Abstract
Background
The parasitic protozoa belonging to Leishmania (L.) donovani complex possess abundant, developmentally regulated cathepsin L-like cysteine proteases. Previously, we have reported the isolation of cysteine protease gene, Ldccys2 from Leishmania (L.) chagasi. Here, we have further characterized this cysteine protease gene and demonstrated its role during infection and survival of Leishmania (L.) chagasi within the U937 macrophage cells.
Results
The amastigote specific Ldccys2 genes of L. (L.) chagasi and L. (L.) donovani have identical gene organization, as determined by southern blots. In vivo expression analyses by Northern blots showed that Ldccys2 is amastigote specific. Western blot using anti-Ldccys2 antibody confirmed the amastigote specific protein expression. Recombinant expression of Ldccys2, a 30 kDA protein, was functionally active in a gelatin assay. Results from Ldccys2 heterozygous knockout mutants showed its role during macrophage infection and in intra-macrophage survival of the parasites. Since attempts to generate null mutants failed, we used antisense RNA inhibition to regulate Ldcccys2 gene expression. Not surprisingly, the results from antisense studies further confirmed the results from heterozygous knockout mutants, reiterating the importance of amastigote specific cysteine proteases in Leishmania infection and pathogenesis.
Conclusions
The study shows that Ldccys2 is a developmentally regulated gene and that Ldccys2 is expressed only in infectious amastigote stages of the parasite. The collective results from both the heterozygous knockout mutants and antisense mRNA inhibition studies shows that Ldccys2 helps in infection and survival of L. (L.) chagasi amastigotes within the macrophage cells. Finally, antisense RNA technique can be used as an alternate approach to gene knockout, for silencing gene expression in L. (L.) chagasi, especially in cases such as this, where a null mutant cannot be achieved by homologous recombination.
Background
Leishmania are the etiological agents of a variety of disease manifestations, collectively termed as leishmaniasis. Visceral leishmaniasis caused by Leishmania (L.) donovani and Leishmania (L.) chagasi is a serious health problem in many tropical and subtropical countries [1-3]. During the digenetic life cycles of Leishmania, it alternates between gut of sand fly vector as an extra cellular promastigote and in the acidic phagolysosome of macrophage as an intracellular amastigote. However, the most intriguing question is the capacity of Leishmania to withstand the hydrolytic conditions of the macrophages, the mechanism of which is still unclear. Thus, identifying the genes expressed specifically in the amastigote stage of the parasites and elucidating their biological function is very important as it would provide new insights into the role of these gene products in the intracellular life cycle of the parasites. Further, this will also help in designing specific drugs and identifying vaccine candidates.
Cysteine proteases play an important role in the infection, replication, development and metabolism of protozoan parasites [4,5]. They have been implicated in the invasion of human erythrocyte by Plasmodium falciparum [6] and considered as virulence factors in the pathogenesis of Entamoeba histolytica [7]. Cysteine protease activity is necessary for the survival of Leishmania (L.) mexicana [8,9] and related protozoan, Trypanosoma cruzi, within the macrophages, in vitro [10]. Knockout studies in L. (L.) mexicana have shown that cysteine proteases not only are virulence factors but also act as modulators of host immune responses [11,12]. Thus, cysteine proteases have become a potential target for chemotherapy and a candidate for vaccine development. Initial studies have confirmed the efficacy of cysteine protease inhibitors in treatment of T. cruzi, P. falciparum and L. (L.) major [13-15]. Immunization with the hybrid protein vaccine, consisting of L. (L.) major cysteine proteases CPB and CPA, partially protected against leishmaniasis [16]. So far, functionally well characterized cysteine proteases are from the New World species of Lesihamania causing the cutaneous forms of leishmaniasis. The members of L .(L.) donovani complex also possess multiple classes of cysteine proteases, which are developmentally regulated [17,18] and are not functionally well characterized. Therefore, there is a need to study the function of these proteases and their role in visceral leishmaniasis.
Studies aimed at defining the function of protozoan parasite components have often used gene disruption approach by homologous recombination. Recently an alternative antisense RNA method was followed that may easily and rapidly answer the complex biological questions. Anti sense RNA approach has been used to study the functions of certain gene products in Entamoeba and Leishmania, to elucidate the functions of cysteine protease [19,20], A2 protein [21] and gp63 [22]. Previously, we have isolated and characterized two distinct cysteine protease cDNA clones Ldccys1 and Ldccys2 from promastigote and amastigote specific cDNA libararies of L. (L.) chagasi [17]. Ldccys1, a member of multi gene family was characterized both in L. (L.) chagasi (Ldccys1) and L. (L.) donovani (Lddcys1) parasites [18]. In the present study, we have characterized the functional role of amastigote specific cysteine protease gene (Ldccys2) of L. (L.) chagasi. We have generated Ldccys2 heterozygous knockout mutants of L. (L.) chagasi by homologous recombination. As an alternative approach, antisense mRNA expression was also employed. Results obtained from both, gene disruption and antisense mRNA expression shows that Ldccys2 plays an important role in the survival of amastigotes within the U937 macrophages.
Results
Ldccys2 is a single copy gene
From our earlier studies it is known that Ldccys2 is a single copy gene in L. (L.) chagasi. In order to compare the genomic organization of Ldccys2 gene in L. (L.) donovani and L. (L.) chagasi, the members of L. (L.) donovani complex, Southern analysis of genomic DNA was performed. Genomic DNA was digested with different restriction enzymes as shown in Figure 1A and probed with Ldccys2 coding region as a probe. Identical hybridizing bands were present in both L. (L.) chagasi and L. (L.) donovani (Figure 1A). PCR amplification and sequencing of Ldccys2 coding region from L. (L.) donovani genomic DNA showed identical nucleotide sequences, indicating that L. (L.) chagasi and L. (L.) donovani had identical Ldccys2 cysteine protease genes.
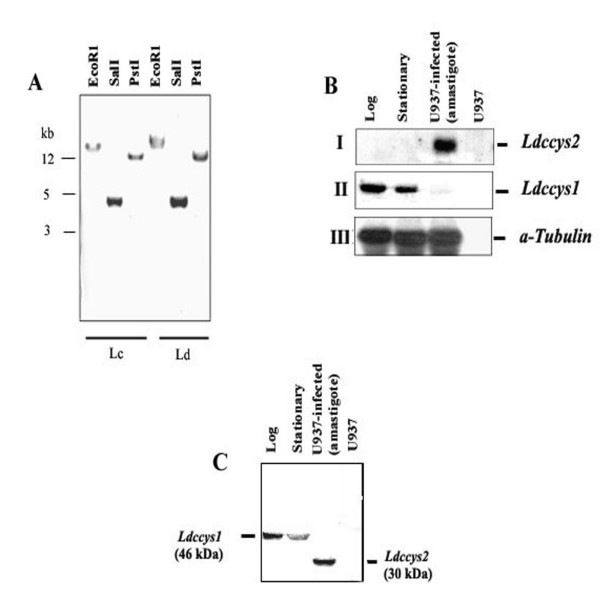
Figure 1.
