Abstract
As the processes of embryogenesis become increasingly well understood, there is growing interest in the development that occurs at later, postembryonic stages. Postembryonic development holds tremendous potential for discoveries of both fundamental and translational importance. Zebrafish, which are small, rapidly and externally developing, and which boast a wealth of genetic resources, are an outstanding model of vertebrate post-embryonic development. Nonetheless, there are specific challenges posed by working with zebrafish at these stages, and this chapter is meant to serve as a primer for those working with larval and juvenile zebrafish. Since accurate staging is critical for high-quality results and experimental reproducibility, we outline best practices for reporting postembryonic developmental progress. Emphasizing the importance of accurate staging, we present new data showing that rates of growth and size–stage relationships can differ even between wild-type strains. Finally, since rapid and uniform development is particularly critical when working at postembryonic stages, we briefly describe methods that we use to achieve high rates of growth and developmental uniformity through postembryonic stages in both wild-type and growth-compromised zebrafish.
INTRODUCTION
Since the 1980s, the zebrafish (Danio rerio) has exploded in popularity as a model organism (Fig. 1). The zebrafish was initially developed as an experimental system because of its suitability for studies of early development: embryos develop rapidly and externally, and are optically transparent; embryos are produced in large, uniform clutches; and genetic screens are straightforward. Many of the advantages at embryonic stages also apply to research focused on postembryonic stages, and the ever-expanding genomic, genetic, and transgenic resources available for zebrafish are equally useful for studies at any stage.
FIGURE 1.
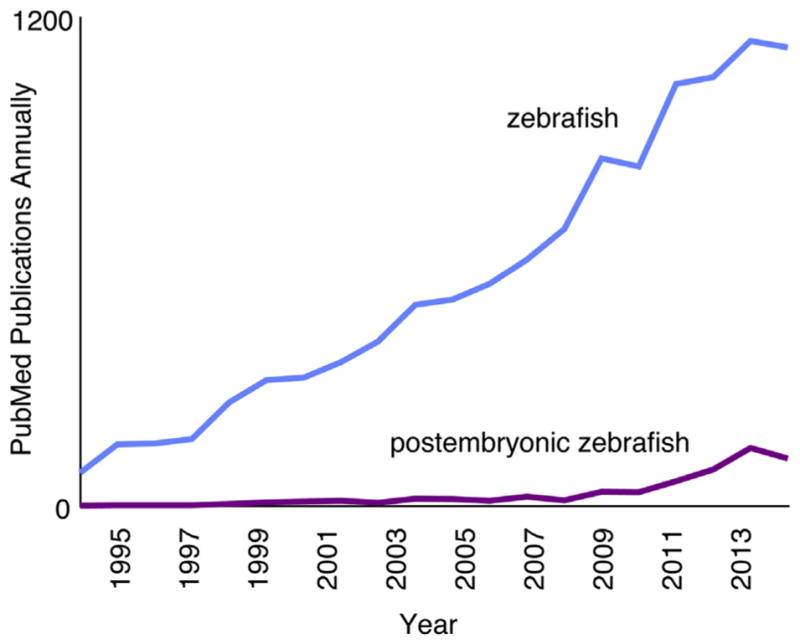
Pubmed papers utilizing zebrafish (blue (light gray in print versions)) and numbers of those papers focusing on postembryonic stages (purple (dark gray in print versions)).
As events of embryogenesis become better understood, many zebrafish researchers have begun to focus on processes that occur after embryogenesis (Fig. 1). Postembryonic development involves major modifications to multiple tissues and organs; many of these changes in zebrafish are similar or identical to those of fetal and neonatal human development. For example, like fetal human skin, the integument of larval zebrafish is colonized by melanocytes, many of which are derived from the peripheral nervous system (Adameyko et al., 2009; Budi, Patterson, & Parichy, 2011); zebrafish scales develop through processes important for dermal bone in mammals (Pasqualetti, Banfi, & Mariotti, 2012; Teck Ho Lee, Thiery, & Carney, 2013); and a conserved suite of mechanisms regulates the development and function both of swim bladders in zebrafish and lungs in mammals (Cass, Servetnick, & McCune, 2013; Longo, Riccio, & McCune, 2013; Zheng et al., 2011). Many other events occurring at postembryonic stages in zebrafish also resemble human fetal development, including modifications to the kidney (Drummond, 2005; Elizondo, Budi, & Parichy, 2010; Gerlach & Wingert, 2013) and gut (Crosnier et al., 2005; Wallace, Akhter, Smith, Lorent, & Pack, 2005); neurogenesis (Kizil, Kaslin, Kroehne, & Brand, 2012; Schmidt, Strahle, & Scholpp, 2013; Zupanc, 2011); ossification of axial and craniofacial bones (Bird & Mabee, 2003; Cubbage & Mabee, 1996; Elizondo et al., 2005; Kimmel, DeLaurier, Ullmann, Dowd, & McFadden, 2010); and continued, but differential, growth across the body (Parichy, Elizondo, Mills, Gordon, & Engeszer, 2009). Research focused on postembryonic stages has further made significant inroads toward understanding the development of the lateral line (Ghysen, Wada, & Dambly-Chaudière, 2014; Thomas, Cruz, Hailey, & Raible, 2015; Wada & Kawakami, 2015), pigment pattern (Kondo and Watanabe, 2015; Parichy & Spiewak, 2015; Singh & Nüsslein-Volhard, 2015), skeleton (Akiva et al., 2015; Eames et al., 2013), heart (Matrone, Wilson, Mullins, Tucker, & Denvir, 2015; Singleman & Holtzman, 2012), microbiome (Burns et al., 2015; Roeselers et al., 2011; Stephens et al., 2015), and lipid stores (Flynn, Trent, & Rawls, 2009; Minchin & Rawls, 2011). Investigating these and other postembryonic processes in zebrafish can lend critical insight into the conserved mechanisms by which postembryonic development occurs in humans.
