Summary
Background
People with HIV-1 in low-income and middle-income countries increasingly need second-line regimens with boosted protease inhibitors. However, data are scarce for treatment response in patients with HIV-1 subtype C (HIV-1C), which is predominant in these regions. We aimed to examine factors associated with virological failure in patients in a standardised national health-care setting.
Methods
We analysed data for participants in InfCare HIV, a prospective national cohort that includes more than 99% of people with HIV in Sweden. We extracted data for the cohort from the InfCare HIV database on Jan 14, 2015. Baseline was initiation of antiretroviral therapy. We used logistic regression to assess factors associated with primary virological failure (failure to suppress HIV-1 within 9 months) in patients with HIV-1B and HIV-1C and calculated odds ratios (OR) for failure. We also used Cox regression models to calculate hazard ratios (HR) for time-to-secondary virological failure (detectable viral load after initial virological suppression). We did homology-based molecular modelling to assess docking.
Findings
We included 1077 patients with HIV-1B and 596 with HIV-1C. In multivariate regression analysis, pre-therapy higher viral load (OR 1.82, 95% CI 1.49–2.21; p<0.0001), subtype C infection (1.75, 1.06–2.88; p=0·028), and boosted protease inhibitor-based regimens (1.55, 1.45–2.11; p=0.004) were associated with increased risk of primary virological failure. Individuals with HIV-1C who were given therapy with boosted protease inhibitors had earlier time-to-secondary virological failure than did those with HIV-1B given similar regimens (adjusted HR 1.92, 95% CI 1.30–2.83; p=0.002). Molecular modelling suggested lower affinity for protease inhibitors to HIV-1C protease than to HIV-1B.
Interpretation
Our findings suggest an increased risk of virological failure in patients with HIV-1C, especially in those on boosted protease inhibitor-based regimens. Future studies should further dissect the biochemical and viral mechanisms of resistance to protease inhibitors in patients with non-B subtypes of HIV-1, including clinical studies to assess the efficacy of boosted protease inhibitor-based regimens in low-income and middle income countries.
Introduction
Most HIV-1 infections occur in people in low-income and middle-income countries (LMICs). HIV-1 subtype C (HIV-1C) causes about half of HIV-1 infections worldwide and causes most HIV infections in LMICs, especially in Ethiopia, India, and sub-Saharan Africa.1 The overwhelming success of antiretroviral therapy for HIV-1B, which is predominant in high-income countries, led WHO to propose a public health approach to scale up therapy in LMICs, although specialist physician management and laboratory monitoring are absent in many of these regions.2
After a decade of scale-up of antiretroviral therapy in LMICs with first-line treatment consisting of two nucleoside reverse transcriptase inhibitors in combination with a non-nucleoside reverse transcriptase inhibitor, increased drug-resistance mutations mean availability of second-line or alternative regimens is imperative. In most LMICs, only 1–2% of eligible patients receive standardized second-line regimens that contain ritonavir-boosted protease inhibitors. Data are scarce for second-line treatment response in patients infected with HIV-1 strains that are prevalent in LMICs. An earlier systemic review and meta-analysis indicated high rates of virological failure in patients on second-line therapies with ritonavir-boosted protease inhibitors in LMICs.3
Although studies from high-income countries report that patients with non-B subtype HIV-1 had responses to antiretroviral therapy similar to those with HIV-1B,4–9 a 2015 study suggested that HIV-1C infection was independently associated with virological failure.10 These studies included only a few patients, typically grouped non-B subtypes together, and were done in diverse geographical and clinical settings. We aimed to investigate the virological and immunological response in patients initiated on treatment regimens based on either ritonavir-boosted protease inhibitors or non-nucleoside reverse transcriptase inhibitors in a standardised national healthcare setting with the specific objective to investigate any difference between patients with HIV-1C and those with HIV-1B. We also used molecular modelling and binding simulations to investigate the mechanisms of differential response by HIV subtypes. Most patients with HIV-1C in the cohort are from LMICs,11 and our study provides insights into the potential factors for virological failure in HIV-1C-dominated epidemics in LMICs.
Methods
Study design and participants
InfCare HIV is prospective national cohort that includes more than 99% of residents living with HIV in Sweden and contains data about most individuals who have died with a confirmed HIV diagnosis since 1983. Patients are recruited from 30 infectious disease clinics across Sweden.11 As of Jan 14, 2015, 10 010 patients were registered in the cohort. Of these, 8117 (81%) patients had been initiated on ART. For this analysis, we extracted data from the database for patients with the following criteria: infection with HIV-1B or HIV-1C, as determined by automated subtyping in REGA version 3 and COMET-HIV followed by maximum likelihood phylogenetic analysis of the pol-region obtained through Sanger sequencing;11,12 age older than 18 years; not pregnant; and started ART after 1996 with two nucleoside reverse transcriptase inhibitor and either one nonnucleoside reverse transcriptase inhibitor or a ritonavir-boosted protease inhibitor. Patients’ information was anonymised and de-identified prior to analysis. The study was approved by regional ethics committees of Stockholm (2005/1167–31/3) and Gothenburg (Diary number 532-11 including amendment 20111118).