Ldccys2 is a single copy gene and is expressed only in amastigotes. (A) Southern blot hybridization of digested genomic DNA from L. (L.) chagasi (Lc) and L. (L.) donovani (Ld). 5 μg of genomic DNA was digested with restriction enzymes as mentioned in the figure and blotted onto Hybond-N membranes. The blot was probed with coding region pf Ldccys2 cDNA clone. (B) Northern blot analyses of L. (L.) chagasi total RNA. Total RNA (10 μg/lane) from promastigotes of logarithmic (lane 1), stationary (lane 2) growth phase, U937 cells (human macrophage cell line) infected with promastigotes for 96 h (lane 3) and uninfected U937 cells (lane 4) were separated on 1.2% (w/v) formaldehyde agarose gel and transferred on to Hybond N+ membrane. Blot was hybridized with PCR amplified DNA fragment containing 3'UTR from Ldccys2 (panel I), Ldccys1 (154 bp, near the polyA region, panel II), and coding region of α-tubulin from L. (L.) chagasi, a kind gift from Dr. M.E. Wilson (panel IV). (C) Western blot analysis of L. (L.) chagasi promastigotes and amastigotes. Equal amounts of proteins from promastigotes of logarithmic (lane 1), stationary (lane 2) phase, U937 cells infected with promastigotes (lane 3) and uninfected U937 cells (lane 4) were separated on 10% (w/v) SDS-PAGE and blotted on to Hybond – P membrane. Western blot analysis was carried out using α-Ldccys2 antibody.
Ldccys2 gene is differentially expressed
To study the expression of Ldccys2 cysteine protease genes, Northern blot analysis was performed using L. (L.) chagasi total RNA from log and stationary phase promastigotes and amastigotes. The 3'UTR region of Ldccys2 used as a probe, hybridized to a 2.5 kb transcript in amastigote (Figure 1BI) unlike the cysteine protease, Ldccys1, which is predominantly expressed in promastigote stages of the parasite (Figure 1 BII). The α-tubulin gene expression was used as an internal control (Figure 1 BIII). Ldccys2 showed similar pattern of expression in L. (L.) donovani (data not shown). It is important to note that this result is in contrast to the earlier report (17), wherein Ldccys2 was identified as promastigote specific cysteine protease gene and Ldccys1 was thought to be amastigote specific. We have confirmed the expression pattern and rectified this error in the following publication (18).
To further confirm the amastigote expression of Ldccys2, Western blot analysis was carried out using extracts from both promastigotes and U937 infected amastigotes of L. (L.) chagasi. The anti-Ldccys2 antibody detected a 30 kDa band in amastigotes alone (Figure 1C, lane 3).
However, a single 46 kDa band seen in log and stationary phase promastigotes (Figure 1C, lanes 1 and 2) represents promastigote specific cysteine protease Ldccys1, due to the cross-reactivity of the antibody. Taken together, the above results show that Ldccys2 is expressed only in the amastigote stage of L. (L.) chagasi.
Recombinant Ldccys2 expressed in insect cells is functionally active
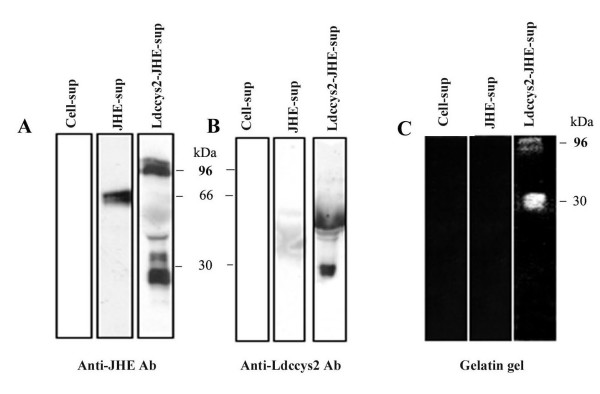
In order to determine the cysteine protease activity encoded by Ldccys2, the predicted coding region was cloned into pIE1/153A.jhe (6hep) vector and transfected into High Five insect cells (invitrogen). The supernatant was harvested and analyzed for the presence of recombinant protein by Western blot analysis using JHE and Ldccys2 antibodies. Western blot analyses with α-JHE antibody (Figure 2A, lane3) detected a band of 96 kDa, corresponding to the fusion protein. The lower molecular mass bands are due to the processed products as a result of autocatalytic activity. Anti- Ldccys2 antibody however, could not detect the recombinant fusion protein (Figure 2B lane 3). The processed products were detected instead, which may be because of inaccessibility of the epitope in the fusion protein. Both the antibodies did not give any background with the negative control (cell extract, Figure 2B and 2C, lane1). Protease activity determined by gelatin SDS-PAGE, showed a band of approximately 30 kDa (Figure 2C, lane 3), confirming the protease activity. Cysteine protease activity was completely inhibited by a specific inhibitor, E-64 at 20 μM concentration (data not shown).
Figure 2.
Expression of recombinant Ldccys2 using insect cell expression system. Western blots showing recombinant expression of Ldccys2 using anti-JHE antibody (A) and duplicate blot probed with anti-Ldccys2 antibody (B). The lanes are labeled appropriately. 5 μg of supernatant from the insect cells expressing recombinant plasmids or the control were separated on 8% SDS-PAGE, blotted onto to PVDF membranes and probed with 1:500 dilution of anti-JHE or 1:100 dilution of anti-Ldccys2 antibody. A horseradish peroxidase conjugated anti-mouse secondary antibody was used and the blot was developed by using ECL chemiluminescent kit. (C) Gelatin assay showing cysteine protease activity of the recombinant protein. The supernatant was separated on a gelatin gel and incubated in a reducing buffer. The gel was then stained with Coomassie Blue to visualize the clearing of gelatin on the gel indicating cysteine protease activity.
