ABSTRACT
Long noncoding RNAs (lncRNAs) are important regulators of various biological processes, including spermatogenesis. Our previous studies have revealed the regulatory loop of mrhl RNA and Wnt signaling, where mrhl RNA negatively regulates Wnt signaling and gets downregulated upon Wnt signaling activation. This downregulation of mrhl RNA is important for the meiotic progression of spermatogonial cells. In our present study, we identified the transcription factor Sox8 as the regulatory link between mrhl RNA expression, Wnt signaling activation, and meiotic progression. In contrast to reports from other groups, we report the expression of Sox8 in germ cells and describe the molecular mechanism of Sox8 regulation by mrhl RNA during differentiation of spermatogonial cells. Binding of mrhl RNA to the Sox8 promoter is accompanied by the assembly of other regulatory factors involving Myc-Max-Mad transcription factors, corepressor Sin3a, and coactivator Pcaf. In the context of Wnt signaling, Sox8 directly regulates the expression of premeiotic and meiotic markers. Prolonged Wnt signaling activation in spermatogonial cells leads to changes in global chromatin architecture and a decrease in levels of stem cell markers.
KEYWORDS: Myc-Mad-Max, Sin3A, Sox8, Wnt signaling, mrhl RNA
INTRODUCTION
The complexity of the eukaryotic genome has been attributed to its multifaceted regulatory networks. Over recent years, significant advancements in high-throughput analyses of the transcriptome have demonstrated the abundance of transcripts that do not code for proteins (noncoding RNAs [ncRNAs]). These ncRNAs are broadly classified as small and long noncoding RNAs. The small ncRNAs, such as microRNA (miRNA) and small interfering RNA (siRNA), play important roles in transcriptional and posttranscriptional gene regulation, while Piwi-associated RNAs (piRNAs) are involved in transposon regulation (1, 2). Another class is that of the long noncoding RNAs (lncRNAs), which are of various sizes between 200 bp and several kilobases (3). The role of lncRNAs in a plethora of functions, for example, dosage compensation (Xist and roX) (4, 5), genomic imprinting (Air and Kcnq1ot1) (6, 7), pluripotency (Evx1as and Hoxb5/6as) (8), cell differentiation and development (Fendrr, Bvht, Miat, Hotair, etc.) (9), nuclear architecture (NEAT1) (10), chromosome segregation (Concr) (11), immune response (lincRNA-EPS, EGOT) (12, 13), etc., have been characterized. lncRNAs bring about such copious functions by employment of diverse mechanisms such as translational inhibition (lincRNA-p21) (14), mRNA degradation (1/2-sbs RNAs) (15), RNA decoys (Gas5) (16), facilitation of recruitment of chromatin modifiers (HOTTIP, HOTAIR, and XIST) (17, 18), regulation of protein activity (Evi2 and Lethe) (19, 20), regulation of the availability of miRNAs by sponging mechanisms (linc-MD1) (21), modulation of splicing (MALAT1) (22), and alteration of protein localization (NRON) (23). In addition to this, various lncRNAs have also been shown to regulate stress responses and disease pathobiology, especially cancer metastasis, emphasizing their importance in cellular homeostasis (24).
Mrhl, one such lncRNA, discovered in our laboratory, is encoded in a meiotic recombination hot spot locus of mouse chromosome 8, within the 15th intron of the phkb gene (25). The primary transcript is 2.4 kb in length, RNA polymerase II (Pol II) transcribed, nucleus restricted, polyadenylated, and unspliced (26). Ddx5/p68 RNA helicase is an interacting partner of mrhl RNA in the nucleus and is required for the regulatory function of mrhl RNA as a negative regulator of Wnt signaling (27). We have also mapped the chromatin occupancy of mrhl RNA and shown that mrhl RNA regulates the expression of several genes, many of which are known to play key roles in spermatogenesis as well as Wnt signaling (28). One such gene is Sox8, which encodes a developmentally important transcription factor, and mrhl RNA interacts with the promoter of Sox8. We have previously shown that Wnt3a ligand triggers Wnt signaling in mouse spermatogonial Gc1-Spg cells (derived from B-type spermatogonia) and at the same time downregulates mrhl RNA gene expression (27). In our most recent study, we have elucidated the molecular mechanism of mrhl RNA downregulation upon activation of Wnt signaling. Ctbp1 acts as a corepressor for the downregulation of mrhl RNA, and this Wnt-mediated downregulation of the RNA is a requisite for the expression of meiotic marker genes and the meiotic commitment of spermatogonial cells (29).
During the course of meiotic progression, mrhl RNA is highly downregulated in spermatocytes compared to spermatogonial cells in the mouse testes. We have observed that overexpression of mrhl RNA in trans abrogates the upregulation of various premeiotic and meiotic marker genes (29), which are vital for the meiotic commitment and progression of spermatogonia. Sox8 has an established role in Sertoli cell function, and it has been shown through knockout studies that the absence of Sox8 leads to defects in sex cord formation (30). In the light of this connection between Wnt signaling, mrhl RNA, and Sox8, we were interested in delineating the detailed molecular mechanism by which mrhl RNA regulates Sox8 during spermatogonial cell differentiation. Here we present a detailed analysis of the promoter of Sox8 and have dissected its regulation by mrhl RNA through interaction with the Myc-Max-Mad transcription factors. The Max-Mad repressor complex with Sin3a as corepressor and the Myc-Max activator complex including Pcaf as a coactivator bring about changes in chromatin to facilitate the transition from a transcriptionally repressed state to an activated state of the Sox8 gene, respectively. Interestingly, the binding site of the Myc-Max-Mad proteins overlaps with the binding region of mrhl RNA on the Sox8 promoter. We also elucidated the role of Sox8 in meiotic progression of spermatogonia and implicate Sox8 in the direct regulation of important premeiotic and meiotic markers in the context of Wnt signaling. We also observed a change in the global chromatin architecture of Gc1-Spg cells upon prolonged Wnt ligand treatment which is corroborated by a gradual decrease in the levels of stem cell markers and a concomitant increase in the expression of differentiation marker genes.
RESULTS
Expression of Sox8 in mouse testicular germ cells.
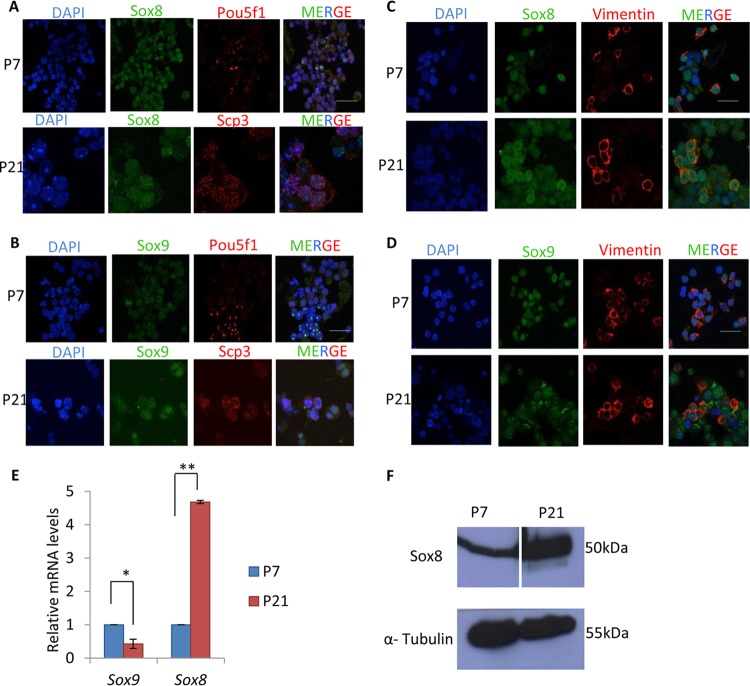
Sox8 is an important transcription factor during spermatogenesis due to its crucial function in Sertoli cells (30, 31). However, there are no reports of Sox8 expression in testicular germ cells. We first checked for its expression by immunofluorescence using testis cellular spread from 7-day-old (P7) and 21-day-old (P21) mice. Different testicular or germ cell types were distinguished based on cell type-specific marker proteins such as Pou5f1 (Oct-4) for spermatogonia, Scp3 for spermatocytes, and vimentin for Sertoli cells. The P7 testes predominantly contain spermatogonia and represent the Wnt-repressed state, while P21 testes which predominantly contain spermatocytes and represent the Wnt-activated state (32). We have demonstrated the suitable use of P7 and P21 mouse testes as an in vivo correlate of Wnt signaling in our previous studies (28, 29). In addition to expression of Sox8, we also looked for that of Sox9 since both belong to group E of Sox proteins.
As can be seen, both Sox8 and Sox9 are expressed in spermatogonial cells as well as in spermatocytes (Fig. 1A and B). We also confirmed the expression pattern in Sertoli cells. Both Sox8 and Sox9 are expressed in Sertoli cells at both stages of testicular development (Fig. 1C and D). Expression of Sox8 was also assessed at the transcriptional level by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 1E) and at the protein level by Western blotting (Fig. 1F), which showed upregulation in the P21 testes compared to the P7 testes. Therefore, these observations showed ubiquitous expression of Sox8 in mouse testicular germ cells.
