Abstract
The harlequin bug, Murgantia histrionica (Hahn), is an agricultural pest in the continental United States, particularly in southern states. Reliable gene sequence data are especially useful to the development of species-specific, environmentally friendly molecular biopesticides and effective biolures for this insect. Here, mRNAs were sampled from whole insects at the 2nd and 4th nymphal instars, as well as sexed adults, and sequenced using Illumina RNA-Seq technology. A global assembly of these data identified 72,540 putative unique transcripts bearing high levels of similarity to transcripts identified in other taxa, with over 99% of conserved single-copy orthologs among insects being detected. Gene ontology and protein family analyses were conducted to explore the functional potential of the harlequin bug’s gene repertoire, and phylogenetic analyses were conducted on gene families germane to xenobiotic detoxification, including glutathione S-transferases, carboxylesterases and cytochrome P450s. Genic content in harlequin bug was compared with that of the closely related invasive pest, the brown marmorated stink bug, Halyomorpha halys (Stål). Quantitative analyses of harlequin bug gene expression levels, experimentally validated using quantitative real-time PCR, identified genes differentially expressed between life stages and/or sexes.
Keywords: harlequin bug, invasive insects, agricultural pest, RNAi targets, insect pheromones, xenobiotic detoxification, transcriptomics
1. Introduction
The harlequin bug, Murgantia histrionica (Hahn) (Hemiptera: Pentatomidae), though native to Mexico and Central America, has been a serious pest of mustard-family crops (Brassicaceae) in the southern USA for ~150 years, and also infests plants in the caper family (Capparaceae) [1,2,3]. It is limited in its northern spread by cold winter temperatures, and through the mid-20th century, there were destructive harlequin bug outbreaks following mild winters [3,4,5,6]. Earlier still, the insect’s unexpected arrival in Confederate states during the Civil War prompted a formal accusation that the Union had introduced it as a deliberate act of biological warfare [7].
Destruction of crop debris in which bugs overwinter, use of soaps and oil emulsions, handpicking, and trap cropping with attractive plants such as early cruciferous crops, were recommended controls in the 19th and early 20th centuries [3]. With the advent of synthetic organic insecticides, harlequin bug was suppressed by broad-spectrum organophosphates and carbamates, and later by pyrethroids and neonicotinoids [3]. However, its recently increasing pest status reflects its tolerance of more selective management tactics for other pests of cole crops, chiefly aphids and caterpillars, and the prevalence of organic vegetable culture. Natural enemies are limited to hymenopterous egg parasitoids and generalist predators [8]. Harlequin bugs sequester glucosinolates from their host plants [9], accounting for their red-and-black aposematic coloration and possibly for the lack of nymphal or adult parasitoids known to attack other pentatomids.
With continued insecticide usage, rising populations under selective management tactics, and limited suppression by natural enemies, the need to develop alternative control mechanisms for this insect pest is becoming increasingly urgent. The development of effective, species-specific and environmentally friendly molecular biopesticides offers a possible alternative (or complement) to chemical pesticides. RNA interference (RNAi) involves the targeting of specific host genes using double-stranded RNA, thereby effecting temporary knockdowns of gene expression and associated gene function [10]. Prerequisite to identifying specific gene targets is the availability of high quality, annotated gene sequence and expression data, which for harlequin bug has heretofore been unavailable.
Pheromone biosynthesis-related genes constitute one such family of potential gene targets. In a laboratory setting, Zahn et al. [11,12] showed that an aggregation pheromone emitted by adult males feeding on a plant host was attractive to both sexes of adults, and concluded that the pheromone was a single isomer of 10,11-epoxy-1-bisabolen-3-ol [4-[3-(3,3-dimethyloxiran-2-yl)-1-methylpropyl]-1-methylcyclohex-2-en-1-ol], dubbed murgantiol. Khrimian et al. [13] showed that the pheromone consisted of two stereoisomers, (3S,6S,7R,10S)- and (3S,6S,7R,10R)-10,11-epoxy-1-bisabolen-3-ol, which occur in a ~1.4:1 ratio in emissions of adult male harlequin bug. Weber et al. [14] demonstrated that, in the field, lures containing a synthetic eight-stereoisomer blend were of comparable attractiveness with pure pheromone components to both sexes in adults, as well as to nymphs, and that presence of a host plant (collard) increased this attractiveness. Cabrera Walsh et al. [15] found that, although harlequin bugs were strongly attracted to host plants by their pheromone lure, they were not retained more on plants with pheromone than those without it, an important consideration for trap cropping and pesticide application strategies.
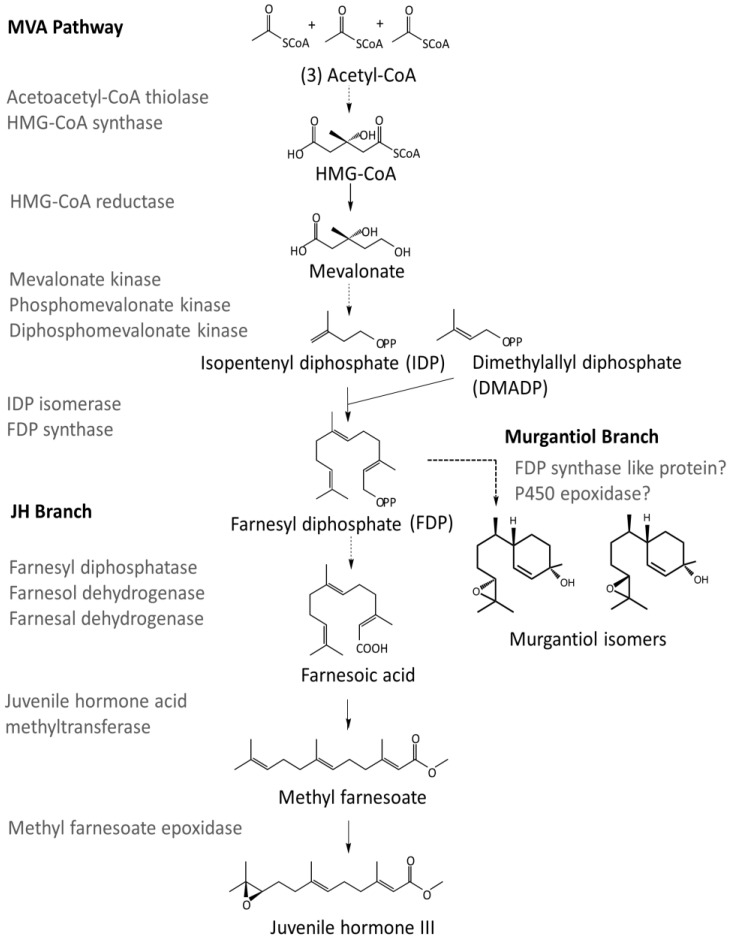
As an epoxybisabolenol, murgantiol is most likely synthesized via the mevalonic acid (MVA) pathway as the core terpenoid biosynthetic pathway in insects [16] (Figure 1). The six enzymatic steps of this pathway convert acetyl-CoA to the universal 5-carbon terpenoid building block, isopentenyl diphosphate (IDP). Enzymatic condensation of two units of IDP with one unit of its isomer dimethylallyl diphosphate (DMADP) gives rise to farnesyl diphosphate (FDP), which is the central 15-carbon precursor in juvenile hormone (JH) biosynthesis (Figure 1). Based on recent findings by Beran et al. [17] on the biosynthesis of sesquiterpene aggregation pheromones in flea beetle, it can be assumed that FDP is also a precursor of murgantiol in a pathway that includes steps catalyzed by isopentenyl diphosphate synthase (IDS) type and cytochrome P450s (CYP) epoxidase enzymes (Figure 1).
Figure 1.
Mevalonate (MVA) and Juvenile Hormone (JH) biosynthetic steps and the proposed pathway for the formation of the aggregation pheromone murgantiol. Multiple enzyme conversions are represented with dashed arrows.
Another class of gene targets involves enzymes used in detoxification of xenobiotic compounds, which includes synthetic insecticides. Glutathione S-transferases (GSTs), carboxylesterases (COEs), and CYPs are three classes of enzymes that have been widely associated with rising levels of insecticide resistance, and their characterization is of urgent concern in maintaining feasible and effective pest management programs. In particular, RNAi-mediated knockdown of these genes may result in more efficacious and/or efficient use of chemical pesticides, thereby reducing the monetary and environmental costs associated with their use in modern agricultural production operations.
In insects, GSTs are typically split into two groups: cytosolic and microsomal. Cytosolic GSTs are ordinarily divided into six subclasses: Delta, Epsilon, Sigma, Omega, Theta and Zeta [18,19]. In particular, Delta and Epsilon groups are unique to insects and have been widely associated with insecticide resistance; although the Delta type are observed among the Insecta in general, the Epsilon subclass appears to be unique to the Holometabola [18]. In contrast to cytosolic GSTs, those of the microsomal type are membrane-associated, and, although they are functionally similar to cytosolic GSTs, they are not currently implicated in insecticide resistance.
Insecticide resistance among hemipterans has been associated with multiple carboxylesterase groupings. Point mutations of acetylcholinesterases, as well as gene duplication and over-expression of E4 and FE4 β-esterases, have been reported in Myzus persicae [20] and Aphis gossypii [21]. The most recent classification of carboxylesterases divides these genes into three major clades [22,23,24]. The first and most basal is the neurodevelopment and cell adhesion clade, which comprises neuroligins, glioactins, neurotactins, acetylcholinesterases and glutacin enzymes. With the exception of acetylcholinesterases, all members of the neurodevelopmental clade are non-catalytic. The second is the hormone and semiochemical processing clade, which contains secreted β-esterases, integument esterases and juvenile hormone esterases. The third clade is associated with dietary and detoxification functions and contains α-esterases.
Cytochrome P450s have a two-fold role in gene-environment interactions, participating both in host biosynthetic pathways, as well as in detoxification of xenobiotic compounds. Biosynthesis of critical endogenous molecules via P450s can be targeted by pesticides. Molting, hormone/pheromone synthesis and turnover, and cuticular hydrocarbon waterproofing may all be targets to exploit for pest control. Conversely, insects have evolved modified P450s to detoxify exogenous chemicals like pesticides, leading to resistance. These two classes of P450s are easily observed in phylogenetic trees. Highly conserved one-to-one orthologs between insect species are parts of pathways to make essential biomolecules like ecdysone (Halloween genes: CYP302, CYP306, CYP307, CYP314, and CYP315; [25]), juvenile hormone (CYP15; [26]), as well as fatty-acid-derived alkanes and alkenes for exoskeleton coating (CYP4G; [27,28]).