Beyond advancing our understanding of normal development, postembryonic zebrafish provide valuable models of human diseases and pathologies, including gastrointestinal (Marjoram & Bagnat, 2015; Yang, Tomkovich, & Jobin, 2014) and metabolic disorders (Asaoka, Terai, Sakaida, & Nishina, 2013; Schlegel & Gut, 2015; Seth, Stemple, & Barroso, 2013); neurodegenerative (Babin, Goizet, & Raldúa, 2014; Lee & Freeman, 2014; Schmid & Haass, 2013), behavioral, and neurological pathologies (Kalueff, Stewart, & Gerlai, 2014; Nguyen et al., 2013; Stewart, Braubach, Spitsbergen, Gerlai, & Kalueff, 2014); cancers (White, Rose, & Zon, 2013; Zhao, Huang, & Ye, 2015); osteoporosis (De Vrieze et al., 2014; Laizé, Gavaia, & Cancela, 2014); and numerous other diseases (Ablain & Zon, 2013; Löhr & Hammerschmidt, 2011; Phillips & Westerfield, 2014; Santoriello & Zon, 2012; Shin & Fishman, 2002). Larval and adult zebrafish are further used to model wound healing (Godwin, 2014; Richardson et al., 2013), regeneration (Coffin, Brignull, Raible, & Rubel, 2014; Gemberling, Bailey, Hyde, & Poss, 2013; Wehner & Weidinger, 2015), and behavior (Fero, Yokogawa, & Burgess, 2011; Norton & Bally-Cuif, 2010; Norton et al., 2011), and are frequently used in pharmacological screening and drug discovery (Ali, Champagne, Spaink, & Richardson, 2011; de Esch, Slieker, Wolterbeek, Woutersen, & de Groot, 2012; Jung et al., 2012; Rennekamp & Peterson, 2015).
There is thus tremendous potential for making discoveries of both fundamental and translational importance using postembryonic zebrafish, but there are specific challenges faced by researchers working with these stages. This chapter aims to serve as a primer for those working with larval and juvenile zebrafish. One major issue in the field of postembryonic zebrafish development is the fact that many studies fail to apply a standard system of staging, making precise replication of experiments challenging or impossible. Such imprecision can further interfere with experimental outcomes when fish used in a study actually represent a range of stages that may differ greatly in developmental state. In this review, we suggest best practices for reliably, reproducibly, and conveniently reporting postembryonic developmental progress. We further provide new data showing that rates of growth and development and relationships between size and stage can differ between wild-type strains, underscoring the importance of accurate staging. Finally, we outline methods for achieving high rates of growth and developmental uniformity through postembryonic stages in both wild-type and genetically or experimentally growth-compromised fish.
1. PART I: POSTEMBRYONIC STAGING
As the field of postembryonic zebrafish research grows and matures, there is a critical need for consensus and community standards regarding staging methods. Only with standardized, uniformly applied staging criteria can experiments and observations be validated, repeated, and compared within and between laboratories. Fish at even slightly different developmental stages may differ greatly in morphology, gene expression, physiology, behavior, and responses to experimental manipulations; inadequate staging can compromise and confound experimental results.
Development can be described along at least three different axes: stage, as defined by attainment of specific developmental milestones; size, as measured on one or more dimensions (eg, standard length, from nose to base of caudal fin); or age, typically days or weeks postfertilization. In the following sections, we consider these three parameters and their utility as indicators of developmental progress. Staging is a flexible and accurate method of reporting developmental state, and we present methods of reporting postembryonic stage and discuss their appropriateness for different contexts. We show that age is unacceptable as an indicator of developmental progress, and that size is an acceptable proxy for development only in certain limited contexts, generally in conjunction with other developmental information. We also present new data comparing the relationships between these three parameters in different wild-type genetic backgrounds; our findings underscore the importance of proper staging and stage reporting. Finally, we discuss methods for customizing staging methods for different uses, and best practices for efficient staging and sample collection.