Procedures
At initiation of therapy and during routine clinical follow-up, plasma HIV-1 RNA concentrations were measured with the COBAS AmpliPrep sample preparation system followed by COBAS Amplicor HIV-1 monitor version 1.5 or COBAS TaqMan HIV-1 version 1.0 or version 2.0 (Roche Molecular Systems, Basel, Switzerland). CD4 cell counts were measured with routine flow cytometry. Genotypic resistance was tested with ViroSeq HIV-1 genotyping system (Abbott, Abbott Park, IL, USA) with drug-resistance mutations defined according to the International Antiviral Society USA 2014 update. Partial pol sequences obtained from the genotyping system were used for phylogenetic study. The Swedish HIV-1C sequences used in this study were pooled with our recently reported HIV-1C sequences from Ethiopia (n=127), India (n=102), and South Africa (n=427), as representative control sequences from east Africa (HIV-1CEA), south and southeast Asia (HIV-1CSSEA), and southern Africa (HIV-1CSA), respectively.13,14 Maximum likelihood phylogenetic analysis was done in FastTree version 2.15 HIV-1C sequences were termed as HIV-1CEA, HIV-1CSA, and HIV-1CSSEA, respectively, if the sequences clustered with the control sequences from Ethiopia, South Africa, and India with more than 70% bootstrap support or being a monophyletic cluster within a large group with control sequences.
In a subset of patients with available data, adherence was assessed with a 7 day recall via a quality-assured electronic questionnaire during a routine clinical visit. Patients were classified as perfectly adherent if they had taken 100% of prescribed doses in the past 7 days without any treatment interruptions.
We defined primary virological failure as when a regimen failed to suppress the viral load within 9 months of the start of ART (i.e, viral load >500 copies per mL if ART started between 1996 and 1998 or >50 copies per mL if started between 1999 and 2015). We defined secondary virological failure as one viral load higher than 500 copies per mL or two consecutive viral loads greater than 50 copies per mL after 9 months on ART. We included a definition of viral load higher than 500 copies per mL because this was frequently considered as therapy failure in routine clinical practice leading to change in therapy after one measurement this high. We assessed CD4 cell count changes as a secondary outcome. We also did molecular investigations. For insight into the molecular mechanism of any possible differential response to protease inhibitors, we investigated structural differences in HIV-1C and HIV-1B proteases. We used Prime version 4.2 of the Schrödinger Suite (Schrödinger, New York, NY, USA), integrated into Maestro (Schrodinger) for homology modelling of consensus HIV-1C and HIV-1B proteases from the Swedish cohort (HIV-1CSE and HIV-1BSE). The crystal structures of South African wild-type HIV-1CZA (Protein Data Bank entry 3U71)16 and HIV-1B (2IEN)17 were used as template structures for the generation of homology models of HIV-1CSE and HIV-1BSE proteases, respectively. The deposited coordinates of HIV-1CZA structure had only one subunit (monomer). We also assessed binding modes of protease inhibitors (darunavir and lopinavir) through docking scores into the structures of the proteases. The dimer of the modelled structure was generated by duplicating the structure of one subunit followed by rotation of the structure by approximately 176° with the crystal structure of HIV-1B protease (Protein Data Bank entry 2IEN) as a guide. The modelled dimer of HIV-1CSE protease was subjected to energy minimisation for 1000 iterations by use of OPLS_2005 force field followed by molecular dynamics simulations for 10,000,000 steps with a 50 fs step size. An averaged structure that contained somewhat closed flap (compared with 3U71) was used to assess docking of darunavir and lopinavir. The docking of these two protease inhibitors individually was done with Induced-Fit-Docking protocol of Schrodinger Suite, which models flexible docking (i.e., optimisation of protein side-chains to obtain best docking pose). The structures with best docking scores were further subjected to molecular dynamic simulation for 10,000,000 iterations using OPLS_2005 force field. All atoms more than 20 Å away from protease inhibitors were constrained to their mean position in all molecular dynamic simulations. Identical parameters such as force field, atom type, grid size, and number of iterations in molecular dynamic simulations were used in all molecular modelling protocols for HIV-1C and HIV-1B proteases. Before docking score was calculated, we tested the docking model by deleting darunavir and lopinavir in the crystal structures of HIV-1B protease bound to darunavir (2IEN)17 and lopinavir and calculated the root mean square deviation between most favourable docked poses (as determined by glide docking score) and the conformation in respective crystal structures.
Statistical analysis
Demographics and the baseline characteristics of patients were compiled with descriptive statistical methods; we used the χ2 test to compare groups (HIV-1B vs HIV-1C) categorical data and used the Mann-Whitney U test to compare continuous data. In univariate and multivariate logistic regression analyses we inputted the following confounding factors as variables: HIV-1 subtype (HIV-1C vs HIV-1B), gender, age of patient and year at first ART initiation, CD4 cell count and log10 viral load at first ART initiation, type of treatment at initiation (non-nucleoside reverse transcriptase inhibitor vs ritonavir-boosted protease inhibitors), year of HIV diagnosis, route of transmission, country of birth (Sweden vs not Sweden), and country of infection (Sweden vs not Sweden). We estimated the effect of each variable on the outcome and summarise with odds ratios (OR)18.