Ldccys2 single allele gene replacement by homologous recombination
In order to investigate the function Ldccys2 gene, we have performed gene replacement by disrupting a 500 bp coding region of the gene with hyg and dhfr-ts cassette as shown in figure 3A. Following transfection, hygromycin B resistant parasites (Ldccys2KO, heterozygous knockout mutants) were selected for further analysis. The PstI restriction enzyme digested genomic DNA from wild type L. (L.) chagasi and Ldccys2 KO were subjected to Southern analysis and results are presented in figure 3B. The disrupted Ldccys2 allele had an additional PstI site that is present within the hyg gene. As shown in the figure 3B, 5' and 3'probes (probes a and b) hybridized to a single 3.5 kb band with wild type genomic DNA. In Ldccys2KO, probe-a hybridized to two bands (3.5 kb and 2.8 kb) and probe-b hybridized to 3.5 kb and 3.0 kb bands. As expected, one of the bands corresponds to the wild type allele whereas the other band corresponded to the disrupted allele. The hyg specific probe-c resulted in no hybridizing band with PstI digested wild type genomic DNA, while in Ldccys2KO, two bands (2.8 kb and 3.0 kb) corresponding to the disrupted allele were detected (Figure 3B, probe c). These results clearly indicate that one of the alleles of the Ldccys2 gene has been replaced by hygromycin gene. Similarly, a second cassette using neomycin gene was used in order to replace the second allele of Ldccys2 gene. Despite numerous attempts, we were unable to obtain a null mutant of Ldccys2. We then checked for any events of rearrangements or change in ploidy levels in the heterozygous mutants. No such detectable events were detected (data not shown).
Figure 3.
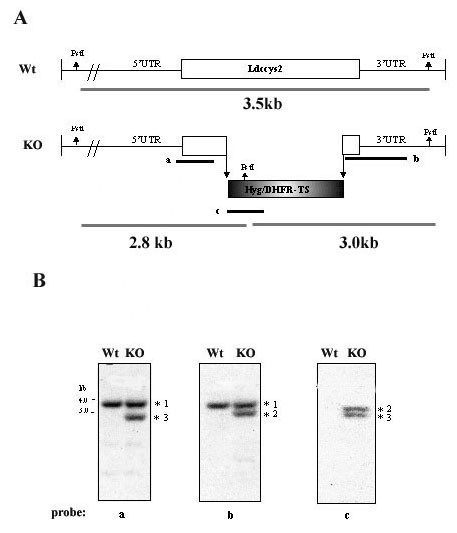
Ldccys2 single allele gene replacement. (A) Schematic representation of the wild type and hyg/dhfr-ts targeted alleles of Ldccys2. The location of the PstI sites used to characterize the hygromycin B resistant recombinants is shown by arrows. The bold lines labelled a, b and c represents the location of different probes used in Southern analyses. (B) Southern analyses of Ldccys2 heterozygous knockout mutants. Genomic DNA (2 μg/lane) from wild type L. (L.) chagasi (WT) and Ldccys2 heterozygous knockout mutants (KO) were digested with PstI and separated on a 0.9% (w/v) agarose gel. The DNA was blotted on to Hybond N+ membrane and hybridized with probes a (i), b (ii) and c (iii). Asterisks: 1- represents wild type allele of Ldccys2 (3.5 kb), 2 and 3- represent disrupted allele (3.0 kb and 2.8 kb). The grey lines indicate the size of expected bands upon PstI digestion.
Ldccys2 single allele knock-out mutants had reduced mRNA and protein levels
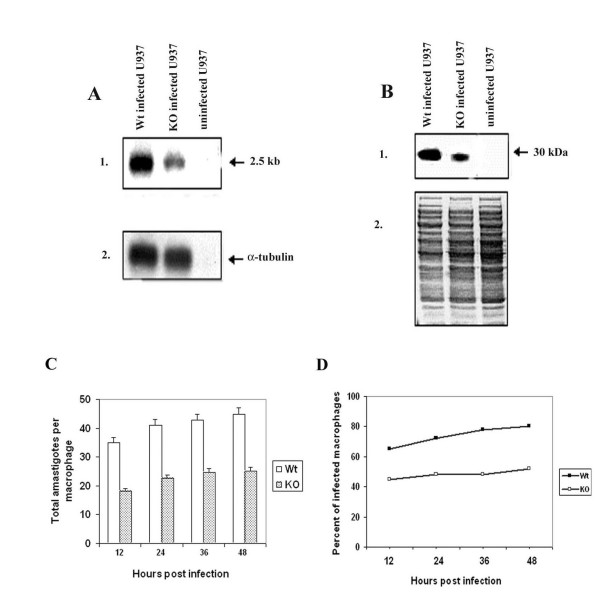
In order to compare the expression of Ldccys2 gene in wild type and heterozygous knockout mutants (Ldccys2 KO), amastigote RNA was isolated from U937 cells infected with wild type as well as Ldccys2KO mutant L. (L.) chagasi promastigotes. Northern blot analysis with Ldccys2 coding region probe detected a single transcript of 2.5 kb from wild type as well as Ldccys2 KO amastigotes (Figure 4A, panel 1). However, the transcript level of Ldccys2 in the Ldccys2KO amastigotes was reduced by 64% compared to that in the wild type L. (L.) chagasi. There was no change in the level of α-tubulin expression either in the Ldccys2KO or in the wild type amastigotes (Figure 4A, panel 2).
Figure 4.
Characterization of L. (L.) chagasi heterozygous knockout mutant amastigotes and intra macrophage survival of Ldccys2KO amastigotes in vitro. (A) Northern blot analysis. Total RNA (10 μg/lane) from U937 cells infected with wild type promastigotes, Ldccys2KO and uninfected U937 cells were separated on 1.2% (w/v) formaldehyde agarose gel and transferred on to Hybond N+ membrane. Blot was hybridized with Ldccys2 coding region DNA probe (1) and α-tubulin gene from L. (L.) chagasi (2). (B) Western blot analysis. Equal amount of lysates from U937 cells infected with wild type promastigotes, Ldccys2KO promastigotes and uninfected U937 cells were separated on 10% (w/v) SDS-PAGE, blotted on to Hybond P membrane. Panel 1 represents the membrane that was probed with α-Ldccys2 antibody and panel 2 is the duplicate gel stained with Coomassie blue. (C) Bar graph showing the number of intracellular amastigotes. U937 macrophage cells were infected at a macrophage to parasite ratio of 1:10. The survival of the amastigotes within the macrophages was evaluated every 12 hours by cytospin and Diff-Quick staining. (D) Graph showing the percent of total macrophages infected at the given time points. For each treatment, 100 infected macrophages were counted. Values represent means ± SEM from three independent experiments.
