strain culture supernatant at 37°C for 6 h. The cells were incubated with CFSE-labeled THP-1 cells at 37°C for 24 h. These cells were washed and the CFSE-labeled THP-1 cells were viewed with a fluorescent microscope. The fluorescent intensity of CFSE-labeled cells was quantified as described in Materials and Methods. The results shown represent the mean ± SEM; n = 5. Data were analyzed using a one-way ANOVA with Dunnett's multiple comparison test. Significant differences from the S. pyogenes SSI-I culture supernatant group are shown: *P < 0.01.
SlaA induced the expression of adhesion molecules in HUVECs
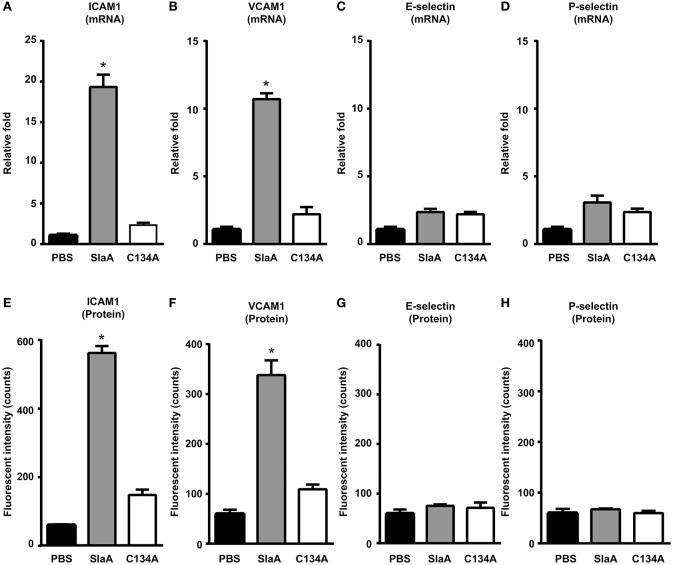
To investigate whether SlaA induced the expression of adhesion molecules in HUVECs, HUVECs were incubated with SlaA. SlaA upregulated the transcription of ICAM1 and VCAM1 mRNAs and the expression of ICAM1 and VCAM1 proteins, but C134A did not (Figures 3A,B,E,F). However, SlaA did not upregulate the transcription of E-selectin and P-selectin mRNAs and the expression of their proteins (Figures 3C,D,G,H).
Figure 3.
SlaA induced the expression of adhesion molecules in HUVECs. HUVECs were incubated with 1 μg/ml SlaA or C134A at 37°C for 3 h (A–D) and 6 h (E–H). The transcription of ICAM1 (A), VCAM1 (B), E-selectin (C), and P-selectin (D) mRNAs in HUVECs was measured by real-time PCR, as described in Materials and Methods. The relative quantity of these mRNAs was normalized to the relative quantity of GAPDH mRNA. The protein expression of ICAM1 (E), VCAM1 (F), E-selectin (G), and P-selectin (H) in HUVECs was analyzed by immunostaining using fluorescently labeled antibodies. The fluorescence intensity of each protein was quantified as described in Materials and Methods. The results shown represent the mean ± SEM; n = 5. Data were analyzed using one-way ANOVA with Dunnett's multiple comparison test. Significant differences from the PBS group are shown: *P < 0.01.
