Abstract
Novel N-3-alkylated 6-anilinouracils have been identified as potent and selective inhibitors of bacterial DNA polymerase IIIC, the enzyme essential for the replication of chromosomal DNA in gram-positive bacteria. A nonradioactive assay measuring the enzymatic activity of the DNA polymerase IIIC in gram-positive bacteria has been assembled. The 6-anilinouracils described inhibited the polymerase IIIC enzyme at concentrations in the nanomolar range in this assay and displayed good in vitro activity (according to their MICs) against staphylococci, streptococci, and enterococci. The MICs of the most potent derivatives were about 4 μg/ml for this panel of bacteria. The 50% effective dose of the best compound (6-[(3-ethyl-4-methylphenyl)amino]-3-{[1-(isoxazol-5-ylcarbonyl)piperidin-4-yl]methyl}uracil) was 10 mg/kg of body weight after intravenous application in a staphylococcal sepsis model in mice, from which in vivo pharmacokinetic data were also acquired.
The antibiotic therapy of nosocomial infections caused by gram-positive bacteria, in particular, Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, and Streptococcus pneumoniae, has become more and more problematic during the last 15 years because of the growing rates of development of resistance of these organisms to traditional antibiotics (26). The identification of novel classes of antibacterial substances that inhibit a new or unexploited target is expected to facilitate novel therapeutic approaches for the treatment of infections caused by multiresistant pathogens. 6-Anilinouracils (AUs) have been described in the literature (1, 9, 24, 25, 27, 28, 31) and in patent applications (3, 7) as a novel antibacterial class showing selective inhibition of DNA polymerase IIIC (Pol IIIC; formerly Pol III), the enzyme required for the replication of chromosomal DNA in gram-positive organisms with low G+C contents (29).
Pol IIIC, encoded by the structural gene polC, is one of the two essential replication-specific DNA polymerases in gram-positive bacteria (10, 12). The polC gene is absent from the eubacteria with high G+C contents, e.g., Mycobacterium, Corynebacterium, and Streptomyces, and the gram-negative eubacteria, e.g., Escherichia, Salmonella, and Haemophilus, as well as eukaryotic cells, but is strongly conserved in a broad group of gram-positive pathogens, including staphylococci, streptococci, and enterococci (29). Pol IIIC has an essential function in chromosomal DNA replication and can be selectively inhibited by the AU family (29).
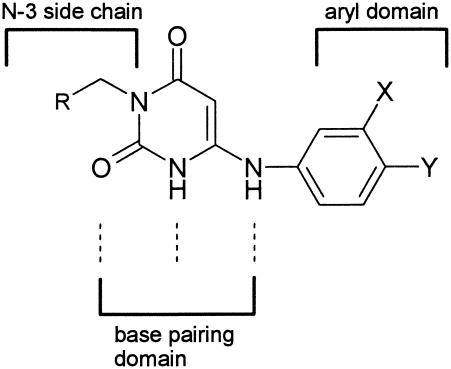
The AU-based inhibitors can be regarded as consisting of three different domains (Fig. 1) (25). The central part of the molecule is the base-pairing domain, which comprises three uracil-specific substituents (the ring NH, the 2-oxo, and the 6-NH). The aryl domain, which consists of an appropriately substituted aryl group, interacts with the Pol IIIC enzyme by binding into a hydrophobic pocket (25). Although they are formally pyrimidines, the AUs inhibit the Pol IIIC enzyme due to their capacity to mimic the guanine moiety of the 2′-deoxyribonucleoside triphosphate guanine, dGTP (25). The unconventional base-pairing domain of the AUs is able to form three hydrogen bonds with the pyrimidine base cytosine (25). The Pol IIIC enzyme, which tries to elongate the primer terminus past a cytosine residue in the DNA template, is inhibited by binding of the aryl domain of the anilinouracils into a unique hydrophobic pocket of the enzyme (25). As a consequence, the inhibitor sequesters the enzyme into a nonproductive, reversible ternary complex with the template primer (25).
FIG. 1.
Important pharmacophoric elements of a generic AU.
The increased inhibitory activities of AU compounds against Pol IIIC and the growth of gram-positive bacteria have been described for N-3-substituted anilinouracils (9, 25). The enhanced antibacterial activities of the compounds with the N-3 side chain alkyl substituents compared to those of N-3-unsubstituted molecules have been described (9, 25). Moreover, more complex aminoalkyl substituents have also been described in this position of the molecule (31).
In this paper we describe the preparation of several N-3-substituted anilinouracils, as well as the effects of these structural variations on antibacterial potency and aqueous solubility. We developed a nonradioactive Pol IIIC assay that enables the determination of the anti-Pol IIIC activities of the novel N-3-substituted anilinouracils. Moreover, a more detailed biological characterization of 6-[(3-ethyl-4-methylphenyl)amino]-3-{[1-(isoxazol-5-ylcarbonyl)piperidin-4-yl]methyl}uracil (EMAIPU;compound 1) is described.
(This work was presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 2003 [A. Kuhl, K. Ehlert, N. Svenstrup, C. Ladel, M. Brands, D. Haebich, T. Lampe, and K. Ziegelbauer, Abstr. 43th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-2155, 2003].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were S. aureus 133, S. pneumoniae 1707/4, and E. faecalis ICB 27159. All strains were subcultured on agar plates containing 5% sheep blood. Liquid cultures were prepared in brain heart infusion (BHI) medium containing 10% bovine serum for S. pneumoniae and E. faecalis at 37°C.
Cloning, expression, and purification of Pol IIIC from S. aureus.
The polC gene was amplified from S. aureus 133 genomic DNA by PCR with the primers SAPOL31 5′-GCGCCATATGGACAGAGCAACAAAAATTTAA-3′ and SAPOLrev 5′-GCGCGGATCCTTACATATCAAATATCGAAA-3′ and transformed into pET15b (Novagen), which provides an N-terminal His tag. The PCR product encoding the polC gene was digested with BamHI and NdeI and ligated into the BamHI-NdeI-digested expression vector, resulting in plasmid pSapolCHis. Upon transformation into Escherichia coli BL21(DE3), the Pol IIIC protein could be expressed as a His tag fusion protein at 18°C for 20 h after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in a soluble form. Briefly, cells were harvested by centrifugation, washed in phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride (PMSF), and resuspended in 50 mM phosphate buffer (pH 8.0) containing 10 mM imidazole, 2 mM β-mercaptoethanol, 1 mM PMSF, and 20% glycerol. The cells were broken with a French press at 12,000 lb/in2, and the cell debris was removed by centrifugation at 27,000 × g for 2 h at 4°C. The supernatant was incubated with Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen) for 1 h at 4°C; placed into a column; and washed with 50 mM phosphate buffer (pH 8.0) containing 20 mM imidazole, 2 mM β-mercaptoethanol, 1 mM PMSF, and 10% glycerol. The protein was eluted in the presence of 100 mM imidazole and was stored in 50% glycerol at −20°C.
DNA Pol IIIC activity.
