Abstract
One hundred forty M phenotype Streptococcus pneumoniae isolates were evaluated by PCR-restriction fragment length polymorphism, serotyping, and pulsed-field gel electrophoresis. Molecular genotyping revealed that the predominant macrolide resistance mechanism in S. pneumoniae in Canada is mef(E) and resistance dissemination is due to both spread of the genetic element MEGA as well as clonal dissemination of penicillin- and/or macrolide-resistant strains.
Low-level resistance to 14- and 15-member macrolides and susceptibility to lincosamides and streptogramin B (M phenotype) in Streptococcus pneumoniae (16) are conferred by the presence of a membrane-bound efflux protein, encoded by the mef gene mef(A) or mef(E) (1, 4, 5, 10, 13). The genetic elements carrying the mef genes, MEGA and Tn1207.1, have recently been described and are well described in the literature (5, 14). Clinical isolates of S. pneumoniae with reduced susceptibility to macrolides may arise through the horizontal acquisition of the genetic element carrying the mef gene or through clonal expansion of resistant strains. Unlike penicillin-resistant S. pneumoniae, the molecular epidemiology of macrolide-resistant S. pneumoniae in Canada has not been examined extensively (9, 12). The aim of this study was to identify the prevalence of mef genes in a large collection of S. pneumoniae strains isolated in Canada from 1997 to 2002 and to determine the genetic relatedness between mef(E)- and mef(A)-carrying isolates.
One hundred forty macrolide-resistant (erythromycin MIC, ≥1 μg/ml) and clindamycin-susceptible (MIC, ≤0.25 μg/ml) (M phenotype) S. pneumoniae clinical isolates were selected from among 6,991 isolates collected between 1997 and 2002 as part of an ongoing annual national surveillance study, the Canadian Respiratory Organism Susceptibility Study (7). Study isolates were collected from medical centers in 9 out of 10 Canadian provinces. Isolates for this study were collected from respiratory tract specimens only and were limited to one isolate per patient.
Erythromycin, clindamycin, and penicillin susceptibilities were determined by the NCCLS M7-A4 broth microdilution method (11). MIC interpretive standards for erythromycin, clindamycin, and penicillin were defined according to the NCCLS breakpoints for 2000 (11). The presence of the mef gene was determined by a previously described PCR assay that did not distinguish between the two variants (15). Discrimination between mef(A) and mef(E) was performed by PCR-restriction fragment length polymorphism analysis according to a previously described protocol (5). The relatedness among mef(A)- and mef(E)-carrying isolates was examined by pulsed-field gel electrophoresis (PFGE) by published methods (9, 12). Genomic DNAs were digested with SmaI prior to electrophoresis with a contour-clamped homogenous electric field apparatus (CHEF DRIII; Bio-Rad Laboratories, Hercules, Calif.). Isolates that differed by one to three bands were considered clonally related (12). DNA patterns were digitized for analysis with Molecular Analyst (Fingerprinting Plus, version 1.12) software. A dendrogram was calculated by the unweighted pair group method with arithmetic averages. Isolates were serotyped by the capsular swelling in antisera (Quellung reaction) from the Statens Serum Institut (Copenhagen, Denmark) according to the manufacturer's instructions.
The distribution of the mef(A) and mef(E) variants of the mef gene among pneumococcal isolates is summarized in Table 1. Among a sample of 140 M phenotype S. pneumoniae isolates, 133 (95%) isolates carried the mef(E) gene and 7 (5%) isolates carried the mef(A) gene. Both mef(E)- and mef(A)-carrying isolates were resistant to erythromycin (MIC, ≥1 μg/ml); however, all mef(A)-carrying isolates were susceptible to other antibiotics, including penicillin (penicillin-susceptible MIC, ≤0.06 μg/ml), while 66% (92 of 140) of mef(E)-carrying isolates demonstrated reduced susceptibility to penicillin (MIC, ≥0.12 μg/ml).
TABLE 1.
Prevalence of mef(E) and mef(A) S. pneumoniae genotypes in Canada between 1997 and 2002
| Yr of isolation | No. (%) of isolates
|
||
|---|---|---|---|
| Total | With mef(E) | With mef(A) | |
| 1997-1998 | 29 | 28 | 1 |
| 1998-1999 | 29 | 28 | 1 |
| 1999-2000 | 27 | 24 | 3 |
| 2000-2001 | 29 | 29 | 0 |
| 2001-2002 | 26 | 24 | 2 |
| Total | 140 | 133 (95) | 7 (5) |
Fourteen unique capsular serotypes were identified among the 140 isolates. Predominant serotypes included 12F (14%), 19F (13%), 23F (12%), and 14 (12%). Seventy-four of 140 (53%) mef-positive S. pneumoniae isolates belonged to serotypes 6B, 9V, 18C, 19F, 14, and 23F. One hundred seven of the 140 (76%) mef-positive S. pneumoniae isolates studied belonged to serotypes 8, 14, 11A, 12F, 18C, 19F, 23F, 6B, and 9V. Nontypeable strains accounted for 18% (25 of 140). Based on the incidence of the particular serotypes among our S. pneumoniae population, we found that the currently available heptavalent and 23-valent pneumococcal vaccinations would provide potential coverage for 53 and 76% of the isolates, respectively. This vaccine coverage was greater for penicillin-resistant isolates, increasing to 77 and 86%, respectively.
