Abstract
The Mitchell’s satyr, Neonympha mitchellii, is an endangered species that is limited to highly isolated habitats in the northern and southern United States. Conservation strategies for isolated endangered species often implement captive breeding and translocation programs for repopulation. However, these programs risk increasing the spread of harmful pathogens, such as the bacterial endosymbiont Wolbachia. Wolbachia can manipulate the host’s reproduction leading to incompatibilities between infected and uninfected hosts. This study uses molecular methods to screen for Wolbachia presence across the distribution of the Mitchell’s satyr and its subspecies, St. Francis satyr, which are both federally listed as endangered and are considered two of the rarest butterflies in North America. The screens confirmed the presence of Wolbachia in the northern and newly discovered southern populations of the Mitchell’s satyr, but not in the St. Francis satyr population. These results combined with previous reports of Wolbachia in N. mitchellii, highlight that Wolbachia infection varies both geographically and temporally in satyr populations. The temporal variance shows the importance of continued monitoring of Wolbachia infection during conservation programs. To reduce the risk of reproductive incompatibilities, it is advised that all individuals collected for conservation purposes be screened for Wolbachia and recommended to avoid the use of infected individuals for captive breeding and translocation programs.
Keywords: endosymbiont, cytoplasmic incompatibility, captive rearing, conservation, nymphalid
1. Introduction
The Mitchell’s satyr, Neonympha mitchellii mitchellii (French), is often cited as one of the rarest butterflies in North America [1,2,3] and was first described from southern Michigan [4]. Until recently, the known distribution of N. m. mitchellii included Indiana and Michigan, with historic, now extinct, populations in Ohio, New Jersey, and Maryland. The St. Francis Satyr, N. mitchellii francisci, is restricted to a single location in Fort Bragg, North Carolina and differs from other N. mitchellii populations in several traits, including male genitalia, wing coloration and habitat [1]. Due to the limited number of extant populations of N. m. mitchellii (17 in Michigan and two in Indiana), its extirpation in three states, and the restriction of N. m. francisci to Fort Bragg, both subspecies were placed on the endangered species list and given federal protection [5].
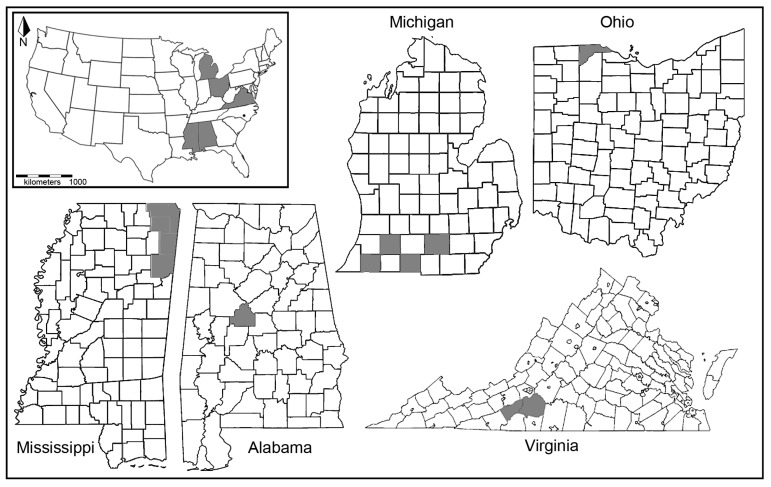
In 2000 and 2001, eight populations of N. m. mitchelli were documented in east-central Alabama [6]. In 2003, a curator of the Mississippi Entomological Museum (MEM), Terence Schiefer, discovered three populations in northeast Mississippi along the Natchez Trace Parkway. In 2010, staff from the Mississippi Museum of Natural History and MEM initiated surveys to determine the distribution of N. m. mitchellii in Mississippi. From 2010 to 2014, surveys of small, open to partially wooded wetlands located Mitchell’s Satyrs at 15 sites across Tishomingo, Itawamba, eastern Prentiss, eastern Alcorn, and Monroe counties in Mississippi (Figure 1). The United States Fish and Wildlife Service now includes these southern populations under the Endangered Species Act.
Figure 1.
State and county distributions of N. mitchellii. Insert of Unites States of America shows states with N. m. mitchelli present in grey, and a black dot for N. m. francisci presence. Grey regions reflect counties N. m. mitchelli were sampled for this study. Black filled circle in North Carolina denotes Fort Bragg that N. m. francisci is present.