After exclusion of patients with primary virological failure, we constructed Kaplan-Meier survival curves and used the log-rank test to assess time-to-secondary virological failure per HIV-1 subtype. We constructed a Cox regression model and adjusted for only variables that were significant at univariate analysis19 to show hazard ratios HR) between HIV1-B and HIV-1C. Martingale and Schoenfeld residuals were used to assess the model assumptions.20,21 We used multivariate logistic regression to assess characteristics associated with the odds of having missed a dose. We adjusted the 7 day pill recall data by route of transmission, subtype (HIV-1B vs HIV-1C), gender, country of birth, and type of treatment. We investigated CD4 cell count change over time in patients with data available with a generalised linear mixed-effects model with a random intercept and random slope adjusted by subtype, gender, age in years, country of birth, country of infection, CD4 cell count and log10 viral load at first ART initiation, time on ART in months, and the CD4 cell count.22
For all analyses, p values lower than 0.05 were regarded as statistically significant. We did all statistical analyses with Stata version 14.0.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
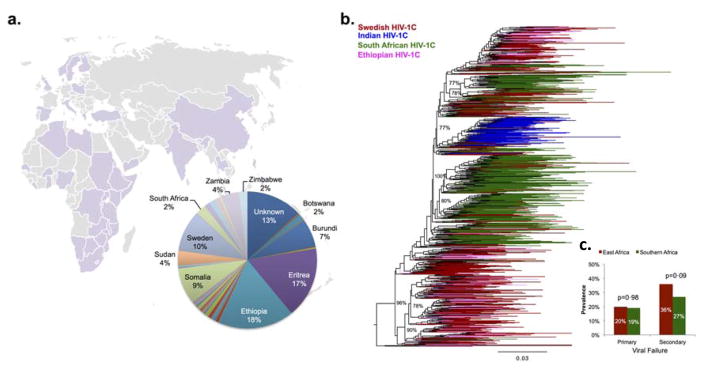
We used data for 1077 people with HIV-1B and 596 people with HIV-1C (table 1). Among the patients who been prescribed ritonavir-boosted protease inhibitors (n=863), regimen base did not differ between those with HIV-1B and those with HIV-1C (appendix p 1). Most individuals with HIV-1C were either born or infected outside of Sweden, and infected mainly through heterosexual sex (table 1). Self-reported country of infection was east African for 56%; individuals with HIV-1C were infected in 46 different countries, mainly in sub-Saharan Africa (figure 1). Despite the fact that pretreatment viral load was higher in patients with HIV-1B than in those with HIV-1C, primary virological failure and secondary virological failure were more common in patients with HIV-1C (table 1). 443 (74%) patients were infected with HIV-1CEA strains, whereas 135 (23%) were infected with HIV-1CSA strains, and 18 (3%) were infected with HIV-1CSSEA strains. No differences were observed in the proportion of patients with primary or secondary virological failure between the HIV-1CEA or HIV-1SA groups (figure 1).
Table 1.
Patient characteristics at initiation of antiretroviral therapy.
| Characteristics | Subtype B | Subtype C | Over All | P-value |
|---|---|---|---|---|
| N (%) | 1077 (64.4) | 596 (35.6) | 1673 (100) | |
| Gender n (%) | ||||
| Female | 95 (8.8) | 326 (54.7) | 421 (25.2) | <0·001 |
| Male | 982 (91·2) | 270 (45.3) | 1252 (74.8) | |
| Median age start ART (IQR) | 40 (33–49) | 37 (31–45) | 39 (32–47) | <0·001 |
| Country of birth n (%) | ||||
| Sweden | 674 (62.6) | 61 (10·2) | 735 (43.9) | <0·001 |
| Non-Swedish | 403 (37.4) | 535 (89.8) | 938 (56.1) | |
| Country of transmission; n (%) | ||||
| Sweden | 659 (65.4) | 58 (10·6) | 717 (46.1) | <0·001 |
| Abroad | 349 (34.6) | 488 (89.4) | 837 (53.9) | |
| Route of transmission | ||||
| PWID | 85 (7.9) | 4 (0·7) | 89 (5.4) | <0·001 |
| Heterosexual | 148 (13.8) | 474 (83) | 622 (37.9) | |
| MSM | 814 (76) | 21 (3.7) | 835 (50·9) | |
| Other | 24 (2.2) | 72 (12.6) | 96 (5.9) | |
| Median year of HIV diagnosis (IQR) | 2007 (2003–2010) | 2008 (2005–2011) | 2009 (2006–2012) | <0·001 |
| Median year of first ART (IQR) | 2009 (2006–2011) | 2010 (2007–2012) | 2009 (2006–2012) | <0·001 |
| Median Log10 VL start ART (IQR) | 5.0 (4.43–5.46) | 4.80 (4.12–5.38) | 4.92 (4.34–5.43) | 0·001 |
| Median CD4 start ART (IQR) | 280 (177–394) | 233 (120–338) | 264 (150–377) | <0·001 |
| Type of treatment | ||||
| NNRTI | 518 (48.1) | 292 (49.1) | 810 (48.4) | 0·72 |
| PI/r | 559 (51·9) | 303 (50·9) | 862 (51·6) | |
| Therapeutic response | ||||
| Primary failure | 172 (16) | 119 (20) | 291 (17.6) | 0·039 |
| Secondary failure | 259 (24) | 199 (39.4) | 458 (27.4) | <0·001 |
Figure 1. Country of infection of patients with HIV-1C (A) and phylogenetic origin of the virus (B, C).
Phylogenetically 74% of the patients were infected with HIV-1C strains representative of east Africa. No intra-HIV-1C differences in primary or secondary viralogical failure were observed (C).
In univariate regression analysis, risk of primary virological failure was higher in patients infected with HIV-1C, patients with higher viral load at the start of ART, and patients who initiated on ritonavir-boosted protease inhibitors (table 2). After adjustment for these factors in multivariate analysis, patients with HIV-1C had nearly twice the risk of primary virological failure than had those with HIV-1B (OR 1.75, 95% CI 1.06–2.9). Patients who initiated treatment on a boosted protease inhibitor-based therapy had higher risk of failure than did those who initiated on a treatment based on non-nucleoside reverse transcriptase inhibitor (table 2). Risk of viral failure was lower in women patients than in men patients. Injection drug use was associated with higher risk of failure; however, numbers were very small. Because HIV-1C infection is more common in individuals infected through heterosexual sex, we did a stratified analysis with only these patients (n=622): outcomes were consistent with the whole population (i.e., higher risk of virological failure with HIV-1C than with HIV-1B; table 2).
