Abstract
In 1996, a set of 20 trinucleotides was identified in genes of both prokaryotes and eukaryotes which has on average the highest occurrence in reading frame compared to its two shifted frames. Furthermore, this set has an interesting mathematical property as is a maximal self-complementary trinucleotide circular code. In 2015, by quantifying the inspection approach used in 1996, the circular code was confirmed in the genes of bacteria and eukaryotes and was also identified in the genes of plasmids and viruses. The method was based on the preferential occurrence of trinucleotides among the three frames at the gene population level. We extend here this definition at the gene level. This new statistical approach considers all the genes, i.e., of large and small lengths, with the same weight for searching the circular code . As a consequence, the concept of circular code, in particular the reading frame retrieval, is directly associated to each gene. At the gene level, the circular code is strengthened in the genes of bacteria, eukaryotes, plasmids, and viruses, and is now also identified in the genes of archaea. The genes of mitochondria and chloroplasts contain a subset of the circular code . Finally, by studying viral genes, the circular code was found in DNA genomes, RNA genomes, double-stranded genomes, and single-stranded genomes.
Keywords: circular code in genes, DNA genes, RNA genes, double-stranded genes, single-stranded genes
1. Introduction
Circular code is a mathematical structure of genes and genomes. This concept initially found for genes is extended for genomes (non-coding regions of eukaryotes) according to recent results. A circular code is a set of words such that any motif from , called motif, allows it to retrieve, maintain, and synchronize the original (construction) frame.
The circular code identified in the genes of bacteria, eukaryotes, plasmids, and viruses [1,2] contains the 20 following trinucleotides
| (1) |
which allows it to both retrieve the reading frame with a window of 13 nucleotides (Figure 3 in [3]) and to code the 12 following amino acids
| (2) |
The current genetic code is not circular. Thus, it cannot retrieve the reading frame. The loss during evolution of this circular code property on the 4-letter alphabet required a complex translation mechanism using 20 amino acids and proteins in current genomes.
motifs from Equation (1) are identified in (i) genes “universally” [1,4]; (ii) tRNAs of prokaryotes and eukaryotes [3,5]; (iii) rRNAs of prokaryotes (16S) and eukaryotes (18S), in particular in the ribosome decoding center where the universally conserved nucleotides G530, A1492, and A1493 are included in the motifs [3,6,7]; and (iv) genomes (non-coding regions of eukaryotes) [4,8].
The motifs of maximal cardinality 20 (composition) in genes with the properties of the circular code, and complementary allow the two reading frames and the four shifted frames to be retrieved by pairing between DNAs-DNAs, DNAs-mRNAs, mRNAs-rRNAs, mRNAs-tRNAs, and rRNAs-tRNAs, as shown with a 3D visualization of the motifs in the ribosome [3,6,7].
The motifs in genomes have a different structure compared to the motifs in genes [8]. Indeed, their cardinality is not maximal (less than 10 for an order of magnitude), their size is longer, and their structure contains repeated trinucleotides. Furthermore, the motifs of minimal cardinality 1 generated with the 20 repeated trinucleotides where (Equation (1)) are very common in the genomes of eukaryotes (e.g., [8,9,10]). Their length can be very large (e.g., , see Figure 1). The repeated trinucleotides are very unstable with mutation rates up to 100,000 times higher than the genomic average mutation rate. Mutation in repeats increases its evolutionary stability.
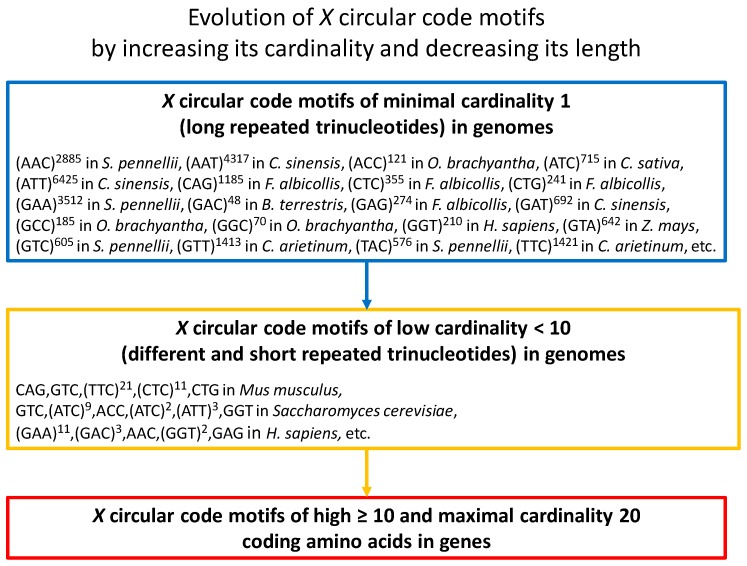
Figure 1.
Model of evolution of the circular code motifs (Equation (1)) by increasing its cardinality (composition) and decreasing its length. Evolution begins with motifs of minimal cardinality 1 (long repeated trinucleotides) in genomes (the examples given are extracted from Table 2 in [8]). Then, the mutations in repeated trinucleotides lead to motifs of low cardinality (different and short repeated trinucleotides) in genomes (the examples given are extracted from Table 4 in [8]) up to motifs of high and maximal cardinality 20 coding the 12 amino acids (Equation (2)).
A model of evolution of the motifs in genes and genomes can be proposed according to the previous works and the recent results [8]. It proposes that the motifs of maximal cardinality 20 in genes have evolved from the motifs of minimal cardinality 1 (repeated trinucleotides) in genomes (Figure 1). An motif of minimal cardinality 1 which is unstable, mutates into an motif of low cardinality containing thus different repeated trinucleotides of short lengths. This evolutionary process continues by increasing the cardinality and decreasing the length of the motifs up to generate the motifs of high and maximal cardinality 20 coding the 12 amino acids (Equation (2)) in genes. The motifs of high cardinality have acquired the protein coding function in addition to the reading frame retrieval. This model suggests that the property of reading frame retrieval has preceded the protein coding function.