The effect of Ldccys2 single allele disruption at protein level was studied using western blot analysis. Equal amounts of lysates from wild type and Ldccys2KO L. (L.) chagasi amastigotes were analyzed using antiserum raised against Ldccys2 protein. Wild type L. (L.) chagasi amastigotes showed a major band of 30 kDa (Figure 4B, panel 1), while Ldccys2KO amastigotes showed an identical sized band, but there was a reduction in amounts by 45%, based on the quantitation of bands. The antibody did not pick up any signal from the negative control (U937 cells). Figure 4B, panel 2 indicates the equal loading of proteins. These results once again, confirm the deletion of one of the alleles of Ldccys2.
Ldccys2 heterozygous knockout mutants exhibit reduced infectivity and intra-macrophage survival of amastigotes
In order to study the effect of Ldccys2 heterozygous knockout mutants on parasite growth, a growth curve analysis was performed. No significant differences were observed between the wildtype parasites and the Ldccys2KO (data not shown). Next, we performed an infection assay with differentiated U937 macrophage cells using with wild type and Ldccys2KO L. (L.) chagasi promastigotes. Interestingly, twelve hours after infection, the number of amastigotes in Ldccys2KO infected macrophages were reduced by half as compared to that of wild type infected macrophage cells. This trend remained same at all the time intervals tested (24, 36 and 48 hours post infection), indicating that the intracellular amastigotes were not able to multiply efficiently (Figure 4C). The percentage of infected macrophages in the wildtype and Ldccys2KO were similar at 12, 24, 36 and 48 hours post infection (Figure 4D), however, there was a clear decrease in the initial infection in case of Ldccys2KO, as compared to the wildtype parasites. The above data suggests that Ldccys2 heterozygous knockout has resulted in reduced levels of Ldccys2 expression and furthermore, has decreased the ability of the amastigotes to infect the macrophages and survive inside the macrophage cells. Taken together, these results suggest that Ldccys2 gene has a significant role in the infection and intramacrophage survival of amastigotes.
Antisense mRNA inhibition of Ldccys2 resulted in reduced macrophage infection and poor amastigote survival within the macrophages
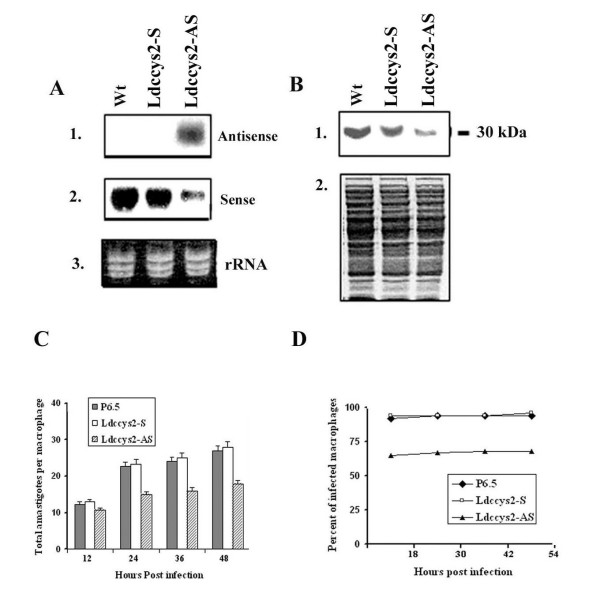
We were unable to generate a complete knock out mutant of Ldccys2 and the heterozygous knockout mutants only partially inhibited the gene expression. Therefore, we used an alternative approach of antisense mRNA inhibition to silence the Ldccys2 gene and to confirm the conclusions derived from Ldccys2 heterozygous knockout mutants studies. Plasmid constructs containing the Ldccys2 antisense and sense sequences were transfected into L. (L.) chagasi promastigotes. The presence of plasmids in these transfectants was confirmed by Southern analysis with Ldccys2 probes (data not shown). Northern blot analyses of amastigotes expressing different plasmid constructs were carried out using specific sense and antisense probes. As expected, Ldccys2 antisense mRNA was expressed only in amastigotes with antisense constructs (Figure 5A, panel 1), and these amastigotes showed a 74% decrease in the Ldccys2 mRNA levels (Figure 5A, panel 2). The amastigotes containing either the sense plasmid or the control plasmid did not show any change in the levels of Ldccys2 mRNA (Figure 5A, panel 2). The stained agarose gel indicates the equal amounts of RNA used in the Northern blots (Figure 5A, panel 3). These results clearly demonstrate that the episomal expression of Ldccys2 antisense mRNA reduces the levels of endogenous Ldccys2 mRNA.
Figure 5.
Expression of sense and antisense Ldccys2 transcripts in L. (L.) chagasi and survival of wild type, sense and antisense Ldccys2 expressing amastigotes in macrophages. (A) Northern blot analyses of amastigotes expressing sense and antisense transcripts. Total RNA (10 μg/lane) isolated from axenic amastigotes of wildtype parasites, transfectants with P6.5/Ldccys2 sense plasmid and P6.5/Ldccys2 antisense plasmid were separated on 1.2% (w/v) formaldehyde agarose gel and blotted on to Hybond N+ membrane. Blots were probed with probes specific for antisense (panel 1) and sense (panel 2) transcripts. Panel 3 represents ethidium bromide stained gel. (B) Western blot analysis of sense and antisense Ldccys2 expressing amastigotes. Equal amounts of total protein extracted from axenically transformed amastigotes with P6.5/Ldccys2 sense plasmid and P6.5/Ldccys2 antisense plasmid and wildtype parasites were separated on 10% (w/v) SDS-PAGE and blotted on to Hybond P membrane. The blot was probed with α-Ldccys2 antibody (panel 1) and a duplicate gel was stained with Coomassie blue (panel2). (C) Bar graph showing total number of amastigotes within the macrophage cells. U937 macrophage cells were infected at a macrophage to parasite ratio of 1:10. The survival of the amstigotes within the macrophages was evaluated every 12 hours by cytospin and Diff-Quick staining.(D) Graph showing the percent of total macrophages infected at the given time points. For each treatment, 100 infected macrophages were counted. Values represent means ± SEM from three independent experiments.
We next performed Western blot analysis of transfectants expressing sense and antisense Ldccys2 mRNA using α-Ldccys2 antibody. Amastigotes expressing antisense mRNA showed a significant reduction (by 76%) in the levels of Ldccys2, however, the expression of sense mRNA did not increase the levels of Ldccys2 compared to amastigotes containing the control plasmid (Figure 5B, panel1). Equal loading of the protein is depicted by the Coomassie stained gel (Figure 5B, panel 2). The results further confirm that Ldccys2 antisense mRNA expression does inhibit the endogenous levels of Ldccys2.