DNA Pol IIIC activity was assayed by an enzymatic coupled assay containing activated (partially digested with DNase I) calf thymus DNA as the template-primer DNA and deoxynucleoside triphosphates (dNTPs) as substrates. The reaction mixture contained 5 μl (25.0 A260 U/ml) of activated calf thymus DNA (Amersham); dATP, dTTP, dGTP, and dCTP at 20 μM each; 20 μM adenosine 5′-phosphosulfate; and 60 μM luciferin in 50 μl of 50 mM Tris-Cl (pH 7.5)-5 mM dithiothreitol-10 mM MgCl2-30 mM NaCl-0.1 mg of bovine serum albumin per ml-10% glycerol. The reaction was started by the addition of the Pol IIIC enzyme preparation. The reaction mixture was incubated for 30 min at 30°C prior to the addition of 20 μl of 0.01 mg of ATP sulfurylase (7.2 U/mg) per ml from baker's yeast (Sigma). The mixture was further incubated for 15 min at 30°C. Upon addition of 20 μl of 0.3 μg of firefly luciferase (Promega) per ml, luminescence can be measured for 1 min in a luminometer. For determination of the Km values of the nucleotides, the nucleotide of interest was used at different concentrations, ranging from 5 to 50 μM, whereas the remaining nucleotides were used in excess concentrations of 500 μM each. For the determination of the anti-Pol IIIC activities of the described N-3-substituted anilinouracils, dGTP as the competitive dNTP was omitted from the enzymatic assay (25). Test compounds were dissolved in dimethyl sulfoxide to a final concentration not higher than 2%.
MIC determinations.
MICs were determined by the broth microdilution method with an inoculum of 5 × 105 CFU/ml in BHI medium. Growth was read after 18 h of incubation at 37°C. For S. pneumoniae and E. faecalis, 10% bovine serum was added to the medium and incubation was performed under microaerophilic conditions. The test compounds were dissolved in dimethyl sulfoxide and diluted to a concentration not greater than 2.5% dimethyl sulfoxide.
Kill curves.
For the time-kill assays, log-phase cultures in BHI medium were diluted to 105 CFU/ml and treated with different concentrations of the test compound. Cultures were sampled at 0, 2, 4, 6, and 24 h of incubation at 37°C. Samples were serially diluted, plated on BHI agar, and incubated for 24 h at 37°C to determine the colony counts.
Resistance development.
Resistance development was investigated by serial passages of S. aureus 133 in 10 ml of BHI medium containing the test compounds at a concentrations of one-half the MIC, the MIC, and two times the MIC. Cells grown in the presence of the highest concentration of compound 1 after overnight incubation at 37°C were used as the inoculum for the next passage and were diluted 1:100 into fresh BHI medium containing further increasing concentrations of compound 1.
Metabolic incorporation assay.
A cell culture of Bacillus subtilis 168 was grown aerobically to the logarithmic growth phase (optical density at 535 nm, 0.1 to 0.2) at 37°C in Belitsky medium supplemented with 1 μM l-leucine (23). After the cells were diluted into fresh medium to an optical density at 535 nm of 0.02, each 1.25 ml of culture was labeled separately with 25 kBq each of l-[4,5-3H]leucine (5.11 TBq/mmol), [5,6-3H]uridine (1.48 TBq/mmol), [methyl-1′,2′-3H]thymidine (4.48 TBq/mmol), and N-acetyl-d-[1-3H]glucosamine (185 GBq/mmol). After 5 min, compound 1 (MIC, 2 μg/ml) was added to the cultures at concentrations corresponding to one-fourth the MIC, the MIC, and four times the MIC for up to 60 min. Samples of 0.1 ml were withdrawn at regular intervals and precipitated with 6% perchloric acid in a 96-well multiscreen filter plate (pore size, 0.45 μm; Millipore) to separate the radioactive metabolites from the radioactivity incorporated into high-molecular-weight material. The precipitates were washed with 0.1 ml of ethanol, and the filter plates were dried prior to the addition of 30 μl of scintillation cocktail (Ultima Gold; Packard). Radioactivity was determined in a 1450 Microbeta scintillation counter (Wallac). All radiochemicals were purchased from Amersham Pharmacia Biotech, Little Chalfont, United Kingdom.
Cytotoxicity.
Cytotoxicity was determined by measuring the metabolic (mitochondrial dehydrogenase) activities of different cell lines after 3 days of incubation with serial dilutions of the compounds. All permanent cell lines were obtained from the American Type Culture Collection (Manassas, Va.). The cells were cultured in RPMI 1640 medium (Gibco, Paisley, United Kingdom) supplemented with 10% fetal calf serum (Seromed, D), 1% glutamine, and 1% penicillin-streptomycin (Gibco). The metabolic activity was determined with an EZ4U kit (Biomedica, Vienna, Austria), which contains a soluble tetrazolium salt, according to the procedures of the manufacturer. Briefly, after the 3-day incubation with the compounds, 25 μl of the EZ4U reagent was added to the cells. The absorbance at 450 nm was determined after 4 h of incubation at 37°C in a 5% CO2 atmosphere with a SpectraFluor microplate reader (Tecan). The 50% effective concentrations (EC50s; the concentration at which 50% of the cells were killed compared to the number of control cells) were determined with GraphPad Prism software by calculation of dose-response curves by using sigmoidal curve fit. No-effect concentrations (NOECs; the concentration at which no differences in killing between treated and control cells were observed) were estimated by means of the calculated curves.
Murine sepsis model.
CFW1 mice (weight, 20 g; six mice per group) were infected with a single intraperitoneal (i.p.) injection of S. aureus 133 (0.25 ml containing 10% mucin per mouse; 106 CFU/mouse). At 30 min after infection the mice were treated intravenously (i.v.) with 0.1 ml of test compound dissolved in 2% dimethyl sulfoxide-12% Solutol at a concentration sufficient to give a dose of 10 mg/kg of body weight. The mice were monitored over a 5-day period, and the results are expressed as the number of surviving mice.
Pharmacokinetics.
The animals used were female CFW1 mice (weight, 18 to 25 g; n = 3), male Wistar rats (weight, 175 to 225 g; n = 3), and female beagle dogs (weight, 9 to 12 kg; n = 2).
For the animal studies, the compound was dissolved in 10% ethanol, 20% Solutol HS15, and 70% water. The concentration of the solution was between 0.5 and 1 mg/ml. A volume of 2 ml/kg was administered to the mice and the rats. For the dogs, the vehicle was 10% ethanol and 60% polyethylene glycol, and the volume was 0.5 ml/kg. The formulation of the test compound was given as a single i.v. administration via a caudal vein (mice and rats) or a cephalic vein (dog). The i.v. doses were given either as a bolus injection (mice and rats) or as a short infusion over 5 min (dogs).
For the rats, blood samples were drawn from the right jugular vein through an implanted cannula while the rats were conscious. For the mice, blood was obtained by exsanguination while the mice were under anesthesia. For the dogs, blood samples were obtained from a punctured jugular vein while the dogs were conscious. The blood was collected and placed into heparinized syringes or vials. Plasma was obtained by centrifugation of blood samples. The plasma was stored below −15°C before further analysis.
LC/MS/MS.