Resistance has been associated with numerous cytochrome P450s, often members of “gene blooms”, which are large expansions of P450s in tandem duplication arrays on chromosomes. These are not highly conserved or even limited to one CYP clan. Almost any P450 family can become adapted to detoxify a pesticide [29,30,31]. Resistance may not only be due to pesticide inactivation, but it may be caused by blocking pesticide entry via thickening of the cuticular hydrocarbon barrier [32]. On the biocontrol side, entomopathogenic fungi kill insects by using P450s like CYP52X1 to degrade and penetrate the hydrocarbon coating on insects [33].
Mating in many if not most insects is dependent on pheromone signaling. The volatile chemicals must be synthesized, released and rapidly turned over at the receptors in antennae. P450s are actively involved in both areas [34,35,36]. Even the status of the queen in social insects, such as the termite Cryptotermes secundus, depends on molecules made by P450s [37]. Disruption of these signals by P450 inhibitors may also constitute an effective pesticidal mode of action.
In this study, we provide the first large scale assembly of transcriptomic sequences in the important agricultural pest, Murgantia histrionica (Hahn). We also identify a number of pheromone synthesis- and insecticide resistance-related genes with potential utility in the development of molecular biopesticides and/or transgenic organisms for insect biocontrol.
2. Materials and Methods
Field-collected harlequin bugs were cultured for several generations in a laboratory setting without exposure to insecticides. Original collections were by hand from their host plants in gardens and small farms within 80 km of Beltsville, MD. They were reared under 16:8 L:D photoperiod on collards in a greenhouse with temperature 18 to 28 °C. Messenger RNA populations were extracted from whole-insect preparations from 2nd and 4th instar nymphs, as well as female and male adults. For 2nd instar nymphs, 15 insects were pooled per bioreplicate, whereas five insects were pooled for all other samples. Only one bioreplicate was submitted for PE100 transcriptome sequencing (i.e., paired-end reads, 100 bp apiece) on an Illumina HiSeq instrument: RNA libraries were prepared using the TruSeq RNA v2 kit per manufacturer’s protocol (Illumina, San Diego, CA, USA). Total RNA quality was checked using an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA), and enrichment for mRNA was done using poly-dT beads. Libraries were resuspended in Qiagen EB buffer (Qiagen, Germantown, MD, USA) and a Fragment Analyzer NGS (Advanced Analytical, Ankeny, IA, USA) was used to check library size and quality; library concentration was checked using a Qubit fluorometer (ThermoFisher Scientific/Life Technologies, Waltham, MA, USA).
Raw sequencing volumes achieved are shown in Table 1, and these sequencing data were uploaded to the NCBI Sequence Read Archive (SRA) division under the BioProject accession identifier, PRJNA302154. Raw reads were normalized in a sample-specific manner using utilities supplied with the Trinity RNA-Seq assembly software package, version 2.2.0 [38]. A global transcriptome assembly was produced using Trinity: default parameter settings were used, with the exceptions that the read normalization (as here applied to the set of pooled reads) and Trimmomatic-based quality control submodules were invoked as pre-processing steps [39]. The resulting putative unique transcripts (PUTs) were post-processed using PRINSEQ-lite [40], which in particular was used to clip any residual poly-A/T tails. Resultant PUTs having lengths of at least 200 bp were then compared with the UniVec database using the NCBI C++ Toolkit’s vecscreen utility [41]—transcripts having hits flagged as “Strong” and/or “Moderate” were purged. Residual transcripts were checked for contaminants and low-quality elements using the NCBI Transcriptome Shotgun Assembly (TSA) division’s internal screening system, and assessed for completeness with respect to known single-copy orthologs generally conserved among insects using the BUSCO pipeline [42]. The final assembly was also made publicly available under the aforementioned BioProject accession.
Table 1.
Sequencing data volumes achieved, pre- and post-normalization. A paired-end sequencing strategy was used, with a targeted read length of 100 bp.
| Raw Sequence Data | Data Post-Normalization | |||
|---|---|---|---|---|
| read pairs | bases | read pairs | bases | |
| 2nd Instar | 213,817,714 | 42,763,542,800 | 13,268,461 | 2,653,692,200 |
| 4th Instar | 222,117,185 | 44,423,437,000 | 15,199,439 | 3,039,887,800 |
| Female | 205,127,388 | 41,025,477,600 | 11,786,560 | 2,357,312,000 |
| Male | 218,215,787 | 43,643,157,400 | 14,191,526 | 2,838,305,200 |
| Totals | 859,278,074 | 171,855,614,800 | 54,445,986 | 10,889,197,200 |
NCBI-vetted PUTs were compared with the NR protein database using Blastx [43] with default parameter settings. PUTs were partitioned into gold, silver and bronze tiers accordingly to similarity with existing sequence data as described previously [44,45]: Briefly, each gold-tier entry involved a PUT at least 300 nucleotide residues long exhibiting a single high-scoring segment pair in its Blastx hit against a reference protein of at least 100 amino acids; at least 75% of aligned residues had to be positively similar, with the ratio of hit length to subject sequence length being at least 90%. Silver-tier PUTs were a minimum of 100 nucleotide residues long with a hit spanning at least 75% of an NR reference protein sequence’s length. Bronze-tier entries involved PUTs at least 100 nucleotides long with hits covering at least 30% of the NR protein’s length.
Protein family analyses were conducted using HMMER3 with default settings in conjunction with the Pfam-A (version 30) database [46,47]. Each PUT could be associated with zero, one or more distinct Pfams. Inferred translational products identified in the gold-tier PUT subset were analyzed for Pfam family content. An additional analysis was performed using the complete set of PUT sequences by comparing the longest ORF present in each (considered across all six frames) with Pfam-A. Annotations based on gene ontology (GO) terms were determined using the pfam2go mapping table made available by Pfam, for each of the ontology’s cellular component, biological process and molecular function aspects. Fine-grained GO terms were resolved to their respective penultimate ancestral nodes in the directed acyclic graph (DAG) used to represent the ontology, thereby providing a more abstract view of the functional capacity inherent in the harlequin bug transcriptome—more specifically, these GO-Slim terms were recovered through traversals of “is_a” paths in the DAG using custom scripts.
Isoform-level expression amounts were estimated using the RSEM package and were recorded using the Transcripts per Million (TPM) measure [48]. Three contrasts were made based on these abundance estimates: 2nd vs. 4th instar nymphs, female vs. male adults, and nymphs vs. adults—reads from 2nd and 4th instar nymphs were pooled to compose the “nymphal” group, and those from adult males and females for the “adults” set. A listing of all PUTs exhibiting at least a two-fold expression differential is presented in Table S1. Validation of gene expression differences was performed on a subset of 16 transcripts using quantitative real-time PCR, incorporating three biological replicates and three technical replicates. Primers were designed using PrimerPlex version 2.62 (PREMIER Biosoft, Palo Alto, CA, USA); these data are also presented in Table S1. Following manufacturer protocol, cDNA was synthesized using SuperScriptTM II Reverse Transcriptase (ThermoFisher Scientific). Primers targeting harlequin bug 18S rRNA (forward: 5′-TTTTATCCAGAAAATCCCGATCA-3′, reverse: 5′-ACAACAAGTCCTCCGAAAAACC-3′) were used to control for RNA quantity, and those targeting harlequin bug elongation factor 1-alpha (forward: 5′-CGAGAAAGAGGCTCAGGAGA-3′, reverse: 5′-TCAGCCTGAGAAGTCCCTGT-3′) were used as an internal reference for gene expression. Real-time PCR was ran on an ABI 7500 instrument using Power SYBR Green master mix (ThermoFisher Scientific), as follows: 40 cycles of 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min; followed by 95 °C for 15 s, 60 °C for 1 min, 95 °C for 30 s, and finally 60 °C for 15 s.
To identify harlequin bug transcripts involved in insecticide resistance, proteins identified in the Gnomon-based annotation of the brown marmorated stink bug (Halyomorpha halys) genome as being carboxylesterases (COE) or glutathione-S-transferases (GST) were retrieved. Blastp was used to search the full H. halys inferred protein set to identify any COE or GST proteins that had not been properly labeled as such by automated methods. Following manual scrutiny, these refined sets were used as Blastp queries against the harlequin bug data—in particular, H. halys sequences were compared against the longest ORF present in each harlequin bug transcript to test for homology. Inclusion of an M. histrionica gene in the COE or GST gene family required its top high-scoring segment pair to exhibit a bit score of at least 75, a subject sequence length of at least 100 amino acid residues, a hit length to subject sequence length ratio of 0.90 or greater, a hit length to query sequence length ratio of 0.75 or greater, and a ratio of positively similar aligned residues to hit length of at least 0.80. Independently for each of these gene families, combined M. histrionica and H. halys protein sets were multiply aligned using the MUSCLE program [49], from which a maximum likelihood-based phylogeny was generated using the method of Le and Gascuel [50] as implemented in PhyML [51] using 100 bootstrap replicates. Phylogenies were visually rendered using the R package, phytools [52]. For both gene family analyses, GenomeThreader [53] was used to align M. histrionica proteins to the brown marmorated stink bug genome (available at https://i5k.nal.usda.gov/Halyomorpha_halys) in an effort to elucidate patterns of molecular evolution across this taxonomic divide.
The full PUT set was batch tBlastn searched with each cytochrome P450 subfamily from Halyomorpha halys (brown marmorated stink bug; 48 sequences). The top 50 hits for each search were kept using an expect value of 10. Blast results were imported into a spreadsheet and duplicate accessions were removed by filtering for unique entries only. In total, 478 PUTs were identified. These sequences were batch Blastx searched against 9125 named insect P450 sequences and the best hit was retained with percent identity and alignment length. The results were sorted by best blast hit which clustered closely related sequences together. The longest member of each group was translated and used to search the 478 PUTs for exact or near-exact matches. These sequences were binned into a collection from a single gene. Additional P450 PUTs were identified by HMMER searches of Pfam-A and Blastx searches of the NCBI protein database, NR. These were mostly short (i.e., <175 aa) fragments.
To determine whether harlequin bug harbors an iflavirus similar to that observed in the brown marmorated stink bug [54], raw RNA-Seq reads were mapped against the viral genome sequence available under NCBI Accession number KF699344.1 using bowtie2 [55]. PUTs potentially associated with organisms of the genera Pantoea, Erwinia and Nosema, which correspond to known pentatomid symbionts [45,56,57], were identified through Blastx comparisons with representative sequences present in the NR database. These comparisons involved the full set of assembled PUTs rather than those that had been explicitly vetted by the NCBI TSA, a vetting which involves removal of putative contaminant sequences, comprising microbial symbionts.