Since 1996, all the statistical analyses studying the preferential occurrence of trinucleotides among the three frames were done at the gene population level (kingdoms, taxonomic groups, genomes). We extend here the method from [1] at the gene level. This new approach is important as all the genes, i.e., of large and small lengths, are now considered with the same weight in the statistical definition for searching the circular code . As a consequence, the concept of circular code, in particular the reading frame retrieval, is directly associated to each gene. Thus, at the gene level, the circular code is searched here in the genes of bacteria, archaea, eukaryotes, plasmids, viruses, and eukaryotic organelles, i.e., mitochondria and chloroplasts. Finally, genes of double-stranded DNA and RNA viruses, and single-stranded DNA and RNA viruses are also analysed with this approach in order to assign a genetic information unit (DNA or RNA, double-stranded or single-stranded) to the circular code .
2. Method
2.1. Definitions
We recall a few definitions without detailed explanations (i.e., without figures and examples) for understanding the main properties of the trinucleotide circular code identified in genes [1,2].
Notation 1.
Let us denote the nucleotide 4-letter alphabet where A stands for Adenine, C stands for Cytosine, G stands for Guanine, and T stands for Thymine. The trinucleotide set over is denoted by . The set of non-empty words (words, respectively) over is denoted by (, respectively).
Notation 2.
Genes have three frames . By convention here, the reading frame is set up by a start trinucleotide , and the frames and are the reading frame shifted by one and two nucleotides in the direction (to the right), respectively.
Two biological maps are involved in gene coding.
Definition 1.
According to the complementary property of the DNA double helix, the nucleotide complementarity map is defined by , , , and . According to the complementary and antiparallel properties of the DNA double helix, the trinucleotide complementarity map is defined by for all . By extension to a trinucleotide set , the set complementarity map , being the set of all subsets of , is defined by , e.g., .
Definition 2.
The trinucleotide circular permutation map is defined by for all . denotes the 2nd iterate of . By extension to a trinucleotide set , the set circular permutation map is defined by , e.g., and .
Definition 3.
A set is a code if, for each , , the condition implies and for .
Definition 4.
Any non-empty subset of the code is a code and called trinucleotide code .
Definition 5.
A trinucleotide code is self-complementary if, for each , , i.e., .
Definition 6.
A trinucleotide code is circular if, for each , , , , the conditions and imply , (empty word), and for .
The proofs to decide whether a code is circular or not are based on the flower automaton [2], the necklace 5LDCN (Letter Diletter Continued Necklace) [11], the necklace LDCCN (Letter Diletter Continued Closed Necklace) with [12], and the graph theory [13].
Definition 7.
A trinucleotide circular code is self-complementary if , , and are trinucleotide circular codes such that (self-complementary), , and ( and are complementary).
The trinucleotide set (Equation (1)) coding the reading frame () in genes is a maximal (20 trinucleotides) self-complementary trinucleotide circular code [2] where the circular code coding the frame contains the 20 following trinucleotides
| (3) |
and the circular code coding the frame contains the 20 following trinucleotides
| (4) |
The trinucleotide circular codes and are related by the permutation map, i.e., and , and by the complementary map, i.e., and [14].
Several classes of methods were developed for identifying the circular code in genes over the last 20 years: frequency methods [2,15,16], correlation function [17], covering capability function [18], and occurrence probability of a complementary/permutation (CP) trinucleotide set at the gene population level [1].
The class of the 216 self-complementary trinucleotide circular codes (Definition 7; [2]; list given in Tables 4a, 5a, and 6a in [19]; [20]) is included in a larger class of codes by relaxing the circularity property which was defined in [1]:
Definition 8.
A trinucleotide code is self-complementary if (self-complementary), , and ( and are complementary) where and .
The statistical approach developed analyses the self-complementary codes (Definition 8) for searching the particular circular code .
2.2. Gene Kingdoms
Gene kingdoms of bacteria , archaea , plasmids , eukaryotes , chromosomes of eukaryotes , mitochondria , chloroplasts , viruses , and its five taxonomic double-stranded DNA viruses , double-stranded RNA viruses , single-stranded DNA viruses , single-stranded RNA viruses , and retro-transcribing viruses are obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/genome/browse/, May 2016) (Table 1). Computer tests exclude genes when (i) their nucleotides do not belong to the alphabet ; (ii) they do not begin with a start trinucleotide ; (iii) they do not end with a stop trinucleotide ; and (iv) their lengths are not modulo 3. In order to have an order of magnitude of data acquisition (details in Table 1), the kingdom of bacteria contains 15,735,053 genes and 5,222,267,667 trinucleotides (7,851,762 genes and 2,481,566,882 trinucleotides in [1]), i.e., a trinucleotide increase of about 110%, and the kingdom of eukaryotes contains 4,356,391 genes and 2,406,844,838 trinucleotides (1,662,579 genes and 824,825,761 trinucleotides in [1]), i.e., a trinucleotide increase of about 192%. The gene kingdoms , , , , and have gene and trinucleotide data that are significantly lower (less than 1 million trinucleotides) than the other gene kingdoms (Table 1).
Table 1.
Kingdoms of genes extracted from the GenBank database (http:// www.ncbi.nlm.nih.gov/genome/browse/, May 2016) with their symbol and their numbers of genomes, genes, and trinucleotides.
| Kingdom | (Symbol) | Nb of Genomes | Nb of Genes | Nb of Trinucleotides |
|---|---|---|---|---|
| Bacteria | 7039 | 15,735,053 | 5,222,267,667 | |
| Archaea | 182 | 282,802 | 81,460,549 | |
| Plasmids | 2319 | 575,760 | 159,169,387 | |
| Eukaryotes | 190 | 4,356,391 | 2,406,844,838 | |
| Chromosomes of eukaryotes | 2979 | 4,356,391 | 2,406,844,838 | |
| Mitochondria | 228 | 3347 | 862,327 | |
| Chloroplasts | 39 | 3192 | 925,303 | |
| Viruses | 5217 | 299,401 | 66,677,580 | |
| Double-stranded DNA viruses | 2480 | 259,696 | 59,239,700 | |
| Double-stranded RNA viruses | 211 | 1061 | 783,020 | |
| Single-stranded DNA viruses | 715 | 3291 | 802,405 | |
| Single-stranded RNA viruses | 1257 | 5093 | 4,406,365 | |
| Retro-transcribing viruses | 137 | 560 | 289,447 |
2.3. Preferential Frame of a Trinucleotide in a Gene
The method developed in [1] for identifying the circular code in genes determined the preferential frame of trinucleotides at the gene population level (kingdoms, taxonomic groups, genomes), i.e., after summing the trinucleotide frequencies of all genes in a kingdom. We extend this method at the gene level, i.e., the preferential frame of trinucleotides among the three frames is determined for each gene. There is no sum of trinucleotide frequencies of all genes in a kingdom. Thus, all the genes, i.e., of large and small lengths, have the same weight in respect to the preferential frame.