Plasma drug concentrations were quantified by a liquid chromatography (LC)-mass spectrometry (MS) (LC/MS/MS) method. A total of 10 μl of internal standard and 1 ml of acetonitrile were added to 0.1 ml of plasma. Plasma proteins were precipitated by shaking. The supernatant was evaporated under nitrogen and reconstituted in 300 μl of the mobile phase, which consisted of 50% 10 mM ammonium acetate (pH 3.5) and 50% acetonitrile. Twenty microliters of the sample was injected onto a Luna C8 column (50 by 3 mm; 5 μm; Phenomenex, Torrance, Calif.). Separation was performed with an HP1100 high-pressure liquid chromatography (HPLC) instrument (Agilent Technologies, Palo Alto, Calif.). The mobile phases consisted of acetonitrile (mobile phase B) and 10 mM ammonium acetate-formic acid (pH 3.5) (mobile phase A). The flow rate was 0.8 ml/min. The following gradient was used: from 0 to 2.0 min, 10% mobile phase B; from 2.0 to 2.1 min, 90% mobile phase B; from 2.1 to 4.0 min, 90% mobile phase B; from 4.0 to 4.1 min, 10% mobile phase B; and from 4.1 to 5.0 min, 10% mobile phase B. The HPLC column effluent was analyzed directly on an API 3000 instrument (PE SCIEX, Toronto, Ontario, Canada) with turbo ion spray ionization (positive mode). A parent-product ion combination of m/z 438.4 to 229.1 was used to monitor multiple ions. A standard curve was generated from the mean of three replicates, which were made by spiking an equal volume of plasma from untreated animals with increasing amounts of drug (1 to 1,000 ng/ml). The amount of drug was quantified by comparison of its peak area ratio in a given sample to the standard curve.
To determine the partitioning to erythrocytes and blood, 10 μl of the test substance (200 ng in acetonitrile) was added to 1 ml of blood and the mixture was incubated for 30 min at room temperature. Plasma was prepared by centrifugation and analyzed by LC/MS/MS, as described above.
The level of protein binding of compound 1 to plasma proteins was determined by the erythrocyte partitioning method described by Schuhmacher et al. (19). Pharmacokinetic data from the concentration-time data were determined by noncompartmental analysis with the KINCALC software package (version 2.33; Bayer HealthCare). The allometric scaling (correlation with body weight) of the pharmacokinetic parameters was performed as described by Boxenbaum (6). After logarithmic/logarithmic transformation, the parameters were fitted to the equation y = a · BWb, where BW is body weight and a and b are the allometric coefficient and the allometric exponent, respectively.
RESULTS
Development of a homogeneous enzymatic assay monitoring the activity of S. aureus Pol IIIC.
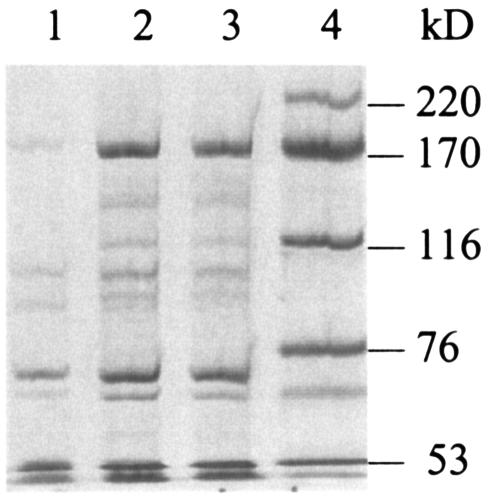
Descriptions of the cloning and expression of the S. aureus Pol IIIC enzyme in E. coli and purification of the enzyme by classical methods have been reported previously (13, 17). In order to facilitate efficient purification of enzymatically active protein in sufficient amounts, we cloned the polC gene amplified from S. aureus into pET15b, which provided an N-terminal His tag. The Pol IIIC protein could be expressed as a His tag fusion protein in a soluble and enzymatically active form. However, it was important to avoid high salt concentrations during purification to preserve the enzymatic activity of S. aureus Pol IIIC, which resulted in only partially purified His-tagged Pol IIIC protein after Ni-NTA affinity chromatography (Fig. 2).
FIG. 2.
SDS-PAGE (7.5% polyacrylamide gel) analysis of the purification of His-tagged Pol IIIC from S. aureus 133 with Coomassie blue staining. Fractions that eluted from a Ni-NTA-agarose column by using 100 mM imidazole are shown. Lane 1, uninduced control; lanes 2 and 3, expressed Pol IIIC (∼166-kDa) protein in the presence of 1 mM IPTG; lane 4, molecular mass marker.
Pol IIIC activity has commonly been measured in a heterogeneous radioactive assay that incorporates radiolabeled thymidine into acid-precipitable DNA (25). A homogeneous nonradioactive assay would facilitate the identification of potential Pol IIIC inhibitors from huge compound libraries in a short time. An enzymatic method for continous monitoring of DNA polymerase activity has been described by Nyren and Lundin (15) and Nyren (16). The DNA polymerase reaction leads to the formation of PPi (template DNA + dNTPs → replicated template DNA + PPi [Pol IIIC reaction]). The PPi can be converted into ATP by the enzyme ATP sulfurylase by using adenosine 5′-phosphosulfate as a substrate (PPi + adenosine 5′-phosphosulfate → ATP + SO4 [ATP-sulfurylase reaction]). The resulting ATP is used for the production of bioluminescence, which can easily be measured in a luminometer. This reaction is catalyzed by the well-known enzyme firefly luciferase with luciferin as the substrate (ATP + O2 + luciferin → AMP + PPi + light + CO2 [luciferase reaction]).
We obtained linear kinetics using the described enzymatically coupled nonradioactive assay with low concentrations of DNA Pol IIIC from S. aureus 133. Figure 3 shows the rate of PPi production measured as bioluminescence as a function of time and DNA Pol IIIC concentrations. The apparent Km values of the recombinant S. aureus Pol IIIC in this assay were 0.3 μM for dCTP and dGTP, 1.5 μM for dTTP, and 0.7 μM for dATP.
FIG. 3.
Time course of S. aureus Pol IIIC activity. The assay was performed as described in Materials and Methods in the absence of dGTP, and bioluminescence is shown in relative light units (RLU) per minute.