Aspirin inhibited the expression of adhesion molecules
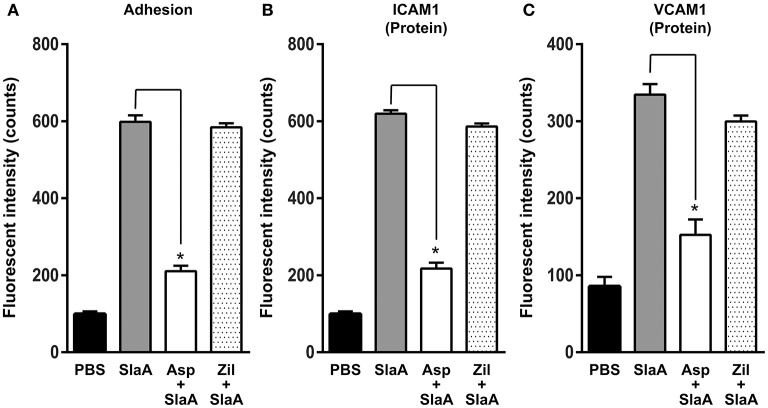
Phospholipase A2 is a key enzyme in the arachidonic acid cascade. We investigated the effect of arachidonic acid cascade inhibitors such as aspirin (cyclooxygenase 2 inhibitor) and zileuton (5-lipoxygenase inhibitor) on the adhesion of THP-1 cells to HUVECs and the expression of ICAM1 or VCAM1. HUVECs were treated with 5 mM aspirin or 50 μM zileuton at 37°C for 60 min. Aspirin inhibited the adhesion of THP-1 cells to HUVECs and the expression of ICAM1 or VCAM1, but zileuton did not (Figure 4).
Figure 4.
Aspirin inhibited the expression of adhesion molecules. HUVECs were pre-incubated with 5 mM aspirin and 50 μM zileuton at 37°C for 60 min, and then the cells were incubated with SlaA at 37°C for 6 h. (A) The treated cells were incubated with CFSE-labeled THP-1 cells at 37°C for 24 h. Adhesion of THP-1 cells to HUVECs (A) and the expression of ICAM1 (B) and VCAM1 (C) on the cells were analyzed as described in Materials and Methods. The results shown represent the mean ± SEM; n = 4. Data were analyzed using one-way ANOVA with Dunnett's multiple comparison test. Significant differences from the SlaA-treated group are shown: *P < 0.01.
SlaA upregulated the transcription of ICAM1 and VCAM1 mRNAs in mouse Aorta
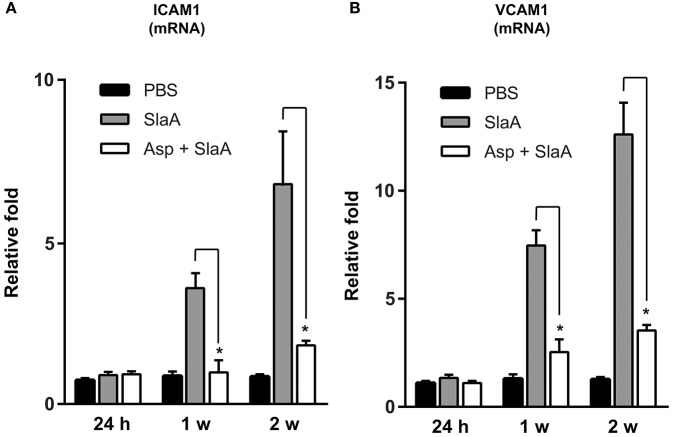
We investigated whether SlaA stimulated the transcription of ICAM1 and VCAM1 mRNAs in mouse aorta. The mice were intravenously administered 30 μg/kg SlaA every 3 days. SlaA upregulated the transcription of ICAM1 and VCAM1 mRNAs at the aorta at 1 and 2 weeks after the initial injection, but did not at 24 h. The upregulation of these mRNAs in the aorta of mice injected with SlaA was inhibited by i.p. administration of 100 mg/kg aspirin (Figures 5A,B).
Figure 5.
SlaA upregulated the transcription of ICAM1 and VCAM1 mRNAs in mouse aorta. Balb/c mice (five mice each) were injected i.v. with 30 μg/kg SlaA or PBS every 3 days for the indicated periods. Aspirin (100 mg/kg) and its vehicle (0.5% CMC) were administered i.p. 3 h prior to the injection of SlaA. The transcription of ICAM1 (A) and VCAM1 (B) mRNAs in the vascular endothelium of aorta was measured by real-time PCR as described in Materials and Methods. The relative quantities of these mRNAs were normalized to the level of GAPDH mRNA. Data were analyzed using one-way ANOVA. Significant differences from the SlaA-treated group are shown: *P < 0.01.