Survival of L. (L.) chagasi amastigotes within the macrophages was assessed in vitro, using U937 cells. Results from three individual experiments are pooled and the collective data is presented in figure 5C. Results showed that the amastigotes carrying either the control plasmid (P6.5) or expressing Ldccys2 sense mRNA did not have any effect on their capacity to infect and survive inside the macrophages. The number of amastigotes per macrophage cell remained the same at initial stages of infection (data not shown). However, amastigotes expressing Ldccys2 antisense mRNA showed significant reduction (by almost 50%) in terms of the initial infection and intra macrophage survival starting from 12 hours post infection and remained consistent up to 48 hours after infection (Figure 5D). The percentage of total infected macrophages is shown in figure 5D. Consistent with the results from the heterozygous knockout mutants, the Ldccys2 antisense transfectants had reduced levels of infection when compared to that of the wildtype and control plasmids. These results further underline the importance of Ldccys2 in infection of macrophages and subsequent survival of amastigotes within the macrophage cells. Taken together, these results indicate that the amastigote specific cysteine protease, Ldccys2 plays an important role in intramacrophage survival of L. (L.) chagasi amastigotes.
Discussion
In this study, we have shown that amastigote cysteine protease, Ldccys2 necessary for macrophage infection and for survival of L. (L.) chagasi amastigotes inside the macrophage cells using two different approaches, gene disruption and antisense mRNA expression. Leishmania (L.) chagasi cysteine protease, Ldccys2 is a single copy gene [18] and is expressed only in amastigote stage of the parasite. L. (L.) donovani also possesses an identical gene. Expression studies performed demonstrates that Ldccys2 is expressed only in amastigotes of L. (L.) chagasi and L. (L.) donovani. A similar single copy gene in L. (L.) mexicana (Lmcpa) has a significant sequence identity (80%) to Ldccys2. Lmcpa gene of L. (L.) mexicana which is the closest homolog of Ldccys2 is differentially expressed in amastigotes [23], whereas the Ldccys2 gene of L. (L.) chagasi is expressed specifically in the amastigote stage. In addition, the major amastigote specific cysteine proteinase gene of L. (L.) mexicana, cpb2.8 is a multicopy gene, located within the lmcpb cluster [24] and there is no significant sequence identity (34%) between Ldccys2 and lmcpb2.8. Therefore, gene organization and expression of amastigote specific cysteine proteases in L. (L.) mexicana and L. (L.) donovani complex are not identical. Interestingly, Llacys1, an amastigote specific cysteine protease from L. (L.) amazonensis was identified recently, which has high homology (88%) with Ldccys2 and like Ldccys2, is only expressed in amastigote stages [25]. In order to confirm the protease activity of amastigote cysteine protease, full-length Ldccys2 was expressed as JHE-fusion protein in an insect cell expression system. The recombinant protein hydrolysed gelatin in activity gels.
Recently, functions of many genes have been established in Leishmania by following gene disruption approach [26,27]. To study the biological role of Ldccys2 cysteine protease, we have followed targeted gene disruption using hyg gene as the selectable marker. Genomic Southern analyses of the hygromycin B resistant clones using specific probes confirmed the replacement of 500 bp region within the ORF of the Ldccys2 by hyg/dhfr-ts. Northern and Western blot analyses Ldccys2KO amastigotes showed a clear decrease in the endogenous levels of Ldccys2 compared to that of wild type L. (L.) chagasi amastigotes (Figure 4), demonstrating the disruption of one of the Ldccys2 alleles. Despite our repeated effort using different selectable marker genes, we failed to create a null mutant of Ldccys2 in L. (L.) chagasi. Since gene amplification and increase in ploidy level has been reported for Leishmania species [28], we analyzed for changes in ploidy level and or gene amplification and found no such events occurring in these heterozygous knockout mutants. This suggests that Ldccys2 is an important gene necessary for the initiation of macrophage infection and survival of amastigotes within the macrophages. Therefore we used the Ldccys2 heterozygous knockout parasites for further functional studies. The U937 macrophage cells infected with Ldccys2 heterozygous knockout mutants showed a significant decrease in the initial infection and survival within the macrophage cells, compared to wild type (Figure 4). This trend remained consistent from 12 hours up to 48 hours after infection. The decrease in initiation of infection and subsequent survival of amastigotes may explain the role of Ldccys2 in amastigote metabolism and its survival inside the macrophages [4]. Consistent with our results, null mutants of the lmcpb cluster in L. (L.) mexicana exhibited reduction in macrophage infectivity and survival [23]. However, lack of Ldccys2 null mutant prevented us from a detailed study on the role of Ldccys2 in survival and pathogenesis, using animal models.
The lack of Ldccys2 null mutants prompted us to use an alternative approach of antisense mRNA inhibtion in order to confirm the results obtained from Ldccys2 heterozygous knockout studies. Using antisense RNA technique it has become possible to investigate the contribution of cysteine proteases to virulence mechanism of E. histolytica [19,20] and A2- protein in the survival of L. (L.) donovani amastigotes in macrophages [21]. Furthermore, episomal expression of sense and anti sense mRNA of L. (L.) amazonensis gp63using Leishmania- specific P6.5 vector provided evidence for a role both in binding of macrophages and the intracellular survival and replication [22]. Not surprisingly, L. (L.) chagasi amastigotes expressing Ldccys2 antisense mRNA has clearly down regulated the endogenous Ldccys2 mRNA levels (Figure 5). However, a small amount of Ldccys2 mRNA is still present in antisense transfectants, which may be due to the incomplete suppression, which is inherent to this approach. Infection of U937 macrophage cells with transfectants expressing Ldccys2 antisense mRNA showed a significant decrease in amastigote survival inside the macrophage cells, as compared to amastigotes expressing either sense Ldccys2 mRNA or the control plasmid (Figure 5). This is in agreement with the results obtained from Ldccys2 heterozygous knockout mutants, reaffirming the role of Ldccys2 in initiation of intracellular infection, and subsequent survival and multiplication of amastigotes. However, the actual effect of antisense mRNA expression may be greater, since reversal of antisense effects is expected from the release of the selective pressure necessary to avoid drug toxicity to macrophages [21]. This is especially true in the assay for intracellular survival and replication, since it requires prolonged period of incubation. The transient nature of the antisense suppression in the absence of selection prevented us from taking up animal studies that also require prolonged infection with these transfectants.