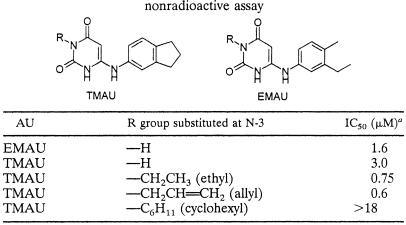
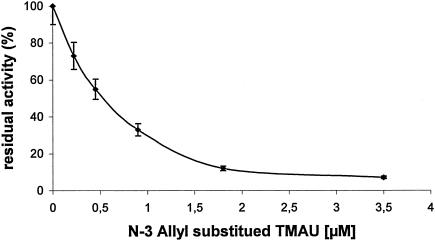
The inhibitory activities of various anilinouracil-based compounds against the S. aureus enzyme were measured in order to validate the homogeneous nonradioactive Pol IIIC assay described here in comparison to the published radioactive assay. Since the anilinouracil-based compounds act as competitive inhibitors of dGTP (25), this nucleotide was usually omitted from the DNA Pol IIIC assay. In the absence of dGTP, the enzymatic activity of the S. aureus Pol IIIC enzyme was lowered by ∼30% compared to the activity measured in the presence of all dNTPs. As can be seen from Table 1, 0.75 μM was detected as the 50% inhibitory concentration (IC50) of the ethyl-substituted compound 6-([3,4-trimethylene]anilino)uracil (TMAU), which has an IC50 of 0.38 μM in the radioactive assay, as published elsewhere (25). Similarly, the IC50s of various published AU compounds determined by the homogeneous enzymatic coupled assay described here were found to be in the same range as those obtained by the radioactive assay (Table 1) (25). As an example the profile of the inhibitory activity of allyl-substituted TMAU against the Pol IIIC enzyme is shown in Fig. 4. IC50s for N-3-substituted anilinouracils determined in the absence of dGTP were highly reproducible; for instance, a standard deviation of 13% was obtained for the IC50 of 0.75 μM for ethyl-substituted TMAU in 10 independent experiments. Moreover, at high concentrations (∼20 μM) of this compound, complete inhibition of the luminescence signal to the background level was observed, indicating that the enzymatic activity measured was due only to Pol IIIC activity and was not caused by other polymerase or pyrophosphate-producing enzymatic activities present in the enzyme preparation, which could be only partially purified without a loss of activity (Fig. 2).
TABLE 1.
IC50 of EMAU and N-3-substituted TMAUs for the Pol IIIC enzyme from S. aureus 133 determined by the nonradioactive assay
IC50, the concentration of a test compound that inhibited the Pol IIIC enzyme activity by 50% compared to the activity of an untreated control.
FIG. 4.
Inhibition of the S. aureus Pol IIIC enzyme by TMAU with an allyl side chain substitution at N-3. An IC50 of 0.6 μM was determined. Bioluminescence of ∼187,000 relative light units/min was measured for the uninhibited Pol IIIC enzyme.
The IC50s of the N-3-substituted anilinouracils determined by the nonradioactive assay were clearly due to their specific inhibitory mode of action on the polymerase activity and were not influenced by the inhibitory activities of the compounds used in the coupled assay system, i.e., ATP sulfurylase and firefly luciferase. This could be demonstrated by using poly(dA) as a template primer and dTTP as the nucleotide substrate for the S. aureus Pol IIIC enzyme. None of the compounds used in the assay system showed inhibitory effects up to a concentration of 50 μM under the assay conditions used compared to the results for an untreated control.
A relatively high background luminescence of ∼16% was measured in the absence of the Pol IIIC enzyme in comparison to the enzymatic activity of the untreated control S. aureus Pol IIIC enzyme; i.e., a signal-to-noise ratio of 6 was obtained by using the assay conditions described for N-3-substituted anilinouracils in the absence of dGTP. The reason for this luminescence was the fact that dATP as a substrate nucleotide for the polymerase reaction could also be used as a substrate by the firefly luciferase. For data analysis the background luminescence values were subtracted from the values obtained in the presence of the Pol IIIC enzyme.
Inhibition of S. aureus Pol IIIC and activities against gram-positive cocci of newly synthesized anilinouracil-based compounds.
The N-3 position of the anilinouracils is described as the only position in the molecule in which a substitution can be made without adversely affecting the inhibition of the bacterial polymerase (25, 27), and consequently, we chose to focus our efforts on optimization of potency by variation of the substituents at this position. Among the anilinouracil-based Pol IIIC inhibitors described in the literature, those with substitutions of 3-ethyl-4-methylanilinouracil (EMAU) and TMAU residues at the aryl site of the molecule were described as the most potent compounds (25, 27). Therefore, in the attempt to enhance the antibacterial activities of the compounds in this class, these two aryl-substituted amino uracils were used as templates in combination with broad variations of the N-3 side chain. Some of the most potent newly identified compounds are shown in Table 2. The anti-Pol IIIC activities (IC50s) could be significantly improved by variation of the N-3 side chain (Table 2) compared to the activities of previously described alkyl-substituted compounds, which reportedly exhibited IC50s of 0.09 to 1.3 μM (25). By introducing more polar functional groups in the N-3 side chain, we achieved a dual goal: first, the antibacterial efficacy as well as the target activity could be improved, and second, the aqueous solubility was significantly enhanced. For example, the IC50 of compound 4 for the S. aureus Pol IIIC enzyme was 8 nM. In general, N-3-(4-piperidylmethyl)anilinouracil amides prepared from five-membered-ring aromatic heterocyclic carboxylic acids were potent inhibitors of Pol IIIC (Table 2).
TABLE 2.
IC50 for Pol IIIC from S. aureus and in vitro activities of the N-3-substituted anilinouracils against the indicated species
Detailed characterization of EMAIPU (compound 1).
On the basis of the relatively high level of activity in vitro of compound 1 against different gram-positive cocci (MICs, 4 to 8 μg/ml) (Table 2) as well as its favorable aqueous solubility of 90 mg/liter at pH 6.5 in phosphate-buffered saline, this compound was selected for in-depth characterization of its in vitro as well as its in vivo properties.
Killing curve.
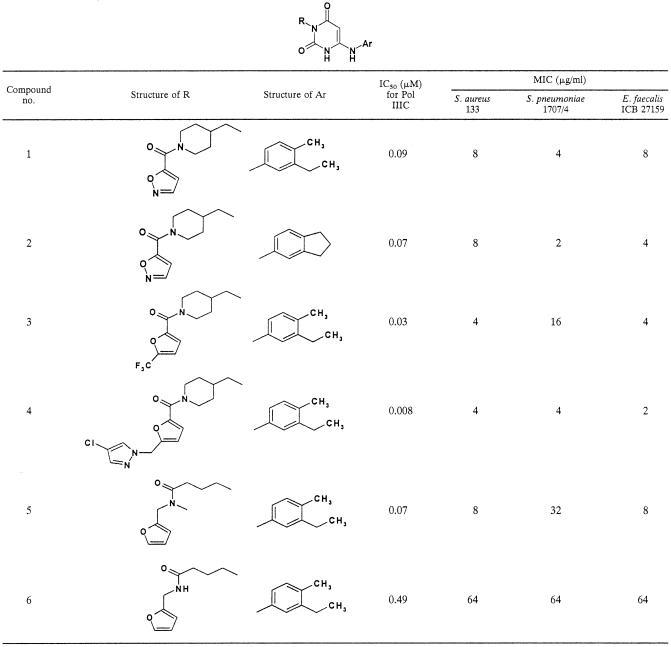
The bactericidal properties of the AU compound class were described by Daly et al. (9). Time-kill studies with compound 1 were performed with S. aureus 133. As shown in Fig. 5, with compound 1 only a 1-log-unit drop in the number of CFU per milliliter was observed within 24 h at eight times the MIC, indicating that compound 1 shows a bacteriostatic killing effect against this staphylococcal strain.
FIG. 5.