Discussion
As shown in Table 1, SlaA is known to be mainly released from M3 strains of S. pyogenes, and is encoded on a prophage genome. The serotype M3 GAS strains of S. pyogenes expressing SlaA and some M3 GAS strains have been shown to be particularly invasive (Beres et al., 2002, 2004; Banks et al., 2004; Nagiec et al., 2004). We also detected the slaA gene in the SSI-1 strain, which is a serotype M3 strain derived from a patient with an invasive infection (Terao et al., 2002), and the SlaA protein in the culture supernatant of S. pyogenes co-cultured with HUVECs. Recently, there has been an increase in infectious diseases induced by S. pyogenes carrying the slaA gene. Previous reports described that SlaA contributed to the colonization and infection of S. pyogenes in pharyngeal epithelial cells (Sitkiewicz et al., 2006, 2007). In addition, Sitkiewicz et al. (2006) reported that the ΔslaA isogenic mutant strain caused significantly less morbidity than the wild-type strain. However, the pathogenesis of SlaA is still not completely understood. In the present study, we have shown that SlaA from S. pyogenes induced the expression of ICAM1 and VCAM1 in HUVECs, resulting in the enhanced adhesion of THP-1 cells to HUVECs. In addition, SlaA also stimulated the expression of ICAM1 and VCAM1 in mouse aorta, and the pre-administration of aspirin inhibited these events.
As cytosolic and secretory phospholipase A2 in mammalian cells is known to be related to the expression of ICAM1 (Thommesen et al., 1998; Zhu et al., 1999; Barnett et al., 2001; Yu et al., 2012), we examined whether SlaA induced the expression of adhesion molecules such as ICAM1 in HUVECs. SlaA upregulated the transcription of ICAM1 and VCAM1 mRNAs and the expression of ICAM1 and VCAM1 proteins, resulting in the adhesion of THP-1 cells. On the other hand, C134A did not stimulate these events, suggesting that the PLA2 activity of SlaA is essential for them. The expression of E-selectin and P-selectin was not detected under our experimental conditions. In addition, aspirin inhibited the expression of ICAM1 and VCAM1 in HUVECs treated with SlaA, but zileuton did not. This suggested that the arachidonic acid cascade via cyclooxygenase 2 plays an important role in the expression of adhesion molecules in HUVECs treated with SlaA. The culture supernatant of ΔslaA isogenic mutant strain did not result in complete absence of the adhesion of THP-1 cells to HUVECs. This suggests that other secretory proteins besides SlaA participate in the adhesion of monocytes to endothelial cells.
Consequently, we suggested that the SlaA-induced expression of ICAM-1 and VCAM-1 may contribute to the leukostasis, vascular injury, and capillary leakage characteristics of invading S. pyogenes infection. Further studies are needed to clarify the mechanism behind the inflammation of endothelial cells treated with SlaA.
Author contributions
Conceived and designed the experiments: MO and YT. Performed the experiments: MO, HD, MK, and TI. Preparation of the materials: MY and SK. Analyzed the data: MO, TI, and TM. Wrote the paper: MO and YT.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tatsuya Kimura for providing technical assistance.