Both subspecies of this butterfly are associated with the sedge-dominated edges of wetlands, where an open canopy is present. Several species of sedges (Carex spp.) have been recorded as larval host plants [7,8,9,10]. Northern populations of N. m. mitchellii inhabit prairie fens, a relatively stable habitat, whereas the southern populations are generally associated with ephemeral habitats such as open to partially wooded, small wetlands near streams, along the wooded borders, or on the edges of wetlands associated with beaver activity [6,9]. In North Carolina, the constant interruption of plant community succession by ordnance use, prescribed fire, and beaver impoundments has resulted in the persistence of suitable habitat for N. m. francisci at Fort Bragg [10]. Northern populations of N. m. mitchellii are univoltine with just one generation per year, whereas southern populations and those of N. m. francisci are bivoltine, having two generations per year. In Mississippi, the first flight period is during early to mid-June, followed by a second flight period in late August. The two subspecies also show clear population genetic differences, however the recently discovered southern populations were genetically indistinguishable from N. m. francisci at one mitochondrial and five nuclear markers [11]. Based on this, the recommendation has been for both subspecies to continue to be managed as separate endangered species.
Captive breeding and translocation programs have become common management strategies for repopulating endangered species that have highly fragmented distributions [12]. However, these strategies can greatly increase the risk of disease transmission. In Danaus plexippus, monarchs, the translocation of mass bred individuals for commercial trade can spread spores of the parasite Ophryocystis elektroscirrha, which can have lethal effects [13]. In many insects, the transmission of bacterial endosymbionts can also have major impacts on the host population, which can be a concern for conservation. Wolbachia (Rickettsiales: Rickettsiaceae) is a maternally inherited bacterial endosymbiont that has a range of hosts from phyla Arthropoda to Nematoda, with as many as 75% of all insect species harboring Wolbachia [14,15]. Wolbachia has evolved mechanisms to increase its transmission that has major impacts of their host’s reproduction. As a result, host populations often experience highly distorted sex ratios and cytoplasmic incompatibilities (CI) between infected and uninfected individuals [16]. The introduction of infected captive individuals during restoration efforts can result in CI with individuals uninfected or carrying a different Wolbachia strain. The introduced CI will further reduce the number of reproducing host individuals and increase the chance of extirpation [17]. Therefore, it is pertinent that conservation strategies for arthropods involve screens for Wolbachia in captive rearing and translocation efforts.
Captive rearing has been a key strategy in the conservation of N. m. mitchellii and in 2014 Wolbachia was first reported in N. m. mitchellii [18,19]. This previous study by Hamm et al. [19] tested for Wolbachia in N. m. mitchellii from a single county in Michigan, and ~11% of the individuals tested positive. Here, we assay for the presence of Wolbachia in 63 N. m. mitchellii sampled across its range including the captive breeding population, 4 N. m. francisci, and 10 Megisto cymela (little wood satyr) whose distribution overlaps with N. mitchellii. We use these to determine (1) if the captive rearing population shows evidence of Wolbachia; (2) if Wolbachia remains present in the Michigan populations; (3) if additional northern populations show presence of Wolbachia; (4) if Wolbachia is present in the newly discovered populations in Mississippi and Alabama and (5) if Wolbachia is also present in N. m. fransicii. The results of this study will identify populations susceptible for Wolbachia induced CI and directly inform conservation managers on best practices for the collection and release N. mitchelli during captive rearing and translocation programs.
2. Materials and Methods
2.1. Specimen Acquisition
Whole body samples were obtained from 76 satyr butterflies across four populations: (i) 35 Neonympha mitchellii mitchellii samples from southern populations in Alabama, Mississippi, and Virginia; (ii) 27 N. m. mitchellii samples from northern populations in Ohio and Michigan; (iii) four Neonympha mitchellii francisci from Fort Bragg; and (iv) 10 samples of Megisto cymela. All samples were collected from wild populations except the 19 Ohio samples, which were obtained from a captive breeding colony at the Toledo Zoo. Five of the samples from the Toledo Zoo were obtained in the form of wing snips or whole wings. Specific locations of collection sites have been omitted from the manuscript for the safety of the endangered butterflies, but a map of counties that were sampled for this study is provided in Figure 1.