Killing curve of EMAIPU (compound 1) (MIC, 8 μg/ml) for S. aureus 133. The compound was tested, as indicated, at half (0.5×), two times (2×), and eight times (8×) its MIC.
Selective inhibition of DNA biosynthesis.
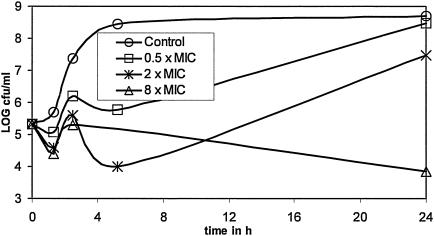
The ability of compound 1 to selectively inhibit DNA synthesis was investigated by the incorporation of radiolabeled metabolic precursors into whole cells of B. subtilis 168. As shown in Fig. 6, compound 1 selectively inhibited DNA synthesis, as measured by determination of the level of incorporation of labeled thymidine at one-fourth the MIC. Even at a concentration of four times the MIC, the syntheses of other macromolecules, i.e., RNA, protein, and peptidoglycan, remained unaffected, demonstrating the highly specific mode of action of the compound. This result confirms similar previous results with this compound class (1).
FIG. 6.
Mode-of-action studies for EMAIPU (compound 1) by the incorporation of radiolabeled metabolic precursors (as indicated) into whole cells of B. subtilis 168 (MIC, 2 μg/ml). Labeling was performed at one-fourth the MIC, the MIC, and four times the MIC. —, control; ▴, EMAIPU at 0.5 μg/ml; •, EMAIPU at 2 μg/ml; ▪, EMAIPU at 8 μg/ml.
Resistance development.
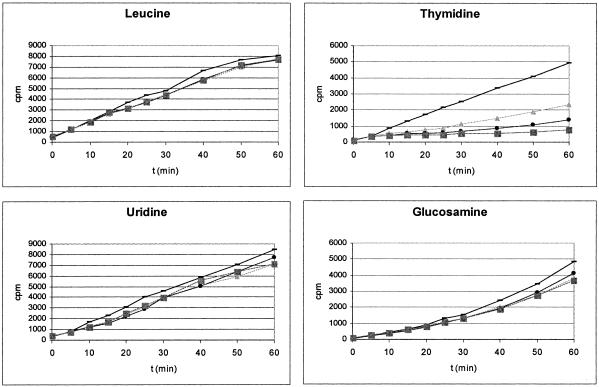
Resistance development is regarded as a critical property for a novel antibacterial compound class. The rate of resistance development was slower for compound 1 than for rifampin and moxifloxacin with strain S. aureus 133, as determined by serial passages in the presence of increasing concentrations (in terms of the fold increase in the MICs) of compound 1 (Fig. 7). However, regarding the relatively weak MIC of 8 μg/ml of compound 1 for this strain, a resistant isolate for which the MIC increased at least 32 times, to >256 μg/ml, was obtained after four passages, whereas the MIC of moxifloxacin increased from 0.03 μg/ml to 2 μg/ml within this number of passages. The MICs of the whole anilinouracil compound class for the compound 1-resistant S. aureus strain that was isolated were increased, and these increases occurred independently of the N-3 side chain substitution. However, no cross-resistance to other known antibiotics was observed. To investigate the resistance mechanism of this strain, the polC gene was amplified by PCR and sequenced. In this strain the phenylalanine at position 1261 of the Pol IIIC enzyme was found to be replaced by a leucine residue. The mutation was caused by a transversion of C3843G. The results show that resistance to compound 1 was due to a target mutation, which might lower the binding affinity of compound 1 to the Pol IIIC enzyme.
FIG. 7.
Development of S. aureus 133 resistance to EMAIPU (compound 1; ▪), determined as described in Material and Methods, in comparison to the development of resistance to rifampin (•) and moxifloxacin (▴).
In vitro cytotoxicity.
To estimate the cytotoxicity of compound 1 in in vivo experiments, eukaryotic cell lines, including human cells, were incubated in the presence of compound 1, as described in Material and Methods. As shown in Table 3, compound 1 showed no severe cytotoxic side effects in this in vitro assay. The EC50 was >100 μg/ml for all cell lines tested, which is generally regarded as uncritical (trovafloxacin, a quinolone antibiotic, and cycloheximide, a known cytotoxic agent, were included for comparison; Table 3). The results are consistent with the selective inhibition of the Pol IIIC enzyme, which is absent from eukaryotic cells.
TABLE 3.
In vitro cytotoxicity of EMAIPU (compound 1) compared to those of cycloheximide and trovafloxacin against different eukaryotic cell lines
| Compound and parameter | P388 (mouse monocyte) | THP-1 (human monocyte) | U937 (human macrophage) | CHO (hamster ovary) | HUH7 (human hepatocyte) |
|---|---|---|---|---|---|
| Cycloheximide | |||||
| EC50 (μg/ml) | 0.06 | 0.11 | 0.28 | 3.59 | 0.08 |
| NOEC (μg/ml) | <0.1 | <0.1 | <0.1 | 1.0 | <0.1 |
| Compound 1 | |||||
| EC50 (μg/ml) | >100 | >100 | >100 | >100 | >100 |
| NOEC (μg/ml) | 31.6 | 31.6 | 31.6 | >100 | >100 |
| Trovafloxacin | |||||
| EC50 (μg/ml) | 7.55 | 24.29 | 5.42 | 23.84 | 24.29 |
| NOEC (μg/ml) | 1.0 | 3.16 | 1.0 | 1.0 | 3.16 |
In vivo efficacy in a murine sepsis model.
On the basis of the in vitro activity of compound 1 against S. aureus 133 (Table 2), its in vivo efficacy was investigated in a murine sepsis model with this strain. As mentioned above the improved, solubility of 90 mg of compound 1 per liter (calculated log P, 1.35 [where P is the distribution coefficient between water and n-octanol of compound 1]) enabled an i.v. application route. Groups of mice (n = 6) were infected i.p., as described in Materials and Methods, and 30 min thereafter were treated i.v. with 10 mg of compound 1 per kg. Compound 1 protected the mice from lethal staphylococcal sepsis; i.e., 50% survival could be observed after administration of the described dosage compared to the rate of survival for the untreated controls, none of which survived. Moreover, an ED100 of 50 mg/kg was found for compound 1 in a sepsis model with E. faecalis ICB 27159 (5 × 107 CFU/mouse) after i.p. administration at 15 min postinfection.
Pharmakokinetic properties.
In order to evaluate the pharmacokinetic properties of compound 1 in different species, the drug was administered i.v. to mice, rats, and dogs. Plasma clearances (CLplasmas) of 1.58, 1.32, and 0.65 liters/(h · kg) were observed for the mice, rats, and dogs, respectively. The elimination half-life (t1/2) and volume of distribution at steady state (Vss) increased from the mice to the dogs (Table 4). The unbound fractions of drug in plasma (fus) were determined to be 1.1% for the mice, 2.6% for the rats, and 2.5% for the dogs. By application of allometric scaling, a CLplasma of 0.53 liters/(h · kg), a Vss of 1.88 liters/kg, and a mean residence time (MRT) after i.v. administration (MRTi.v.) of 3.5 h were predicted for humans.