Footnotes
Funding. This research was supported by JSPS KAKENHI Grant Numbers 15H05017, 16K15785, 26293390, and the Takeda Science Foundation.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00300/full#supplementary-material
References
- Banks D. J., Beres S. B., Musser J. M. (2002). The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10, 515–521. 10.1016/S0966-842X(02)02461-7 [DOI] [PubMed] [Google Scholar]
- Banks D. J., Porcella S. F., Barbian K. D., Beres S. B., Philips L. E., Voyich J. M., et al. (2004). Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190, 727–738. 10.1086/422697 [DOI] [PubMed] [Google Scholar]
- Barnett C. C., Jr., Moore E. E., Silliman C. C., Abdalla E. K., Partrick D. A., Curley S. A. (2001). Cytosolic phospholipase A(2)-mediated ICAM-1 expression is calcium dependent. J. Surg. Res. 99, 307–310. 10.1006/jsre.2001.6188 [DOI] [PubMed] [Google Scholar]
- Beres S. B., Musser J. M. (2007). Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS ONE 2:e800. 10.1371/journal.pone.0000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S. B., Sylva G. L., Barbian K. D., Lei B., Hoff J. S., Mammarella N. D., et al. (2002). Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. U.S.A. 99, 10078–10083. 10.1073/pnas.152298499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S. B., Sylva G. L., Sturdevant D. E., Granville C. N., Liu M., Ricklefs S. M., et al. (2004). Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc. Natl. Acad. Sci. U.S.A. 101, 11833–11838. 10.1073/pnas.0404163101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M. W. (2000). Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511. 10.1128/CMR.13.3.470-511.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti V. A. (2000). Gram-Positive Pathogens. Washington, DC: ASM Press. [Google Scholar]
- Honda-Ogawa M., Ogawa T., Terao Y., Sumitomo T., Nakata M., Ikebe K., et al. (2013). Cysteine proteinase from Streptococcus pyogenes enables to evade innate immunity via degradation of complement factors. J. Biol. Chem. 288, 15854–15864. 10.1074/jbc.M113.469106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa M., Oda M., Domon H., Saitoh I., Hayasaki H., Terao Y. (2016). Streptococcus pyogenes CAMP factor attenuates phagocytic activity of RAW 264.7 cells. Microbes Infect. 18, 118–127. 10.1016/j.micinf.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Li Z., Sakota V., Jackson D., Franklin A. R., Beall B. (2003). Array of M protein gene subtypes in 1064 recent invasive group A Streptococcus isolates recovered from the active bacterial core surveillance. J. Infect. Dis. 188, 1587–1592. 10.1086/379050 [DOI] [PubMed] [Google Scholar]
- Moses A. E., Hidalgo-Grass C., Dan-Goor M., Jaffe J., Shetzigovsky I., Ravins M., et al. (2003). Emm typing of M nontypeable invasive group A Streptococcal isolates in Israel. J. Clin. Microbiol. 41, 4655–4659. 10.1128/JCM.41.10.4655-4659.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. P., Low D. E., Green K. A., Simor A. E., Loeb M., Gregson D., et al. (2003). Clinical and epidemiologic features of group a streptococcal pneumonia in Ontario, Canada. Arch. Intern. Med. 163, 467–472. 10.1001/archinte.163.4.467 [DOI] [PubMed] [Google Scholar]
- Murakami J., Kawabata S., Terao Y., Kikuchi K., Totsuka K., Tamaru A., et al. (2002). Distribution of emm genotypes and superantigen genes of Streptococcus pyogenes isolated in Japan from 1994 to 1999. Epidemiol. Infect. 128, 397–404. 10.1017/S0950268802006854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Krause R. M. (1998). Emerging Infections. San Diego, CA: Academic; Press, 185–218. [Google Scholar]