FIG 1.
Expression of Sox8 and Sox9 in mouse testes. The Sox8 and α-tubulin Western blots are from different gels. For the Sox8 blot, the space separating P7 and P21 is due to a protein marker lane that was excised. (A and B) Expression of Sox8 (A) and Sox9 (B) in 7-day-old mouse testis with Pou5f1 as a marker for spermatogonial cells and in 21-day-old mouse testis with Scp3 as a marker for spermatocytes. (C and D) Expression of Sox8 (C) and Sox9 (D) in 7-day-old and 21-day-old mouse testis with vimentin as a marker for Sertoli cells. (E) Expression analysis of Sox8 and Sox9 in P7 and P21 mouse testes. (F) Western blot showing Sox8 expression in P7 and P21 mouse testes. Data in panel E are plotted as means ± standard deviations (SD) (n = 4). **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student's t test). Bars, 25 μm.
Sox8 expression is upregulated in germ cells and Sertoli cells upon Wnt signaling activation.
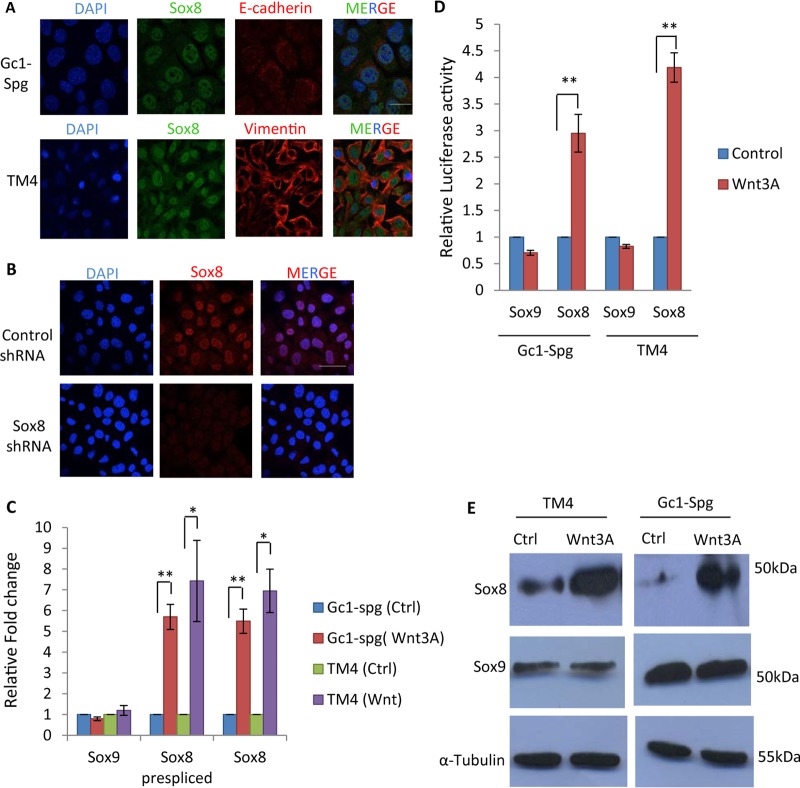
It is known that Wnt signaling can downregulate mrhl RNA expression and that mrhl RNA binds to the promoter of Sox8 and plays a role in the expression of Sox8 (28). Sox8 expression was also revalidated in the B-type spermatogonial Gc1-Spg cell line and the Sertoli TM4 cell line (Fig. 2A). The specificity of the Sox8 antibody used in the present study was confirmed by immunofluorescence upon Sox8 silencing (Fig. 2B). We examined the expression of Sox8 and Sox9 (mRNA and protein levels) upon Wnt signaling activation in the Gc1-Spg and TM4 cell lines. In addition to scoring for mature transcript, we also scored for the prespliced primary transcript of Sox8 by using the intron-exon junction primers (Table 1) to unequivocally establish that the upregulation is due to transcriptional activation and not the stability of mature mRNA. Wnt signaling activation leads to upregulation of Sox8 expression in the spermatogonial Gc1-Spg cell line as well as in the Sertoli TM4 cell line, while expression of Sox9 was not significantly perturbed (Fig. 2C). The transcriptional upregulation of Sox8 is regulated at the level of the promoter as demonstrated by the increase in luciferase activity upon Wnt signaling activation in Gc1-Spg and TM4 cells (Fig. 2D). Even at the protein level, Sox8 expression was upregulated in both cell lines but Sox9 expression was not (Fig. 2E). Expression of Sox8 showed 5-fold to 6-fold upregulation upon Wnt3A activation in Gc1-Spg cells at both the RNA and protein levels.
FIG 2.
Upregulation of Sox8 upon Wnt3a conditioned medium treatment in Gc1-Spg and TM4 cells. (A) Expression and localization of Sox8 in Gc1-Spg and TM4 cell lines. (B) Immunofluorescence analysis in Gc1-Spg cells upon Sox8 silencing to show the specificity of the antibody used. (C) Expression analysis of Sox8 and Sox9 in Wnt3A conditioned medium-treated Gc1-Spg and TM4. Ctrl, control. (D) Luciferase assay for Sox8 and Sox9 promoter in control and Wnt3A conditioned medium-treated Gc1-Spg cells. (E) Western blot showing upregulation of Sox8 in Wnt3A conditioned medium-treated Gc1-Spg and TM4 cells. Data in panels C and D are plotted as means ± SD (n = 4). **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student t test). Bar (A and B), 25 μm.
TABLE 1.
List of primers used for RT-qPCR, ChIP-qPCR, and nucleosome mapping (NM1 to NM19)
| Gene or locus (purpose) | Forward primer | Reverse primer |
|---|---|---|
| Actba (RT-qPCR) | AGGTCATCACTATTGGCAACG | TACTCCTGCTTGCTGATCCAC |
| Sox9 (RT-qPCR) | TGCTGAACGAGAGCGAGAAGAGAC | GGACCCTGAGATTGCCCAGAGTG |
| Sox8 primary (RT-qPCR) | CTGTGTTTTCTGTAGCTTGCTG | CCTGGAGCCCACCTGTGTG |
| Sox8 (RT-qPCR) | GCTGTGGCGCTTGCTGAG | CTGTGTGGTGGTCACTGTG |
| Mrhl (RT-qPCR) | TGAGGACCATGGCTGGACTCT | AGATGCAGTTTCCAATGTCCAAAT |
| c-Myc (RT-qPCR) | CTAGTGCTGCATGAGGAGAC | CTCTTGGCAGCTGGATAGTC |
| Max (RT-qPCR) | GATAACGATGACATCGAGGTG | TTGCTCCAGAAGAGCATTCTG |
| Mad (RT-qPCR) | CTGTCCACCAAATAGACCAG | TGCATGCTGCCTCGCTCG |
| Stra-8 (RT-qPCR) | CGTGGCAAGTTTCCTGGACAAG | GGCTCTGGTTCCTGGTTTAATGG |
| Lhx8 (RT-qPCR) | GTCTGGAGATAGTTGGCTCGAGT | GGATGGTAGGCTTTGTAAACTAG |
| c-Kit (RT-qPCR) | CCCGACGCAACTTCCTTA | CGCTTCTGCCTGCTCTTC |
| Mtl-5 (RT-qPCR) | CGTCTGGGAGCTGCTAAA | GGAGGTCCTGAGAACTTGG |
| Hspa2 (RT-qPCR) | CTACGTGGCCTTCACTGACA | GGTCAGGATGGACACATCGA |
| Actb (ChIP-qPCR) | TCCACAAGGGCGGAGGCTAT | GGGTTTTATAGGACGCCACA |
| Ccnd1 (ChIP-qPCR) | CCATTCTCCCGGTTTAAGAACAG | AGCCTTCGTAGATATGCAAATCG |
| Cad (ChIP-qPCR) | CACACAAGACTTCCAGCTCAC | TGTAGTCAATAATACAAATTAGTAC |
| NM1 | GATCCCAGACCTGAAGGTTAG | GTGGTCTCTAATATCTCCATCTC |
| NM2 | GAGATGGAGATATTAGAGACCAC | ACCTGTTTCCCAAGCCCGTG |
| NM3 | CACGGGCTTGGGAAACAGGT | CTTGTCTGGGTGGCATAGAG |
| NM4 | CTCTATGCCACCCAGACAAG | CGAAACTCAAAGGAGGCGATG |
| NM5 | CATCGCCTCCTTTGAGTTTCG | GAGGCGAGAGCCAAGCCTG |
| NM6 | CAGGCTTGGCTCTCGCCTC | CCTAGAGAGAAGGCGGAGAG |
| NM7 | CTCTCCGCCTTCTCTCTAGG | CTGGGTGGTCTGGCGGAG |
| NM8 | CTCCGCCAGACCACCCAG | GTCCTTCCGGGTATGACCTG |
| NM9 | CAGGTCATACCCGGAAGGAC | CTGGCAGTGACTGCTGAGTC |
| NM10 | GACTCAGCAGTCACTGCCAG | GCTAGACAGAGGTGGGAGG |
| NM11 | CCTCCCACCTCTGTCTAGC | CCAGGTGACCCTGGTAACTC |
| NM12 | GAGTTACCAGGGTCACCTGG | GCAACCAAACCCGGGTTCTG |
| NM13 | CAGAACCCGGGTTTGGTTGC | AAGGTTCCTGAGTCCAACATC |
| NM14 | GATGTTGGACTCAGGAACCTT | GCAGCACGGCTTGACTTTCG |
| NM15 | CGAAAGTCAAGCCGTGCTGC | GGTCAGAGGGCTAAGGGTG |
| NM16 | CACCCTTAGCCCTCTGACC | TCCGTACCACGTGGGCCAG |
| NM17 | TGGCCCACGTGGTACGGAG | AATCCAAAGAAGCCTTGCGAC |
| NM18 | TCGCAAGGCTTCTTTGGATTC | TCCGGGTTGGCAGGTCCTC |
| NM19 | AGGACCTGCCAACCCGGAC | CAGTTCTGGGACCCGCAAC |
| Mutant Tcf4 binding site | GTACATCGCCTCCGCTGAGTTTCGCGGGG | CCCCGCGAAACTCAGCGGAGGCGATGTAC |
| Zfp113 (ChIP-qPCR) | ACCACAACTTCCACCTCACC | GGGTCTGTAACAGGGTCCAA |
| Pou5f1 (RT-qPCR) | GACACCTGGCTTCAGACTTC | TCAGGCTGCAAAGTCTCCAC |
| Sox2 (RT-qPCR) | AGTGGTACGTTAGGCGCTTC | GACCACGAAAACGGTCTTGC |
| Nanog (RT-qPCR) | TGATTCAGAAGGGCTCAGCAC | AAGGCTGCAGAAAGTCCTCC |
| Scp3 (RT-qPCR) | AAATCTGGGAAGCCACCTTTG | CAATTTTCTGGTTACTGGCTTTG |
| Dmc1 (RT-qPCR) | GCGGCTACTCAGGTGGAAAG | AGTCGTGACAACATCTGGGC |
| Spo11 (RT-qPCR) | TGCTGTGCCGACTAACATTC | CGTAGGGATCTGCATCGAC |
| Sox8 (bp −139) | CGAAAGTCAAGCCGTGCTGC | AATCCAAAGAAGCCTTGCGAC |
| Sox8 (bp −500 to −700) | CTCTCCGCCTTCTCTCTAGG | CTGGCAGTGACTGCTGAGTC |
| Sox8 (bp −800) | CACGGGCTTGGGAAACAGGT | GAGGCGAGAGCCAAGCCTG |
Beta-actin gene.