TABLE 4.
Pharmacokinetic parameters for compound 1 (EMAIPU) in mice, rats, and dogs
| Parametera | Female CFW1 mice | Male Wistar rats | Male beagle dogs |
|---|---|---|---|
| Doseb (mg/kg) | 1 | 2 | 1 |
| t1/2 (h) | 0.53 | 0.89 (1.17) | 1.54 (1.08) |
| MRT (h) | 0.35 | 0.57 (1.09) | 1.55 (1.04) |
| Vss (liter/kg) | 0.55 | 0.75 (1.27) | 1.01 (1.23) |
| CLplasma [liters/(h · kg)] | 1.58 | 1.32 (1.26) | 0.65 (1.19) |
| CLplasma/CLblood | 1.36 | 1.21 | 1.22 |
| CLblood [liters/(h · kg)] | 2.15 | 1.60 | 0.79 |
| fu (%) | 1.1 | 2.6 | 2.5 |
For plasma clearance, the equation y = 1.02·BW0.848 (r2 = 0.999) was used; for volume of distribution, the equation y = 1.23·BW1.098 (r2 = 0.999) was used. The values of the pharmacokinetic parameters are geometric means for the mice and geometric means (standard deviations) for the rats and dogs.
The doses were administered as i.v. bolus injections for the mice and rats and as a 5-min i.v. infusion for the dogs.
DISCUSSION
In order to identify novel antibacterial agents that inhibit the Pol IIIC enzyme we performed for the first time a nonradioactive and homogeneous enzyme assay by omitting filtration steps. The enzymatic activity of Pol IIIC from S. aureus was measured in an enzymatically coupled assay; i.e., the PPi produced from the DNA polymerization reaction was converted into bioluminescence with the enzymes ATP sulfurylase and firefly luciferase. Using this assay, we determined that the Km values for the dNTPs were similar to those for the B. subtilis Pol IIIC, as determined in the heterogeneous radioactive assay (14). The IC50s of various published AU compounds determined by the homogeneous enzymatic coupled assay described here were found to be in the same range as those determined by the radioactive assay (25). Moreover, we also found that AU compounds containing increasing numbers of carbon atoms in the aliphatic side chains at N-3 had increased anti-Pol IIIC activities. Addition of a cyclohexyl group significantly reduced the inhibitory activity against the Pol IIIC enzyme. The results reported here are in full agreement with those presented in earlier reports (25), suggesting that the assay described here might be suitable for the characterization of newly synthesized anilinouracil-based compounds.
In a recently published report, Yang et al. (30) described a homogeneous scintillation proximity assay with DNA Pol III from Streptococcus pyogenes. Those investigators used synthetic double-stranded oligonucleotides as substrates, although the Pol III activity was considerably lower (30). They described inconsistent results for inhibition of DNA Pol IIIC by anilinouracils when activated calf-thymus DNA was used as the substrate (30). Yang et al. (30) pointed out that the sequence and the concentration of activated calf thymus DNA are unknown and might be the reason for the inconsistent inhibition measurements obtained with anilinouracils. However, here we found reproducible IC50s of the anilinouracils when we used activated calf thymus DNA as the substrate, and these values were also comparable to those determined by the published radioactive heterogeneous Pol IIIC assay (25).
Previous work with the anilinouracil compound class has primarily focused on the optimization of the anilino substituent; among the anilinouracil-based Pol IIIC inhibitors described in the literature, those with 3-ethyl-4-methylanilino- and 3,4-trimethyleneanilino substitutions at the aryl site of the molecule have been described as the most potent (25); these moieties were kept constant when we varied the substituents at other positions. Despite a great deal of work to establish the structure-activity relationship (SAR) of uracil ring substitutions, so far the N-3 position appears to be the only place in the molecule where substitutions can be made without a loss of activity (25, 27). Among the series of linear N-alkyl substituents, the potency seems to correlate with lipophilicity; thus, the highest activity was achieved with N-3 butyl-substituted TMAU (25), whereas the addition of a benzyl group at N-3 significantly reduced the anti-Pol IIIC activity (25), seemingly indicating an unfavorable steric interaction. A recent report (31) has described the more hydrophilic N-3-piperidinylbutyl derivative as a potent Pol IIIC inhibitor.
Our work with this compound class was based on the hypothesis that an energetically favorable interaction with the Pol IIIC enzyme could be achieved by elaboration of the N-3 substituent, which in turn would lead to improved antibacterial potency.
Indeed, many of the resulting compounds showed significant inhibitory activities against the Pol IIIC target in the low nanomolar range. Several structural classes with improved potencies were identified, and although each structural subclass seemed to have a distinct SAR, an overall SAR did not emerge. Zhi et al. (31) also observed the lack of steric limits for bulky substituents at this position and assumed only a low-level interaction of this group with the Pol IIIC enzyme.
Nevertheless, the anti-Pol IIIC activity could be significantly influenced by variation of the N-3 side chain. It is known that the formation of a hydrogen-bonded complex between an AU molecule and a cytosine base in the DNA template is the first interaction in the inhibitory mechanism of the AU compound class (25). This could be demonstrated by the fact that polymerization of a poly(dA)-oligo(dT) DNA template-primer containing no cytosine bases in the template could not be inhibited by anilinouracils (25). It might be that the newly identified N-3 substituents with improved anti-Pol IIIC activities enhance the base-pairing faculties of the AU compounds.
The MICs of the compounds described here for gram-positive cocci were found to be, at best, in the range of 4 to 8 μg/ml, which is in agreement with data from recent reports describing aminoalkyl substituents at position 3 of the uracil ring (31). It is noteworthy that the MICs of the investigated derivatives somehow seemed to be limited, regardless of the anti-Pol IIIC activity measured in the enzymatic assay; i.e., although the anti-Pol IIIC activities of the compounds could be significantly improved, the antibacterial activities (MICs) of the compounds could not be improved beyond a comparatively modest MIC of 2 μg/ml. Although these activities are much improved relative to those of previously reported anilinouracils, they are still far from the potencies of fluoroquinolones and modern beta-lactam antibiotics. Apparently, the anti-Pol IIIC activity does not linearly correlate with the MICs, and factors other than the activity against the target alone contribute to the antibacterial activities, and physicochemial parameters are of importance. Zhi et al. (31) also observed this variation in antibacterial activity, despite strong inhibition of Pol IIIC.
The apparently inherent minimum MICs of the AU compounds might be explained by one of the following three hypotheses or a combination thereof. (i) The level of penetration of the AU molecules into a bacterial cell might be somehow limited. (ii) The IC50s were determined in the absence of dGTP, which is present in the bacterial cell. Indeed, addition of dGTP to the enzymatic Pol IIIC assay increased the IC50s of the AU compounds (data not shown), which is consistent with the competitive mode of action of this compound class (25). (iii) It has been reported that the S. aureus Pol IIIC activity is lowered from ∼480 to ∼120 nucleotides/s in in vitro assays, in which the accessory proteins present in the bacterial cell, e.g., the β sliding clamp, are lacking (11). Thus, it might be that the molecules are able to inhibit the slower activity of the Pol IIIC enzyme in the in vitro assay but are not potent enough to inhibit the enzymatic activity to the same extent inside a bacterial cell in the presence of dGTP.