Table 2.
Factors associated with primary viral treatment failure (multivariate logistic regression model).
| All patients (n= 1673) | Heterosexuals (n= 622) | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | P-value | Multivariate | P-value | Univariate | P-value | Multivariate | P-value | |
| Subtype | ||||||||
| HIV-1B | 1 | 1 | 1 | |||||
| HIV-1C | 1·31 (1·01; 1·70) | 0·039 | 1·75 (1·06; 2.9) | 0·028 | 1·48 (0·88; 2.50) | 0·143 | 2·26 (1·13; 4.52) | 0·021 |
| Gender | ||||||||
| Male | 1 | 1 | 1 | 1 | ||||
| Female | 0·75 (0·55; 1·02) | 0·07 | 0·49 (0·32; 0·75) | 0·001 | 0·51 (0·33; 0·77) | 0·001 | 0·42 (0·25; 0·70) | 0·001 |
| Age in years | 1 (0·99; 1·02) | 0·488 | 0·99 (0·97; 1·01) | 0·216 | 1·00 (0·98; 1·02) | 0·723 | 0·99 (0·96; 1·01) | 0·365 |
| Year start ART | 0·97 (0·94; 1·00) | 0·095 | 0·98 (0·92; 1·04) | 0·464 | 0·97 (0·92; 1·02) | 0·273 | 1·04 (0·94– 1·15) | 0·443 |
| CD4 start ART | 0·99 (0·99; 1) | <0·001 | 0·99 (0·99; 1) | 0·265 | 0·99 (0·99; 1·00) | 0·026 | 0·99 (0·99; 1) | 0·402 |
| Log VL start ART | 1·94 (1·63; 2.30) | <0·001 | 1·82 (1·49; 2.21) | <0·001 | 1·87 (1·42; 2.46) | <0·001 | 1·83 (1·33; 2·53) | <0·001 |
| Type of treatment | ||||||||
| NNRTI | 1 | 1 | 1 | 1 | ||||
| PI/r | 1·81 (1·40; 2.35) | <0·001 | 1·55 (1·15; 2.11) | 0·004 | 1·54 (1·01; 2.36) | 0·047 | 1·81 (1·09; 2.99) | 0·020 |
| Year HIV diagnosis | 1 (0·98; 1·02) | 0·878 | 1·02 (0·98; 1·07) | 0·256 | 0·99 (0·95; 1·03) | 0·713 | 0·97 (0·9; 1·04) | 0·383 |
| Route of transmission | ||||||||
| MSM | 1 | 1 | ||||||
| PWID | 2.08 (1·25; 3.44) | 0·005 | 2.52 (1·43; 4.44) | 0·001 | ||||
| Heterosexual | 1·2 (0·90; 1·58) | 0·212 | 1·11 (0·66; 1·87) | 0·684 | ||||
| Other | 1·39 (0·81; 2.37) | 0·231 | 1·34 (0·64; 2.80) | 0·442 | ||||
| Country of birth | ||||||||
| Sweden | 1 | 1 | 1 | 1 | ||||
| Abroad | 1·09 (0·84; 1·40) | 0·529 | 0·91 (0·63; 1·32) | 0·62 | 1·1 (0·66; 1·86) | 0·709 | 0·81 (0·38; 1·73) | 0·593 |
| Country of infection | ||||||||
| Sweden | 1 | 1 | 1 | 1 | ||||
| Other | 1·14 (0·87; 1·48) | 0·35 | 1·12 (0·77; 1·62) | 0.565 | 1·45 (0·79; 2.66) | 0·233 | 1·18 (0·57; 2·44) | 0·650 |
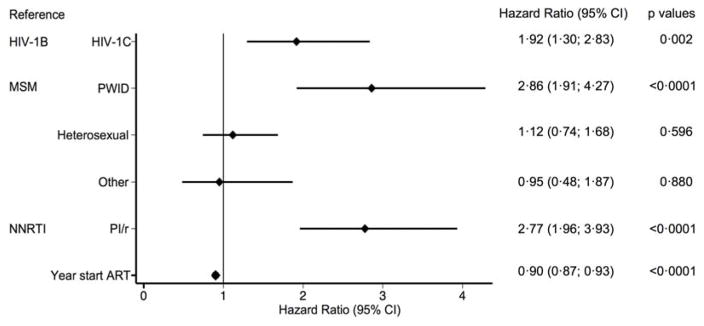
In our analysis of time-to-secondary virological failure (n=223), we adjusted for the following variables: HIV-1 subtype, type of treatment, route of transmission, and year of start of ART, with type of combination ART regimen as a time-dependent variable. Patients with HIV-1C had earlier virological failure than did those with HIV-1B (figure 2). Patients on regimens based on ritonavir-boosted protease inhibitors also had earlier failure than did those on treatment based on a non-nucleoside reverse transcriptase inhibitor (figure 2). In patients who initiated therapy with ritonavir-boosted protease inhibitors, those with HIV-1C had earlier virological failure than those with HIV-1B (1.79, 1.26–2.86; p=0.014); a difference not present between patients initiated on therapy based on a non-nucleoside reverse transcriptase inhibitor (1.98, 094–4.17; p=0.072). A Cox regression analysis of time-to-secondary virological failure that adjusted for all variables had similar findings (appendix).