Role of mrhl RNA and the Tcf4 binding site present in the Sox8 promoter in Wnt-mediated upregulation of Sox8.
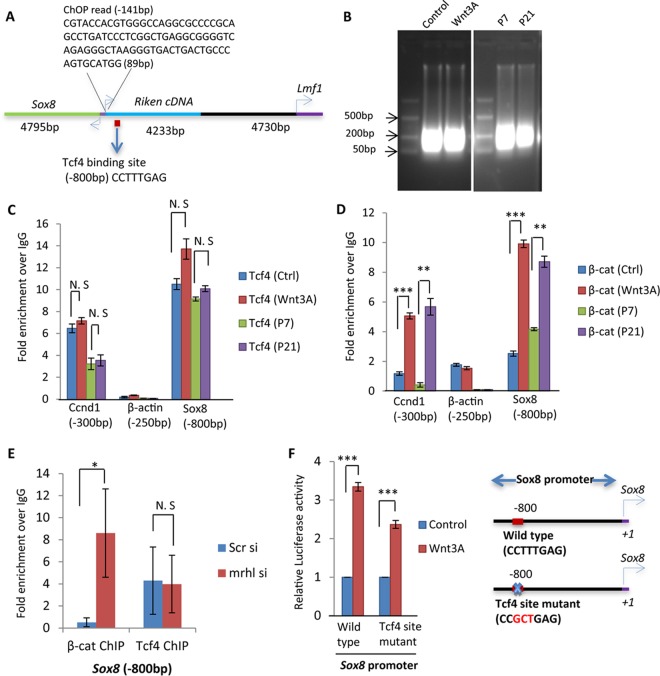
Sox8 has an interesting genomic architecture and is transcribed from a bidirectional promoter in mouse (Fig. 3A). The binding region of mrhl RNA is located at around 141 bp upstream of the transcriptional start site (TSS) and stretches to around 89 bases on the chromatin. Bioinformatic analysis using the MAPPER search engine for computational identification of putative transcription factor binding sites revealed a Tcf4 binding site (CTTTGA) at around 800 bp upstream of TSS in the Sox8 promoter. Tcf4 is a key transcription factor of Wnt signaling and brings about activation or repression of various genes by recruitment of β-catenin and associated coactivators and corepressors (33). We explored the possibility of Tcf4 and β-catenin being the regulators of Sox8 upon Wnt signaling activation. For this purpose, we examined the occupancy of Tcf4 and β-catenin on the Sox8 promoter by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR). For the ChIP experiments, the DNA was sonicated to be enriched in the size range of 100 to 300 bp (Fig. 3B). We found that Tcf4 constitutively occupies the site (Fig. 3C) whereas β-catenin is recruited only upon Wnt activation (Fig. 3D) in Gc1-Spg cells. The Ccnd1 gene promoter was used as a positive control, and the β-actin gene promoter was used as a negative control for both Tcf4 and β-catenin ChIP. The results were also validated in P7 and P21 mouse testicular cells. To determine whether Tcf4 and β-catenin binding to the Sox8 promoter is directly dependent on mrhl, we performed mrhl RNA silencing and checked the occupancy of Tcf4 and β-catenin on the Sox8 promoter (Fig. 3E). Occupancy of TCF4 was unchanged, while there was a significant increase in the occupancy of β-catenin at the Sox8 promoter upon silencing of mrhl RNA. Given that the site is occupied by β-catenin upon Wnt activation, the functionality of the site was checked next. We performed a luciferase reporter assay with the Sox8 promoter containing either the wild-type or mutated Tcf4 binding site, the latter of which was mutated from CTTTGA to CGCTGA, using site-directed mutagenesis. Even though there was a reduction in the Sox8 promoter function upon mutation (Fig. 3F), the reduction was not as drastic as would be expected if the Tcf4 binding site were the sole regulator of Sox8 gene expression.
FIG 3.
Analysis of the role of the Tcf4 binding site in the Sox8 promoter in Wnt-mediated upregulation of Sox8. (A) Genomic architecture of Sox8. The bidirectional promoter of Sox8, its neighboring genes, and locations of the Tcf4 binding site and of the mrhl RNA binding site are depicted. (B) Agarose gel (1.5%) showing the size range of sonicated DNA used for ChIP experiments. (C and D) Tcf4 (C) and β-catenin (β-cat) (D) ChIP, showing their occupancy on the Sox8 gene promoter containing the Tcf4 binding site (bp −800) selectively upon Wnt signaling activation. P7 testes represent the Wnt-repressed state, while P21 testes represent the Wnt-activated state. Ccnd1 (bp −300) is used as a positive control, whereas the β-actin gene (bp −250) is used as a negative control. (E) Occupancy of Tcf4 and β-catenin on the Sox8 gene promoter containing the Tcf4 binding site (bp −800) upon silencing of mrhl RNA. (F) Luciferase assay in Gc1-Spg cells treated with control medium or Wnt3A conditioned medium using plasmid constructs containing the 1-kb upstream promoter region of Sox8 with either the wild-type Tcf4 binding site or a mutant Tcf4 binding site. Data in panels C to F are plotted as means ± SD, n = 4. ***, P < 0.001; **, P < 0.01; *, P < 0.05 (two-tailed Student t test). N. S, not significant.
Wnt signaling downregulates mrhl RNA, which is in contrast to the expression of Sox8 seen upon Wnt signaling activation. To test the role of mrhl RNA in Sox8 regulation, mrhl RNA was overexpressed and silenced under control and Wnt conditions. It was observed that Sox8 expression was upregulated under control conditions when mrhl RNA was silenced whereas the upregulation of Sox8 was abrogated under Wnt conditions when mrhl RNA was overexpressed in trans (Fig. 4A). The results were also corroborated by a luciferase reporter assay (Fig. 4B). The drastic change in the expression of Sox8 upon mrhl RNA silencing or overexpression clearly establishes mrhl RNA as the regulator of the Sox8 gene.
FIG 4.