- Nagiec M. J., Lei B., Parker S. K., Vasil M. L., Matsumoto M., Ireland R. M., et al. (2004). Analysis of a novel prophage-encoded group A Streptococcus extracellular phospholipase A(2). J. Biol. Chem. 279, 45909–45918. 10.1074/jbc.M405434200 [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Kurokawa K., Yamashita A., Nakata M., Tomiyasu Y., Okahashi N., et al. (2003). Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13, 1042–1055. 10.1101/gr.1096703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa T., Furukawa N., Aikawa C., Watanabe T., Haobam B., Kurokawa K., et al. (2011). CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS ONE 6:e19543. 10.1371/journal.pone.0019543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K. L., Beall B., Barrett N. L., Cieslak P. R., Reingold A., Farley M. M., et al. (2002). Epidemiology of invasive group a streptococcus disease in the United States, 1995-1999. Clin. Infect. Dis. 35, 268–276. 10.1086/341409 [DOI] [PubMed] [Google Scholar]
- Pearson J. A., Tyler M. I., Retson K. V., Howden M. E. (1993). Studies on the subunit structure of textilotoxin, a potent presynaptic neurotoxin from the venom of the Australian common brown snake (Pseudonaja textilis). 3. The complete amino-acid sequences of all the subunits. Biochim. Biophys. Acta 1161, 223–229. 10.1016/0167-4838(93)90217-F [DOI] [PubMed] [Google Scholar]
- Schmitz F. J., Beyer A., Charpentier E., Normark B. H., Schade M., Fluit A. C., et al. (2003). Toxin-gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J. Infect. Dis. 188, 1578–1586. 10.1086/379230 [DOI] [PubMed] [Google Scholar]
- Sharkawy A., Low D. E., Saginur R., Gregson D., Schwartz B., Jessamine P., et al. (2002). Severe group a streptococcal soft-tissue infections in Ontario: 1992-1996. Clin. Infect. Dis. 34, 454–460. 10.1086/338466 [DOI] [PubMed] [Google Scholar]
- Sitkiewicz I., Nagiec M. J., Sumby P., Butler S. D., Cywes-Bentley C., Musser J. M. (2006). Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc. Natl. Acad. Sci. U.S.A. 103, 16009–16014. 10.1073/pnas.0607669103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkiewicz I., Stockbauer K. E., Musser J. M. (2007). Secreted bacterial phospholipase A2 enzymes: better living through phospholipolysis. Trends Microbiol. 15, 63–69. 10.1016/j.tim.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2001a). Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45, 101–113. 10.1006/plas.2000.1510 [DOI] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2001b). Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46, 140–148. 10.1006/plas.2000.1510 [DOI] [PubMed] [Google Scholar]
- Terao Y., Kawabata S., Nakata M., Nakagawa I., Hamada S. (2002). Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes associated with isolates from toxic shock-like syndrome patients. J. Biol. Chem. 277, 47428–47435. 10.1074/jbc.M209133200 [DOI] [PubMed] [Google Scholar]
- Terao Y., Mori Y., Yamaguchi M., Shimizu Y., Ooe K., Hamada S., et al. (2008). Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J. Biol. Chem. 283, 6253–6260. 10.1074/jbc.M704821200 [DOI] [PubMed] [Google Scholar]
- Terao Y., Yamaguchi M., Hamada S., Kawabata S. (2006). Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 281, 14215–14223. 10.1074/jbc.M513408200 [DOI] [PubMed] [Google Scholar]
- Thommesen L., Sjursen W., Gasvik K., Hanssen W., Brekke O. L., Skattebol L., et al. (1998). Selective inhibitors of cytosolic or secretory phospholipase A2 block TNF-induced activation of transcription factor nuclear factor-κ B and expression of ICAM-1. J. Immunol. 161, 3421–3430. [PubMed] [Google Scholar]
- Yokote K., Morisaki N., Zenibayashi M., Ueda S., Kanzaki T., Saito Y., et al. (1993). The phospholipase-A2 reaction leads to increased monocyte adhesion of endothelial cells via the expression of adhesion molecules. Eur. J. Biochem. 217, 723–729. 10.1111/j.1432-1033.1993.tb18298.x [DOI] [PubMed] [Google Scholar]
- Yu J. A., Sadaria M. R., Meng X., Mitra S., Ao L., Fullerton D. A., et al. (2012). Lung cancer cell invasion and expression of intercellular adhesion molecule-1 (ICAM-1) are attenuated by secretory phospholipase A(2) inhibition. J. Thorac. Cardiovasc. Surg. 143, 405–411. 10.1016/j.jtcvs.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Zhu X., Munoz N. M., Kim K. P., Sano H., Cho W., Leff A. R. (1999). Cytosolic phospholipase A2 activation is essential for beta 1 and beta 2 integrin-dependent adhesion of human eosinophils. J. Immunol. 163, 3423–3429. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.