Figure 2. Analysis of time-to-secondary virological failure.
NNRTI=non-nucleoside reverse transcriptase inhibitor. PI/r=ritonavir-boosted protease inhibitor.
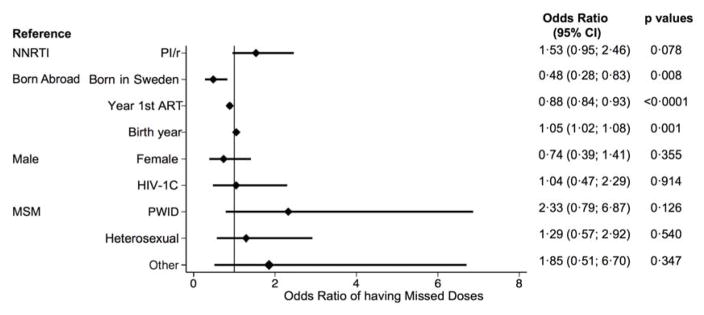
We had 7 day adherence data for 616 (57%) patients with HIV-1B and 213 (36%) with HIV-1C. 729 (88%) of the 829 patients reported being perfectly adherent in the past 7 days. Multivariate logistic regression showed no difference in odds of having missed doses between the two types of treatment or the two HIV-1 subtypes (figure 3).
Figure 3. Logistic regression model of odds of having missed one dose in the past 7 days.
NNRTI=non-nucleoside reverse transcriptase inhibitor. PI/r=ritonavir-boosted protease inhibitor.
We did analysis of CD4 cell increase (gain) among 1173 patients with 9842 CD4 cell counts measured during viral suppression until viral failure or to end of first-line therapy or to end of the study period (Jan 14, 2015). There was slightly lower gain in patients with HIV-1C than in those with HIV-1B (table 3). Baseline CD4 cell count, log10 viral load, and age at initiation, and country of infection also had a significant effect on the CD4 cell count gain over time (table 3).
Table 3. CD4+ T-cell change over time analysed with an adjusted generalized linear mixed-effects model with a random intercept and random slope.
The analysis was performed among 1173 patients and 9842 CD4+ T-cell counts, during viral suppression until viral failure or to end of first line therapy or to end of the study period (14 Jan 2015).
| Subtype | p-value | |
|---|---|---|
| HIV-1B | Ref | |
| HIV-1C | −43.05 (−65.53; −20·58) | <0·001 |
| Gender | ||
| Male | Ref | |
| Female | −18.96 (−40·53; 2.61) | 0·085 |
| Age in years | −1·31 (−2.06; −0·56) | 0·001 |
| Country of birth | ||
| Abroad | Ref. | |
| Sweden | −3.92(−22.52; 14.69) | 0·680 |
| Country of infection | ||
| Infected abroad | Ref | |
| Sweden | 34.41 (15.86; 52.96) | <0·001 |
| CD4+ T-cell count | 0·85 (0·81; 0·90) | <0·001 |
| Log10 viral load | 18·47 (8·96; 27·97) | <0·001 |
| Time on ART in Months | 3·47 (3·35; 3·59) | <0·001 |
| CD4 T-cell gain/months | ||
| HIV-1B | Ref | |
| HIV-1C | −0·27 (−0·50; −0·04) | 0·02 |
| Intercept | 142·93 (81·86; 203·99) | <0·001 |
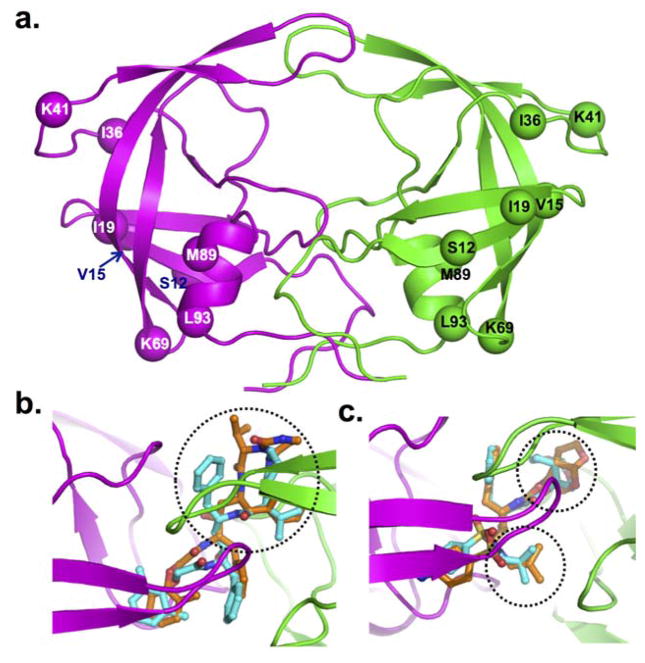
Sequence analysis of the HIV-1C protease from individuals who had not taken ritonavir-boosted protease inhibitors for the first time identified naturally occurring polymorphisms at eight positions: Thr12Ser, Ile15Val, Leu19Ile, Met36Ile, Arg41Lys, His69Lys, Leu89Met, and Ile93Leu (compared with the consensus HIV-1B used in Stanford HIV database; figure 4). Similar results were observed when Stanford University HIV Drug Resistance Database sequences were analysed (data not shown). No specific pattern was observed in the reverse transcriptase sequences. 99 sequences were available from patients with secondary virological failure. 16 (44%) of 36 people for whom reverse transcriptase inhibitors failed had at least one drug-resistance mutation compared with two (3%) of 62 for whom treatment with boosted protease inhibitors failed. Major protease-inhibitor drug-resistance mutations were identified in two patients with HIV-1B (Met46Leu and Leu90Met) and one with HIV-1C (Met46Leu).