3. Results
Assembly of the full RNA-Seq dataset generated in this study produced 526,403 putative unique transcripts (PUTs) comprising 427,532,460 bases. Following data cleaning routines, a total of 523,102 NCBI-vetted PUTs, containing 425,689,726 assembled bases, was made available under BioProject accession number PRJNA302154. A total of 95.7% of conserved single-copy orthologs in the Insecta class were fully represented in this assembly, with 3.6% fragmented and only 0.7% absent. Sorting these sequences on the basis of extrinsic homology information resulted in the labeling of 19,303 gold-tier, 19,755 silver-tier and 33,482 bronze-tier PUTs. Pfam family analysis of translation products inferred from gold-tier PUT sequences identified 4518 unique families in total. Assaying Pfam family presence among longest ORFs present in the comprehensive PUT collection resulted in detection of 5352 distinct Pfam families in total. Table 2 presents the 25 most abundant Pfam entries detected in both datasets. These described a fairly typical array of protein family content, evincing presence of genes associated with cell signaling, DNA binding and innate insect immunity.
Table 2.
The 25 most abundant Pfam families encountered in each of the gold-tier and complete harlequin bug PUT datasets.
| Gold-Tier PUTs (Trimmed per Homology Info) | All PUT Sequences (Untrimmed) | ||||
|---|---|---|---|---|---|
| Pfam hit | Pfam Description | Counts | Pfam hit | Pfam Description | Counts |
| PF00069.23 | Protein kinase domain | 511 | PF00078.25 | Reverse transcriptase (RNA-dependent DNA polymerase) | 3272 |
| PF07714.15 | Protein tyrosine kinase | 504 | PF00096.24 | Zinc finger, C2H2 type | 1424 |
| PF13894.4 | C2H2-type zinc finger | 383 | PF13894.4 | C2H2-type zinc finger | 1292 |
| PF00096.24 | Zinc finger, C2H2 type | 381 | PF13465.4 | Zinc-finger double domain | 1262 |
| PF13857.4 | Ankyrin repeats (many copies) | 324 | PF00069.23 | Protein kinase domain | 1256 |
| PF13465.4 | Zinc-finger double domain | 323 | PF12796.5 | Ankyrin repeats (3 copies) | 1245 |
| PF12796.5 | Ankyrin repeats (3 copies) | 323 | PF13637.4 | Ankyrin repeats (many copies) | 1209 |
| PF00400.30 | WD domain, G-beta repeat | 323 | PF13857.4 | Ankyrin repeats (many copies) | 1203 |
| PF13637.4 | Ankyrin repeats (many copies) | 319 | PF07714.15 | Protein tyrosine kinase | 1199 |
| PF00023.28 | Ankyrin repeat | 316 | PF07690.14 | Major Facilitator Superfamily | 1183 |
| PF13606.4 | Ankyrin repeat | 313 | PF00023.28 | Ankyrin repeat | 1171 |
| PF07690.14 | Major Facilitator Superfamily | 288 | PF13606.4 | Ankyrin repeat | 1154 |
| PF12894.5 | Anaphase-promoting complex subunit 4 WD40 domain | 280 | PF13909.4 | C2H2-type zinc-finger domain | 956 |
| PF13927.4 | Immunoglobulin domain | 268 | PF01359.16 | Transposase (partial DDE domain) | 844 |
| PF14531.4 | Kinase-like | 266 | PF00400.30 | WD domain, G-beta repeat | 834 |
| PF13895.4 | Immunoglobulin domain | 261 | PF13927.4 | Immunoglobulin domain | 741 |
| PF00047.23 | Immunoglobulin domain | 259 | PF00083.22 | Sugar (and other) transporter | 734 |
| PF07679.14 | Immunoglobulin I-set domain | 257 | PF07679.14 | Immunoglobulin I-set domain | 676 |
| PF01926.21 | 50S ribosome-binding GTPase | 256 | PF00047.23 | Immunoglobulin domain | 636 |
| PF07686.15 | Immunoglobulin V-set domain | 255 | PF13895.4 | Immunoglobulin domain | 635 |
| PF08477.11 | Ras of Complex, Roc, domain of DAPkinase | 244 | PF00005.25 | ABC transporter | 620 |
| PF00071.20 | Ras family | 238 | PF00076.20 | RNA recognition motif (a.k.a. RRM, RBD, or RNP domain) | 611 |
| PF00076.20 | RNA recognition motif (a.k.a. RRM, RBD, or RNP domain) | 235 | PF00067.20 | Cytochrome P450 | 590 |
| PF12799.5 | Leucine Rich repeats (2 copies) | 234 | PF00665.24 | Integrase core domain | 578 |
| PF00025.19 | ADP-ribosylation factor family | 211 | PF07686.15 | Immunoglobulin V-set domain | 546 |
Analysis of the complete PUT dataset yielded 552 distinct GO terms associated with the ontology’s Biological Process aspect, 199 with Cellular Component and 627 with Molecular Function. Analysis on the gold-tier PUT subset resulted in identification of 463, 162 and 554 distinct terms corresponding to the Biological Process, Cellular Component and Molecular Function aspects, respectively. Supplementary Figure S1 presents the 10 most frequently encountered GO terms seen within each of these dimensions, for each set of PUTs; no appreciable differences were evident with which to distinguish functional capacity of the complete PUT set relative to the gold-tier subset. Analogous to its full-ontology counterpart, Supplementary Figure S2 presents the five most abundant GO-Slim terms observed. Similarly, the distribution of GO-Slim terms between the two datasets appeared equitable. Of the PUTs that exhibited the greatest fold changes in expression levels between 2nd and 4th instar nymphs, the transcripts seemingly most abundantly up-regulated in the former relative to the latter comprised a cuticular protein, perhaps associated with the molting process (see Table 3). Those seemingly more abundantly expressed among 4th instar nymphs were usually digestion or stress-response related. Transcripts predominantly expressed in females included a juvenile hormone acid O-methyltransferase and a muscle component, troponin C. Transcripts that suggested male-dominant expression patterns included an odorant binding protein, which may be involved in mate recognition. No clear functional trend seemed evident among genes whose fold changes suggested more abundant expression in adults relative to nymphs, but in the reverse case, transcripts were often associated with cuticular tissues. In all three comparisons, transcripts with the greatest perturbations in expression fold changes included a number of distinct uncharacterized proteins. Gene expression patterns for a subset of these genes were validated experimentally using quantitative real-time PCR, and these results generally corroborated findings suggested by RNA-Seq data (see Figure 2).
Table 3.
Gold-tier transcripts exhibiting the greatest gene expression fold changes within each of the three harlequin bug mRNA population comparisons performed. Expression levels were conveyed using the Transcripts Per Million (TPM) measure, and binary logarithms were used to rank sample-specific ratios of these amounts. A floor on TPM values of 5.0 was imposed.
| 2nd Instar Expression Relative to 4th Instar | ||||
| log2FC | Direction | 2nd Instar | 4th Instar | NR Gene |
| 5.1284 | up | 253.24 | 7.24 | adult-specific cuticular protein ACP-20-like |
| 4.6366 | up | 1055.89 | 42.45 | uncharacterized protein LOC106688964 |
| 3.6726 | up | 366.36 | 28.73 | tubulin beta-1 chain |
| 3.5047 | up | 380.01 | 33.48 | protein takeout-like |
| 3.3727 | up | 192.25 | 18.56 | GTP cyclohydrolase 1 isoform X1 |
| 5.3563 | down | 8.17 | 334.68 | heat shock 70 kDa protein cognate 4 |
| 5.3279 | down | 7.17 | 287.99 | acyl-CoA Delta(11) desaturase-like isoform X1 |
| 5.2056 | down | 6.33 | 233.59 | uncharacterized protein LOC106679388 |
| 4.7751 | down | 9.21 | 252.18 | lysosomal alpha-mannosidase |
| 4.6475 | down | 6.05 | 151.63 | uncharacterized protein LOC106683993 |
| Female Expression Relative to Male | ||||
| log2FC | Direction | Female | Male | NR Gene |
| 4.1357 | up | 375.11 | 21.34 | heat shock 70 kDa protein cognate 4 |
| 3.5908 | up | 104.22 | 8.65 | JH acid O-methyltransferase-like isoform X1 |
| 2.8439 | up | 88.45 | 12.32 | uncharacterized protein LOC106679932 |
| 2.7760 | up | 124.18 | 18.13 | troponin C, isoform 1-like |
| 2.7262 | up | 34.21 | 5.17 | adult-specific cuticular protein ACP-20-like |
| 3.9320 | down | 570.39 | 8706.01 | putative odorant-binding protein 4 |
| 2.7080 | down | 5.58 | 36.46 | E3 ubiquitin-protein ligase TRIM37-like isoform X2 |
| 2.2543 | down | 6.29 | 30.01 | exocyst complex component 7 |
| 1.9412 | down | 6.51 | 25.00 | DDB1- and CUL4-associated factor 6-like |
| 1.9045 | down | 7.18 | 26.88 | exosome complex component CSL4 |
| Adult Expression Relative to Nymph | ||||
| log2FC | Direction | Adult | Nymph | NR Gene |
| 3.8701 | up | 5288.23 | 361.66 | putative odorant-binding protein 4 |
| 3.7040 | up | 105.04 | 8.06 | inositol monophosphatase 2-like |
| 3.4889 | up | 98.80 | 8.80 | uncharacterized protein LOC106686819 |
| 3.4091 | up | 78.93 | 7.43 | uncharacterized protein LOC106691169 |
| 3.0628 | up | 264.22 | 31.62 | NADP-dependent malic enzyme-like isoform X3 |
| 6.0853 | down | 44.80 | 3041.89 | uncharacterized protein LOC106688633 |
| 4.3486 | down | 5.41 | 110.22 | pro-resilin-like |
| 3.7376 | down | 13.83 | 184.48 | probable antibacterial peptide |
| 3.6876 | down | 9.00 | 115.96 | cuticle protein 18.6, isoform B |
| 3.2451 | down | 11.07 | 104.96 | GTP cyclohydrolase 1 isoform X1 |
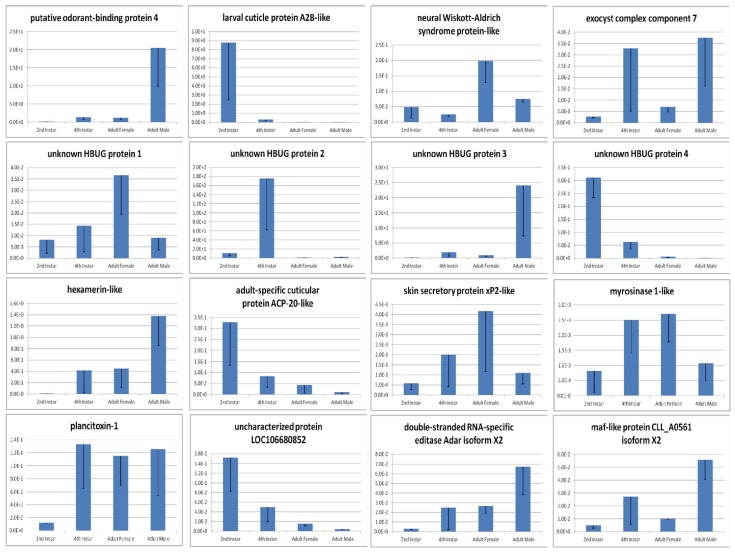
Figure 2.