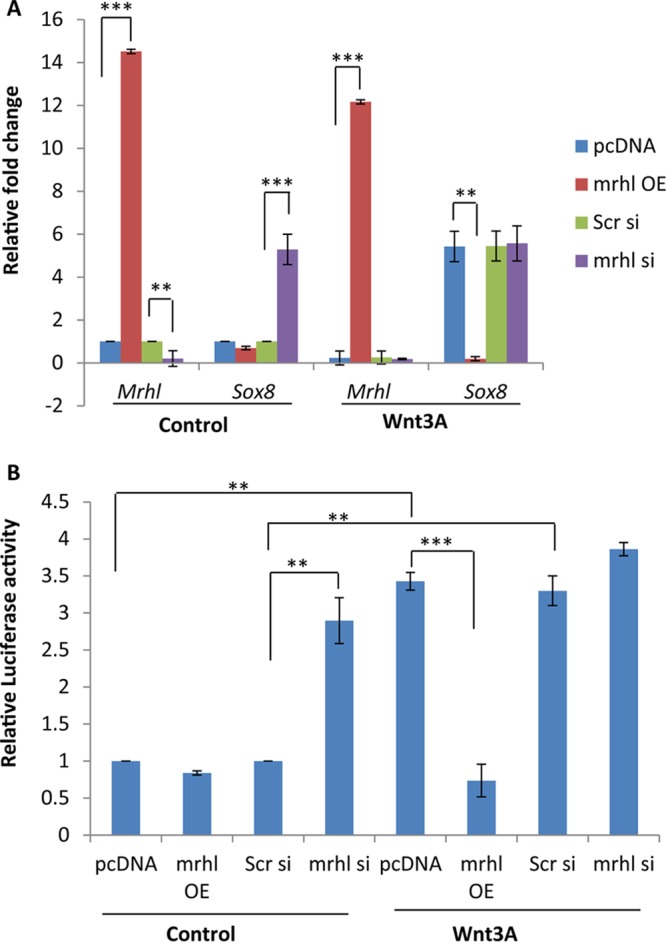
Analysis of the role of mrhl RNA in Wnt-mediated upregulation of Sox8. (A) Expression analysis of mrhl RNA and Sox8 in control and Wnt3A conditioned medium-treated Gc1-Spg upon mrhl RNA overexpression (OE) and mrhl RNA silencing (si). pcDNA, vector control. (B) Luciferase assay for the Sox8 promoter in control and Wnt3A conditioned medium-treated Gc1-Spg upon mrhl RNA overexpression and mrhl RNA silencing. Data are plotted as means ± SD, n = 4. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student t test).
c-Myc binds to the Sox8 promoter at the mrhl RNA binding site upon Wnt signaling activation.
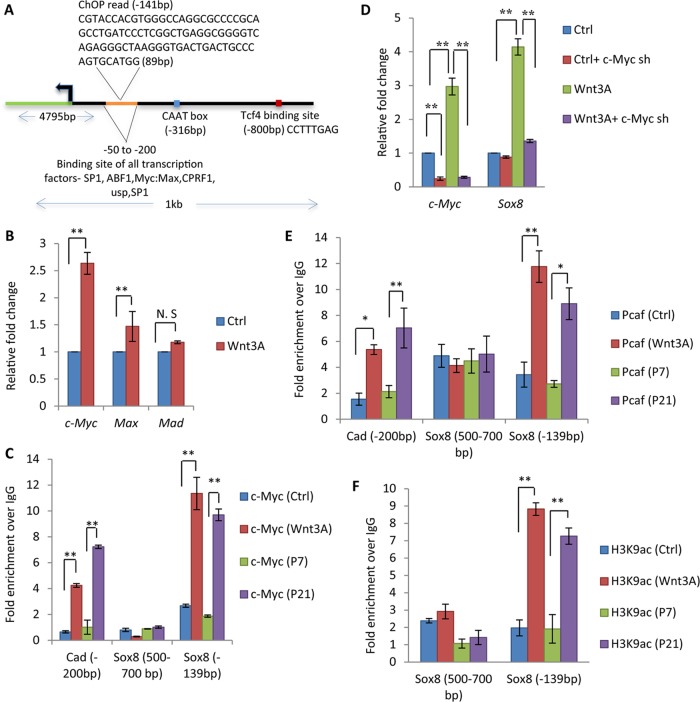
Bioinformatic analysis of the Sox8 gene promoter also revealed binding sites for five putative transcription factors, among which Myc-Max-Mad was a promising candidate (Fig. 5A). ABF1 and CPRF1 are not expressed in mammalian germ cells, while SP1 is a ubiquitous chromatin remodeller and ultraspiracle encodes a protein expressed in Drosophila. Interestingly, it was observed that the binding site of all the transcription factors, including Myc-Max-Mad, overlapped the site where mrhl RNA binds to the Sox8 promoter, indicating the importance of this region in Sox8 gene regulation.
FIG 5.
c-Myc binds to the Sox8 promoter at the Myc-Max-Mad binding site upon Wnt signaling activation. (A) The architecture of the 1-kb upstream promoter of the Sox8 gene with the CAAT box, Tcf4 binding site, mrhl RNA binding site, and putative transcription factors is identified along with the binding sites. (B) Expression analysis of c-Myc, Max, and Mad in Gc1-Spg cells (control, Wnt3A conditioned medium treated). (C) c-Myc ChIP in Gc1-Spg cells and mouse testis with Cad as a positive control for c-Myc occupancy. The region from bp −500 to −700 in the Sox8 promoter was used as a negative control. (D) Expression analysis of c-Myc and Sox8 upon c-Myc silencing in control and Wnt3A conditioned medium-treated Gc1-Spg. (E) ChIP showing recruitment of c-Myc-associated coactivator Pcaf at the Myc-Max-Mad binding site in Wnt3A conditioned medium-treated Gc1-Spg cells and in P21 mouse testes. (F) Increased level of H3K9ac at the Myc-Max-Mad binding site upon Wnt activation in Gc1-Spg cells and in P21 mouse testes. Data are plotted as means ± SD, n = 4. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student t test). N. S, not significant.
There were earlier reports showing upregulation of c-Myc upon Wnt activation (34). We checked for the same in our model system, and it was observed that c-Myc is indeed upregulated upon Wnt signaling activation in Gc1-Spg cells whereas there was no significant effect on the expression of Max or Mad genes (Fig. 5B). Next, we checked for the occupancy of c-Myc on the Sox8 promoter upon Wnt activation. As shown in Fig. 5C, c-Myc exhibits a strong occupancy on the Sox8 promoter upon Wnt signaling activation both in Gc1-Spg cells and in mouse testis (P21), establishing it to be a candidate responsible for the Wnt-mediated upregulation of Sox8. The Carbamoyl-Phosphate Synthetase 2, Aspartate Transcarbamylase and Dihydroorotase (cad) gene promoter was used as a positive control, and the bp −500 to −700 region of the Sox8 gene promoter was used as a negative control. Since c-Myc occupies the Sox8 gene promoter only upon the cue of Wnt signaling, we went ahead with silencing the expression of c-Myc using short hairpin RNA (shRNA) in Gc1-Spg cells. Analysis of Sox8 gene expression upon c-Myc silencing showed abrogation of the Wnt-mediated upregulation of Sox8 gene expression (Fig. 5D). It is known that c-Myc recruits the histone acetyltransferase (HAT) Pcaf, which leads to an increase in the level of histone activation mark H3K9ac, which in turn facilitates the transcriptional activation (35). Therefore, we also scored for the occupancy of Pcaf and observed an increase in Pcaf occupancy at the Myc-Max-Mad binding site but not in the region from bp −500 to −700 on the Sox8 promoter in both Wnt3A-treated Gc1-Spg cells and P21 mouse testis (Fig. 5E). Consistent with the recruitment of Pcaf, there was also an increase in the level of the H3K9ac mark at the Sox8 promoter (Fig. 5F).
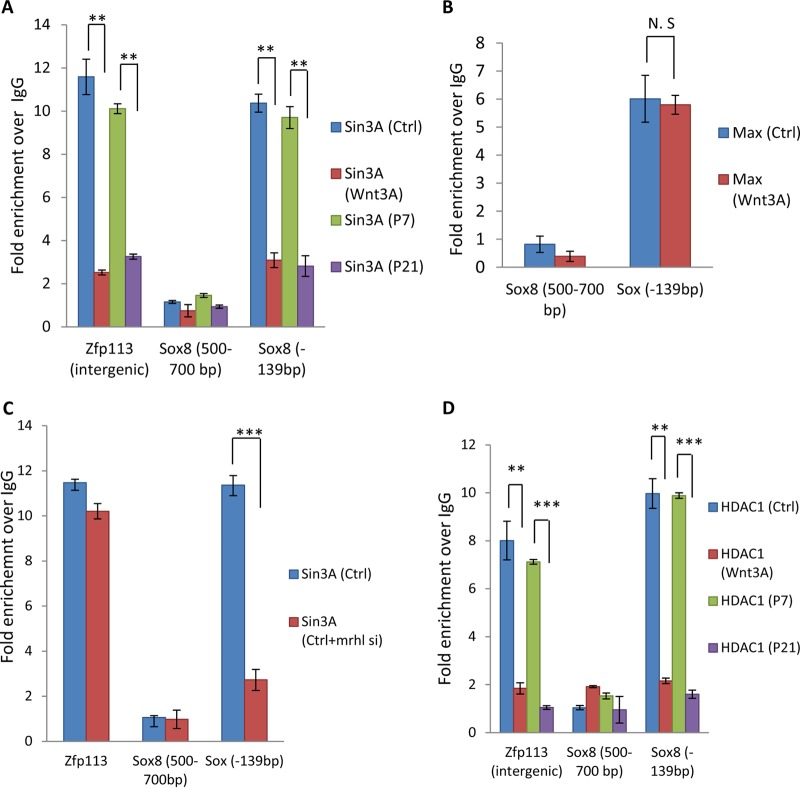
Sin3A is bound at the Myc-Max-Mad binding site on the Sox8 promoter to keep it in a repressed state.