Figure 4. Molecular model of HIV-1C protease dimer.
The backbone of protease is shown in secondary structure representation: α-helices as helical ribbon, β strands as flat arrows, and unordered structure as thin tubes (A). The two dimers are coloured magenta and green. The Cα atoms of the residues that are not conserved between HIV-1B and HIV-1C proteases are shown as solid balls. Superposition in HIV-1B and HIV-1C is shown for lopinavir (B) and darunavir (C). The protease inhibitors are rendered in balls-and-stick (cyan carbons in HIV-1B, and orange carbons in HIV-1C) in two proteases. The other atoms are coloured by atom type (red for oxygen, blue for nitrogen, and yellow for sulphur). The ribbon diagram shows the backbone of HIV-1C proteases. The difference in conformation of different moieties for the drugs in the two proteases is circled by dotted lines.
The overall modelled structure of HIV-1CSE (figure 4) was similar to that of HIV-1BSE but differed significantly from the reported monomeric crystal structure of South African HIV-1CZA protease (PDB entry 3U71). The major difference existed in the conformation of flap region. The molecular dynamics simulations showed high flexibility in the conformation of the flap region of HIV-1CSE protease. When testing our docking model in the crystal structures of HIV-1B protease bound to darunavir (Protein Data Base entry 2IEN)17 and lopinavir (1MUI)23 the root-mean-square deviation between most favourable docked poses (as determined by glide docking score) for lopinavir or darunavir and the conformation in respective crystal structures were less than 0.7 Å. In our analysis, lopinavir bound to HIV-1C protease in significantly different conformation than it did to HIV-1B protease (figure 4). The major difference was seen in the conformation of 1,3-diazacyclohexa-2-one moiety of lopinavir. This change significantly affected the binding of lopinavir to HIV-1CZA protease compared with HIV-1BSE protease as reflected by glide docking score of −8.07 versus −8.9, respectively. The superposed structures of darunavir in HIV-1CSE and in HIV-1BSE showed two major differences between the subtypes: in the position of sulphonyl moiety and the acyclic isobutyl ring (figure 4). These changes significantly affected the binding of darunavir to HIV-1CZA protease (docking score of −7.81) compared with HIV-1BSE proteases (−8.11). These data, although based on the modelled structures, suggest that differences exist between HIV-1C and HIV-1B molecules in the binding of protease inhibitors to their proteases.
Discussion
In this analysis of a prospective cohort, patients with HIV-1C who received ART regimens based on ritonavir-boosted protease inhibitors had a significantly increased primary and secondary virological failure compared with those with HIV-1B. In the patients in whom therapy failed, a shorter time-to-secondary virological failure was found in those with HIV-1C than in those with HIV-1B, especially in those on protease inhibitors. The poorer treatment outcome occurred in the patients with HIV-1C despite developed clinical care, modern laboratory monitoring, and focused adherence support at highly HIV-specialised infectious disease clinics in a high-income country (Sweden). Naturally occurring poly morphisms in HIV-1C protease might affect the binding of at least some protease inhibitors, potentially contributing to the differences we observed.
Studies from high-income countries of the viral and immunological responses to ART in patients with non-B subtype HIV-1 have suggested a similar outcome to those with HIV-1B,4,5,7–9 although the Swiss HIV cohort has reported better viral response in white people with non-B subtype HIV-1 than in those with HIV-1B.6 However, the small number of people with non-B subtypes, which were grouped together despite potential biological differences, in these studies is likely to lead to an over simplification of the data.5 A study from the UK comparing HIV-1B, HIV-1A1, HIV-1C, HIV-1D, and HIV-1CRF02_AG reported more frequent viral rebound in patients with HIV-1D.8 A more recent clinical trial indicated that patients with HIV-1C were more prone to virological failure.10 However, the inclusion of few patients from diverse geographical regions and socio medico-economic backgrounds could have biased the outcome.10 Also, data obtained through clinical trials do not always reflect the real-world situation, which might be especially true in LMICs.
Our findings of increased virological failure in patients with HIV-1C cannot be explained by differences in adherence between the HIV-1 subtypes or regimen types (figure 3); moreover, clinical care was harmonised between the different categories of patients and all were treated at highly experienced HIV specialised infectious disease clinics with an almost 100% linkage and retention to care.24 Our analysis does include the early generation protease inhibitors. However, most patients were taking ritonavir-boosted lopinavir, ritonavir-boosted atazanavir, or ritonavir-boosted darunavir, which are the still available for use in LMICs.
When analysing pretherapy protease sequences we found naturally occurring polymorphisms at eight positions without any major protease inhibitor drug-resistant mutations. These polymorphisms have also been reported in the HIV-1CSA proteases from South African consensus sequences,16 investigators of which claimed that the polymorphism in the position 36 might affect the stability of the hinge region of the protease.16 Our molecular dynamics simulations showed high flexibility in the conformation of the flap region of HIV-1CSE protease compared with HIV-1BSE protease, which is consistent with this previous finding.16 Earlier structural and biochemical studies posit that increased flexibility is likely to contribute to less protease inhibitor susceptibility in HIV-1CSA protease.25,26 Our molecular docking analyses further corroborate this hypothesis because both lopinavir and darunavir showed lower binding affinity to HIV-1CSE protease than to HIV-1BSE protease.