Consider a gene kingdom listed in Table 1. Let be the occurrence frequency of a trinucleotide in a frame of a gene belonging to a kingdom . Thus, there are trinucleotide occurrence frequencies in the three frames of a gene . Then, the preferential frame of a trinucleotide in a gene is the frame of maximal occurrence frequency among the three frames of
| (5) |
The three frequencies of a given trinucleotide are computed in the three frames 0, 1, and 2 of a gene. Then, the preferential frame of the trinucleotide in this gene is the frame associated to its highest trinucleotide frequency.
Remark 1.
In [1], the three occurrence frequencies of a trinucleotide in the three frames computed in a gene kingdom , always have different values, thus a unique preferential frame can be assigned to the trinucleotide. At the gene level, particularly for genes of small lengths, a trinucleotide may have an identical occurrence frequency in two or three frames . In this case, two or three preferential frames are assigned to the trinucleotide . If a trinucleotide is absent in a gene , mainly for genes of very small lengths, then no preferential frame is attributed to .
The indicator function is 1 if the preferential frame of a trinucleotide is equal to the frame of a gene , and 0 otherwise
| (6) |
where is defined in Equation (5).
2.4. Number of Preferential Frames of a Trinucleotide in a Gene Kingdom
The number of preferential frames of a trinucleotide for each frame in a gene kingdom is simply obtained by summing for all genes in
| (7) |
where is defined in Equation (6).
2.5. Occurrence Probability of a Complementary/Permutation Trinucleotide Set in a Gene Kingdom
In order to study the self-complementary codes (Definition 8) including the class of circular codes, and in particular the circular code , Equation (7) for a trinucleotide is expanded to a set of six trinucleotides involving the complementarity map and the permutation map simultaneously, precisely with in frame 0, in frame 1, in frame 2, and . is called a complementary and permutation (CP) trinucleotide set and is completely defined by the trinucleotide .
Remark 2.
and (proof obvious).
When the trinucleotide is given then the trinucleotide is also known. Thus, there are CP trinucleotide sets noted where with in frame 0, in frame 1, and in frame 2. A maximal (20 trinucleotides) self-complementary code is identified with the first 10 values of the numbers (defined below). Precisely, the code has 20 trinucleotides in frame 0, 20 trinucleotides in frame 1, and 20 trinucleotides in frame 2 with (self-complementary), , and ( and are complementary). There are self-complementary trinucleotide codes, and among them only 216 are circular [2,20].
Notation 3.
A CP trinucleotide set belongs to the self-complementary trinucleotide circular code , i.e., , if , i.e., if the trinucleotide and its complementary trinucleotide belong to . Ten CP trinucleotide sets among 30 belong to the circular code , i.e., such that 10 sets with and .
Notation 4.
In order to facilitate the reading of Table 2, the 30 CP trinucleotide sets are presented in the following way (i) the first 10 sets belong to the circular code (with , and ) and are in lexicographical order with respect to the trinucleotide (in bold), and (ii) the 20 remaining sets are in lexicographical order with respect to the trinucleotide (in italics).
Table 2.
Identification of the maximal self-complementary trinucleotide circular code in gene kingdoms of bacteria , archaea , plasmids , eukaryotes , chromosomes of eukaryotes , mitochondria , chloroplasts , viruses , and its five taxonomic groups: double-stranded DNA viruses , double-stranded RNA viruses , single-stranded DNA viruses , single-stranded RNA viruses , and retro-transcribing viruses (Table 1). Occurrence probability (%) of the 30 complementary and permutation (CP) trinucleotide sets with in frame 0, in frame 1, in frame 2, in a gene kingdom computed according to Equation (9) and its rank , the 1st rank being associated to the highest value of and the 30th rank, to the lowest value of . The 20 trinucleotides of the self-complementary circular code are in bold, the 20 trinucleotides of the circular code are in italics, and the 20 trinucleotides of the circular code are both in bold and italics. The first 10 CP sets belong to the circular code ( with and ) and are in lexicographical order with respect to the trinucleotide in bold, and the 20 remaining CP sets are in lexicographical order with respect to the trinucleotide in italics. The numbers in italics occurring with the CP sets are associated with the two trinucleotides of which do not occur preferentially in the gene kingdom.