Quantitative real-time PCR validation of select harlequin bug transcript expression patterns.
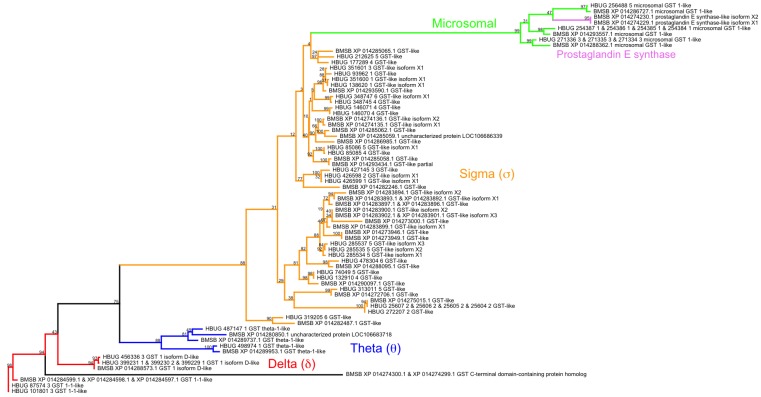
Forty-one H. halys and 44 M. histrionica glutathione S-transferase associated transcripts encode 35 and 34 unique protein sequences, respectively. A phylogeny of these genes’ unique translation products is shown in Figure 3. Four distinct clades corresponding to GST class were observed: Theta, Delta/Epsilon, Sigma, and microsomal (which was positioned within the Sigma clade). Although members of the Delta/Epsilon classes are not distinguished in annotations made available by the NCBI, the observation that Epsilon-type GSTs are exclusive to the Holometabola [18] suggests genes from these two Paraneopteran species are of the Delta type. The majority of protein sequences were placed in the Sigma clade. Two H. halys prostaglandin E synthase isoforms were observed and subsumed under the phylogeny’s microsomal GST glade; no harlequin bug homologs to these enzymes were evident, however. Thirty-one of the 44 unique M. histrionica GST associated protein sequences could be reliably splice-aligned to the H. halys genome, but only 19 of these aligned to unique loci. After further accounting for isoforms, an additional 14 GST homologs were identified: one Theta, two Delta, eight Sigma and three microsomal GSTs. Homologs of GSTs from the Zeta and Omega subclasses were not apparent in these data.
Figure 3.
Maximum likelihood-based glutathione S-transferase phylogeny for H. halys and M. histrionica proteins. Four distinct clades corresponding to GST class were observed: delta (red), theta (blue), sigma (brown) and microsomal (green). Two prostaglandin E synthase isoforms (pink) were placed in the microsomal GST clade. Bootstrap support (100 replicates) is indicated on branches.
Ninety H. halys and 88 M. histrionica carboxylesterase associated transcripts encode 82 and 70 unique translation sequences, respectively. A maximum likelihood phylogeny of these unique translation products is shown in Supplementary Figure S3. A preponderance of sequences were phylogenetically positioned in a clade containing carboxylesterases proper, comprising carboxylesterase 4A-like, carboxylesterase 5A-like and β-esterase clades. The large β-esterase clade contains venom carboxylesterase 6-like enzymes, as well as E4 and FE4 esterases. Acetylcholinesterase-, neurotactin- and neuroligin-associated sequences were all basal to the large carboxylesterase clade.
Fifty-five of the 70 unique M. histrionica carboxylesterase associated transcripts map to the H. halys genome, suggesting two acetylcholinesterase (four mapped transcripts), three neuroligin (11 mapped transcripts), one neurotactin (three mapped transcripts) and 19 carboxylesterase (37 mapped transcripts) homologs. All fifteen unique, unmapped M. histrionica carboxylesterase associated transcripts are located within the carboxylesterase clade, perhaps suggesting a lineage-specific expansion of these genes in harlequin bug (see Supplementary Figure S3).
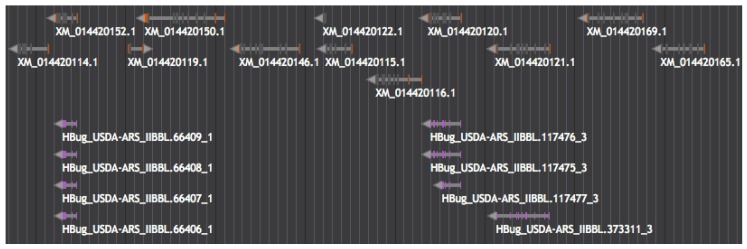
Some H. halys scaffolds contain multiple esterase genes in close proximity, specifically scaffolds 261 and 2592, which comprise tandem arrays of eleven and six esterase genes, respectively. On both scaffolds 261 and 2592, M. histrionica esterase proteins mapped to three distinct, homologous esterase H. halys loci, suggesting a potentially elevated esterase gene copy number in this latter species relative to the former (see Figure 4).
Figure 4.
Segment of scaffold 261 from the Halyomorpha halys genome assembly harboring an array of eleven E4/FE4 esterase genes. The H. halys COE genes are show in the top track, in grey (note that gene model XM_014420119.1 appears to correspond to an unrelated lipase gene) a total of six unique M. histrionica proteins (bottom track, purple) could be aligned to this chromosomal region, and these associated with three distinct COE loci present in the H. halys genome sequence.
H. halys scaffolds 261, 2592, 159, 1429 and 116 contain, respectively, 11, 6, 6, 4 and 2 carboxylesterase genes in close proximity. On both scaffolds 261 and 2592, M. histrionica carboxylesterase associated transcripts map to three distinct, homologous H. halys carboxylesterase associated loci (see Figure 4). All other M. histrionica carboxylesterase associated transcripts map to H. halys scaffolds containing just one carboxylesterase associated loci.
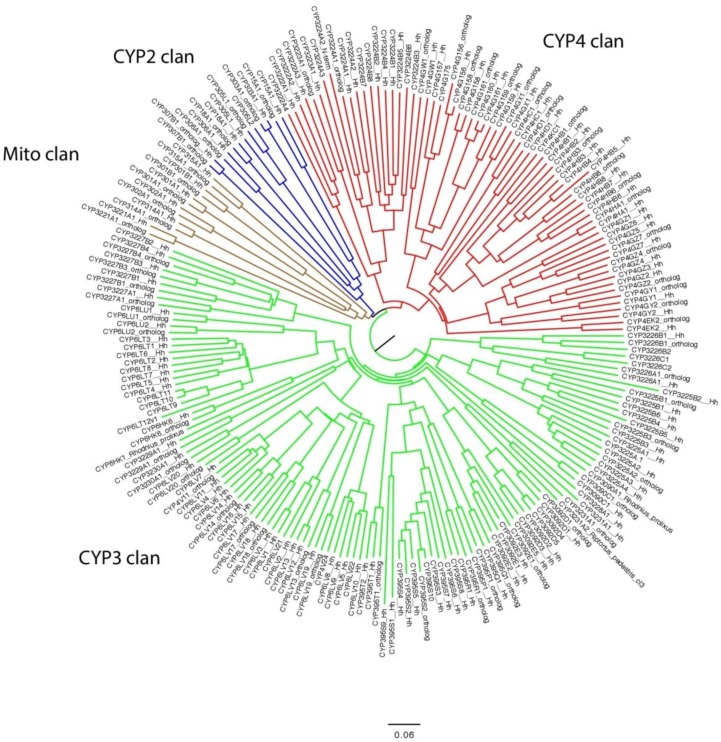
A total of 133 novel cytochrome P450 sequences in M. histrionica were assigned names based on their relationship to named P450s from other insects. Many were orthologs to P450s of Halyomorpha halys. Every P450 family in brown marmorated stink bug was found in harlequin bug except CYP3228A1. This sequence was searched for specifically and found as a pseudogene fragment. An NJ tree was made using Clustal Omega using midpoint rooting (Figure 5). The four insect clans are present. The CYP2 clan and the mito clans are small, containing all of the Halloween genes and some other highly conserved genes like CYP15, CYP18, CYP301 and CYP305. The CYP3 clan is large containing about half of the sequences with a gene bloom in the CYP6LV subfamily. The CYP4 clan has about one quarter of the total sequences. These two clans contain the least conserved genes with the exception of the CYP4G genes (cuticular hydrocarbon synthesis).
Figure 5.
Cytochrome P450s from H. halys (126 sequences) and M. histrionica (86 sequences) plus two from Rhodnius prolixus and one from Riptortus pedestris were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) at EBI. An NJ midpoint-rooted tree was made. The tree was drawn using Figtree v1.3.1 and labeled in Adobe Illustrator CC ver17.0.0. P450 clans are colored. Sequences less than 175 amino acids were not included.
A number of P450-encoding transcripts exhibited possible sex-preferential gene expression patterns. Five exhibited gene expression patterns suggestive of female-specific expression among adult insects, and seven suggested a male-dominant pattern (see Table 4). Table 4 also presents the seven PUTs having expression values of at least 5 TPM in both male and female imagoes, and exhibited at least a two-fold expression differential per RNA-Seq data—among these, six (~86%) suggested a male-dominant pattern.
Table 4.