Only two major protease inhibitor drug-resistance mutations were found in patients with HIV-1B for whom treatment failed but not in those with HIV-1C, which is congruent with the fact that mutations causing resistance to protease inhibitors are seldom seen at short-term virological failure.27–32 However, routine population sequencing was used, which precludes the identification of resistance mutations in the minor viral quasispecies, a region we have reported to contain a higher amount of drug-resistance mutations to reverse transcriptase inhibitors in east African patients with HIV-1C than in Indian and white patients with HIV-1C.12 Moreover, a 2013 study reported that sequence changes in the gp41 cytoplasmic tail and uncleaved gag can confer resistance to protease inhibitors; these regions are not included in current clinical assays.33 Thus, we cannot exclude that non-identified drug-resistance mutations could have contributed to the difference in outcome between the HIV-1 subtypes.
This study has limitations and strengths. Most patients with HIV-1C were born in Africa; therefore we cannot exclude that social and ethnic factors affected the outcome. However, most of the population reported being perfectly adherent and there was no recognised difference in adherence between the patients with HIV-1B and those with HIV-1C. A major strength of our study is that all patients received care at HIV specialised infectious disease clinics in a high-income country where linkage and retention in care is almost 100% and where patients receive standardised monitoring with modern laboratory access and adherence support by nurses and counsellors.24 Another strength of our study is that we included a large number of patients with HIV-1C and did not pool all non-subtype B into one category. Therefore, our results of a higher virological failure rate in patients with HIV-1C are of concern and might be aggravated in treatment settings that use a public health approach without any viral load monitoring or genetic resistance testing analysis.
In conclusion, we show that patients with HIV-1C given ritonavir-boosted protease inhibitors had higher risk of primary and secondary virological failure than did those with HIV-1B. Our findings also support biochemical and structural predictions of less susceptibility to protease inhibitors for HIV-1C molecules. As LMICs are poised to scale up second-line ART containing ritonavir-boosted protease inhibitors, a concern is that protease inhibitors will be less efficient in patients with HIV-1C. Therefore, increased understanding is needed for the biochemical and viral mechanisms of resistance to protease inhibitors and therapy dynamics in HIV-1C and other non-B subtypes. Studies of new drugs such as integrase inhibitors should be done in patients with HIV-1C and other non-B subtypes, which are responsible for the greatest global HIV burden.
Supplementary Material
Acknowledgments
We were funded by grants from Karolinska Institutet Research Foundation (2014fobi41250), Swedish Research Council (2012–3476), Stockholm County Council (2013–0042), and Swedish Physicians against AIDS grant (Foa2013–0014 and Fob2015–009). The molecular modelling was supported by a grant from National Institute of Health (grant number P50 GM103368) and University of Missouri (Mizzou Advantage; grant number DKE13) We thank the steering committee of the Swedish InfCare HIV cohort, which provided the data, and thank all participating clinics. We also thank Michele Santacatterina for his assistance in statistical coding.
Footnotes
Contributors
UN conceived of the study. UN and AH designed the study. AH, VS, KS, and UN generated and analysed data. VS and AS did clinical interpretation. UN wrote the first draft, which was corrected by AH, VS, KS, and AS.
Declaration of interests
We declare no competing interests.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S WHO-UNAIDS Network for HIV Isolation and Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–89. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilks CF, Crowley S, Ekpini R, et al. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006;368:505–10. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 3.Ajose O, Mookerjee S, Mills EJ, Boulle A, Ford N. Treatment outcomes of patients on second-line antiretroviral therapy in resource-limited settings: a systematic review and meta-analysis. AIDS. 2012;26:929–38. doi: 10.1097/QAD.0b013e328351f5b2. [DOI] [PubMed] [Google Scholar]
- 4.Bannister WP, Ruiz L, Loveday C, et al. HIV-1 subtypes and response to combination antiretroviral therapy in Europe. Antivir Ther. 2006;11:707–15. [PubMed] [Google Scholar]
- 5.Geretti AM, Harrison L, Green H, et al. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis. 2009;48:1296–305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- 6.Scherrer AU, Ledergerber B, von Wyl V, et al. Improved virological outcome in white patients infected with HIV-1 non-B subtypes compared to subtype B. Clin Infect Dis. 2011;53:1143–52. doi: 10.1093/cid/cir669. [DOI] [PubMed] [Google Scholar]
- 7.Chaix ML, Seng R, Frange P, et al. Increasing HIV-1 non-B subtype primary infections in patients in France and effect of HIV subtypes on virological and immunological responses to combined antiretroviral therapy. Clin Infect Dis. 2013;56:880–87. doi: 10.1093/cid/cis999. [DOI] [PubMed] [Google Scholar]
- 8.Easterbrook PJ, Smith M, Mullen J, et al. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc. 2010;13:4. doi: 10.1186/1758-2652-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dierynck I, De Meyer S, Lathouwers E, et al. In vitro susceptibility and virological outcome to darunavir and lopinavir are independent of HIV type-1 subtype in treatment-naive patients. Antivir Ther. 2010;15:1161–69. doi: 10.3851/IMP1697. [DOI] [PubMed] [Google Scholar]