| AAC | GTT | 6.1 | 6 | 7.4 | 4 | 6.0 | 4 | 8.5 | 3 | 8.4 | 3 | 4.4 | 9 | 4.9 | 9 | 6.6 | 3 | 6.8 | 4 | 6.4 | 3 | 5.9 | 1 | 7.0 | 3 | 5.0 | 5 | |
| AAT | ATT | 7.4 | 2 | 5.3 | 8 | 7.3 | 2 | 8.7 | 2 | 8.7 | 2 | 3.4 | 14 | 5.8 | 1 | 6.6 | 2 | 7.5 | 2 | 5.9 | 4 | 5.6 | 2 | 6.3 | 4 | 5.3 | 4 | |
| ACC | GGT | 5.1 | 9 | 3.9 | 12 | 5.1 | 8 | 5.1 | 8 | 4.7 | 10 | 5.7 | 4 | 5.2 | 3 | 4.9 | 8 | 4.9 | 9 | 4.6 | 8 | 4.6 | 5 | 5.3 | 7 | 3.7 | 9 | |
| ATC | GAT | 6.5 | 4 | 6.7 | 5 | 6.2 | 3 | 8.1 | 4 | 8.2 | 4 | 3.6 | 13 | 4.6 | 14 | 6.5 | 4 | 6.7 | 5 | 6.4 | 2 | 5.4 | 4 | 7.1 | 2 | 6.0 | 2 | |
| CAG | CTG | 4.6 | 10 | 3.7 | 13 | 3.9 | 10 | 4.2 | 10 | 4.9 | 9 | 0.7 | 30 | 0.0 | 30 | 2.9 | 15 | 3.8 | 10 | 2.4 | 18 | 2.8 | 19 | 1.5 | 24 | 2.9 | 18 | |
| CTC | GAG | 6.2 | 5 | 7.5 | 3 | 5.9 | 6 | 7.0 | 5 | 7.5 | 5 | 2.6 | 18 | 0.4 | 27 | 5.6 | 6 | 6.3 | 6 | 5.3 | 7 | 4.1 | 10 | 5.4 | 6 | 4.9 | 6 | |
| GAA | TTC | 5.8 | 7 | 5.3 | 7 | 5.5 | 7 | 5.0 | 9 | 5.2 | 7 | 6.3 | 2 | 4.9 | 7 | 5.2 | 7 | 5.6 | 7 | 5.3 | 6 | 4.6 | 6 | 4.9 | 8 | 5.4 | 3 | |
| GAC | GTC | 8.2 | 1 | 9.7 | 1 | 7.8 | 1 | 9.0 | 1 | 9.1 | 1 | 5.8 | 3 | 0.6 | 26 | 7.2 | 1 | 8.0 | 1 | 7.3 | 1 | 5.4 | 3 | 7.3 | 1 | 6.4 | 1 | |
| GCC | GGC | 6.7 | 3 | 8.2 | 2 | 6.0 | 5 | 5.7 | 6 | 5.2 | 8 | 7.1 | 1 | 5.3 | 2 | 5.9 | 5 | 7.0 | 3 | 5.5 | 5 | 4.3 | 8 | 5.7 | 5 | 4.7 | 7 | |
| GTA | TAC | 5.4 | 8 | 6.6 | 6 | 5.0 | 9 | 5.4 | 7 | 5.7 | 6 | 4.6 | 8 | 4.8 | 11 | 4.7 | 9 | 5.3 | 8 | 4.6 | 9 | 4.0 | 11 | 4.5 | 10 | 4.3 | 8 | |
| AAG | CTT | 3.0 | 13 | 4.1 | 11 | 2.9 | 16 | 3.4 | 14 | 3.5 | 13 | 1.4 | 27 | 3.2 | 18 | 3.3 | 12 | 3.0 | 14 | 3.2 | 13 | 3.6 | 12 | 3.6 | 13 | 2.9 | 16 | |
| ACA | TGT | 1.1 | 26 | 1.4 | 20 | 1.1 | 26 | 0.3 | 30 | 0.2 | 30 | 4.3 | 10 | 2.9 | 19 | 1.2 | 27 | 0.9 | 26 | 1.3 | 28 | 1.4 | 28 | 1.5 | 26 | 1.5 | 29 | |
| ACG | CGT | 1.6 | 22 | 0.3 | 28 | 1.8 | 21 | 0.6 | 26 | 0.6 | 25 | 1.9 | 24 | 5.0 | 4 | 1.8 | 22 | 1.4 | 23 | 1.6 | 25 | 3.2 | 16 | 1.6 | 22 | 2.1 | 24 | |
| ACT | AGT | 2.9 | 15 | 1.3 | 22 | 3.3 | 13 | 3.0 | 15 | 2.5 | 15 | 2.1 | 22 | 4.9 | 8 | 3.6 | 11 | 3.1 | 13 | 3.7 | 11 | 4.3 | 7 | 4.1 | 11 | 3.4 | 14 | |
| AGC | GCT | 2.9 | 14 | 3.4 | 14 | 3.1 | 15 | 3.4 | 13 | 3.1 | 14 | 3.9 | 12 | 4.9 | 6 | 4.0 | 10 | 3.6 | 11 | 4.2 | 10 | 4.3 | 9 | 4.6 | 9 | 3.5 | 13 | |
| AGG | CCT | 2.3 | 19 | 1.5 | 19 | 2.5 | 19 | 2.4 | 16 | 2.0 | 17 | 2.2 | 21 | 4.8 | 13 | 2.7 | 17 | 2.5 | 17 | 2.6 | 17 | 3.5 | 13 | 2.7 | 17 | 2.4 | 22 | |
| ATA | TAT | 1.6 | 21 | 4.1 | 10 | 1.6 | 24 | 0.8 | 23 | 0.9 | 23 | 4.2 | 11 | 2.7 | 20 | 2.3 | 19 | 1.7 | 20 | 2.9 | 16 | 2.8 | 21 | 2.7 | 16 | 2.9 | 17 | |
| ATG | CAT | 3.1 | 12 | 2.5 | 16 | 3.2 | 14 | 1.3 | 20 | 1.4 | 19 | 1.8 | 26 | 4.8 | 10 | 2.5 | 18 | 2.6 | 15 | 2.3 | 19 | 3.1 | 17 | 1.8 | 20 | 2.7 | 20 | |
| CCA | TGG | 1.6 | 23 | 1.3 | 21 | 1.8 | 20 | 1.1 | 22 | 0.8 | 24 | 2.8 | 16 | 4.5 | 15 | 2.0 | 21 | 1.6 | 22 | 2.3 | 20 | 2.6 | 22 | 1.8 | 19 | 2.8 | 19 | |
| CCG | CGG | 0.6 | 28 | 0.1 | 29 | 0.7 | 28 | 0.8 | 24 | 1.0 | 22 | 0.8 | 29 | 0.6 | 25 | 1.5 | 26 | 0.8 | 28 | 1.4 | 27 | 2.8 | 20 | 1.5 | 25 | 2.3 | 23 | |
| GCG | CGC | 2.7 | 17 | 1.7 | 18 | 3.4 | 11 | 3.5 | 12 | 3.9 | 12 | 2.0 | 23 | 4.2 | 17 | 2.9 | 16 | 2.4 | 18 | 3.2 | 15 | 3.4 | 14 | 3.0 | 14 | 3.6 | 12 | |
| GTG | CAC | 3.3 | 11 | 4.7 | 9 | 3.4 | 12 | 3.8 | 11 | 4.5 | 11 | 1.8 | 25 | 0.3 | 28 | 3.3 | 13 | 3.5 | 12 | 3.2 | 14 | 3.2 | 15 | 2.9 | 15 | 3.7 | 10 | |
| TAG | CTA | 1.7 | 20 | 2.1 | 17 | 1.6 | 22 | 1.6 | 19 | 1.8 | 18 | 3.0 | 15 | 0.3 | 29 | 1.7 | 24 | 1.6 | 21 | 2.1 | 22 | 1.8 | 26 | 1.6 | 23 | 2.0 | 25 | |
| TCA | TGA | 0.4 | 29 | 0.8 | 25 | 0.6 | 29 | 0.6 | 25 | 0.4 | 28 | 4.8 | 7 | 0.7 | 24 | 0.7 | 30 | 0.6 | 30 | 1.0 | 29 | 0.9 | 30 | 0.8 | 30 | 0.9 | 30 | |
| TCC | GGA | 1.5 | 24 | 1.0 | 24 | 1.6 | 23 | 0.6 | 27 | 0.6 | 26 | 5.0 | 6 | 4.8 | 12 | 1.7 | 23 | 1.1 | 25 | 1.9 | 23 | 2.4 | 23 | 2.0 | 18 | 2.7 | 21 | |
| TCG | CGA | 0.2 | 30 | 0.1 | 30 | 0.4 | 30 | 0.4 | 29 | 0.3 | 29 | 2.6 | 17 | 4.3 | 16 | 1.0 | 29 | 0.7 | 29 | 0.8 | 30 | 1.7 | 27 | 1.0 | 28 | 1.7 | 28 | |
| TCT | AGA | 1.2 | 25 | 0.6 | 27 | 1.5 | 25 | 1.6 | 18 | 1.3 | 21 | 2.4 | 19 | 2.1 | 22 | 1.6 | 25 | 1.4 | 24 | 1.8 | 24 | 2.0 | 25 | 1.7 | 21 | 1.7 | 27 | |
| TGC | GCA | 2.5 | 18 | 3.0 | 15 | 2.8 | 18 | 2.4 | 17 | 2.0 | 16 | 5.2 | 5 | 4.9 | 5 | 3.0 | 14 | 2.6 | 16 | 3.4 | 12 | 2.9 | 18 | 3.7 | 12 | 3.2 | 15 | |