2.2. Sample Preparation
The 72 whole body genomic DNA (gDNA) samples were isolated using a DNA extraction kit, Qiagen DNeasy. GDNA extractions for the five wing snips were conducted using mechanical homogenization with a sterile pellet mixer in 10 nm Tris HCl, 1 nm Ethylenediaminetetraacetic acid (EDTA), pH = 8 (TE) and left overnight at room temperature [20]. All gDNA samples were stored at −20 °C.
2.3. Wolbachia Screens
Two concurrent Polymerase Chain Reaction (PCR) based assays were required to confirm the absence/presence of Wolbachia [17]. The first PCR assayed was a control to confirm DNA extraction was successful, by amplifying the arthropod-specific 28s rRNA gene. Primers used for the 28s PCRs were (28sF3633: 5′ TAC CGT GAG GGA AAG TTG AAA 3′; 28sR4076: 5′ AGA CTC CTT GGT CCG TGT TT 3′) [21]. Those samples that did not amplify for 28s were not used in subsequent analyses. To assay for Wolbachia presence, we used Wolbachia specific primers to amplify the 16s rRNA gene (WSpec), (WSpecF: 5′CAT ACC TAT TCG AAG GGA TAG 3′ and WSpecR: 5′AGC TTC GAG TGA AAC CAA TTC 3′) [15,22]. Standard PCR protocols were followed using 10 μm forward and reverse primers, 10 mM dNTP PCR grade Mix (Invitrogen), and Taq DNA polymerase with 10× Standard Taq Buffer (New England Biolabs) with the suggested routine thermocycling conditions for Taq (New England Biolabs) and a 52 °C annealing temperature. PCR products were visualized alongside a 100 base pair ladder on a 1% agarose gel run at 100 volts for one hour.
2.4. Wolbachia Confirmation
Samples that tested positive for PCR amplification of Wspec were cleaned with ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) to remove excess nucleotides. Forward and reverse cycle sequencing reactions were run separately using BigDye Terminators (Applied Biosystems) and the Wspec primers. Dye terminator removal and sequencing was conducted at Georgia Genomics Institute. Chromatogram files for each sequence were cleaned by removing low quality and aligned using Sequencher version 5.5.1 (Gene Codes, Ann Arbor, MI, USA). Consensus sequences were generated from the forward and reverse sequences for each sample. To confirm the amplified 16s fragments were from Wolbachia, the consensus sequences were used to conduct TBLASTX [23] sequence similarity comparisons against the National Center for Biotechnology Information (NCBI) nucleotide database restricted to sequences from (i) bacteria only and (ii) Wolbachia pipientis only. The sequence for the 16s fragment isolated from N. mitchellii and M. cymela is available at NCBI genbank (Accession # MF002138 and MF002139).
3. Results and Discussion
3.1. Wolbachia Presence
For all 76 samples tested, the 28s arthropod rRNA gene was successfully amplified. Of these 76 samples, only 11 showed amplification of the Wolbachia 16s ribosomal RNA gene. Sequence of the 16s PCR product from positive N. m. mitchellii showed the greatest similarity to Wolbachia strains isolated from insects (Drosophila simulans or to Maculinea teleius, the Large Scale Blue Butterfly). The 16s sequences alone were not sufficient to distinguish between Wolbachia strains found in insects [24].
3.2. Wolbachia Demographics in N. mitchellii
Wolbachia infection was variable among the northern and southern N. m. mitchellii populations (Table 1, Figure 1). Our results re-confirm the presence of Wolbachia in Michigan populations [19]. Two of eight individuals sampled in Michigan tested positive for Wolbachia presence. One of these individuals was collected from Jackson county, which is the same county Hamm et al. [19] first reported Wolbachia presence among ~11% of the tested N. m. mitchellii. This suggests that Wolbachia infections can remain variable in N. m. mitchellii populations without being lost or spread among all individuals. We report the first evidence of Wolbachia outside Jackson Co., in Cass Co., Michigan. Unfortunately, only a single individual was available for genetic testing from Cass Co., but four individuals from two other counties in Michigan were all negative for Wolbachia. Collectively, these findings suggest there are likely barriers to Wolbachia transmission in the Michigan N. m. mitchellii populations, which could be the result of incompatibilities between infected and uninfected individuals.
Table 1.