The out come of this study shows that Ldccys2 is expressed only in amastigote stage of the parasite. Both the heterozygous knockout mutants and antisense mRNA expression shows that Ldccys2 is important for infection of macrophages and survival of L. (L.) chagasi amastigotes inside the macrophage cells. Based on the data obtained in this study, there is a clear justification for targeting the Ldccys2 gene in L (L.). chagasi and carrying out further studies in animal models. Currently, we are working towards generating null mutants of Ldccys2 in L. (L.) chagasi and to further understand the role of this protein in the pathogenesis of L. (L.) chagasi parasites.
Methods
Parasite culture
L. (L.) chagasi strain MHOM/BR/74/PP75 promastigotes were grown in HOMEM, pH 7.4 (Gibco BRL) supplemented with 10% (v/v) heat inactivated calf serum, at 26°C. Promastigote cultures were seeded at 1 × 106parasites/ml and harvested in logarithmic or stationary growth phase, as defined by cell concentration. L. (L.) chagasi promastigotes were transformed to axenic amastigotes using the standardized protocol as described earlier [29]. All the transfectants were periodically converted to amastigotes by infecting U937 macrophage cells in order to ensure the property of virulence.
RNA isolation and northern hybridization
Total RNA was isolated using trizol (GibcoBRL) as recommended by the manufacturer. Promastigote RNA was obtained from logarithmic and stationary growth phase parasites of L. (L.) chagasi. To isolate amastigote RNA, human macrophage cell line (U937, ATCC) was infected with promastigote parasites, which were subsequently converted into intracellular amastigotes [29]. RNA (10 μg/lane) was separated on 1.2% (w/v) formaldehyde agarose gels and transferred onto Hybond N+ membrane (Amersham Pharmacia). Standard procedures were followed to perform Southern and Northern blot hybridizations. Using Quick Prime Labeling Kit (Amersham Pharmacia Biotech), probes representing specific region of Ldccys2 cDNA clone was labeled with [α32P] dCTP. The sense and antisense probes were generated by 5'end-labeling the specific sense and antisense oligonuecleotide primers with γ-32P ATP, using T4 polynucleotide kinase according to the standard protocol.
Polyclonal antibody production against Ldccys2
Direct DNA immunization [30] was used to generate antibody against Ldccys2. Briefly, the pro-mature region of the gene was PCR amplified from the Ldccys2 cDNA clone using specific 5' (5'CAGACAGGATCCG CGGCCGCCATGGACGACTTCATTGCC 3') and 3' primers (5' GGCCGCGGATCCGCG GCCGCCTATGAGGTGTTGGAGTCGTC 3') and the PCR product was sub-cloned into BamHI site of pcDNA 3.1 (Invitrogen). After confirmation by sequencing the plasmid constructs were amplified in E. coli and DNA was isolated using endotoxin free maxi prep kit (Qiagen). 50 μg of plasmid DNA in 100 μl of saline was injected into the hind limb quadriceps muscles of BALB/c mice. All the mice were boosted after 14 days and tested for antibody response seven days after the boost. Mice with positive response were boosted one more time and after a week sera was collected and stored at -20°C for further use.
Western blot analysis
Denaturing gel electrophoresis was carried out according to the standard protocols. Following SDS-PAGE, proteins were transferred onto to Hybond P membrane (Amersham Pharmacia Biotech) using semi-dry transblot apparatus (BioRad). Membranes were blocked for 2 hours with 5% (w/v) skim milk solution and probed overnight at 4°C with 1:200 dilution of anti-Ldccys2 antiserum. Membranes were washed three times with phosphate buffered saline containing 0.1% (v/v) Tween and incubated with 1:5000 dilution of anti-mouse horseradish peroxidase linked secondary antibody (Molecular Probes). Enhanced chemiluminescent kit (Amersham Pharmacia Biotech) was used for detection.
Expression of cysteine protease genes
In order to express Ldccys2 cysteine protease gene, the coding region was amplified by PCR. The digested PCR product was cloned in-frame into unique NotI site of pIE1/153A.jhe (6hep) vector [31] and confirmed by sequencing. The fusion of the Cysteine proteases to the c-terminus of Juvenile hormone esterase (JHE) directs the secretion into the culture supernatant of transfected insect cells. Culture of insect cells and procurement of samples containing recombinant protein from transfected insect cells are described earlier [32]. Briefly, High Five insect cells (Invitrogen) were seeded into6-well culture plates (35 mm diameter) at a density of 5 × 105 cells/ml (2 ml/well) and transfected for 5 h with 0.5 ml of transfection solution containing 30 μg/ml lipofectin (Life Technologies) and 6 μg/ml total plasmid DNA in basal IPL-41 medium. Then the transfection solution was removed, the cells were rinsed with basal media and 2 ml IPL-41 + 10% FBS was added and incubated for sixty hours. The supernatant was then harvested and stored at -20°C for analysis. Enzyme activity was analyzed on gelatin SDS-PAGE containing 0.2% gelatin and 8% (w/v) acrylamide as described previously [17]. For inhibition studies, gels were pre-incubated with 20 μM E-64 (Sigma), a Cysteine protease specific inhibitor. Gels were stained with Coomassie Brilliant Blue for half hour and destained with 30% Methanol for 2 hours, to visualize the bands. Bands were photographed using Diamed apparatus (Bio-Rad).
Ldccys2 gene disruption by homologous recombination
Gene replacement vector, pXLdccys2-HygKO was designed to replace 500 bp mature region of Ldccys2 coding region and constructed as outlined below (Figure 2b). The sense primer Ldccys2 HindIII (5' GCTCTCAAGCTTGCTCACGCATCCGCCGC-3') and the antisense primer Ldccys2 Xho I (5'GAAGGCCTCGAGCGAGCCGCACATTCCCTG-3') were used to amplify a 500 bp region from Ldccys2 cDNA and cloned between HindIII – XhoI sites in pX63-Hyg (kindly provided by Dr. S. M. Beverley) for the 5' end homologous recombination. A 450 bp 3' end homologous recombination fragment was amplified using the sense primer Ldccys2 SmaI (5'CCGGAGCCCGGGCCCACGGCGCTTGTGCAG-3') and antisense primer Ldccys2 BglII (5' ACTGTCAGATCTGCTGTGCGCCAGATCGCG-3') and cloned between SmaI and BglII sites of pX63-Hyg. The plasmid construct pXLdccys2-HygKO was confirmed by sequencing and digested with HindIII and BglII. The resulting 3.8 kb linear fragment containing hygromycin phospotransferase/dihydrofolate reductase-thymidylate synthase (hyg/dhfr-ts) gene sequence and the homologous recombination fragment was purified and used for transfection.