M. histrionica PUTs encoding cytochrome P450 enzymes that appear to have sex-preferential expression patterns. Those among the most predominantly expressed in female adults are highlighted in yellow; those more predominantly expressed in males are highlighted in orange; and those having expression values of at least 5 TPM in both sex-specific samples, as well as at least a two-fold expression differential, are presented with grey highlighting.
| ♂ (TPM) | ♀ (TPM) | |log2(♂:♀)| | Direction | HBUG PUT Id | Best NR hit |
| 0.03 | 18.87 | 9.2969 | - | 472510 | XP_014270541.1 (CYP4GZ4) |
| 0.07 | 24.35 | 8.4424 | - | 473495 | XP_014286441.1 (CYP6LT5) |
| 0.40 | 72.44 | 7.5006 | - | 419699 | XP_014273208.1 (CYP6LT7) |
| 0.06 | 8.73 | 7.1849 | - | 103772 | XP_014286439.1 (CYP6LT3) |
| 0.27 | 19.68 | 6.1876 | - | 103769 | XP_014286439.1 (CYP6LT3) |
| 6.35 | 0.00 | undefined | + | 478727 | XP_014293876.1 (CYP307B1) |
| 4.71 | 0.00 | undefined | + | 173857 | XP_014285590.1 (CYP3226B1) |
| 9.95 | 0.01 | 9.9586 | + | 298833 | XP_014293876.1 (CYP307B1) |
| 4.30 | 0.07 | 5.9408 | + | 137898 | XP_014288222.1 (CYP3227B4) |
| 3.26 | 0.15 | 4.4418 | + | 521147 | XP_014276563.1 (CYP3225B3) |
| 5.85 | 0.61 | 3.2616 | + | 428429 | XP_014279285.1 (CYP302A1) |
| 38.09 | 6.24 | 2.6098 | + | 103771 | XP_014286439.1 (CYP6LT3) |
| 21.99 | 6.92 | 1.6680 | + | 504543 | XP_014284935.1 (CYP4HB7) |
| 15.69 | 5.20 | 1.5933 | + | 456532 | XP_014285589.1 (CYP3226B1) |
| 25.99 | 8.88 | 1.5493 | + | 486339 | XP_014285590.1 (CYP3226B1) |
| 14.45 | 5.10 | 1.5025 | + | 316472 | XP_014274999.1 (CYP3224A2) |
| 15.06 | 5.51 | 1.4506 | + | 165088 | XP_014271425.1 (CYP6LU1) |
| 13.48 | 6.26 | 1.1066 | + | 212119 | XP_014284933.1 (CYP4HB6) |
| 7.51 | 15.33 | 1.0295 | - | 504542 | XP_014284935.1 (CYP4HB7) |
In searching for M. histrionica terpene biosynthetic genes, a total of 19 sequences were identified using query sequences for the MVA and JH biosynthetic pathways from various insects (Table 5). These sequences were orthologs to respective genes in H. halys and included transcripts of all six genes of the MVA pathway (acetoacetyl-CoA thiolase, HMG-CoA synthase, HMG-CoA reductase, mevalonate kinase, phosphomevalonate kinase and diphosphomevalonate decarboxylase) and an IDP isomerase. Furthermore, two IDS homologs of FDP synthase-like genes were observed, one of which exhibited higher expression levels in adults and male bugs (Table 6). In the JH pathway branch, multiple isoforms of farnesol dehydrogenase (3), farnesal dehydrogenase (2), and farnesoic acid methyltransferase (3) were found, but expression values for several of these isoforms were low at all stages of development (Table 6). A single transcript for methyl farnesoate epoxidase was identified; however, no homolog of farnesyl diphosphatase could be observed. While transcripts of MVA pathway genes did not show major developmental or sex specific expression differences, more distinct expression patterns were found for isoforms of JH biosynthetic genes (farnesol dehydrogenase, farnesal dehydrogenase, farnesoic acid methyltransferases) dependent on instar, nymphal/adult stage or sex. As in M. histrionica, MVA pathway gene orthologs in H. halys were equally expressed among sexes and developmental stages, while expression profiles of orthologs in JH biosynthesis differed dependent on the isoform. These expression patterns were, however, not consistent with those found in harlequin bug. Interestingly, a total of four FDP synthase-like transcripts were detected in H. halys as opposed to only two such sequences in M. histrionica.
Table 5.
M. histrionica PUTs encoding terpene biosynthesis-related genes (no genes for ubiquinone or dolichol biosynthesis were considered).
| Enzyme Name | Query Sequences (multiple taxa) | Transcript Identified (M. histrionica) |
Blastx Support (H. halys) |
|---|---|---|---|
| Acetoacetyl-CoA thiolase | XM_014419845,XM_014386017,XM_015512081,AK403218 | HBug_USDA-ARS_IIBBL.267134 | XP_014294739.1 |
| HBug_USDA-ARS_IIBBL.43170 | XP_014277769.1 | ||
| HMG-CoA synthase | XM_014416338,X70034,AB733009 | HBug_USDA-ARS_IIBBL.375421 | XP_014277503.1 |
| HMG-CoA reductase | X70034,XM_014424783,XM_014391838,XM_015521221 | HBug_USDA-ARS_IIBBL.421664 | XP_014280269.1 |
| Mevalonate kinase | XM_014416757,XM_014391202,GEDC01029638,XM_012431690 | HBug_USDA-ARS_IIBBL.227208 | XP_014272243.1 |
| Phosphomevalonate kinase | XM_014416475,XM_014398812,GECZ01001991,GEBQ01010256 | HBug_USDA-ARS_IIBBL.270718 | XP_014271961.1 |
| Diphosphomevalonate decarboxylase | XM_018479978,XM_014434730,XM_014399537,GEBQ01002905 | HBug_USDA-ARS_IIBBL.428020 | XP_014290216.1 |
| IDP Isomerase | XM_014415973,XP_014247428,GECU01023093,AK417896 | HBug_USDA-ARS_IIBBL.92242 | XP_014271459.1 |
| FDP Synthase | XP_014289225 | HBug_USDA-ARS_IIBBL.420512 | XP_014276183.1 |
| HBug_USDA-ARS_IIBBL.414919 | XP_014276401.1 | ||
| Farnesyl diphosphatase | NP_572760.1 | No homologs detected | |
| Farnesol dehydrogenase | XP_014292348 | HBug_USDA-ARS_IIBBL.414590 | XP_014286519.1 |
| HBug_USDA-ARS_IIBBL.376500 | XP_014286525.1 | ||
| HBug_USDA-ARS_IIBBL.328207 | XP_014286524.1 | ||
| Farnesal dehydrogenase | KC243495 | HBug_USDA-ARS_IIBBL.79640 | XP_014292700.1 |
| HBug_USDA-ARS_IIBBL.14716 | XP_014272618.1 | ||
| Juvenile hormone acid methyltransferase | XP_014293044,XP_001651876 | HBug_USDA-ARS_IIBBL.485486 | XP_014290953.1 |
| HBug_USDA-ARS_IIBBL.519494 | XP_014293044.1 | ||
| HBug_USDA-ARS_IIBBL.346622 | XP_014283772.1 | ||
| Methyl farnesoate epoxidase | XP_014283057 | HBug_USDA-ARS_IIBBL.517163 | XP_014283057.1 |
Table 6.
Gene expression levels for Murgantia histrionica and Halyomorpha halys genes associated with terpene biosynthesis. RSEM-calculated expression values are conveyed in units of Transcripts per Million (TPM), and comparisons correspond to binary logs of TPM ratios.
| Harlequin Bug | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2nd | 4th | 4th:2nd | ♂ | ♀ | ♀ : ♂ | Nymphs | Adults | Adults:Nymphs | Overall | Transcript ID |
| Acetoacetyl-CoA thiolase | 176.92 | 131.53 | -0.43 | 211.58 | 344.29 | 0.70 | 154.39 | 267.85 | 0.79 | 211.99 | HBug_USDA-ARS_IIBBL.267134 |
| Acetoacetyl-CoA thiolase | 41.01 | 28.64 | -0.52 | 21.84 | 33.13 | 0.60 | 34.61 | 26.66 | -0.38 | 30.56 | HBug_USDA-ARS_IIBBL.43170 |
| HMG-CoA reductase | 15.83 | 12.42 | -0.35 | 29.97 | 22.95 | -0.39 | 14.10 | 27.05 | 0.94 | 20.60 | HBug_USDA-ARS_IIBBL.421664 |
| HMG-CoA synthase | 0.53 | 0.60 | 0.18 | 0.27 | 0.45 | 0.74 | 0.57 | 0.35 | -0.70 | 0.45 | HBug_USDA-ARS_IIBBL.375421 |
| Mevalonate kinase | 0.11 | 0.28 | 1.35 | 0.05 | 0.33 | 2.72 | 0.22 | 0.13 | -0.76 | 0.19 | HBug_USDA-ARS_IIBBL.227208 |
| Phosphomevalonate kinase | 4.51 | 2.76 | -0.71 | 6.08 | 3.08 | -0.98 | 3.59 | 4.76 | 0.41 | 4.15 | HBug_USDA-ARS_IIBBL.270718 |
| Diphosphomevalonate decarboxylase | 0.05 | 0.12 | 1.26 | 0.13 | 0.11 | -0.24 | 0.09 | 0.14 | 0.64 | 0.15 | HBug_USDA-ARS_IIBBL.428020 |
| IDP Isomerase | 20.22 | 10.48 | -0.95 | 15.04 | 13.73 | -0.13 | 15.32 | 14.53 | -0.08 | 14.92 | HBug_USDA-ARS_IIBBL.92242 |
| FDP Synthase | 3.44 | 0.41 | -3.07 | 46.44 | 3.51 | -3.73 | 1.92 | 28.48 | 3.89 | 15.29 | HBug_USDA-ARS_IIBBL.420512 |
| FDP Synthase | 25.04 | 23.37 | -0.10 | 32.47 | 31.56 | -0.04 | 23.79 | 32.17 | 0.44 | 27.98 | HBug_USDA-ARS_IIBBL.414919 |
| Farnesol dehydrogenase | 0.16 | 0.15 | -0.09 | 0.00 | 0.00 | undefined | 0.16 | 0.00 | - ∞ | 0.08 | HBug_USDA-ARS_IIBBL.414590 |
| Farnesol dehydrogenase | 0.00 | 0.13 | + ∞ | 0.07 | 0.45 | 2.68 | 0.03 | 0.15 | 2.32 | 0.09 | HBug_USDA-ARS_IIBBL.328207 |
| Farnesol dehydrogenase | 13.27 | 9.88 | -0.43 | 7.78 | 7.54 | -0.05 | 11.57 | 7.81 | -0.57 | 9.65 | HBug_USDA-ARS_IIBBL.376500 |
| Farnesal dehydrogenase | 0.29 | 0.20 | -0.54 | 8.14 | 12.84 | 0.66 | 0.24 | 10.14 | 5.40 | 5.20 | HBug_USDA-ARS_IIBBL.14716 |
| Farnesal dehydrogenase | 3.29 | 9.14 | 1.47 | 4.99 | 5.77 | 0.21 | 6.24 | 5.30 | -0.24 | 5.78 | HBug_USDA-ARS_IIBBL.79640 |
| Juvenile hormone acid methyltransferase | 0.04 | 0.00 | - ∞ | 0.12 | 0.03 | -2.00 | 0.00 | 0.09 | + ∞ | 0.00 | HBug_USDA-ARS_IIBBL.519494 |
| Juvenile hormone acid methyltransferase | 3.04 | 10.78 | 1.83 | 4.97 | 64.51 | 3.70 | 6.87 | 30.65 | 2.16 | 17.85 | HBug_USDA-ARS_IIBBL.485486 |
| Juvenile hormone acid methyltransferase | 0.03 | 1.22 | 5.35 | 0.09 | 0.28 | 1.64 | 0.68 | 0.17 | -2.00 | 0.45 | HBug_USDA-ARS_IIBBL.346622 |
| Methyl farnesoate epoxidase | 0.39 | 0.12 | -1.70 | 0.18 | 0.14 | -0.36 | 0.25 | 0.17 | -0.56 | 0.21 | HBug_USDA-ARS_IIBBL.517163 |