Wolbachia presence in N. mitchelli populations.
| Taxa | State | County | Number Tested | Number Positive for Wolbachia |
|---|---|---|---|---|
| Neonympha mitchelli mitchelli | AL | Bibb | 11 | - |
| MS | Alcorn | 1 | - | |
| MS | Prentis | 4 | 1 | |
| MS | Tishomingo | 6 | - | |
| MS | Itawamba | 1 | - | |
| MS | Monroe | 8 | 4 | |
| VA | Floyd | 2 | - | |
| VA | Franklin | 2 | - | |
| MI | Branch | 2 | - | |
| MI | Cass | 1 | 1 | |
| MI | Jackson | 3 | 1 | |
| MI | Kalamazoo | 2 | - | |
| OH | Toledo Zoo | 19 | - | |
| Neonympha mitchelli francisci | NC | Fort Bragg | 4 | - |
| Megisto cymela | AL | Baldwin | 2 | 1 |
| MS | Harrison | 2 | 1 | |
| MS | Tishomingo | 3 | 1 | |
| MS | Wilkininson | 1 | - | |
| TX | Blanco | 1 | 1 | |
| VA | Franklin | 1 | - |
We confirmed that Wolbachia is not present in any of the 19 individuals from the Toledo Zoo captive rearing colony. If Wolbachia were present in the Zoo population at a similar frequency as the Michigan populations (~11%–25%), then at least one or a few individuals would be expected to test positive for Wolbachia. The lack of Wolbachia presence in the 19 individuals strongly suggests Wolbachia is either absent or present at negligible levels in the Toledo Zoo captive rearing colony.
Southern populations of N. m. mitchelli samples tested positive for the presence of Wolbachia. However, at no single southern location (e.g., county) were all N.m. mitchellii individuals positive for Wolbachia. Five of the 20 individuals collected from Mississippi tested positive for Wolbachia (Table 1). Four of the five positive samples were collected from the same area in Monroe County, on the same day. The additional positive individual was collected in Prentis Co., over 300 km from the collections in Monroe Co. Three other individuals collected in Prentis were all negative for Wolbachia. Eight additional individuals collected across four different counties also tested negative for Wolbachia. Again, these results suggest there may be limited transmission of Wolbachia within and between local N. m. mitchellii populations.
In Alabama and Virginia populations, no N. m. mitchellii were positive for Wolbachia. In Alabama, individuals included in this study were only sampled from a single county, however all 11 tested individuals were negative for Wolbachia (Table 1). In Virginia, four individuals collected from two counties were all negative. These results suggest that Wolbachia infection varies within both northern and southern N. m. mitchellii populations. Including more N. m. mitchellii individuals from each population and sampling additional loci from Wolbachia is needed to accurately estimate the frequency of Wolbachia infection and identify potential strain incompatibilities. However, Wolbachia was repeatedly found in southern populations of the little brown satyr, M. cymela, despite only sampling 1–3 individuals per county. In Mississippi, Wolbachia was only present in a portion of the M. cymela individuals (Table 1), which was very similar to Wolbachia presence in Mississippi populations of N. m. mitchellii. Unlike N. m. mitchellii, M. cymela in Alabama and Virginia tested positive for Wolbachia. This confirms that Wolbachia is present in these regions; however, there appears to be limited transmission between these satyr species. Wolbachia presence was also confirmed in a Texas population of M. cymela, demonstrating the broad range of Wolbachia presence among the satyrs.
None of the N. m. francisci samples were positive in this study. These results are in stark contract with the study by Hamm et al. [19], which found that all four N.m. francisci tested were positive for Wolbachia. Instead, here we found that the four individuals we sampled were negative for Wolbachia. The lack of Wolbachia found in our samples suggests that Wolbachia presence may vary temporally and no longer be present in N. m. francisci. Alternatively, if the two studies are considered collectively, regardless of when collected, then 50% of the St. Francis would be estimated to be infected with Wolbachia. These alternative explanations are difficult to discern with the small sample sizes of St. Francis samples in each study, however there is only a 6.25% chance of sampling four positive (or four negative) individuals from a population with 50% infection. Regardless, our results confirm that Wolbachia infection is not ubiquitous throughout the range of N. m. francisci.