Episomal expression of Ldccys2 specific sense and antisense mRNA
The Ldccys2 cDNA clone was isolated from an amastigote specific cDNA library of L. (L.) chagasi. Ldccys2 was earlier reported as promastigote specific cDNA clone [17]. However, it was further verified to be amastigote specific gene (18). The coding region (1 kb) of Ldccys2 was inserted at BamHI site of Leishmania specific vector P6.5 [22], a kind gift from Dr. K.P.Chang. Cloning was done in both sense and antisense orientations with reference to that of N-acetylglucosamine 1-phosphate transferase (nagt), which was used as the selective marker for tunicamycin resistance. Plasmid constructs were confirmed by sequencing. P6.5 plasmid and the sense and antisense plasmid constructs were amplified in E. coli and isolated with a DNA maxi prep kit (Qiagen) and the purified DNA samples were used for transfection.
DNA transfection
All the transfections were carried out using L. (L.) chagasi promastigotes. Briefly, 4 × 107 log phase promastigotes were pelleted, washed twice with phosphate buffered saline and resuspended in 0.4 ml of electroporation buffer in a 0.2 cm cuvette. Twenty-five microgram of P6.5 plasmid DNA constructs was used for episomal expression, whereas for homologous recombination 5 μg of linearized DNA fragment was transfected. DNA was electroporated using BioRad Gene Pulser Unit at 0.45 kV, capacitance of 25 μF and resistance of 250 ohms. Following electroporation, cells were resuspended in drug-free media for 24 hours. Parasites with P6.5 plasmid DNA constructs were selected for tunicamycin resistance at 5, 10 and 20 μg/ml tunicamycin (Sigma). Parasites transfected with DNA fragment for homologous recombination were selected for hygromycin B (GibcoBRL) resistance at 100 μg/ml.
In vitro assay for intra macrophage survival
The U937 macrophage cells were infected with L. (L.) chagasi stationary promastigotes at a host to parasite ratio of 1:10 [33]. Briefly, U937 suspension cells (10 6 cells/ml) in 10 ml RPMI 1640 medium supplemented with 10% (v/v) heat inactivated fetal bovine serum, 2 mM L-glutamine and 50 μg gentamycin (Gibco BRL) were allowed to grow at 37°C for 3 days in 10% (v/v) CO2. The cells were then induced to adhere with 7.5 ng/ml of phorbol myristate acetate (PMA) and allowed to recover for 72 hours. The macrophage cells were then infected with stationary phase promastigotes and allowed to infect for 6 hours. The unbound parasites were washed away with RPMI medium and the infected macrophage cells were fed with fresh RPMI medium and left for 4 days. At every 12 hours interval up to 72 hours, cells were scraped and Diff-Quick stained slides were prepared for microscopic counting,
Abbreviations
Hyg, hygromycin; JHE, juvenile hormone esterase; kb, kilobase; KO, knock-out; ORF, open reading frame; UTR, untranslated region
Authors' contributions
VM carried out all the plasmid constructs for gene disruption and antisense mRNA inhibiton studies and performed the Southern and Northern blot analyses shown in Figures 1, 3, 4 & 6; wrote parts of the manuscript. ASK performed all the transfections, western blots and macrophage assays shown in Figures 1, 2, 4, 5, 6 and 7; wrote parts of the manuscript. LG supervised the experiments and gave laboratory support, and critically read the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This investigation received financial support from the Natural Sciences and Engineering Council (NSERC) of Canada, Canadian Institute of Health Research (CIHR) and UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases to L.G. We would like to thank Dr. K.P. Chang, Chicago Medical School for providing the Leishmania specific P6.5 plasmid.
Contributor Information
Vasanthakrishna Mundodi, Email: mundodi@uthscsa.edu.
Ashwini S Kucknoor, Email: kucknoor@uthscsa.edu.
Lashitew Gedamu, Email: lgedamu@ucalgary.ca.
References
- Badaro R, Jones TC, Lorenzo R. A prospective study of visceral leishmaniasis in an endemic area of Brazil. J Infect Dis. 1986;154:639–649. doi: 10.1093/infdis/154.4.639. [DOI] [PubMed] [Google Scholar]
- Evans TG, Teixeira MJ, McAuliffe IT. Epidemology of visceral leishmaniasis in Northeast Brazil. J Infect Dis. 1992;166:1124–1132. doi: 10.1093/infdis/166.5.1124. [DOI] [PubMed] [Google Scholar]
- Tselentis Y, Gilkas A, Chaniotis B. Kala-azar in Athens basin. Lancet. 1994:1635. doi: 10.1016/S0140-6736(94)93085-6. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Brooks DR, Coombs GH. Roles of cysteine proteinases of trypanosomes and Leishmania in host-parasite interactions. Curr Opin Microbiol. 1998;1:445–460. doi: 10.1016/S1369-5274(98)80065-9. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Coombs GH, Alexander J. Cysteine peptidases as virulence factors of Leishmania. Curr Opin Microbiol. 2004;7:375–381. doi: 10.1016/j.mib.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Mayer R, Picard I, Lawton P, Grellier P, Barrault C, Monsigny M, Schrevel J. Peptide derivatives specific for a Plasmodium falciparum proteinase inhibit the erythrocyte invasion by merozoites. J Med Chem. 1991;34:3029–3035. doi: 10.1021/jm00114a011. [DOI] [PubMed] [Google Scholar]
- Que X, Reed SL. Cysteine proteinases and the pathogenesis of amebiasis. Clin Microbiol Rev. 2000;13:196–206. doi: 10.1128/CMR.13.2.196-206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs GH, Baxter J. Inihibition of Leishmania amastigote growth by antipain and leupeptin. Ann Trop Med Parasitol. 1984;78:21–24. doi: 10.1080/00034983.1984.11811768. [DOI] [PubMed] [Google Scholar]
- Denise H, McNeil K, Brooks DR, Alexander J, Coombs GH, Mottram JC. Expression of multiple CPB genes encoding cysteine proteases is required for Leishmania mexicana virulence in vivo. Infect Immun. 2003;71:3190–3195. doi: 10.1128/IAI.71.6.3190-3195.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G, Andrews N, Mills AA, Engel JC, Smith R, McKerrow JH. Peptide- fluromethyl ketones arrest intracellular replication and intracellular transmission of Trypanosoma cruzi. Mol Biochem Parasitol. 1993;58:17–24. doi: 10.1016/0166-6851(93)90086-D. [DOI] [PubMed] [Google Scholar]