Among the different N-3-substituted compounds, EMAIPU (compound 1) not only showed moderate activity against S. aureus (Table 2) but also showed a much increased aqueous solubility compared to those of N-unsubstituted derivatives. This improved solubility enabled i.v. application of compound 1 in a murine sepsis model, which is the first time that a compound in this class has been able to be administered by this route; previous in vivo experiments had been conducted with i.p. dosing. We found an ED100 of 50 mg/kg in a murine enterococcal sepsis model and an ED50 of 10 mg/kg in a murine staphylococcal sepsis model. Tarantino et al. (25) reported that an AU compound with a hydroxypropyl N-3 side chain substitution had an ED100 of 10 mg/kg after i.p. administration in a murine staphylococcal sepsis model, which is in the same range as our results. Pyrazolo[3,4-d]pyrimidine-based inhibitors of Pol IIIC with improved aqueous solubilities have also been described, but no animal studies were reported (2).
The pharmacokinetics of compound 1 (EMAIPU) in mice showed a low to moderate CLplasma and a moderate Vss. This results in an MRTi.v. of 0.35 h. Compared to the values of the pharmacokinetic parameters for another marketed antibiotic, such as linezolid, which possesses a CLplasma of 1.0 liters/h · kg and an MRTi.v. of 0.8 h, the values of the pharamacokinetic parameters for EMAIPU are in an acceptable range. Therefore, the good pharmacokinetic properties of compound 1 support the positive findings obtained with the murine sepsis model. For the antibacterial agent linezolid, CLplasma decreases (in relation to the total amount of blood in the liver) and Vss is maintained at a constant level. This results in increased MRTi.v.s of 1.3 and 5.3 h for rats and dogs, respectively (21). By extrapolation of these data to humans by allometric scaling, the MRTi.v. for humans is estimated to be 7 h (5). This is in good agreement with the measured value of 8 h for MRTi.v. after administration of a single i.v. dose (22). Other marketed antibiotics, e.g., moxifloxacin, show an even more pronounced increase in MRTi.v. in species with increasing body weight. The MRTi.v. of moxifloxacin increased from 0.9 h in mice to 13 h in humans (20). Extrapolation of the values obtained for compound 1 (EMAIPU) to the values that would be obtained for humans reveals less favorable pharmacokinetic properties, in particular, less favorable CLplasma and MRTi.v. values. This issue should be considered during the optimization of anilinouracil-based compounds.
The rate of development of resistance to compound 1, as determined by serial passages in S. aureus, was found to be slower than that to rifampin. Nevertheless, a highly resistant S. aureus strain could be isolated after four passages in the presence of compound 1. The mutation that caused resistance to compound 1 was located at position 1261 of the Pol IIIC enzyme, which resulted in a change of phenylalanine to leucine. This conserved phenylalanine was also changed into a leucine residue in different gram-positive organisms resistant to 3(4′-hydroxybutyl)-6-(3′-ethyl-4′-methylanilino) uracil (HBEMAU) (8). A Pol IIIC enzyme from B. subtilis resistant to p-hydroxyphenyl azouracil has been described to be the result of a mutation in azp-12, which contains a single base change that results in the replacement of the serine at position 1175 by alanine (18). The region of this mutation is described as the C-terminal domain of the Pol IIIC enzyme required for dNTP binding and as the known aryl-binding region in B. subtilis (4, 18). The Pol IIIC enzymes from resistant organisms displayed the same polymerase activities as their wild-type counterparts, but they showed higher Ki values for AU compounds (8). Thus, resistance to the AU compound class is based on the lower binding affinities of the inhibitory molecules to the target enzyme. The fact that resistance to anilinouracils containing different N-3 side chains but the same aryl domain was caused by the exchange of a conserved phenylalanine residue within the aryl-binding domain of the Pol IIIC enzyme further supports the assumption that the N-3 substituents investigated so far do not play a crucial role in enzyme binding.
Low frequencies of resistance to HBEMAU were observed (8), which seems to be in accordance with the fact that probably only a single allowable mutation leads to resistance to the anilinouracil-based compound class. However, to estimate the clinical relevance of this resistance phenomenon, more detailed studies with more strains should be performed.
Other key properties of compounds of the AU class, e.g., their specific inhibition of DNA biosynthesis, have been preserved in the newly synthesized compounds, and the results for these properties obtained here are comparable to those reported in previous studies (1, 9, 25).
In summary, by using an approach to optimize the antibacterial activities of compounds within the AU compound class by making variations at position 3 of the uracil ring, a number of derivatives, including EMAIPU (compound l), showing promising in vitro and in vivo antibacterial efficacies have been identified. The enhanced aqueous solubility facilitated i.v. administration in a mouse sepsis model. The anilinouracils are a promising novel antibacterial compound class, but further improvement of their antibacterial potencies would be required for this class to be competitive in a clinical setting. Exploration of the novel derivatives belonging to this class may, it is hoped, lead to the identification of novel antibiotics to combat multiresistant gram-positive pathogens.
Acknowledgments
We gratefully acknowledge the contributions of S. Bartel, M. Bauser, J. Keldenich, I. Knezevic, and H.-O. Werling.
REFERENCES
- 1.Ali, A., S. D. Aster, D. W. Graham, G. F. Patel, G. E. Taylor, R. L. Tolman, R. E. Painter, L. L. Silver, K. Young, K. Ellsworth, W. Geissler, and G. S. Harris. 2001. Design and synthesis of novel antimicrobial agents with inhibitory activity against DNA polymerase III. Bioorg. Med. Chem. Lett. 11:2185-2188. [DOI] [PubMed] [Google Scholar]
- 2.Ali, A., G. E. Taylor, K. Ellsworth, G. Harris, R. Painter, L. L. Silver, and K. Young. 2003. Novel pyrazolo[3,4-d]pyrimidine-based inhibitors of Staphylococcus aureus DNA polymerase III: design, synthesis, and biological evaluation. J. Med. Chem. 46:1824-1830. [DOI] [PubMed] [Google Scholar]
- 3.Ali, A., G. E. Taylor and D. W. Graham. April. 2001. Gram-positive selective antibacterial compounds, compositions containing such compounds and methods of treatment. Patent WO 01/29010.
- 4.Barnes, M. H., R. A. Hammond, C. C. Kennedy, S. L. Mack, and N. C. Brown. 1992. Localization of the exonuclease and polymerase domains of Bacillus subtilis DNA polymerase III. Gene 111:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Bhamidipati, R. K., P. V. Dravid, R. Mullangi, and N. R. Srinivas. 2004. Prediction of clinical pharmacokinetic parameters of linezolid using animal data by allometric scaling: applicability for the development of novel oxazolidinones. Xenobiotica 34:571-579. [DOI] [PubMed] [Google Scholar]
- 6.Boxenbaum, H. 1982. Interspecies scaling, allometry, physiological time and the ground plan of pharmacokinetics. J. Pharmacokinet. Biopharm. 10:201-227. [DOI] [PubMed] [Google Scholar]
- 7.Brown, N. C., and G. E. Wright. March. 1996. Antibiotic compounds and methods to treat gram-positive bacterial and mycoplasmal infections. Patent WO 96/06614.