4. Conclusions
The confirmation of Wolbachia presence in northern and southern populations of N. m. mitchellii could pose major concern for captive breeding and translocation programs. Genetic differences between N. m. mitchellii populations and population size estimates suggest that there has been little historical migration between the northern and southern populations due to habitat loss and fragmentation [7,8,11]. If Wolbachia transmission also decreased with the Mitchell satyr populations, there is a risk of the evolution of incompatibilities between strains infecting northern versus southern populations. Therefore, we recommend that live N. m. mitchellii individuals that show the presence of Wolbachia not be translocated between northern and southern populations. To reduce the possibility of CI, we also recommend avoiding any populations with Wolbachia positive individuals for captive breeding and translocation programs. The difference in Wolbachia presence between the present study and Hamm et al. [19] suggests that Wolbachia can vary temporally and that all collected samples for conservation purposes should be screened, regardless of previous Wolbachia presence at the collection locations. The use of wing snip sampling can allow for more individuals within a population to be tested without sacrificing individuals, but due to the small sample size of wing snips and low numbers of infection in this study, further investigations should be conducted to discern the method’s effectiveness in other species of concern. We urge researchers and conservation managers to use caution during survey and collection efforts, in order to minimize the potential spread and introduction of Wolbachia. Conservation efforts for the Mitchell’s satyr and St. Francis satyr would benefit from further studies of potential CI between infected and uninfected individuals and a detailed examination of Wolbachia strain variation among the populations and subspecies.
Acknowledgments
Funding for this work was provided by United States Fish and Wildlife Service (JoVonn Hill, Jennifer Seltzer and Brian A. Counterman) and Texas Ecolabs (Brian A. Counterman and Jennifer Fenner). We thank Georgia Genomics Facility for providing sequencing service. We thank Nick Haddad for providing St. Francis satyr samples. We thank the following people and agencies for assistance with sample permits: Mississippi Department of Wildlife, Fisheries, and Parks (MS Permit # 0517131); Kathy Lunceford, U.S. Fish and Wildlife Service (MS Permit # TE 198846-1, TE 198846-2); Lisa McGinnis, Natchez Trace Parkway (MS Permit # NATR-2012-SCI-0006); Rick Myers, Department of Conservation and Recreation (VA Permit # DNH-SWR01-13); Deb Carter, U.S. Fish and Wildlife Service (VA Permit # TE 697823); Sarah Burgess, Nature Conservancy; Lori Sargent, Michigan Department of Natural Resources (MI Endangered Species Permit #2039); Lisa Mandell, U.S. Fish and Wildlife Service (MI Permit # TE03501B-0); Tameka Dandridge, U.S. Fish and Wildlife Service, East Lansing Field Office; Daria Hyde, Michigan Natural Features Inventory; Paul Hartfield, U.S. Fish and Wildlife Service, Mississippi Field Office; Steve Roble, Virginia Department of Conservation and Recreation.
Author Contributions
Brian A. Counterman, Jennifer Fenner and JoVonn Hill conceived and designed the experiments; JoVonn Hill, Jennifer Seltzer, Jennifer Fenner, Heather Sullivan, Scott Peyton, Peter Tolson and Ryan Walsh acquired samples; Jennifer Fenner performed the experiments; Jennifer Fenner and Brian A. Counterman analyzed the data; Brian A. Counterman contributed reagents/materials/analysis tools; Jennifer Fenner and Brian A. Counterman wrote the manuscript with assistance from JoVonn Hill and Jennifer Seltzer.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Parshall D.K., Kral T.W. A new subspecies of Neonympha mitchellii (French) (Satyridae) from North Carolina. J. Lepid. Soc. 1989;42:114–119. [Google Scholar]
- 2.Michigan Department of Natural Resources Mitchell’s Satyr. [ (accessed on 25 October 2011)]; Available online: http://www.michigan.gov/dnr/0,1607,7-153-10370_12145_12204-33013--,00.html.