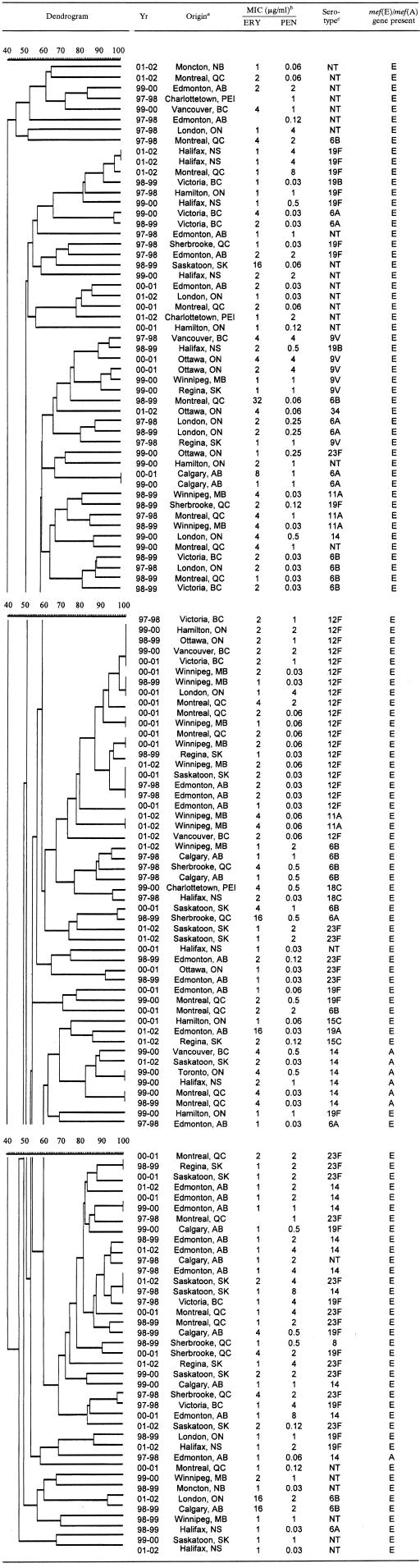
All S. pneumoniae strains were typeable by PFGE; the results are summarized in Table 2. Molecular analysis by PFGE with SmaI-restricted chromosomal DNA revealed 127 distinct DNA profiles among 140 macrolide-resistant S. pneumoniae isolates. One hundred twenty-two unique genotypes were found among the 133 mef(E)-carrying isolates. Dendrogram analysis of the mef(E)-carrying isolates identified 19 clusters (≥85% genetic relatedness), each containing between 2 and 11 isolates, which accounted for 47% (63 of 133) of the mef(E) S. pneumoniae isolates. The majority of the mef(E) isolates within clusters were coresistant to penicillin. Among the 19 clusters, 13 (68%) demonstrated cluster-specific serotypes. Isolates within these 13 clusters belonged to serotype 6B (3 clusters), 6A (2 clusters), 12F (2 clusters), 23F (2 clusters), 11A (1 cluster), 9V (1 cluster), 18C (1 cluster), or 14 (1 cluster).
TABLE 2.
Dendrogram depicting the genetic relatedness of 140 clinical S. pneumoniae isolates on the basis of PFGE results
Province: AB, Alberta; BC, British Columbia; MB, Manitoba; NB, New Brunswick; NS, Nova Scotia; ON, Ontario; QC, Quebec; SK, Saskatchewan.
ERY, erythromycin; PEN, penicillin.
NT, nontypeable.
Among the seven mef(A)-carrying isolates, five unique genotypes were found. Dendrogram analysis identified one cluster (≥80% genetic relatedness) which accounted for 86% (six of seven) of the mef(A) S. pneumoniae isolates. All seven mef(A)-carrying isolates belonged to serotype 14, and all were susceptible to penicillin.
The higher prevalence of the mef(E) variant found among the macrolide-resistant S. pneumoniae population in Canada adds to the conclusion that the mef(E) gene is more prevalent in North America than in Europe (1, 4-6, 10, 13). It has been proposed that the incidence of the two variants occurs as a result of the carriage rates of Streptococcus pyogenes and viridans group streptococci, which carry mef(A) and mef(E), respectively (6). The low incidence of mef(A)-positive S. pyogenes isolates might explain the low incidence of mef(A) among mef-positive S. pneumoniae isolates in Canada (8); however, as the incidence of macrolide-resistant mef(A)-carrying S. pyogenes isolates appears to be increasing, it might affect the incidence of the mef(A) gene in the S. pneumoniae population (8). A low incidence of the mef(A) gene in the S. pneumoniae population might also be due to greater ability of the MEGA mef(E)-containing element to spread horizontally in S. pneumoniae compared to the Tn1207.1 mef(A)-containing element, which has been referred to as “defective” (5, 14).