- 8.Butler, M. M., D. J. Skow, R. O. Stephenson, P. T. Lyden, W. A. LaMarr, and K. A. Foster. 2002. Low frequencies of resistance among Staphylococcus and Enterococcus species to the bactericidal DNA polymerase inhibitor N(3)-hydroxybutyl 6-(3′-ethyl-4′-methylanilino) uracil. Antimicrob. Agents Chemother. 46:3770-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, J. S., T. J. Giehl, N. C. Brown, C. Zhi, G. E. Wright, and R. T. Ellison. 2000. In vitro antimicrobial activities of novel anilinouracils which selectively inhibit DNA polymerase III of gram-positive bacteria. Antimicrob. Agents Chemother. 44:2217-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dervyn, E., C. Suski, R. Daniel, C. Bruand, J. Chapuis, J. Errington, L. Janniere, and S. D. Ehrlich. 2001. Two essential DNA polymerases at the replication fork. Science 294:1716-1719. [DOI] [PubMed] [Google Scholar]
- 11.Gass, K. B., R. L. Low, and N. R. Cozzarelli. 1973. Inhibition of a DNA polymerase from Bacillus subtilis by hydroxyphenylazopyrimidines. Proc. Natl. Acad. Sci. USA 70:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue, R., C. Kaito, M. Tanabe, K. Kamura, N. Akimitsu, and K. Sekimizu. 2001. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet. Genomics 266:564-571. [DOI] [PubMed] [Google Scholar]
- 13.Klemperer, N., D. Zhang, M. Skangalis, and M. O'Donnell. 2000. Cross-utilization of the β sliding clamp by replicative polymerases of evolutionary divergent organisms. J. Biol. Chem. 275:26136-26143. [DOI] [PubMed] [Google Scholar]
- 14.Low, R. L., S. A. Rashbaum, and N. R. Cozzarelli. 1976. Purification and characterization of DNA polymerase III from Bacillus subtilis. J. Biol. Chem. 251:1311-1325. [PubMed] [Google Scholar]
- 15.Nyren, P., and A. Lundin. 1985. Enzymatic method for continuous monitoring of inorganic pyrophosphate synthesis. Anal. Biochem. 151:504-509. [DOI] [PubMed] [Google Scholar]
- 16.Nyren, P. 1987. Enzymatic method for continuous monitoring of DNA polymerase activity. Anal. Biochem. 167:235-238. [DOI] [PubMed] [Google Scholar]
- 17.Pacitti, D. F., M. H. Barnes, D. H. Li, and N. C. Brown. 1995. Characterization and overexpression of the gene encoding Staphylococcus aureus DNA polymerase III. Gene 165:51-56. [DOI] [PubMed] [Google Scholar]
- 18.Sanjanwala, B., and T. Ganesan. 1991. Genetic structure and domains of DNA polymerase III of B. subtilis. Mol. Gen. Genet. 226:467-472. [DOI] [PubMed] [Google Scholar]
- 19.Schuhmacher, J., K. Bühner, and A. Witt-Laido. 2000. Determination of the free fraction and relative free fraction of drugs strongly bound to plasma proteins. J. Pharm. Sci. 89:1008-1021. [DOI] [PubMed] [Google Scholar]
- 20.Siefert, H. M., A. Domdey-Bette, K. Henninger, F. Hucke, C. Kohlsdorfer, and H. H. Stass. 1999. Pharmacokinetics of the 8-methoxyquinolone, moxifloxacin: a comparison in humans and other mammalian species. J. Antimicrob. Chemother. 43:69-76. [DOI] [PubMed] [Google Scholar]
- 21.Slatter, J. G., L. A. Adams, E. C. Bush, K. Chiba, P. T. Daley-Yates, K. L. Feenstra, S. Koike, N. Ozawa, G. W. Peng, J. P. Sams, M. R. Schuette, and S. Yamazaki. 2002. Pharmacokinetics, toxicokinetics, distribution, metabolism and excretion of linezolid in mouse, rat and dog. Xenobiotica 32:907-924. [DOI] [PubMed] [Google Scholar]
- 22.Slatter, J. G., D. J. Stalker, K. L. Feenstra, I. R. Welshman, J. B. Bruss, J. P. Sams, M. G. Johnson, P. E. Sanders, M. J. Hauer, P. E. Fagerness, R. P. Stryd, G. W. Peng, and E. M. Shobe. 2001. Pharmacokinetics, metabolism, and excretion of linezolid following an oral dose of [14C]linezolid to healthy human subjects. Drug Metab. Dispos. 29:1136-1145. [PubMed] [Google Scholar]
- 23.Stulke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 24.Tarantino, P. M., C. Zhi, J. J. Gambino, G. E. Wright, and N. C. Brown. 1999. 6-Anilinouracil-based inhibitors of B. subtilis DNA polymerase III. Antipolymerase and antimicrobial structure-activity relationships based on substitution at uracil N3. J. Med. Chem. 42:2035-2040. [DOI] [PubMed] [Google Scholar]
- 25.Tarantino, P. M., C. Zhi, G. E. Wright, and N. C. Brown. 1999. Inhibitors of DNA polymerase III as novel antimicrobial agents against gram-positive eubacteria. Antimicrob. Agents Chemother. 43:1982-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomasz, A. 1994. Multiple-antibiotic-resistant pathogenic bacteria. N. Engl. J. Med. 330:1247-1251. [DOI] [PubMed] [Google Scholar]
- 27.Trantolo, D. J., G. E. Wright, and N. C. Brown. 1986. Inhibitors of B. subtilis DNA polymerase III. Influence of modifications in the pyrimidine ring of anilino- and (benzylamino)pyrimidines. J. Med. Chem. 29:676-681. [DOI] [PubMed] [Google Scholar]
- 28.Wright, G. E., and N. C. Brown. 1980. Inhibitors of Bacillus subtilis DNA polymerase III. 6-Anilinouracils and 6-(alkylamino)uracils. J. Med. Chem. 23:34-38. [DOI] [PubMed] [Google Scholar]
- 29.Wright, G. E., and N. C. Brown. 1999. DNA polymerase III: a new target for antibiotic developement. Curr. Opin. Anti-Infect. Investig. Drugs 1:45-48. [Google Scholar]
- 30.Yang, F., I. B. Dicker, M. G. Kurilla, and D. L. Pompliano. 2002. PolC-type polymerase III of Streptococcus pyogenes and its use in screening for chemical inhibitors. Anal. Biochem. 304:110-116. [DOI] [PubMed] [Google Scholar]
- 31.Zhi, C., Z. Y. Long, J. Gambino, W. C. Xu, N. C. Brown, M. Barnes, M. M. Butler, W. LaMarr, and G. E. Wright. 2003. Synthesis of substituted 6-anilinouracils and their inhibition of DNA polymerase IIIC and gram-positive bacterial growth. J. Med. Chem. 46:2731-2739. [DOI] [PubMed] [Google Scholar]