- 10.Kantor R, Smeaton L, Vardhanabhuti S, et al. Pretreatment HIV drug resistance and HIV-1 subtype C are independently associated with virologic failure: results from the multinational PEARLS (ACTG A5175) Clinical Trial. Clin Infect Dis. 2015;60:1541–49. doi: 10.1093/cid/civ102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neogi U, Haggblom A, Santacatterina M, et al. Temporal trends in the Swedish HIV-1 epidemic: increase in non-B subtypes and recombinant forms over three decades. PLoS One. 2014;9:e99390. doi: 10.1371/journal.pone.0099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekici H, Rao SD, Sonnerborg A, Ramprasad VL, Gupta R, Neogi U. Cost-efficient HIV-1 drug resistance surveillance using multiplexed high-throughput amplicon sequencing: implications for use in low- and middle-income countries. J Antimicrob Chemother. 2014;69:3349–55. doi: 10.1093/jac/dku278. [DOI] [PubMed] [Google Scholar]
- 13.Neogi U, Haggblom A, Singh K, et al. Factors influencing the efficacy of rilpivirine in HIV-1 subtype C in low- and middle-income countries. J Antimicrob Chemother. 2016;71:367–71. doi: 10.1093/jac/dkv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neogi U, Engelbrecht S, Claassen M, et al. Mutational heterogeneity in p6 Gag late assembly (L) domains in HIV-1 subtype C viruses from South Africa. AIDS Res Hum Retroviruses. 2016;32:80–84. doi: 10.1089/AID.2015.0266. [DOI] [PubMed] [Google Scholar]
- 15.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naicker P, Achilonu I, Fanucchi S, et al. Structural insights into the South African HIV-1 subtype C protease: impact of hinge region dynamics and flap flexibility in drug resistance. J Biomol Struct Dyn. 2013;31:1370–80. doi: 10.1080/07391102.2012.736774. [DOI] [PubMed] [Google Scholar]
- 17.Tie Y, Boross PI, Wang YF, et al. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J Mol Biol. 2004;338:341–52. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 18.Homer D, Lemeshow S, Sturdivant R. Applied logistic regression. 3. Hoboken, NJ: Wiley; 2013. [Google Scholar]
- 19.Kleinbaum DG, Klein M, editors. Survival analysis: a self-learning text. New York, NY: Springer-Verlag; 2005. Competing risks survival analysis; pp. 391–461. [Google Scholar]
- 20.Mellors JW, Kingsley LA, Rinaldo CR, Jr, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–79. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 21.Therneau TM, Grambsch PM, Fleming TR. Martingale-based residuals for survival models. Biometrika. 1990;77:147–60. [Google Scholar]
- 22.Fitzmaurice GM, Laird NM, Ware JH. Applied longitudinal analysis. Hoboken, NJ: John Wiley & Sons; 2012. [Google Scholar]
- 23.Stoll V, Qin W, Stewart KD, et al. X-ray crystallographic structure of ABT-378 (lopinavir) bound to HIV-1 protease. Bioorg Med Chem. 2002;10:2803–06. doi: 10.1016/s0968-0896(02)00051-2. [DOI] [PubMed] [Google Scholar]
- 24.European Centre for Disease Prevention and Control. Thematic report: HIV continuum of care. [accessed Sept 20, 2015];Monitoring implementation of the Dublin Declaration on Partnership to Fight HIV/AIDS in Europe and Central Asia: 2014 progress report. 2015 [Google Scholar]
- 25.Mosebi S, Morris L, Dirr HW, Sayed Y. Active-site mutations in the South African human immunodeficiency virus type 1 subtype C protease have a significant impact on clinical inhibitor binding: kinetic and thermodynamic study. J Virol. 2008;82:11476–79. doi: 10.1128/JVI.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velazquez-Campoy A, Vega S, Fleming E, et al. Protease inhibition in African subtypes of HIV-1. AIDS Rev. 2003;5:165–71. [PubMed] [Google Scholar]
- 27.Hill A, McBride A, Sawyer AW, Clumeck N, Gupta RK. Resistance at virological failure using boosted protease inhibitors versus nonnucleoside reverse transcriptase inhibitors as first-line antiretroviral therapy--implications for sustained efficacy of ART in resource-limited settings. J Infect Dis. 2013;207(suppl 2):S78–84. doi: 10.1093/infdis/jit112. [DOI] [PubMed] [Google Scholar]
- 28.Levison JH, Orrell C, Gallien S, et al. Virologic failure of protease inhibitor-based second-line antiretroviral therapy without resistance in a large HIV treatment program in South Africa. PLoS One. 2012;7:e32144. doi: 10.1371/journal.pone.0032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. Protease inhibitor resistance is uncommon in HIV-1 subtype C infected patients on failing second-line lopinavir/r-containing antiretroviral therapy in South Africa. AIDS Res Treat. 2011;2011:769627. doi: 10.1155/2011/769627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallego O, de Mendoza C, Perez-Elias MJ, et al. Drug resistance in patients experiencing early virological failure under a triple combination including indinavir. AIDS. 2001;15:1701–06. doi: 10.1097/00002030-200109070-00014. [DOI] [PubMed] [Google Scholar]
- 31.Taiwo B, Zheng L, Gallien S, et al. Efficacy of a nucleoside-sparing regimen of darunavir/ritonavir plus raltegravir in treatment-naive HIV-1-infected patients (ACTG A5262) AIDS. 2011;25:2113–22. doi: 10.1097/QAD.0b013e32834bbaa9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland KA, Parry CM, McCormick A, et al. Evidence for reduced drug susceptibility without emergence of major protease mutations following protease inhibitor monotherapy failure in the SARA trial. PLoS One. 2015;10:e0137834. doi: 10.1371/journal.pone.0137834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabi SA, Laird GM, Durand CM, et al. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest. 2013;123:3848–60. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.