It is known that when Max heterodimerizes with c-Myc it acts as an activator whereas when it heterodimerizes with Mad it acts as a repressor (36). Our data clearly show the importance of Myc-Max in Sox8 gene upregulation upon Wnt activation. Therefore, we were curious to see whether the Max-Mad complex was involved in maintaining Sox8 in a repressed state under control conditions. Sin3A is the major corepressor interacting with the Max-Mad complex (37, 38). We carried out Sin3A ChIP at the Myc-Max-Mad binding site in the Sox8 promoter with Zfp113 as a positive control (Fig. 6A). Sin3A is present at the site under control conditions, and upon Wnt signaling activation the occupancy of Sin3A is depleted in Gc1-Spg cells and also in the mouse testicular cells, indicating its role in maintaining Sox8 in a repressed state. Given that Max is expressed under control conditions and upon Wnt activation, we also carried out Max ChIP as an internal control (Fig. 6B). The importance of mrhl RNA in repression of Sox8 has been established, as shown above (Fig. 4). To study the role of mrhl RNA in the recruitment of Sin3A on the Sox8 gene promoter, we carried out Sin3A ChIP upon mrhl RNA silencing; as can be seen from Fig. 6C, mrhl RNA plays an important role in the establishment of the repressive chromatin involving Sin3A. We also checked for the occupancy of histone deacetylase 1 (HDAC1) as it is the known chromatin modifier recruited by Sin3A (37). HDAC1 is present at the Myc-Max-Mad binding site under control conditions in Gc1-Spg cells but is depleted upon Wnt signaling activation, and the result was also corroborated in vivo in the mouse testicular cells (Fig. 6D).
FIG 6.
Sin3A is bound at the Myc-Max-Mad binding site on the Sox8 promoter to keep it in a repressed state. (A) ChIP showing the presence of Max at the Myc-Max-Mad binding site on the Sox8 promoter in both control and Wnt3A conditioned medium-treated Gc1-Spg cells. (B) ChIP in Gc1-Spg cells with Zfp113 as a positive control, showing the occupancy of Sin3A in control Gc1-Spg cells but not in Wnt3A conditioned medium-treated Gc1-Spg cells. The region from bp −500 to −700 in the Sox8 promoter is used as a negative control. (C) Sin3A ChIP upon mrhl RNA silencing. (D) ChIP showing the presence of HDAC1 at the Myc-Max-Mad binding site on the Sox8 promoter in control Gc1-Spg cells but not in Wnt3A conditioned medium-treated Gc1-Spg cells. Data are plotted as means ± SD, n = 4. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student t test). N. S, not significant.
Changes in histone modification marks and nucleosome dynamics on the Sox8 promoter upon Wnt signaling activation.
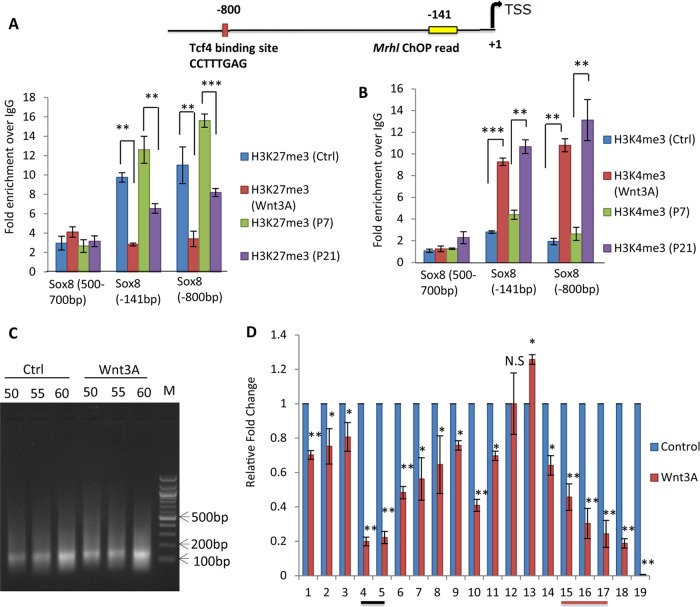
So far, we have established that Sox8 is upregulated upon Wnt signaling activation and that the removal of mrhl RNA from the Sox8 locus is important for this transcriptional activation. The Tcf4 binding site is occupied by β-catenin upon Wnt activation but is minimally active. Next, we were interested in exploring the chromatin changes taking place in the promoter region to facilitate this transcriptional activation. We looked at the histone activation and repression marks at both the sites where mrhl RNA physically binds to the chromatin and the Tcf4 binding site. There was a decrease in the level of repression mark H3K27me3 at both the sites upon Wnt signaling activation in Gc1-Spg cells and also in P21 mouse testicular cells (Fig. 7A). An increase in the level of activation mark H3K4me3 was also observed at these sites both in vitro and in vivo (Fig. 7B). Nucleosome depletion is a common phenomenon and occurs primarily at transcriptionally active promoters to facilitate the binding of the transcriptional machinery and various coactivators. Therefore, the nucleosome position was mapped by histone H3 ChIP followed by qPCR at every 50-bp interval on the Sox8 promoter in control and Wnt3A conditioned medium (CM)-treated Gc1-Spg cells. Mononucleosomes were isolated by micrococcal nuclease digestion (Fig. 7C) and then were subjected to H3 ChIP followed by qPCR to identify the regions of chromatin bound to nucleosomes. Nucleosome depletion was indeed observed at the mrhl RNA binding site as well as the Tcf4 binding site (Fig. 7D).
FIG 7.
Changes in histone modification marks and nucleosome dynamics upon Sox8 promoter Wnt signaling activation. (A and B) Levels of repression of mark H3K27me3 (A) and activation of mark H3K4me3 (B) at the Tcf4 binding site (bp −800) and the mrhl RNA binding site (bp −141) on the Sox8 promoter in Gc1-Spg cells (control and Wnt3A conditioned medium treated) and mouse testes (P7 and P21). (C) Micrococcal nuclease digestion of chromatin for different time intervals (50, 55, and 60 min) from control and Wnt3A conditioned medium-treated Gc1-Spg cells to obtain mononucleosomes. (D) Change in nucleosome dynamics on the 1-kb upstream Sox8 promoter upon Wnt signaling activation. Numbers 1 to 19 represent every 50-bp interval scored for in the 1-kb upstream Sox8 gene promoter, with 19 being adjacent to the TSS. The black line represents the Tcf4 binding site, and the red line represents chromatin oligonucleotide affinity purification (ChOP) read results. Data are plotted as means ± SD, n = 4. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student t test). N. S, not significant.
Role of Sox8 and Wnt signaling in meiotic commitment and progression of spermatogonial cells.
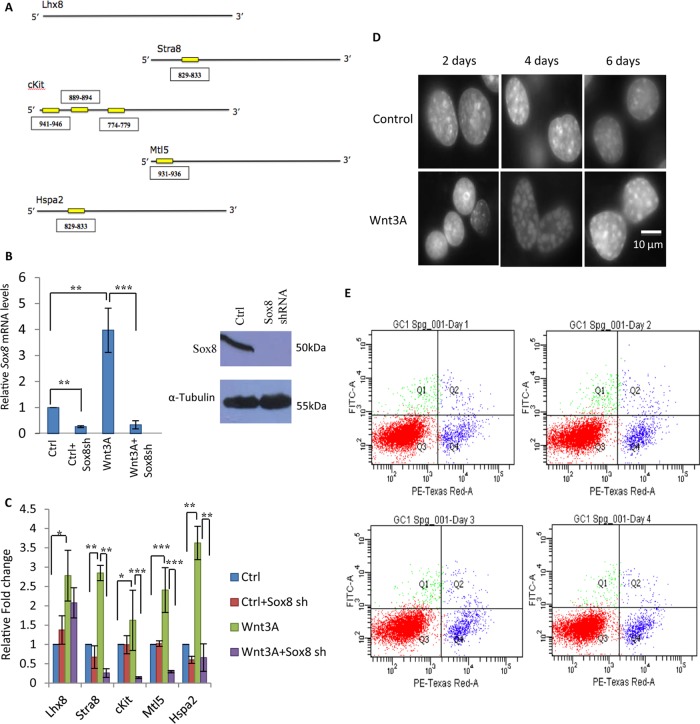
We showed earlier that Wnt-mediated downregulation of mrhl RNA is essential for upregulation of various premeiotic and meiotic marker genes in spermatogonial cells, thus paving the path for the differentiation of spermatogonia to spermatocytes (29). Now that we have established that mrhl RNA can also regulate Sox8 expression, it is possible that Wnt-mediated mrhl RNA downregulation leads to an upregulation of Sox8 which in turn leads to activation of these meiotic commitment marker genes. When the Sox8 binding site was mapped on the promoter of these genes, we observed that all of the genes, with the exception of the Lhx8 gene, possess a Sox8 binding site in the 1-kb upstream sequence as shown in Fig. 8A. The functionality of Sox8 in the regulation of these genes was checked by silencing Sox8 gene expression using the shRNA approach in Gc1-Spg cells (Fig. 8B). When the Wnt signaling-mediated upregulation of Sox8 was perturbed, it also abrogated the upregulation of these premeiotic and meiotic marker genes (Fig. 8C). This clearly establishes a significant role for Sox8 in germ cells and also raises the possibility of Sox8 being important for spermatogonial differentiation.