| Brown Marmorated Stink Bug | |||||||||||
| Annotation | 2nd | 4th | 4th:2nd | ♂ | ♀ | ♀ : ♂ | Nymphs | Adults | Adults:Nymphs | Overall | Protein / RNA IDs |
| Acetoacetyl-CoA thiolase | 175.06 | 168.71 | -0.05 | 793.94 | 942.05 | 0.25 | 171.71 | 864.84 | 2.33 | 452.10 | XP_014294739.1 / XM_014439253.1 |
| Acetoacetyl-CoA thiolase | 70.48 | 65.24 | -0.11 | 147.12 | 185.97 | 0.34 | 67.72 | 165.71 | 1.29 | 107.36 | XP_014275331.1 / XM_014419845.1 |
| HMG-CoA reductase | 18.12 | 21.68 | 0.26 | 44.27 | 25.96 | -0.77 | 20.00 | 35.52 | 0.83 | 26.28 | XP_014280269.1 / XM_014424783.1 |
| HMG-CoA synthase | 0.00 | 0.00 | undefined | 0.44 | 1.40 | 1.67 | 0.00 | 1.28 | + ∞ | 0.54 | XP_014277503.1 / XM_014422017.1 |
| Mevalonate kinase | 8.00 | 8.54 | 0.09 | 12.13 | 7.82 | -0.63 | 8.29 | 10.07 | 0.28 | 9.01 | XP_014272243.1 / XM_014416757.1 |
| Phosphomevalonate kinase | 5.97 | 4.46 | -0.42 | 7.02 | 4.80 | -0.55 | 5.17 | 5.96 | 0.21 | 5.49 | XP_014271961.1 / XM_014416475.1 |
| Diphosphomevalonate decarboxylase | 3.96 | 3.44 | -0.20 | 4.90 | 3.98 | -0.30 | 3.72 | 4.46 | 0.26 | 4.04 | XP_014290216.1 / XM_014434740.1 |
| IDP Isomerase | 13.51 | 10.56 | -0.36 | 11.89 | 14.15 | 0.25 | 11.96 | 12.97 | 0.12 | 12.37 | XP_014271459.1 / XM_014415973.1 |
| FDP Synthase | 3.62 | 8.08 | 1.16 | 15.70 | 18.66 | 0.25 | 5.97 | 17.12 | 1.52 | 10.48 | XP_014276183.1 / XM_014420697.1 |
| FDP Synthase | 12.52 | 14.96 | 0.26 | 13.13 | 15.78 | 0.27 | 13.80 | 14.40 | 0.06 | 14.04 | XP_014276401.1 / XM_014420915.1 |
| FDP synthase | 0.04 | 0.16 | 2.00 | 0.80 | 0.04 | -4.32 | 0.10 | 0.44 | 2.14 | 0.24 | XP_014289203.1 / XM_014433717.1 |
| FDP synthase | 0.29 | 0.29 | 0.00 | 0.35 | 0.22 | -0.67 | 0.29 | 0.28 | -0.05 | 0.29 | XP_014289225.1 / XM_014433739.1 |
| Farnesol dehydrogenase | 204.63 | 77.01 | -1.41 | 35.70 | 46.26 | 0.37 | 137.30 | 40.68 | -1.75 | 98.17 | XP_014286519.1 / XM_014431033.1 |
| Farnesol dehydrogenase | 78.56 | 47.27 | -0.73 | 43.79 | 43.94 | 0.00 | 62.05 | 43.86 | -0.50 | 54.69 | XP_014286524.1 / XM_014431038.1 |
| Farnesol dehydrogenase | 107.93 | 53.10 | -1.02 | 44.15 | 27.32 | -0.69 | 79.01 | 36.10 | -1.13 | 61.65 | XP_014286525.1 / XM_014431039.1 |
| Farnesal dehydrogenase | 28.66 | 94.12 | 1.72 | 35.72 | 49.72 | 0.48 | 63.19 | 42.42 | -0.57 | 54.79 | XP_014272618.1 / XM_014417132.1 |
| Farnesal dehydrogenase | 49.42 | 54.55 | 0.14 | 57.72 | 65.03 | 0.17 | 52.13 | 61.22 | 0.23 | 55.81 | XP_014292700.1 / XM_014437214.1 |
| Juvenile hormone acid methyltransferase | 0.13 | 0.06 | -1.12 | 0.22 | 34.99 | 7.31 | 0.09 | 16.85 | 7.55 | 6.87 | XP_014293044.1 / XM_014437558.1 |
| Juvenile hormone acid methyltransferase | 46.90 | 45.31 | -0.05 | 22.24 | 79.28 | 1.83 | 46.06 | 49.52 | 0.10 | 47.46 | XP_014290953.1 / XM_014435467.1 |
| Juvenile hormone acid methyltransferase | 0.38 | 0.34 | -0.16 | 0.79 | 0.18 | -2.13 | 0.36 | 0.50 | 0.47 | 0.41 | XP_014283772.1 / XM_014428286.1 |
| Methyl farnesoate epoxidase | 1.15 | 0.59 | -0.96 | 0.16 | 0.26 | 0.70 | 0.86 | 0.21 | -2.03 | 0.59 | XP_014283057.1 / XM_014427571.1 |
Transcripts appearing to encode proteins involved in RNAi-related processes were detected, including such RISC-associated components as Dicer, Loquacious and Argonaute, as well as homologs of Aubergine, Tarbp2, various RNA helicases and PIWI-containing proteins. No PUTs with evident homology to sid-1 or sid-2 were detected, nor were any apparent RNA-dependent RNA polymerases observed.
No evidence of an iflavirus homologous to that previously observed in the brown marmorated stink bug was apparent in harlequin bug, and only very weak evidence supporting the possible presence of a Nosema sp. microbe was present: only seven PUTs corresponded to genes observed in the Nosema genus, and these were predominantly associated with retrotransposable elements. In contrast, 653 and 67 PUTs exhibited high levels of sequence similarity with known Pantoea and Erwinia genes, respectively, consistent with the notion that these genera likely comprise the primary microbial endosymbionts of M. histrionica, as well as several other pentatomids [57,58].
4. Discussion
The increasing importance of stink bug pests is concurrent with the reduction in foliar insecticides in major, predominantly transgenic field crops, and the invasion of several major stink bug pests into new continents. Panizzi et al. [59] listed 25 “important” pest species in 17 genera, and about 41 “less important” pests in 16 additional genera, infesting a wide variety of agronomic and horticultural crops and other useful plants. Since that review, two of those pests hitherto considered less important, H. halys and Bagrada hilaris, have become vastly more important following continental-scale invasions, and a number of other species are potentially serious threats for invasion [59,60]. Complicating management is the fact that several predatory pentatomid species in the subfamily Asopinae are important and widespread natural enemies in agroecosystems [61]. All of these trends have prompted increasing interest in new management tools as well as avoidance of insecticide resistance in this important group.
The current study addressed the harlequin bug gene space from both quantitative and qualitative perspectives. That only one biological replicate’s transcriptome was sequenced implies that any inferred fold changes in gene expression per RNA-Seq data are at most suggestive of differential expression (DE), but these cannot support the notion of true DE with any statistical significance. However, a subset of transcripts exhibiting high fold changes was further probed using quantitative real-time PCR combined with rigorous statistical analyses—for these cases, DE could be asserted, and these findings represent compelling targets on which to perform RNAi-based reverse genetics. The lack of biological replication among RNA-Seq data per se has no impact on qualitative results presented herein; in particular, these qualitative findings afford useful insights into the inherent enzymatic capacity of the harlequin bug to detoxify xenobiotic compounds and to synthesize terpenoid compounds.
M. histrionica appears to possess four distinct Delta-class GST genes, although these resolve to two pairs, each of which contain two genes exhibiting very high levels of sequence similarity. Whether these correspond to recent duplication events or to potential RNA sequencing and assembly artifacts will require exploring the genome of M. histrionica, which is beyond the scope of this study. The H. halys genome seems to encode two Delta-type GSTs. These counts are lower than what has been seen in other hemipterans: M. persicae has eight and Acyrthosiphon pisum has ten such genes [62]. They are also lower than what has been observed in the Dipterans Drosophila melanogaster (11) and Anopheles gambiae (12) [63,64], although slightly higher than that seen in the Hymenopteran species, Apis mellifera (one; [23]). The low count of Delta-type GSTs in both harlequin bug and brown marmorated stink bug makes them appealing as knockdown/knockout targets for transcriptional disruption. Their potential utility may, however, be overshadowed by the generally highly conserved nature of genes from the Delta (as well as Epsilon) cytosolic GST subtypes, thereby incurring risks of off-target effects in practical applications. Transcriptome sequencing and analysis of closely related insect taxa should help clarify the extent to which such off-target gene silencing risks might manifest in unanticipated ecological damage.
Some H. halys scaffolds contain multiple β-esterase genes in close proximity, specifically scaffolds 261 and 2592, which comprise tandem arrays of eleven and six β-esterase genes, respectively. On both scaffolds 261 and 2592, M. histrionica β-esterase proteins mapped to three distinct, homologous β-esterase H. halys loci. This may suggest a potentially elevated β-esterase gene copy number in H. halys when compared to M. histrionica. However, fifteen unique M. histrionica carboxylesterase-associated protein sequences that did not map to the H. halys genome via GenomeThreader analysis are all located within the carboxylesterase clade (Supplementary Figure S3). These unmapped M. histrionica carboxylesterase-associated protein sequences may represent multiple M. histrionica-specific carboxylesterases. Currently, with only M. histrionica transcript data, it is not possible to identify novel gene duplication or to identify divergent homologs.