Genotyping of 133 mef(E)-carrying S. pneumoniae isolates showed that approximately half of the isolates were genetically related and the other half remained genetically unrelated (Table 2). This indicates that macrolide resistance associated with the genetic element MEGA is a result of both clonal dissemination (vertical) as well as spread of the genetic element (horizontal). Further analysis showed that the majority of the isolates that are genetically related (cluster) were also resistant to penicillin, while the majority of the genetically unrelated mef(E)-carrying isolates remained susceptible to penicillin, suggesting that penicillin resistance is driving the clonal spread of the MEGA element. Genotyping of seven mef(A)-carrying S. pneumoniae isolates demonstrated genetic relatedness among these isolates. As the isolates are not related in terms of date and location of isolation, the presence of a single cluster containing six of the seven mef(A) strains indicates that resistance due to the genetic element Tn1207.1 is occurring through the expansion of a single penicillin-susceptible serotype 14 clone that has acquired the mef(A) gene. These PFGE patterns are similar to those of other investigators who found that mef(E) strains did not appear to be related by PFGE, while mef(A) strains were genetically indistinguishable (5).
In conclusion, although both mef(E) and mef(A) genes were present in Canadian isolates of S. pneumoniae, the majority of isolates screened were mef(E). This is in contrast to the European studies that reported mef(A) as the major efflux gene among their S. pneumoniae isolates (1, 4, 5, 10, 13). Similar to the findings of others (5), all mef(A) isolates found in our study belonged to serotype 14, and unlike some studies (1, 10) that identified mef(A) isolates scattered over seven different serotypes (23F,19A,3,6B,15B,33A, and 9), no other serotypes were found in our mef(A) S. pneumoniae isolates. The mef(E)-carrying isolates, in concordance with other studies (1, 5, 10), were more scattered (over 14 serotypes).
Because mef(A) and mef(E) in pneumococci appear to originate from different essentially invariant elements, acquired from group A and viridans group streptococci, respectively, and because acquisition of either gene may have implications regarding streptococcal physiology and antibiotic resistance, particularly penicillin, it remains important for mef(E) and mef(A) to be considered independently and to continue to document their horizontal and vertical spread within S. pneumoniae as this may lead to a better understanding of the spread of macrolide-resistant S. pneumoniae. In addition, since both mef(E) and mef(A) have been found in S. pyogenes and Streptococcus agalactiae in at least one study (2, 3), it would be interesting to see whether a similar mixed occurrence of mef(A) and mef(E) is also present in Canadian M phenotype strains of S. pyogenes and viridans group streptococci and this may lead to a better understanding of the dissemination of macrolide resistance in Canada.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpin, C., M. H. Canron, P. Noury, and C. Quentin. 1999. Emergence of mefA and mefE genes in beta-haemolytic streptococci and pneumococci in France. J. Antimicrob. Chemother. 44:133-134. [DOI] [PubMed] [Google Scholar]
- 3.Arpin, C., H. Daube, F. Tessier, and C. Quentin. 1999. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 43:944-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogaert, D., P. W. M. Hermans, I. N. Grivea, G. S. Katopodis, T. J. Mitchell, M. Sluijter, R. De Groot, N. G. Beratis, and G. A. Syrogiannopoulos. 2003. Molecular epidemiology of penicillin-susceptible non-β-lactam-resistant Streptococcus pneumoniae isolates from Greek children. J. Clin. Microbiol. 41:5633-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 7.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibilities to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz, K. C., A. J. McGeer, C. L. Duncan, A. Ashi-Sulaiman, B. M. Willey, A. Sarabia, J. McCann, S. Pong-Porter, Y. Rzayev, J. S. de Azavedo, and D. E. Low. 2003. Emergence of macrolide resistance in throat culture isolates of group A streptococci in Ontario, Canada, in 2001. Antimicrob. Agents Chemother. 47:2370-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louie, M., L. Louie, G. Papia, J. Talbot, M. Lovgren, and A. E. Simor. 1999. Molecular analysis of the genetic variation among penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae serotypes in Canada. J. Infect. Dis. 179:892-900. [DOI] [PubMed] [Google Scholar]
- 10.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCCLS. 2000. Methods for antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2003. Molecular epidemiology of penicillin-resistant and ciprofloxacin-resistant Streptococcus pneumoniae in Canada. Antimicrob. Agents Chemother. 47:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oster, P., A. Zanchi, S. Cresti, M. Lattanzi, F. Montagnani, C. Cellesi, and G. M. Rossolini. 1999. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob. Agents Chemother. 43:2510-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]