FIG 8.
Role of Sox8 and Wnt signaling in meiotic progression and commitment of spermatogonial cells. (A) Sox8 binding site in the 1-kb upstream promoters of important premeiotic and meiotic marker genes. (B) Expression analysis of Sox8 by RT-qPCR and Western blotting upon Sox8 silencing by shRNA. (C) Expression analysis of premeiotic markers (Stra8, Lhx8, and c-kit) and meiotic markers (Mtl5 and Hspa2) upon Wnt signaling activation as well as upon Sox8 silencing in control and Wnt3A conditioned medium-treated Gc1-Spg cells. (D) Change in global chromatin architecture in mouse spermatogonial Gc1-Spg cells upon treatment with Wnt3A conditioned medium for different time durations (2, 4, and 6 days). (E) Apoptosis assay of Gc1-Spg cells upon prolonged Wnt3A treatment (24, 48, 72, and 96 h). PE, R-phycoerythrin.
Given that the many factors which play a vital role in differentiation of spermatogonial stem cells to spermatocytes do so under the veil of Wnt signaling, we went ahead to explore the significance of Wnt signaling independently initiating this differentiation process. First, we studied the cell morphology changes of the Gc1-Spg cell line, which is a B-type spermatogonial cell line, upon prolonged Wnt3A conditioned medium treatment. As can be seen from Fig. 8D, there was a visible increase in number of DAPI (4′,6-diamidino-2-phenylindole) intense chromatin foci after 4 to 6 days of Wnt3A conditioned medium treatment. This change, however, was not due to apoptosis or compromise in the health of the cells (Fig. 8E). We therefore went ahead to score for the Pou5f1, Sox2, and Nanog spermatogonial stem cell markers. There was a gradual decrease in the expression levels of all the three stem cell markers upon prolonged Wnt3A conditioned medium treatment of Gc1-Spg cells (Fig. 9A). Next, we scored for the spermatogonial cell differentiation markers—Stra8, Scp3, Dmc1, and Spo11. Stra8 is a key molecule for this differentiation, which is known to be regulated by retinoic acid (39). As can be seen in Fig. 9B and C, Stra8 expression levels drastically increased upon Wnt signaling activation in Gc1-Spg cells for meiotic commitment of spermatogonia. Furthermore, the other three meiotic marker genes encoding Scp3, Dmc1, and Spo11, involved in chromosome pairing and recombination, were also upregulated upon Wnt signaling activation in Gc1-Spg cells (Fig. 9B). Western blot analysis also showed the gradual decrease in the level of Pou5f1 and an increase in the level of Stra8 upon prolonged Wnt3A treatment (Fig. 9C).
FIG 9.
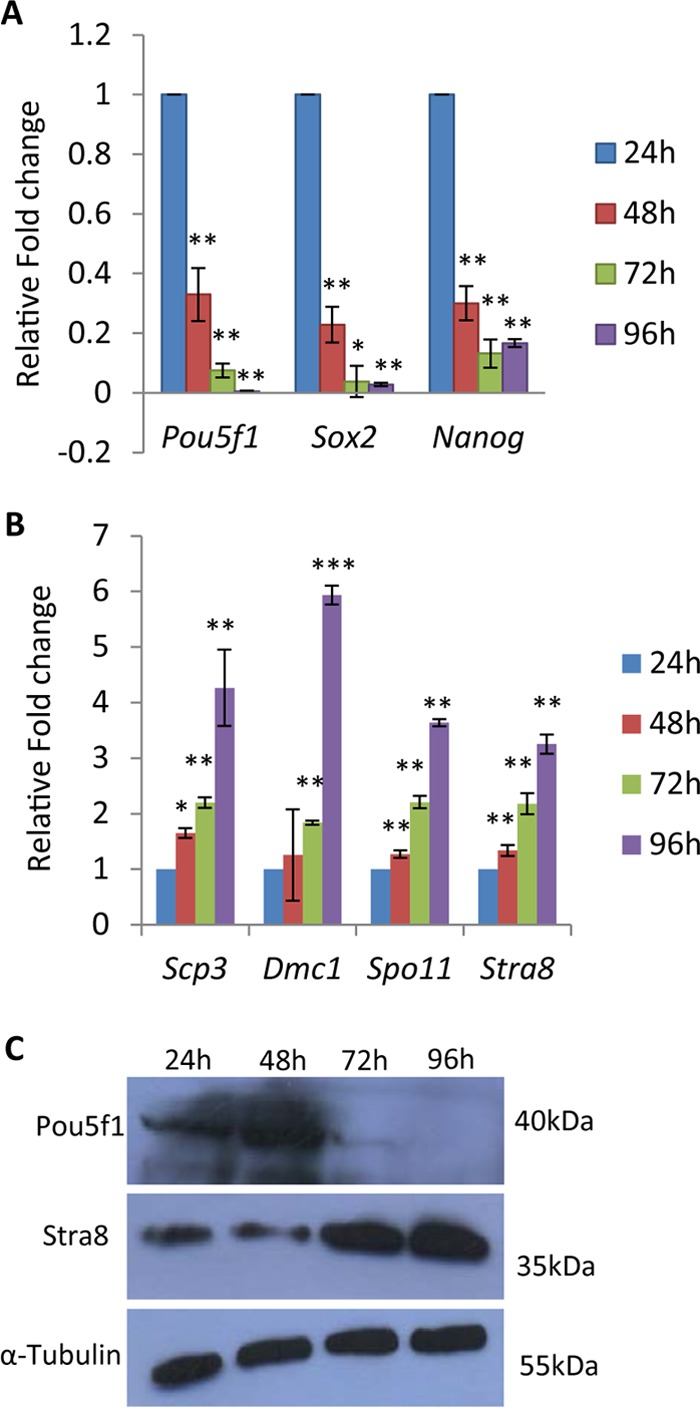
(A and B) Expression analysis of spermatogonial stem cell markers (Pou5f1, Sox2, and Nanog) (A) and meiotic differentiation markers (Scp3, Dmc1, Spo11, and Stra8) (B) upon prolonged (24-, 48-, 72-, and 96-h) Wnt3A treatment in Gc1-Spg cells. (C) Western blot analysis of Pou5f1 and Stra8 upon prolonged (24, 48, 72, and 96 h) Wnt3A treatment in Gc1-Spg cells. The significance values in panels A and B are represented with respect to 24-h Wnt3A treatment. Data in panels A and B are plotted as means ± SD, n = 4. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed Student's t test).
DISCUSSION
An earlier report from our laboratory showed that mrhl RNA physically associates with and regulates 37 genomic loci, 1 of which is the Sox8 genomic locus where the RNA binds to its promoter region (28). In the present study, we explored the regulation of Sox8 by mrhl RNA and identified possible players in the Wnt signaling-mediated regulation of Sox8.
The members of the Sox (sry-related HMG box) group of transcription factors play an important role in development, particularly in cell fate specification and differentiation; among these, Sox8, Sox9, and Sox10 belong to subgroup E. The members of the Sox E group of proteins are involved in neural crest development. Sox9 and Sox10 have been very well studied in terms of their biological roles. Sox8 and Sox9 have overlapping functions, and both of them are expressed in Sertoli cells and are implicated in sex cord development. One of the earlier studies had shown that Sox8 is not expressed in testicular germ cells and had implicated it in having a role only in Sertoli cell function (30). However, since we did observe in our previous studies that Sox8 is expressed in Gc1-Spg cells (B-type spermatogonial cells), we wanted to confirm that it is indeed expressed in testicular germ cells. All the initial experiments carried out in the present study demonstrated that Sox8 is expressed in both spermatogonia and spermatocytes. Since the earlier expression studies were done using minimal histochemistry, it is possible that the Sox8 expression was not detected due to epitope masking in the testicular sections.