Inspection of genome sequence organization hints towards an increase in copy number of H. halys β-esterase genes through tandem gene duplication, which may have been effected by chromosomal slippage during DNA replication, for example. Results considered herein suggest that tandem duplication of β-esterase genes is also present in M. histrionica, albeit to a lesser extent than observed in H. halys. Whether such shared duplications occurred in a common ancestor or were realized as independent events subsequent to speciation remains uncertain, and will require genome sequence analysis of a broader taxonomic sampling of pentatomid species to resolve.
It is possible that β-esterase gene duplication is a common method of generating insecticide resistance in the Hemiptera. Gene duplication and over expression of β-esterases has been observed as one of the primary insecticide resistance mechanisms in M. persicae [65,66]. The presence of numerous carboxylesterase genes being actively transcribed in cultures of H. halys and M. histrionica, neither of which have been exposed to insecticides for at least 30 and 10 generations, respectively, suggests these constitute a standing defense against insecticide and/or other toxin exposure. Future investigation of differential expression between insecticide exposed and non-exposed groups could elucidate gene expression regulation associated with insecticide exposure. Identification of up regulated genes in response to insecticide exposure could help narrow the field of potential β-esterase gene targets, as well as identify regulation mechanisms that may also be of interest for combatting insecticide resistance.
Harlequin bug cytochrome P450s are most similar to brown marmorated stink bug (BMSB) P450s. BMSB has 128 named P450s in 26 families and 11 fragments that were too short to name. Harlequin bug has 87 named P450s and 17 fragments in 25 families. Sixty-three (72%) were considered orthologs. The differences occurred in gene clusters where there was not a 1:1 relationship between the species. The BMSB had more P450s in some large gene blooms like CYP6LV (20:9) and CYP6LT (8:4). One CYP3 clan family CYP3228A1 was found in BMSB but not in harlequin bug. Only two new subfamilies were present in harlequin bug CYP3226C and CYP4KC1. The harlequin bug data are based on transcriptome sequences. If some genes were not expressed they would not be represented. This may explain some of the differences. The large number of genes in the CYP3 (43 genes) and CYP4 (30 genes) clans provides the raw material for developing pesticide resistance. The genes in the CYP2 and mito clans might be good targets to block molting (Halloween genes and CYP18) and juvenile hormone synthesis (CYP15). Inhibition of the CYP4G P450s could act to block or reduce cuticular hydrocarbon synthesis, though this subfamily has six genes so inhibition might be difficult to achieve. Differential P450 expression in antennae could identify potential pheromone metabolizing P450s. Although the mating process in M. histrionica is not exclusively controlled by olfaction, but also via behavioral cues including visual and vibrational signals, inhibitors of P450s may nonetheless have utility in disrupting successful mating.
In the core MVA terpenoid biosynthetic pathway of M. histrionica and H. halys, all enzymes except for acetoacetyl-CoA synthase appear to be represented by single-copy genes, which is consistent with MVA pathways from other insects (e.g., [67,68,69]). On the contrary, enzymes involved in the conversion of FDP to farnesoic acid in JH biosynthesis are known to be encoded by families of genes with expression in different tissues based on the importance of farnesol and farnesal homeostasis for a variety of cellular functions [70]. Accordingly, we found isoforms of M. histrionica and H. halys farnesol and farnesal dehydrogenase; however, we were unable to identify orthologs of farnesyl diphosphatase. Furthermore, in agreement with findings from other insects [70], harlequin bug and brown marmorated stink bug methyl farnesoate epoxidase appears to be represented by a single-copy gene transcript; unexpectedly, we found several potential isoforms for juvenile hormone acid methyltransferase. To build a biosynthetic branch for the formation of murgantiol, it is possible that the pathway is initiated by one of the two identified FDP synthases because of the higher male specific expression of this gene (at least for M. histrionica). It can further be assumed that P450 enzymes with similarity to methyl farnesoate epoxidase are involved in the final epoxidation step in murgantiol biosynthesis because of the same position of the epoxy group at C10-C11 in both pheromone and JH molecules. Further comparative expression profiling paired with gene functional analysis will be required to elucidate the murgantiol biosynthetic pathway in M. histrionica.
5. Conclusions
This study generated the first high-volume collection of transcriptome sequence data for the important insect pest Murgantia histrionica. A number of gene targets for transcriptional disruption were identified, which will require further experimental study in an in vivo context. These genes include elements of three families implicated in the development of resistance to synthetic insecticides (glutathione S-transferases, carboxyesterases and cytochrome P450s), as well as genes involved pheromone biosynthesis pathways. These data will be of further utility in studies addressing additional gene families, in performing comparative transcriptomics among various insect species, and in supporting gene annotation efforts for any possible genome sequencing efforts made specifically for the harlequin bug.
Acknowledgments
The authors thank Megan Herlihy for assistance with insect rearing and Ashot Khrimian for helpful discussions. Comments from two anonymous reviewers substantially improved this manuscript. This work was supported in part by USDA NIFA award 2016-67013-24759.
Supplementary Materials
The following are available online at http://www.mdpi.com/2075-4450/8/2/55/s1, Supplementary Figure S1 (GO term abundances (full ontology)), Figure S2 (GO-Slim term abundances) and Figure S3 (Maximum likelihood based carboxylesterase phylogeny for Halyomorpha halys and Murgantia histrionica proteins); and Table S1: Gene expression levels of putative unique transcripts exhibiting at least two-fold expression differentials among three comparisons.
Author Contributions
D.E.G. and M.E.S. conceived and designed the experiments; M.E.S., J.H.R., D.R.N. and D.K. performed the experiments; M.E.S., J.H.R., B.L. and J.L. analyzed the data; D.C.W. maintained a M. histrionica culture and contributed staged insects from this colony; and M.E.S., J.H.R., D.R.N., D.C.W., D.T. and D.E.G. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Aldrich J.R., Avery J.W., Lee C.J., Graf J.C., Harrison D.J., Bin F. Semiochemistry of cabbage bugs (Heteroptera: Pentatomidae: Eurydema and Murgantia) J. Entomol. Sci. 1996;31:172–182. [Google Scholar]
- 2.Chittenden F.H. Harlequin Cabbage Bug and Its Control. USDA Farmers’ Bulletins (U.S. Department of Agriculture); Washington, DC, USA: 1920. [Google Scholar]
- 3.Wallingford A.K., Kuhar T.P., Schultz P.B., Freeman J.H. Harlequin bug biology and pest management in brassicaceous crops. J. Integr. Pest Manag. 2011;2:H1–H4. doi: 10.1603/IPM10015. [DOI] [Google Scholar]
- 4.Walker H.G., Anderson L.D. Report on the control of the harlequin bug, Murgantia histrionica Hahn, with notes on the severity of an outbreak of this insect in 1932. J. Econ. Entomol. 1933;26:129–135. doi: 10.1093/jee/26.1.129. [DOI] [Google Scholar]
- 5.DiMeglio A.S., Wallingford A.K., Weber D.C., Kuhar T.P., Mullins D. Supercooling points of Murgantia histrionica (Hemiptera: Pentatomidae) and field mortality in the mid-Atlantic United States following lethal low temperatures. Environ. Entomol. 2016;45:1294–1299. doi: 10.1093/ee/nvw091. [DOI] [PubMed] [Google Scholar]
- 6.Koch R.L., Pezzini D.T., Michel A.P., Hunt T.E. Identification, Biology, Impacts, and Management of Stink Bugs (Hemiptera: Heteroptera: Pentatomidae) of Soybean and Corn in the Midwestern United States. J. Integr. Pest Manag. 2017;8:11. doi: 10.1093/jipm/pmx004. [DOI] [Google Scholar]
- 7.Lockwood J.A. Six-Legged Soldiers: Using Insects as Weapons of War. Oxford University Press; Oxford, NY, USA: 2010. [Google Scholar]
- 8.Capinera J.L. Harlequin Bug, Murgantia histrionica (Hahn) (Hemiptera: Pentatomidae) In: Capinera J.L., editor. Encyclopedia of Entomology. Springer; Dordrecht, The Netherlands: 2008. pp. 1766–1768. [Google Scholar]
- 9.Aliabadi A., Renwick J.A.A., Whitman D.W. Sequestration of glucosinolates by harlequin bug Murgantia histrionica. J. Chem. Ecol. 2002;28:1749–1762. doi: 10.1023/A:1020505016637. [DOI] [PubMed] [Google Scholar]
- 10.Joga M.R., Zotti M.J., Smagghe G., Christiaens O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016;7:553. doi: 10.3389/fphys.2016.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn D.K., Moreira J.A., Millar J.G. Identification, synthesis, and bioassay of a male-specific aggregation pheromone from the harlequin bug, Murgantia histrionica. J. Chem. Ecol. 2008;34:238–251. doi: 10.1007/s10886-007-9415-x. [DOI] [PubMed] [Google Scholar]
- 12.Zahn D.K., Moreira J.A., Millar J.G. Erratum to: Identification, synthesis, and bioassay of a male-specific aggregation pheromone from the harlequin bug, Murgantia histrionica. J. Chem. Ecol. 2012;38:126. doi: 10.1007/s10886-012-0061-6. [DOI] [PubMed] [Google Scholar]
- 13.Khrimian A., Shirali S., Vermillion K.E., Siegler M.A., Guzman F., Chauhan K., Aldrich J.R., Weber D.C. Determination of the stereochemistry of the aggregation pheromone of harlequin bug, Murgantia histrionica. J. Chem. Ecol. 2014;40:1260–1268. doi: 10.1007/s10886-014-0521-2. [DOI] [PubMed] [Google Scholar]
- 14.Weber D.C., Walsh G.C., DiMeglio A.S., Athanas M.M., Leskey T.C., Khrimian A. Attractiveness of harlequin bug, Murgantia histrionica, aggregation pheromone: Field response to isomers, ratios, and dose. J. Chem. Ecol. 2014;40:1251–1259. doi: 10.1007/s10886-014-0519-9. [DOI] [PubMed] [Google Scholar]
- 15.Walsh G.C., Dimeglio A.S., Khrimian A., Weber D.C. Marking and retention of harlequin bug, Murgantia histrionica (Hahn) (Hemiptera: Pentatomidae), on pheromone-baited and unbaited plants. J. Pest Sci. 2016;89:21–29. doi: 10.1007/s10340-015-0663-1. [DOI] [Google Scholar]