- 3.Unites States Fish and Wildlife Service; [(accessed on 25 October 2011)]. Mitchell’s Satyr Range wide Habitat Conservation Plan. Available online: http://www.fws.gov/midwest/endangered/permits/hcp/mitchellsatyr/index.html. [Google Scholar]
- 4.French G.H. A new species of Neonympha. Can. Entomol. 1889;21:25–27. doi: 10.4039/Ent2125-2. [DOI] [Google Scholar]
- 5.Casebere L., Denny G., Metzler E., Reznicek T., Schweitzer D., Shuey J., Sferra N., Walker S., Wilsmann L., Rabe M. Mitchell’s Satyr Butterfly Neonympa mitchellii mitchellii Recovery Plan. U.S. Department of the Interior Fish and Wildlife Service; Falls Church, VA, USA: 1998. [Google Scholar]
- 6.Hart B. A Survey for the Mitchell’s Satyr (Neonympha mitchellii French) in the National Forests of Alabama Final Report. Report for the US Fish and Wildlife Service; Falls Church, VA, USA: 2004. [Google Scholar]
- 7.Goldstein P.Z., Hall S., Hart B., Roble S.M., Shuey J. Evaluation of Relationships and Conservation Status Within the Neonympha mitchellii Complex (Lepidoptera: Nymphalidae) Report for US Fish and Wildlife Service; Falls Church, VA, USA: 2004. [Google Scholar]
- 8.Hamm C.A. Development of polymorphic anonymous nuclear DNA markers for the endangered Mitchell’s satyr butterfly, Neonympha mitchellii mitchellii (Lepidoptera: Nymphalidae) Conserv. Genet. Resour. 2012;4:127–128. doi: 10.1007/s12686-011-9490-8. [DOI] [Google Scholar]
- 9.McAlpine W.S., Hubbell S.P., Pliske T.E. The distribution, habits, and life history of Euptychia mitchellii (Satyridae) J. Lep. Soc. 1960;14:209–226. [Google Scholar]
- 10.Kuefler D., Haddad N.M., Hall S., Hudgens B., Bartel B., Hoffman E. Distribution, population structure and habitat use of the endangered Saint Francis Satyr butterfly, Neonympha mitchellii francisci. Am. Midl. Nat. 2008;159:298–320. doi: 10.1674/0003-0031(2008)159[298:DPSAHU]2.0.CO;2. [DOI] [Google Scholar]
- 11.Hamm C.A., Rademacher V., Landis D.A., Williams B.L. Conservation genetics and the implication for recovery of the endangered Mitchell’s Satyr butterfly, Neonympha mitchellii mitchellii. J. Hered. 2013;105:19–27. doi: 10.1093/jhered/est073. [DOI] [PubMed] [Google Scholar]
- 12.Crone E.E., Pickering D., Schultz C.B. Can captive rearing promote recovery of endangered butterflies? An assessment in the face of uncertainty. Biol. Conserv. 2007;139:103–112. doi: 10.1016/j.biocon.2007.06.007. [DOI] [Google Scholar]
- 13.De Roode J.C., Yates A.J., Altizer S. Virulence-transmission trade-offs and population divergence in virulence in a naturally occurring butterfly parasite. PNAS. 2008;105:7489–7494. doi: 10.1073/pnas.0710909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeyaprakash A., Hoy M.A. Long PCR improves Wolbachia amplification: Wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 15.Werren J.H., Windsor D.M. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. R. Soc. Lon. B Biol. Sci. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werren J.H., Baldo L., Clark M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 17.Nice C.C., Gompert Z., Forister M.L., Fordyce J.A. An unseen foe in arthropod conservation efforts: The case of Wolbachia infections in the Karner blue butterfly. Biol. Conserv. 2009;142:3137–3146. doi: 10.1016/j.biocon.2009.08.020. [DOI] [Google Scholar]
- 18.Tolson P. Rearing Mitchell’s satyr at the Toldeo Zoo-a first step towards eventual reintroduction in secure habitats. News J. Lepid. Soc. 2008;50:42–43. [Google Scholar]
- 19.Hamm C.A., Handley C.A., Pike A., Forister M.L., Fordyce J.A., Nice C.C. Wolbachia infection and Lepidoptera of conservation concern. J. Insect Sci. 2014;14:1–8. doi: 10.1673/031.014.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose O.C., Brookes M.I., Mallet J.L.B. A quick and simple nonlethal method for extracting DNA from butterfly wings. Mol. Ecol. 1994;3:275. doi: 10.1111/j.1365-294X.1994.tb00064.x. [DOI] [Google Scholar]
- 21.Morse J.G., Rugman-Jones P.F., Watson G.W., Robinson L.J., Bi J.L., Stouthamer R. High levels of exotic armored scales on imported avocados raise concerns regarding USDA-APHIS’ phytosanitary risk assessment. J. Econ. Entomol. 2009;102:855–867. doi: 10.1603/029.102.0303. [DOI] [PubMed] [Google Scholar]
- 22.Simões P., Mialdea G., Reiss D., Sagot M.F., Charlat S. Wolbachia detection: An assessment of standard PCR Protocols. Mol. Ecol. Resour. 2011;11:567–572. doi: 10.1111/j.1755-0998.2010.02955.x. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Baldo L., Hotopp J.C.D., Jolley K.A., Bordenstein S.R., Biber S.A., Choudhury R.R., Hayashi C., Maiden M.C., Tettelin H., Werren J.H. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]