| TTA | TAA | 0.9 | 27 | 0.6 | 26 | 1.0 | 27 | 0.5 | 28 | 0.4 | 27 | 2.3 | 20 | 1.6 | 23 | 1.1 | 28 | 0.9 | 27 | 1.5 | 26 | 1.4 | 29 | 1.0 | 29 | 1.9 | 26 | |
| TTG | CAA | 2.8 | 16 | 1.2 | 23 | 2.9 | 17 | 1.2 | 21 | 1.4 | 20 | 1.2 | 28 | 2.3 | 21 | 2.1 | 20 | 2.2 | 19 | 2.2 | 21 | 2.3 | 24 | 1.4 | 27 | 3.6 | 11 | |
The occurrence number of a CP trinucleotide set in a gene kingdom is equal to
| (8) |
where is defined in Equation (7).
In order to normalize the numbers which depend on the numbers of genes in a kingdom , we simply define the occurrence probability of a CP trinucleotide set in a gene kingdom as follows
| (9) |
where is defined in Equation (8).
The parameter gives the rank of the values among the 30 CP trinucleotide sets , the 1st rank being associated to the highest value of and the 30th rank, to the lowest value of .
2.6. A Statistical Test to Evaluate the Significance of the Obtained Ranks
In order to evaluate the statistical significance of the ranks of the probabilities (Equation (9)) of the 30 CP trinucleotide sets in a given kingdom , we derive confidence intervals for . If the confidence interval for two probabilities do not overlap, then their associated ranks are assumed to be valid (in the population). The confidence interval for two probabilities is evaluated by using the classical 2-sample z-test which is briefly recalled here.
Let and be the populations associated to the CP trinucleotide sets and of probabilities and , respectively. The probabilities and of and are observed in a given gene kingdom (sample) of size (defined from Equation (8)). The tests carried out in Section 3 are applied on large samples (the size of the smallest sample analysed being with the archaea ). Thus, the assumptions of normality for the variables and of the homogeneity for the variances in the two populations are not needed. The equality is tested against the alternative if they are not equal. Under and with large samples (), (always verified in the tests carried out in Section 3), and and are independent events (realistic hypothesis with kingdoms of large sizes), then
The z-value and the p-value are given for each statistical test carried out in Section 3.
2.7. Explained Example of the Statistical Approach Developed
As an example, we explain the definition of the occurrence probability (Equation (9)) which takes the value of 6.1% (see Table 2) with the CP trinucleotide set with in frame 0, in frame 1 and in frame 2 in the gene kingdom of bacteria (Table 1).
The occurrence frequencies of the 64 trinucleotides are computed in the three frames of each gene belonging to . Then, the preferential frame of each trinucleotide for each gene in is determined according to Equation (5). For example, with the trinucleotide in a gene of , if the frequency of AAC in frame (reading frame) is greater than the two frequencies and of AAC in frames and , i.e., , then the preferential frame of AAC in is 0, i.e., .
The indicator function of each trinucleotide for each gene in is obtained from Equation (6). With the previous example of AAC in the gene of , the indicator function is equal to for the frame and for the frames and .
The number of preferential frames of each trinucleotide for each frame in is computed according to Equation (7). With the previous example of AAC in , the following numbers are obtained: for the frame , for the frame , and for the frame . Thus, the preferential frame of AAC in is 0.
The occurrence number of the 30 CP trinucleotide sets in is determined according to Equation (8). With in , the following numbers are obtained: for the frame , and for the frame , and and for the frame . Then, the occurrence number of in is equal to .
Finally, the occurrence probability of the 30 CP trinucleotide sets in is deduced from Equation (9). With in , the occurrence probability of in is equal to .
3. Results
3.1. Maximal Self-Complementary Circular Code in Genes
This new statistical approach will show that the same set of 20 trinucleotides among sets occurs preferentially in genes (reading frame) of bacteria , archaea , plasmids , eukaryotes , and viruses . This set is the maximal self-complementary circular code defined in Equation (1).