- 16.Bellés X., Martín D., Piulachs M.-D. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu. Rev. Entomol. 2005;50:181–199. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- 17.Beran F., Rahfeld P., Luck K., Nagel R., Vogel H., Wielsch N., Irmisch S., Ramasamy S., Gershenzon J., Heckel D.G., et al. Novel family of terpene synthases evolved from trans-isoprenyl diphosphate synthases in a flea beetle. Proc. Natl. Acad. Sci. USA. 2016;113:2922–2927. doi: 10.1073/pnas.1523468113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman R. Genomic organization of the glutathione S-transferase family in insects. Mol. Phylogenet. Evol. 2011;61:924–932. doi: 10.1016/j.ympev.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Li X., Schuler M.A., Berenbaum M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 20.Moores G.D., Devine G.J., Devonshire A.L. Insecticide-insensitive acetylcholinesterase can enhance esterase-based resistance in Myzus persicae and Myzus nicotianae. Pestic. Biochem. Physiol. 1994;49:114–120. doi: 10.1006/pest.1994.1038. [DOI] [Google Scholar]
- 21.Moores G.D., Gao X., Denholm I., Devonshire A.L. Characterisation of insensitive acetylcholinesterase in insecticide-resistant cotton aphids, Aphis gossypii Glover (Homoptera: Aphididae) Pestic. Biochem. Physiol. 1996;56:102–110. doi: 10.1006/pest.1996.0064. [DOI] [Google Scholar]
- 22.Oakeshott J.G., Johnson R.M., Berenbaum M.R., Ranson H., Cristino A.S., Claudianos C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol. Biol. 2010;19(Suppl. 1):147–163. doi: 10.1111/j.1365-2583.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 23.Claudianos C., Ranson H., Johnson R.M., Biswas S., Schuler M.A., Berenbaum M.R., Feyereisen R., Oakeshott J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakeshott J.G., Devonshire A.L., Claudianos C., Sutherland T.D., Horne I., Campbell P.M., Ollis D.L., Russell R.J. Comparing the organophosphorus and carbamate insecticide resistance mutations in cholin- and carboxyl-esterases. Chem. Biol. Interact. 2005;157–158:269–275. doi: 10.1016/j.cbi.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 25.Rewitz K.F., O’Connor M.B., Gilbert L.I. Molecular evolution of the insect Halloween family of cytochrome P450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 2007;37:741–753. doi: 10.1016/j.ibmb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Qu Z., Kenny N.J., Lam H.M., Chan T.F., Chu K.H., Bendena W.G., Tobe S.S., Hui J.H.L. How did arthropod sesquiterpenoids and ecdysteroids arise? Comparison of hormonal pathway genes in noninsect arthropod genomes. Genome Biol. Evol. 2015;7:1951–1959. doi: 10.1093/gbe/evv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Y., Tittiger C., Wicker-Thomas C., Le Goff G., Young S., Wajnberg E., Fricaux T., Taquet N., Blomquist G.J., Feyereisen R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. USA. 2012;109:14858–14863. doi: 10.1073/pnas.1208650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Z., Zhang X., Wang Y., Moussian B., Zhu K.Y., Li S., Ma E., Zhang J. LmCYP4G102: An oenocyte-specific cytochrome P450 gene required for cuticular waterproofing in the migratory locust, Locusta migratoria. Sci. Rep. 2016;6:29980. doi: 10.1038/srep29980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N., Li M., Gong Y., Liu F., Li T. Cytochrome P450s—Their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015;120:77–81. doi: 10.1016/j.pestbp.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Edi C.V., Djogbénou L., Jenkins A.M., Regna K., Muskavitch M.A.T., Poupardin R., Jones C.M., Essandoh J., Kétoh G.K., Paine M.J.I., et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David J.-P., Ismail H.M., Chandor-Proust A., Paine M.J.I. Role of cytochrome P450s in insecticide resistance: Impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120429. doi: 10.1098/rstb.2012.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balabanidou V., Kampouraki A., MacLean M., Blomquist G.J., Tittiger C., Juárez M.P., Mijailovsky S.J., Chalepakis G., Anthousi A., Lynd A., et al. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 2016;113:9268–9273. doi: 10.1073/pnas.1608295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S., Widemann E., Bernard G., Lesot A., Pinot F., Pedrini N., Keyhani N.O. CYP52X1, representing new cytochrome P450 subfamily, displays fatty acid hydroxylase activity and contributes to virulence and growth on insect cuticular substrates in entomopathogenic fungus Beauveria bassiana. J. Biol. Chem. 2012;287:13477–13486. doi: 10.1074/jbc.M111.338947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maïbèche-Coisne M., Nikonov A.A., Ishida Y., Jacquin-Joly E., Leal W.S. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. USA. 2004;101:11459–11464. doi: 10.1073/pnas.0403537101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomquist G.J., Figueroa-Teran R., Aw M., Song M., Gorzalski A., Abbott N.L., Chang E., Tittiger C. Pheromone production in bark beetles. Insect Biochem. Mol. Biol. 2010;40:699–712. doi: 10.1016/j.ibmb.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 36.He P., Zhang Y.-F., Hong D.-Y., Wang J., Wang X.-L., Zuo L.-H., Tang X.-F., Xu W.-M., He M. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genom. 2017;18:219. doi: 10.1186/s12864-017-3592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann K., Gowin J., Hartfelder K., Korb J. The scent of royalty: A P450 gene signals reproductive status in a social insect. Mol. Biol. Evol. 2014;31:2689–2696. doi: 10.1093/molbev/msu214. [DOI] [PubMed] [Google Scholar]
- 38.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vakatov D. The NCBI C++ Toolkit Book [Internet] National Center for Biotechnology Information; Bethesda, MD, USA: 2004. [Google Scholar]
- 42.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Sparks M.E., Blackburn M.B., Kuhar D., Gundersen-Rindal D.E. Transcriptome of the Lymantria dispar (Gypsy Moth) Larval Midgut in Response to Infection by Bacillus thuringiensis. PLoS ONE. 2013;8:e61190. doi: 10.1371/journal.pone.0061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparks M.E., Shelby K.S., Kuhar D., Gundersen-Rindal D.E. Transcriptome of the invasive brown marmorated stink bug, Halyomorpha halys (Stål) (Heteroptera: Pentatomidae) PLoS ONE. 2014;9:e111646. doi: 10.1371/journal.pone.0111646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 47.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 51.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 52.Revell L.J. Phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 53.Gremme G., Brendel V., Sparks M.E., Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005;47:965–978. doi: 10.1016/j.infsof.2005.09.005. [DOI] [Google Scholar]
- 54.Sparks M.E., Gundersen-Rindal D.E., Harrison R.L. Complete Genome Sequence of a Novel Iflavirus from the Transcriptome of Halyomorpha halys, the Brown Marmorated Stink Bug. Genome Announc. 2013;1:e00910-13. doi: 10.1128/genomeA.00910-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kenyon L.J., Meulia T., Sabree Z.L. Habitat visualization and genomic analysis of “Candidatus Pantoea carbekii,” the primary symbiont of the brown marmorated stink bug. Genome Biol. Evol. 2015;7:620–635. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prado S.S., Almeida R.P.P. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 2009;58:64–69. doi: 10.1007/s00284-008-9267-9. [DOI] [PubMed] [Google Scholar]
- 58.Duron O., Noël V. A wide diversity of Pantoea lineages are engaged in mutualistic symbiosis and cospeciation processes with stinkbugs. Environ. Microbiol. Rep. 2016;8:715–727. doi: 10.1111/1758-2229.12432. [DOI] [PubMed] [Google Scholar]
- 59.Panizzi A.R., McPherson J.E., James D.G., Javahery M., McPherson R.M. Economic importance of stink bugs (Pentatomidae) In: Schaefer C.W., Panizzi A.R., editors. Heteroptera of Economic Importance. CRC Press; Boca Raton, FL, USA: 2000. pp. 421–474. [Google Scholar]
- 60.McPherson J.E. Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management. CRC Press; Boca Raton, FL, USA: 2017. [Google Scholar]
- 61.De Clercq P. Predaceous stinkbugs (Pentatomidae: Asopinae) In: Schaefer C.W., Panizzi A.R., editors. Heteroptera of Economic Importance. CRC Press; Boca Raton, FL, USA: 2000. pp. 737–789. [Google Scholar]
- 62.Ramsey J.S., Rider D.S., Walsh T.K., De Vos M., Gordon K.H. J., Ponnala L., Macmil S.L., Roe B.A., Jander G. Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol. Biol. 2010;19(Suppl. 2):155–164. doi: 10.1111/j.1365-2583.2009.00973.x. [DOI] [PubMed] [Google Scholar]
- 63.Holt R.A., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M.C., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 64.Ranson H., Claudianos C., Ortelli F., Abgrall C., Hemingway J., Sharakhova M.V., Unger M.F., Collins F.H., Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 65.Devonshire A.L., Foster G.N., Sawicki R.M. Peach-potato aphid, Myzus persicae (Sulz.), resistant to organophosphorus and carbamate insecticides on potatoes in Scotland. Plant Pathol. 1977;26:60–62. doi: 10.1111/j.1365-3059.1977.tb01024.x. [DOI] [Google Scholar]
- 66.Devonshire A.L., Sawicki R.M. Insecticide-resistant Myzus persicae as an example of evolution by gene duplication. Nature. 1979;280:140–141. doi: 10.1038/280140a0. [DOI] [Google Scholar]
- 67.Noriega F.G., Ribeiro J.M.C., Koener J.F., Valenzuela J.G., Hernandez-Martinez S., Pham V.M., Feyereisen R. Comparative genomics of insect juvenile hormone biosynthesis. Insect Biochem. Mol. Biol. 2006;36:366–374. doi: 10.1016/j.ibmb.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinjoh T., Kaneko Y., Itoyama K., Mita K., Hiruma K., Shinoda T. Control of juvenile hormone biosynthesis in Bombyx mori: Cloning of the enzymes in the mevalonate pathway and assessment of their developmental expression in the corpora allata. Insect Biochem. Mol. Biol. 2007;37:808–818. doi: 10.1016/j.ibmb.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Nouzova M., Edwards M.J., Mayoral J.G., Noriega F.G. A coordinated expression of biosynthetic enzymes controls the flux of juvenile hormone precursors in the corpora allata of mosquitoes. Insect Biochem. Mol. Biol. 2011;41:660–669. doi: 10.1016/j.ibmb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noriega F.G. Juvenile hormone biosynthesis in insects: What is new, what do we know, and what questions remain? Int. Sch. Res. Notices. 2014 doi: 10.1155/2014/967361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.