- Alexander J, Coombs GH, Mottram JC. Leishmania mexicana cysteine proteinase deficient mutants have attenuated virulence for mice and potentiate a Th1 response. J Immunol. 1998;161:6794–6801. [PubMed] [Google Scholar]
- Buxbaum LU, Denise H, Coombs GH, Alexander J, Mottram JC, Scott P. Cysteine protease B of Leishmania mexicana inhibits host Th1 responses and protective immunity. J Immunol. 2003;171:3711–3717. doi: 10.4049/jimmunol.171.7.3711. [DOI] [PubMed] [Google Scholar]
- McKerrow JH, McGrath ME, Engel JC. The cysteine protease of Trypanosoma cruzi as a model for antiparasitic drug design. Parasitol Today. 1995;11:279–282. doi: 10.1016/0169-4758(95)80039-5. [DOI] [PubMed] [Google Scholar]
- Rosenthal PJ, Kim K, McKerrow JH, Leech JH. Identification of three stage-specific proteinases from Plasmodium palcifarum. J Exp Med. 1987;166:816–821. doi: 10.1084/jem.166.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Himeno K, Ishikawa H, Hisaeda T, Sakai T, Dainichi T, Asao T, Good RA, Katunuma N. Switch of CD4+ T cell differentiation from Th2 to Th1 by treatment with Cathepsin B inhibitor in experimental leishmaniasis. J Immunol. 1998;161:2120–2127. [PubMed] [Google Scholar]
- Zadeh-Vakili A, Taheri T, Doustdari F, Salmanian AH, Rafati S. Immunization with the hybrid protein vaccine consisting of Leishmania major cysteine proteinases Type I (CPB) and Type II (CPA), partially protects against leishmaniasis. Vaccine. 2004;22:1930–1940. doi: 10.1016/j.vaccine.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Omara-Opyene AL, Gedamu L. Molecular cloning, characterization and over expression of two distinct cysteine protease cDNA from Leishmania donovani chagasi. Mol Biochem Parasitol. 1997;90:247–267. doi: 10.1016/S0166-6851(97)00158-8. [DOI] [PubMed] [Google Scholar]
- Mundodi V, Somanna A, Farrell P, Gedamu L. Genomic organization and functional expression of differentially regulated cysteine protease genes of Leishmania donovani complex. Gene. 2002;282:257–265. doi: 10.1016/S0378-1119(01)00851-4. [DOI] [PubMed] [Google Scholar]
- Ankri S, Stolarsky T, Mirelman D. Antisense inhibition of expression of cysteine proteinases in Entamoeba histolytica does not affect cytopathic or hemolytic activity but inhibits phagocytosis. Mol Microbiol. 1998;28:777–785. doi: 10.1046/j.1365-2958.1998.00837.x. [DOI] [PubMed] [Google Scholar]
- Ankri S, Stolarky T, Bracha R, Padilla-Vaca F, Mirelman D. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica – induced formation of liver abscess in hamsters. Infect Immun. 1999;67:421–422. doi: 10.1128/iai.67.1.421-422.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Matlashewski G. Loss of virulence in Leishmania donovani deficient in an amastigote-specific protein, A2. Proc Natl Acad Sci U S A. 1997;94:8807–8811. doi: 10.1073/pnas.94.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DQ, Kolli B, Yadav N, Lu HG, Sachs AG, Peterson DA, Chang KP. Episomal expression of specific sense and antisense mRNAs in Leishmania amazonensis : Modulation of gp63 level in promastigotes and their infection of macrophage in vitro. Infect Immun. 2000;68:80–86. doi: 10.1128/iai.68.1.80-86.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram JC, Robertson CD, Coombs GH, Barry JD. A developmentally regulated cysteine proteinase gene of Leishmania mexicana. Mol Microbiol. 1992;6:1925–1932. doi: 10.1111/j.1365-2958.1992.tb01365.x. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Souza AE, Hutchison JE, Carter R, Frame MJ, Coombs GH. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc Natl Acad Sci U S A. 1996;93:6008–6013. doi: 10.1073/pnas.93.12.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasakosvitsch F, Gentil LG, dos Santos MRM, da Silveiri JF, Barbieri CL. Cloning and characterization of a Cysteine proteinase gene expressed in amastigotes of Leishmania (L.)amazonensis. Int J Parasitol. 2003;33:445–454. doi: 10.1016/S0020-7519(03)00010-9. [DOI] [PubMed] [Google Scholar]
- Papadopoulou B, Roy G, Mourad W, Leblanc E, Quellette M. Changes in folate and pterin metabolism after disruption of the Leishmania H locus short-chain dehydrogenase gene. J Biol Chem. 1994;269:7310–7315. [PubMed] [Google Scholar]
- Papadopoulou B, Roy G, Dey S, Rosen B, Olivier M, Quellette M. Gene disruption of the P-glycoprotein related gene pgpA of Leishmania tarentolae. Biochem Biophys Res Commun. 1996;224:772–778. doi: 10.1006/bbrc.1996.1098. [DOI] [PubMed] [Google Scholar]
- Kundig C, Leblanc E, Papadopoulou B, Ouellette M. Role of the locus and of the resistance gene on gene amplification frequency in methotrexate resistant Leishmania tarentolae. Nucleic Acids Res. 1999;27:3653–3659. doi: 10.1093/nar/27.18.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanna A, Mundodi V, Gedamu L. In vitro cultivation and characterization of Leishmania chagasi amastigote-like forms. Acta Trop. 2002;83:37–42. doi: 10.1016/S0001-706X(02)00054-2. [DOI] [PubMed] [Google Scholar]
- Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A, Hawe LL, Leander KR, Martinez D, Perry HC, Shiver JW, Montogomery DL, Liu MA. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Behie LA, Iatrou K. Secretion of cytoplasmic and nuclear proteins from animal cells using novel secretion modules. Prot Strut Fun Gen. 2000;41:144–153. [PubMed] [Google Scholar]
- Farrell PJ, Lu M, Prevost J, Brown C, Behiw C, Iatrou K. High-level expression of secreted glycoproteins in transformed lepidopteran insect cells using a novel expression vector. Biotechnol Bioeng. 1998;60:6556–6563. doi: 10.1002/(SICI)1097-0290(19981220)60:6<656::AID-BIT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Streit JA, Donelson JE, Agey MW, Wilson ME. Developmental changes in the expression of Leishmania chagasi gp63 and heat shock protein in a human macrophage cell line. Infect Immun. 1996;64:1810–1818. doi: 10.1128/iai.64.5.1810-1818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]