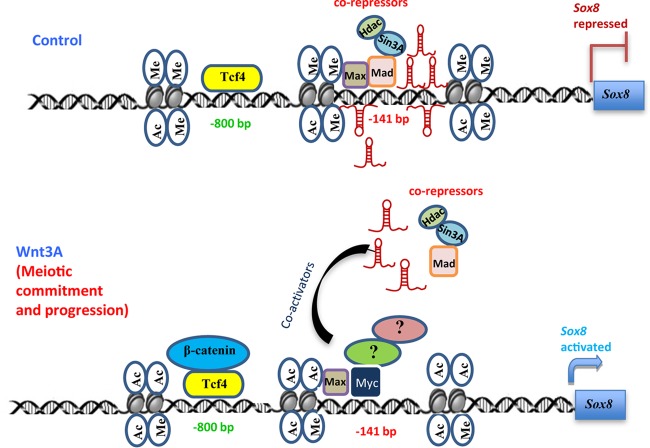
In our present study, we showed that the Wnt signaling-mediated transcriptional regulation of Sox8 involves substantial changes in the chromatin dynamics of the promoter. The Max-Mad complex binds to the site under control conditions and recruits the corepressor complex containing Sin3A and HDAC1 to keep Sox8 in a repressed state, and the presence of mrhl RNA is imperative for this repression. Our experiments clearly suggest that mrhl RNA, along with the corepressors, keeps Sox8 in a repressed state and that the removal of the RNA makes the site available for the recruitment of the coactivators and the activation of Sox8. Multiple transcription factors have binding sites on the Sox8 promoter, and, interestingly, all of them are clustered at the mrhl RNA binding site, showing the importance of this locus in Sox8 promoter regulation. The Myc-Max binding site is present in the Sox8 promoter and overlaps with the mrhl RNA binding site. We observed c-Myc occupancy on the Sox8 promoter upon Wnt activation, and silencing of c-Myc abrogated Wnt-mediated upregulation of Sox8, proving its role in the regulation of Sox8 gene expression as summarized in Fig. 10. The recruitment of c-Myc-related coactivator Pcaf and the increase in the level of H3K9ac at the Myc-Max-Mad binding site further validate the involvement of c-Myc as an activator for the Wnt-mediated upregulation of Sox8 gene expression. The mechanism by which mrhl RNA facilitates the formation of the repressor complex at the Sox8 promoter is only a matter of speculation. It is possible that mrhl RNA recruits Sin3A at the promoter site through a direct interaction. Alternatively, the RNA can influence the heterodimerization of Max-Mad at this locus. It is also not clear how the mrhl RNA interacts with the Sox8 promoter or whether it is through RNA-DNA triplex formation as has been demonstrated previously (40) or through the p68 protein or through yet another uncharacterized bridging protein. We are currently investigating all these possibilities, which should throw light on the detailed molecular mechanism of mrhl RNA-mediated repression of Sox8 gene expression in spermatogonial cells.
FIG 10.
Model summarizing the findings of present study. The model depicts the changes at the proximal promoter region of Sox8 at the mrhl RNA binding site (bp −141) upon Wnt signaling activation with respect to the binding of corepressors and the recruitment of β-catenin at the TCF4 binding site (bp −800). The displaced corepressors could be replaced with unknown (?) coactivators. The changes in histone modifications are also shown (Ac, acetylation; Me, methylation). Upon Wnt activation, the upregulation of Sox8 leads to meiotic commitment and differentiation.
Our results conclusively show that Sox8 is not only expressed in germ cells but also temporally regulated by Wnt signaling activation. It has been reported that Wnt signaling activation paves the path for differentiation of spermatogonia to spermatocytes by upregulation of various premeiotic and meiotic marker genes. We showed earlier that Wnt signaling-mediated downregulation of mrhl RNA is essential for upregulation of these genes (28). Since we knew that mrhl RNA can in turn regulate Sox8, we were interested in seeing the role of Sox8 in regulation of these marker genes and in turn in meiotic progression. We found that silencing of Sox8 abrogates the upregulation of these marker genes, showing that Sox8 can be an important transcription factor in spermatogonial differentiation. We were also interested in studying the role of Wnt signaling in spermatogonial differentiation, as it is the common factor in mrhl RNA downregulation and Sox8 upregulation. Our preliminary data suggest that spermatogonial cells exhibit a global change in chromatin architecture upon prolonged Wnt3A treatment and that the process also leads to a gradual decrease in the levels of spermatogonial stem cell markers and to an increase in the levels of differentiation markers. There is definitely a differentiation program initiated by Wnt signaling, but the molecular details of this very perceivable change are still to be addressed. We note here that B-type spermatogonia are progenitor cells that are primed to meiotic commitment and enter the meiotic prophase upon exposure to the appropriate signal. At present, we believe that Sox8 regulates expression of Stra8, which is a master regulator of a transcription program leading to meiotic commitment and progression (39). In this context, mrhl RNA plays a very important role in regulating expression of the Sox8 transcription factor. We are presently engaged in testing this hypothesis.
In past few years, there have been many studies on lncRNA profiling during different stages of spermatogenesis (41–43). It is clear from such studies that the transcriptome levels of lncRNAs are greater than those of mRNAs during spermatogenesis and that the expression of lncRNAs is also dynamically regulated. Mechanistic details of lncRNA function during spermatogenesis have been provided by only a few studies, including our report of the role of mrhl RNA in regulation of Wnt signaling through p68 RNA helicase (27, 44–46). The present report describes a new dimension in the context of spermatogenesis, with lncRNA regulating the expression of an important transcription factor and such regulation being crucial for meiotic entry or spermatogonial differentiation. It would also be interesting to explore if the whole 2.4-kb RNA or just a part of it is required for its function at the Sox8 locus. Addressing these issues would widen our knowledge of the complex and diverse ways by which lncRNAs can bring about specific spatiotemporal regulation of gene expression.
MATERIALS AND METHODS
Cell line, antibodies, plasmids, and reagents.
Cells of the Gc1-Spg cell line (CRL-2053) and TM4 cell line (CRL-1715) were obtained from ATCC. Cells of the mouse L-control cell line (ATCC CRL-2648) and L-Wnt3A cell line (ATCC CRL-2647) were kind gifts from Jomon Joseph (NCCS, India).
All chemical reagents were of analytical reagent (AR) grade and were purchased from Sigma. Protein G Dynabeads (10003D), protein A-agarose beads (15918-014), an apoptosis kit (V13242), and Lipofectamine 2000 (11668027) were purchased from Invitrogen. DNase I (MO303), T4 DNA ligase (M0202S), and restriction enzymes were purchased from New England BioLabs. A luciferase assay kit was supplied by Promega. Sox8 shRNA (SHCLNG-NM_011447) and Myc-shRNA (SHCLNG-NM_010849) were obtained from Sigma. An Mrhl siRNA pool was obtained from Dharmacon, and the siRNA sequences were described before (27). Real-time PCR was performed using a Corbett rotor gene 2000 cycler (Qiagen).
The list of antibodies used in the present study, with the manufacturers and catalogue numbers indicated in parentheses, is as follows: β-catenin (BD Biosciences; catalog no. 610154), TCF4 (Millipore; catalog no. 05-511), alpha tubulin (Sigma; catalog no. T8203), Hdac1 (Cell Signaling; catalog no. 5356), histone H3 (Abcam; catalog no. ab46765), H3K27me3 (Abcam; catalog no. ab6002), H3K4me3 (Abcam; catalog no. ab12209), H3K9ac (Abcam; catalog no. ab4441), Sox8 (Pierce; catalog no. PA1-28072), Sox9 (Abcam; catalog no. ab3697), E-cadherin (BD Biosciences; catalog no. 610404), vimentin (Abcam; catalog no. ab20346), Pou5f1 (Oct-4) (Abcam; catalog no. ab27985), Stra8 (Abcam; catalog no. ab49602), Sin3A (Abcam; catalog no. ab3479), c-Myc (Santa Cruz; catalog no. 9E10), Pcaf (Sigma; catalog no. P7493), Scp3 (Abcam; catalog no. ab97672), and Max (Cell Signaling; catalog no. 4739).
Preparation of control and Wnt3A conditioned medium (Wnt3A CM).
L-control or L-Wnt3A cells (106 cells) were seeded in 90-mm-diameter culture dishes containing Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). After 48 h, fresh medium was added to the cultured cells and the reaction mixture was incubated for an additional 24 h. The medium was then collected, centrifuged at 500 × g for 5 min, filtered, and stored at −20°C until further use. For treatment of Gc1-Spg cells, the control as well as Wnt3A conditioned medium was diluted 2:1 with DMEM containing 10% FBS.
Cloning of the 1-kb sequence upstream of the Sox8 and Sox9 promoter and luciferase reporter assay.
The 1-kb sequence upstream of the Sox8 and Sox9 genes was cloned in pGL3-Basic vector. Clones were confirmed by DNA sequencing. The Tcf4 binding site on the Sox8 gene promoter (bp −800) was mutated from CTTTGA to CGCTGA using a Stratagene QuikChange site-directed mutagenesis kit (catalog no. 200518). For the luciferase assay, Gc1-Spg cells were transfected with 1 μg of pGL3-Basic vector or 1 μg of a Sox8/Sox9 promoter clone in a six-well plate using Lipofectamine 2000. Transfection with a cytomegalovirus–β-galactosidase (CMV-βGal) plasmid (0.4 μg) was done as an internal control for transfection efficiency. After 24 h, cells were harvested and processed for luciferase assay in accordance with the protocol of the manufacturer (Promega). Luciferase readings for pGL3-Basic as well as for the Sox8/Sox9 promoter plasmid were normalized with the luminometer readings obtained for CMV-βGal.
shRNA- and siRNA-mediated silencing.
All shRNA plasmids were transfected at a concentration of 1.5 μg/ml whereas scrambled and mrhl siRNA pool plasmids were transfected at a concentration of 100 nM using Lipofectamine 2000 at 60% to 70% cell confluence.
Quantitative PCR (qPCR).
RNA isolation was performed using TaKaRa TRIzol in accordance with the manufacturer's protocol. A 2-μg concentration of DNase-treated RNA was used for processing 20-μl reaction mixtures of cDNA using all reagents from Promega. A 2-μl volume of this cDNA was used for each qPCR. β-Actin was used to normalize the threshold cycle (CT) values.