3.1.1. Circular Code in Genes of Bacteria
In the genes of bacteria , the 10 CP trinucleotide sets have occurrence probabilities (Equation (9)) with the 10 highest ranks among 30 (Table 2), i.e., , and leading to the 20 trinucleotides of in frame 0, 20 trinucleotides of in frame 1, and 20 trinucleotides of in frame 2. The highest rank with is related to the complementary pair . The 10th rank with is very significantly greater than the 11th rank with (, , ). The 20 trinucleotides of the circular code are identified in the genes of bacteria:
| (10) |
The same result is obtained at the gene level and the gene population level [1].
3.1.2. Circular Code in Genes of Archaea
In the genes of archaea , the eight CP trinucleotide sets (except and ) have occurrence probabilities with the eight highest ranks among 30 (Table 2). The highest rank with is also related to the complementary pair . The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . As the CP set has a rank with greater than with and with , the two complementary trinucleotides occur preferentially in compared to () and (), however the statistical significance between the ranks and is not confirmed due to the lack of archaeal gene data (see Section 2.2) (, , ). Thus, a subset of of 18 trinucleotides (a non-maximal self-complementary circular code) is identified in the genes of archaea:
| (11) |
Note that the code () is the variant code observed in Deinococcus [1]. The circular code retrieved in the genes of archaea is a new result which was not found in a study of variant codes in archaeal genomes [15].
3.1.3. Circular Code in Genes of Plasmids
In the genes of plasmids , the 10 CP trinucleotide sets have occurrence probabilities with the 10 highest ranks among 30 (Table 2). The highest rank with is again related to the complementary pair . The 10th rank with is very significantly greater than the 11th rank with (, , ). The 20 trinucleotides of the circular code are identified in the genes of plasmids:
| (12) |
The same result is obtained at the gene level and the gene population level [1].
3.1.4. Circular Code in Genes of Eukaryotes
In the genes of eukaryotes , the 10 CP trinucleotide sets have occurrence probabilities with the 10 highest ranks among 30 (Table 2). The highest rank with is again related to the complementary pair . The 10th rank with is significantly greater than the 11th rank with (, , ). The 20 trinucleotides of the circular code are identified in the genes of eukaryotes:
| (13) |
The same result is obtained at the gene level and the gene population level [1].
The subset of of 16 trinucleotides in the genes of Homo sapiens identified at the gene level is also identical to the subset found at the gene population level [1].
3.1.5. Circular Code in Genes of Eukaryotic Chromosomes
The statistical analysis in Section 3.1.4 takes the eukaryotic genome as the genetic information unit. Indeed, Equation (7) with is achieved with eukaryotic genomes (see Table 1). We complete this classical approach by choosing the eukaryotic chromosome as the genetic information unit. Thus, Equation (7) with is performed with eukaryotic chromosomes of genomes (see Table 1).
In the genes of eukaryotic chromosomes , the 10 CP trinucleotide sets have occurrence probabilities with the 10 highest ranks among 30 (Table 2). The highest rank with is again related to the complementary pair . The 10th rank with is very significantly greater than the 11th rank with (, , ). The 20 trinucleotides of the circular code are identified in the genes of eukaryotic chromosomes:
| (14) |
It is a new result which completes the statistical analysis of genes in eukaryotic genomes (Section 3.1.4).
3.1.6. Non-Maximal Circular Code in Genes of Eukaryotic Organelles
The genes of eukaryotic organelles, i.e., mitochondria and chloroplasts, are investigated with this statistical approach. It should also be stressed that the available data have an order of magnitude very significantly lower than the other gene kingdoms studied (less than 1 million trinucleotides for each class of organelles, see Table 1). However, we can already observe some statistical trends with the trinucleotides in the preferential frame.
Non-Maximal Circular Code in Genes of Mitochondria
Surprisingly, in the genes of mitochondria , the four CP trinucleotide sets have occurrence probabilities with the four highest ranks among 30 (Table 2). The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . The CP set with determines that the two complementary trinucleotides () do not occur preferentially in . The CP set with implies that the two complementary trinucleotides () do not occur preferentially in . The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . Thus, a subset of of 12 trinucleotides (a non-maximal self-complementary circular code) is identified in the genes of mitochondria :
| (15) |
This subset is very close to the subset of of 13 trinucleotides previously identified by inspection in mitochondrial genes [21], as has 10 trinucleotides in common.
Non-Maximal Circular Code in Genes of Chloroplasts
In the genes of chloroplasts , the highest occurrences of CP trinucleotide sets again belong to the circular code . The three CP trinucleotide sets have occurrence probabilities with the three highest ranks among 30 (Table 2). The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . The CP set with states that the two complementary trinucleotides () do not occur preferentially in . The CP set with implies that the two complementary trinucleotides () do not occur preferentially in . The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . The CP set with implies that the two complementary trinucleotides () do not occur preferentially in . Thus, a subset of of 10 trinucleotides (a non-maximal self-complementary circular code) is identified in the genes of chloroplasts :
| (16) |
3.1.7. Circular Code in Genes of Viruses
In the genes of viruses , the nine CP trinucleotide sets (except ) have occurrence probabilities with the nine highest ranks among 30 (Table 2). The highest rank with is again related to the complementary pair . The CP set with explains that the two complementary trinucleotides () do not occur preferentially in . Thus, a subset of of 18 trinucleotides (a non-maximal self-complementary circular code) is identified in the genes of viruses:
| (17) |
The statistical method of viral genes at the gene population level [1] could not decide between the two codes and . The statistical analysis at the gene level confirms the code of 18 trinucleotides in the genes of viruses.
3.2. Circular Code Found in DNA and RNA Genomes and in Double-Stranded and Single-Stranded Genomes
The self-complementary property of the circular code has been related since 1996 to the complementary property of the DNA double helix. In order to deepen this idea, we searched with this statistical approach the circular code in five important sub-classes of viral genes using either DNA genome or RNA genome, and either double-stranded genome or single-stranded genome, i.e., in the genes of double-stranded DNA viruses , double-stranded RNA viruses , single-stranded DNA viruses , single-stranded RNA viruses , and retro-transcribing viruses .