Western blotting.
After SDS-PAGE and transfer, the membranes were blocked in 5% skimmed milk for 45 min at room temperature and then incubated with the respective primary antibody dissolved in 1% skimmed milk–0.05% phosphate-buffered saline with Tween 20 (PBST) overnight at 4°C. The membrane was then washed with 0.05% PBST and incubated with the respective secondary antibody dissolved in 1% skimmed milk–0.05% PBST for an hour at room temperature. Washes were then given using 0.05% PBST, and the blot was developed in a dark room using either a Pico ECL kit or a Femto ECL kit from Pierce.
Testis cellular spreads and immunofluorescence.
The preparation of cellular spreads and subsequent immunofluorescence analysis were carried out as previously described (29).
Chromatin immunoprecipitation (ChIP).
Cells were harvested and cross-linked using 1% formaldehyde for 10 min at room temperature followed by quenching using 0.125 M glycine for 5 min. The cross-linked pellet was obtained by centrifugation at 1,000 × g at 4°C for 10 min. The cell pellet was resuspended in 1 ml of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl) and incubated on ice for 15 min. This was followed by sonication of the lysate using a Bioruptor for a total of 35 cycles (30 s on and 30 s off at high pulse). Insoluble material was removed by centrifugation at 13,000 × g at 4°C for 10 min. Sonicated DNA was enriched in the range of 100 to 300 bp. The lysate was incubated with the respective antibodies for immunoprecipitation overnight at 4°C. The immune complexes were allowed to bind to protein G/A Dynabeads for 3 h at 4°C. The beads bound by immune complexes were obtained by magnetic precipitation followed by one wash with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl) and with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl). The immunoprecipitated material was eluted from the beads by adding 400 μl of elution buffer (0.1 M NaHCO3, 1% SDS), and the samples were incubated in an end-to-end rotator at room temperature for 45 min. The eluates were then processed for DNA isolation by the phenol-chloroform method. The fold enrichment over IgG was calculated by considering the CT values obtained for the target DNA as follows:
| (1) |
Microccocal nuclease digestion.
Cells were harvested and pelleted at 2,000 × g for 3 min at 4°C. The pellet was resuspended in lysis buffer (60 mM KCl, 15 mM NaCl, 15 mM Tris, 0.34 M sucrose, 2 mM EDTA, 0.5 mM EGTA, 1 mM dithiothreitol [DTT], 0.03% Triton, 1% glycerol) and incubated on ice for 10 min with regular vortex mixing. The suspension was then centrifuged at 14,000 × g for 10 min at 4°C. The pellet was then resuspended in wash buffer (60 mM KCl, 15 mM NaCl, 15 mM Tris, 0.34 M sucrose, 1 mM DTT) and centrifuged at 14,000 × g for 10 min at 4°C. The pellet was then suspended in MNase buffer (10 mM Tris, 10 mM KCl, 2 mM CaCl2), and micrococcal nuclease enzyme (1.5 units) was added followed by incubation at 37°C for 50 min. The reaction was then stopped by addition of 5 mM EGTA and incubation on ice for 5 min. The digested nuclei were centrifuged at 2,000 × g for 10 min at 4°C. The reaction mixture was then suspended in LSDB 250 buffer (20% glycerol, 50 mM HEPES, 3 mM MgCl2, 250 mM KCl) and was incubated overnight with histone H3 antibody. The beads were blocked with 0.1% bovine serum albumin (BSA) at 4°C for 30 min, after which they were added to the nucleosome suspension and incubated for a further 4 h at 4°C. The beads were then washed with LSDB 250 buffer 3 times (for 5 min at 4°C each time) and eluted as described for ChIP. The eluate was subjected to DNA extraction.
Apoptosis assay.
The apoptosis assay was performed in accordance with the protocol of the manufacturer (Invitrogen).
IACUC approval.
Experiments were performed using mice testes. The institution (JNCASR) has obtained IACUC approval for research involving use of mice.
ACKNOWLEDGMENTS
We thank Jomon Joseph of the National Centre for Cell Sciences (NCCS, India) for providing the mouse L cells. We acknowledge A. Prakash of the Animal Facility, B. S. Suma of the Confocal Imaging Facility, and Y. Swaroopa of the FACS Facility at JNCASR. M.R.S.R. thanks the Department of Science and Technology for J. C. Bose and SERB Distinguished Fellowships.
This work was financially supported by Department of Biotechnology, Govt. of India (Grant Number BT/01/COE/07/09).
We declare that we have no conflicts of interest.
S.K. and B.K. performed the experiments. S.K., V.S.A., and M.R.S.R. designed the experiments, analyzed the data, and wrote the manuscript.
REFERENCES
- 1.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207. [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert SL, Pehrson JR, Sharp PA. 2000. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem 275:36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 5.Park Y, Kelley RL, Hyangyee O, Kuroda MI, Meller VH. 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- 6.Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 7.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 10.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchese FP, Grossi E, Marin-Bejar O, Bharti SK, Raimondi I, Gonzalez J, Martinez-Herrera DJ, Athie A, Amadoz A, Brosh RM Jr, Huarte M. 2016. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell 63:397–407. doi: 10.1016/j.molcel.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA. 2016. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carnero E, Barriocanal M, Prior C, Pablo Unfried J, Segura V, Guruceaga E, Enguita M, Smerdou C, Gastaminza P, Fortes P. 2016. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 17:1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. 2012. LincRNA-p21 suppresses target mRNA translation. Mol Cell 47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. 2 February 2010. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. 2014. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. 2013. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, Zhang S, Zhong F, Liu C, Yin Y, Cai W, Zhang H. 2016. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat Commun 7:11730. doi: 10.1038/ncomms11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatima R, Akhade VS, Pal D, Rao SM. 2015. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther 3:5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishant KT, Ravishankar H, Rao MR. 2004. Characterization of a mouse recombination hot spot locus encoding a novel non-protein-coding RNA. Mol Cell Biol 24:5620–5634. doi: 10.1128/MCB.24.12.5620-5634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganesan G, Rao SM. 2008. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA 14:1399–1410. doi: 10.1261/rna.838308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arun G, Akhade VS, Donakonda S, Rao MR. 2012. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol 32:3140–3152. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akhade VS, Arun G, Donakonda S, Rao MR. 2014. Genome wide chromatin occupancy of mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol 11:1262–1279. doi: 10.1080/15476286.2014.996070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akhade VS, Dighe SN, Kataruka S, Rao MR. 2016. Mechanism of Wnt signaling induced down regulation of mrhl long non-coding RNA in mouse spermatogonial cells. Nucleic Acids Res 44:387–401. doi: 10.1093/nar/gkv1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. 2004. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 31.O'Bryan MK, Takada S, Kennedy CL, Scott G, Harada S, Ray MK, Dai Q, Wilhelm D, de Kretser DM, Eddy EM, Koopman P, Mishina Y. 2008. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol 316:359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. 2009. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 138:151–162. doi: 10.1530/REP-08-0510. [DOI] [PubMed] [Google Scholar]
- 33.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald BT, Tamai K, He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon SB, Wood MA, Cole MD. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol 20:556–562. doi: 10.1128/MCB.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lüscher B. 2001. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 277:1–14. doi: 10.1016/S0378-1119(01)00697-7. [DOI] [PubMed] [Google Scholar]
- 37.Eilers AL, Billin AN, Liu J, Ayer DE. 1999. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem 274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- 38.Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, Eisenman RN, Radhakrishnan I. 2000. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell 103:655–665. doi: 10.1016/S0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 39.Feng CW, Bowles J, Koopman P. 2014. Control of mammalian germ cell entry into meiosis. Mol Cell Endocrinol 382:488–497. doi: 10.1016/j.mce.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. 2015. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun 6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao J, Wu J, Schuster AS, Hennig GW, Yan W. 2013. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod 89:107. doi: 10.1095/biolreprod.113.113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, Li Y, Huang H, Sun F. 2014. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep 4:5966. doi: 10.1038/srep05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran M, Chen B, Li Z, Wu M, Liu X, He C, Zhang S, Li Z. 2016. Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol Reprod 94:77. doi: 10.1095/biolreprod.115.136911. [DOI] [PubMed] [Google Scholar]
- 44.Wen K, Yang L, Xiong T, Di C, Ma D, Wu M, Xue Z, Zhang X, Long L, Zhang W, Zhang J, Bi X, Dai J, Zhang Q, Lu ZJ, Gao G. 2016. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res 26:1233–1244. doi: 10.1101/gr.199547.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoneda R, Satoh Y, Yoshida I, Kawamura S, Kotani T, Kimura AP. 2016. A genomic region transcribed into a long noncoding RNA interacts with the Prss42/Tessp-2 promoter in spermatocytes during mouse spermatogenesis, and its flanking sequences can function as enhancers. Mol Reprod Dev 83:541–557. doi: 10.1002/mrd.22650. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Wang M, Wang M, Wu X, Geng L, Xue Y, Wei X, Jia Y, Wu X. 2016. A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis 7:e2140. doi: 10.1038/cddis.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]