In the genes of double-stranded DNA viruses , the 10 CP trinucleotide sets have occurrence probabilities with the 10 highest ranks among 30 (Table 2). Thus, the circular code is found in :
| (18) |
In the genes of double-stranded RNA viruses , single-stranded RNA viruses , and retro-transcribing viruses , respectively, the nine CP trinucleotide sets (except ) have occurrence probabilities , , and , respectively, with the nine highest ranks , , and , respectively, among 30 (Table 2). Note that the ranks , , and for a given CP trinucleotide set are not identical (Table 2). Thus, by using the reasoning mentioned previously ( with for in , , and ), a subset of of 18 trinucleotides is observed in , , and :
| (19) |
| (20) |
| (21) |
In the genes of single-stranded DNA viruses , the eight CP trinucleotide sets (except and ) have occurrence probabilities with the eight highest ranks among 30 (Table 2). Thus, by using the reasoning as previously mentioned ( with and with ), a subset of of 16 trinucleotides is observed in :
| (22) |
All these results show that the circular code is found almost perfectly in DNA genomes, RNA genomes, double-stranded genomes, and single-stranded genomes. The very few exceptions, either the two trinucleotides or the four trinucleotides for one case, are related to the CP set or the two CP sets having the lowest occurrence among the 10 CP sets .
4. Conclusions
The “universal” occurrence in genes of a same set of 20 trinucleotides, which has in addition the mathematical property to be a circular code, must be confirmed by several statistical approaches and various gene data analyses at different levels: kingdom, taxonomic group, genome, and gene. All the previous approaches have studied and identified the circular code at the gene population level (kingdom, taxonomic group, and genome) [1,2,15,16,17,21]. The statistical approach at the gene level developed here, for the first time since 1996, analyses the preferential occurrence of trinucleotides among the three frames of each gene. This new methodology allows all genes, i.e., of large and small lengths, to be considered with the same weight. As a consequence, the concept of circular code, in particular the reading frame retrieval, is directly associated to each gene. Thus, motifs from the circular code at different locations in a gene may assist the ribosome to maintain and synchronize the reading frame. The number, the cardinality, and the length of motifs in genes may be associated to the length, the function, and the ancestry of genes. This research work is currently under investigation.
At the gene level, the circular code is strengthened in the genes of bacteria, eukaryotes, plasmids, and viruses, and is now also identified in the genes of archaea. In addition to eukaryotic genomes, it is also found in the genes of eukaryotic chromosomes. The genes of mitochondria and chloroplasts contain a subset of the circular code . It should be stressed that some mitochondrial and chloroplast genes lack the stop codon and are excluded from this data acquisition. Such a statistical bias may prevent a proper detection of preferential frames for some trinucleotides in the genes of eukaryotic organelles. The circular code is searched in the large class of self-complementary trinucleotide codes which contains in particular the 216 maximal self-complementary circular codes. Thus, for a basic order of magnitude, the probability to retrieve the same circular code in four independent gene kingdoms (bacteria , plasmids , eukaryotes , double-stranded DNA viruses ) is equal to .
In the genes of the bacterial, eukaryotic, and plasmid kingdoms, 14 among the 47 studied gene taxonomic groups (about 30%) have variant codes [1], i.e., trinucleotide codes which differ from . Seven variant codes are identified. However, all have at least 16 trinucleotides of . Two variant codes (according to the notation in [1]) in cyanobacteria and plasmids of cyanobacteria, and in birds, are self-complementary, without permuted trinucleotides, but are non-circular. Five variant codes in Deinococcus, plasmids of chloroflexi and Deinococcus, mammals, and kinetoplasts, in elusimicrobia and apicomplexans, in fishes, in insects, and in basidiomycetes and plasmids of spirochaetes, are self-complementary circular. Thus, two variant codes and are not circular and do not belong to the set of the 216 maximal self-complementary circular codes [2] having the strong mathematical structure of the dihedral group [20]. The reason could be related to the gene data or to a biological property which remains to be identified. All these variant codes in the genes are identified at the taxonomic group level. However, as the circular code is now also identified at the gene level, variant codes may also be associated with genes belonging to the same genome but with different protein coding functions. This interesting and open problem should be investigated in the future.
A probability measure of the reading frame retrieval () of each trinucleotide of has been introduced in [22] and [23] (Section 2.2 and 1st row of Table 1). The probability of the circular code , i.e., the average probability of the 20 trinucleotides of , is equal to (Result 5 in [22]; 1st row of Table 1 in [23]). This measure can be applied to the non-maximal self-complementary circular codes, precisely to the excluded trinucleotides of archaea (Equation (11)), of mitochondria (Equation (15)), of chloroplasts (Equation (16)), of viruses (Equation (17)), and of single-stranded DNA viruses (Equation (22)). The computation leads to , , , , and . Archaeal genes miss two trinucleotides of which have the lowest values. In contrast, mitochondrial, chloroplast, and viral genes miss trinucleotides of with high values. Thus, the genes in reduced genomes are more flexible in translation, allowing overlap coding by frameshifting in agreement with [24] (and the cited references). However, it should be stressed that this result may vary with the increase of gene data of eukaryotic organelles in the future. The circular code (20 trinucleotides) with the functions of reading frame retrieval and maintenance in regular RNA transcription, may also have, through its bijective transformation codes, the same functions in nucleotide exchanging RNA transcription in mitochondrial genes [23]. Indeed, as the mitochondrial gamma polymerase has bacterial origins (e.g., [25]), mitochondrial polymerization and its associated bijective transformations might use the circular code . However at the translational level, the ribosome might follow the non-maximal self-complementary circular code observed in mitochondrial genes (Equation (15)). A similar explanation could be applied to the chloroplast genes which have also bacterial origins (cyanobacteria).
By a study of viral genes, the circular code is found in DNA genomes, RNA genomes, double-stranded genomes, and single-stranded genomes. Thus, the reading frame retrieval property of could operate for translating DNA and RNA genes, in particular for the “primitive” RNA genes. The property of could be involved for translating the two shifted frames in DNA and RNA genes, in particular for optimizing the genomes of small sizes. The complementarity property of is naturally associated to the double-stranded DNA and RNA genomes. It could also be used to pair single-stranded DNA genomes between them and single-stranded RNA genomes between them. Thus, the and complementary properties of could be involved for translating the three frames (reading frame and its two shifted frames) in one strand and the three frames in the complementary strand of DNA and RNA genes.
In summary, this new statistical approach at the gene level which is applied to massive gene data identifies the maximal self-complementary trinucleotide circular code in the genes of bacteria, archaea, eukaryotes, plasmids, and viruses, which may be involved in translation coding [3].
Acknowledgments
I thank the three reviewers for their advice, and Denise Besch, Svetlana Gorchkova, Elisabeth Michel, Professor Jacques Streith, and Jean-Marc Vassards for their support.
Conflicts of Interest
The author declares no conflict of interest.
References
- 1.Michel C.J. The maximal C3 self-complementary trinucleotide circular code X in genes of bacteria, eukaryotes, plasmids and viruses. J. Theor. Biol. 2015;380:156–177. doi: 10.1016/j.jtbi.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Arquès D.G., Michel C.J. A complementary circular code in the protein coding genes. J. Theor. Biol. 1996;182:45–58. doi: 10.1006/jtbi.1996.0142. [DOI] [PubMed] [Google Scholar]
- 3.Michel C.J. Circular code motifs in transfer and 16S ribosomal RNAs: A possible translation code in genes. Comput. Biol. Chem. 2012;37:24–37. doi: 10.1016/j.compbiolchem.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 4.El Soufi K., Michel C.J. Circular code motifs in genomes of eukaryotes. J. Theor. Biol. 2016;408:198–212. doi: 10.1016/j.jtbi.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Michel C.J. Circular code motifs in transfer RNAs. Comput. Biol. Chem. 2013;45:17–29. doi: 10.1016/j.compbiolchem.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 6.El Soufi K., Michel C.J. Circular code motifs in the ribosome decoding center. Comput. Biol. Chem. 2014;52:9–17. doi: 10.1016/j.compbiolchem.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 7.El Soufi K., Michel C.J. Circular code motifs near the ribosome decoding center. Comput. Biol. Chem. 2015;59:158–176. doi: 10.1016/j.compbiolchem.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 8.El Soufi K., Michel C.J. Unitary circular code motifs in genomes of eukaryotes. Biosystems. 2017 doi: 10.1016/j.biosystems.2017.02.001. in press. [DOI] [PubMed] [Google Scholar]
- 9.Canapa A., Cerioni P.N., Barucca M., Olmo E., Caputo V. A centromeric satellite DNA may be involved in heterochromatin compactness in gobiid fishes. Chromosome Res. 2002;10:297–304. doi: 10.1023/A:1016519708187. [DOI] [PubMed] [Google Scholar]
- 10.Gemayel R., Vinces M.D., Legendre M., Verstrepen K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 11.Pirillo G. A characterization for a set of trinucleotides to be a circular code. In: Pellegrini C., Cerrai P., Freguglia P., Benci V., Israel G., editors. Determinism, Holism, and Complexity. Kluwer Academic Publisher; New York, NY, USA: 2003. [Google Scholar]
- 12.Michel C.J., Pirillo G. Identification of all trinucleotide circular codes. J. Theor. Biol. 2010;34:122–125. doi: 10.1016/j.compbiolchem.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Fimmel E., Michel C.J., Strüngmann L. n-Nucleotide circular codes in graph theory. Philos. Trans. R. Soc. A. 2016;374:20150058. doi: 10.1098/rsta.2015.0058. [DOI] [PubMed] [Google Scholar]
- 14.Bussoli L., Michel C.J., Pirillo G. On conjugation partitions of sets of trinucleotides. Appl. Math. 2012;3:107–112. doi: 10.4236/am.2012.31017. [DOI] [Google Scholar]
- 15.Frey G., Michel C.J. Circular codes in archaeal genomes. J. Theor. Biol. 2003;223:413–431. doi: 10.1016/S0022-5193(03)00119-X. [DOI] [PubMed] [Google Scholar]
- 16.Frey G., Michel C.J. Identification of circular codes in bacterial genomes and their use in a factorization method for retrieving the reading frames of genes. Comput. Biol. Chem. 2006;30:87–101. doi: 10.1016/j.compbiolchem.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Arquès D.G., Michel C.J. A code in the protein coding genes. Biosystems. 1997;44:107–134. doi: 10.1016/S0303-2647(97)00049-X. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez D.L., Giannerini S., Rosa R. Circular codes revisited: a statistical approach. J. Theor. Biol. 2011;275:21–28. doi: 10.1016/j.jtbi.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Michel C.J., Pirillo G., Pirillo M.A. A relation between trinucleotide comma-free codes and trinucleotide circular codes. Theor. Comput. Sci. 2008;401:17–26. doi: 10.1016/j.tcs.2008.02.049. [DOI] [Google Scholar]
- 20.Fimmel E., Giannerini S., Gonzalez D.L., Strüngmann L. Circular codes, symmetries and transformations. J. Math. Biol. 2015;70:1623–1644. doi: 10.1007/s00285-014-0806-7. [DOI] [PubMed] [Google Scholar]
- 21.Arquès D.G., Michel C.J. A circular code in the protein coding genes of mitochondria. J. Theor. Biol. 1997;189:273–290. doi: 10.1006/jtbi.1997.0513. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed A., Frey G., Michel C.J. Essential molecular functions associated with circular code evolution. J. Theor. Biol. 2010;264:613–622. doi: 10.1016/j.jtbi.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Michel C.J., Seligmann H. Bijective transformation circular codes and nucleotide exchanging RNA transcription. Biosystems. 2014;118:39–50. doi: 10.1016/j.biosystems.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Seligmann H. Chimeric mitochondrial peptides from contiguous regular and swinger RNA. Comput. Struct. Biotechnol. J. 2016;14:283–297. doi: 10.1016/j.csbj.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf Y.I., Koonin E.V. Origin of an animal mitochondrial DNA polymerase subunit via lineage-specific acquisition of a glycyl-tRNA synthetase from bacteria of the Thermus-Deinococcus group. Trends Genet. 2001;17:431–433. doi: 10.1016/S0168-9525(01)02370-8. [DOI] [PubMed] [Google Scholar]