1.1 MEANINGFULLY REPORTING DEVELOPMENTAL PROGRESS
Staging is the most direct, reliable, and flexible method for assigning and reporting developmental state. Stage is determined with reference to developmental milestones laid out in a normal table of development, or staging series. An embryonic staging series, produced in the 1990s (Kimmel, Ballard, Kimmel, Ullmann, & Schilling, 1995), helped to bring consistency to embryonic zebrafish research focused on early development. A postembryonic staging series was published in 2009 (Parichy et al., 2009). This postembryonic normal table is simple to use and allows rapid, accurate assignment of stage for a wide variety of experimental contexts. The stages defined may be assessed with only the most basic knowledge of zebrafish anatomy. We describe two conventions for reporting these stages, each with particular advantages to certain applications.
1.1.1 Named stages
A straightforward method of indicating developmental state is simply to report the name of the most mature developmental milestone reached by individuals. The post-embryonic normal table defines 15 stages from early larva to adult, characterized by the appearance of a discrete phenotypic character, eg, the anterior lobe of the swim bladder, caudal fin rays, or scales; nine intermediate stages are illustrated as well (see Table 1).
Table 1.
Milestones in Postembryonic Zebrafish Development (Parichy et al., 2009)
| Milestone | SSL | Defining Characteristic(s) |
|---|---|---|
|
| ||
| pSB, swim bladder inflation | 3.5 | Swim bladder inflates |
| pSB+ | 3.8 | Head shows more anterior mouth position |
| FLe, early flexion | 4.5 | Notochord flexion; condensed mesenchyme pronounced at caudal fin bud |
| CR, caudal fin ray appearance | 4.9 | Rays visible in caudal fin |
| AC, anal fin condensation | 5.4 | Condensed mesenchyme forms at anal fin fold |
| DC, dorsal fin condensation | 5.7 | Condensed mesenchyme forms at dorsal fin fold |
| DC+ | 5.8 | Anal fin fold forms ridges and develops melanophores |
| MMA, metamorphic melanophore appearance | 5.9 | Lightly melanized melanophores present dorsolaterally; 18–20 caudal fin rays present |
| aSB, anterior swim bladder appearance | 6 | Anterior swim bladder inflates (may initially be very small) |
| aSB+ | 6.1 | Cleft develops in the caudal fin |
| AR, anal fin ray appearance | 6.2 | Rays visible in anal fin |
| AR+ | 6.3 | Dorsal fin fold forms ridges and develops melanophores |
| DR, Dorsal fin ray appearance | 6.4 | Rays visible in dorsal fin |
| DR+ | 6.6 | 6–8 dorsal fin rays visible |
| PB, pelvic fin bud appearance | 7.2 | Bud of pelvic fin protrudes from the ventral body wall |
| PB+ | 7.6 | Pelvic fin bud acquires a more oblong shape |
| PR, pelvic fin ray | 8.6 | Rays visible in pelvic fin |
| PR+ | 9.2 | Barbel bud appears |
| SP, squamation onset posterior | 9.8 | Scales visible on the tail |
| SA, squamation through anterior | 10.4 | Scales visible anterior to the dorsal fin |
| J, juvenile | 11 | Scales extend anteriorly to head |
| J+ | 13 | Melanophores completely cleared from primary interstripe |
| J++ | 16 | Secondary dorsal and ventral stripes formed |
| A, adult | 26 | Sexually mature; secondary sex characters evident |
The standardized standard length corresponding to each milestone is listed as SSL (which is also the average size of WT(WA) fish at that stage). Note that this table should be used as a coarse reference only, and actual staging should be performed in comparison to the images of reference individuals in Figures 32–57 of Parichy et al. (2009).
1.1.2 Standardized standard length
Another method of reporting stage is to report Standardized Standard Length (SSL) as defined in the postembryonic normal table. SSLs were based originally on logistic regression analyses of stage transition sizes in a reference population of zebrafish (though no statistics are required for their use!). In this approach, fish are simply matched to images and descriptions in the postembryonic normal table, and stages are reported as the corresponding SSL, even if the actual size of the specimen differs somewhat from the archetypal size owing to individual genetic, environmental, or strain-specific variation. For example, fish that have acquired pelvic fin buds (stage PB), could be reported as “7.2 SSL.” This convention also allows development to be parsed more finely than with named stages, since researchers may report intermediate SSLs between milestone-denoted stages. For example, if fish are well past pelvic fin bud appearance (PB/7.2 SSL) but have not quite developed pelvic fin rays (PR/8.6 SSL), they might be reported as “8.0 SSL,” ideally with exemplar images or descriptions to define this intermediate developmental state. Note that “mm” is dispensed within this reporting convention to further indicate reference to archetypal SSL rather than actual specimen length.
The use of a named stage or SSL, rather than absolute standard length (SL), is valuable because relationships between stage and size vary with rearing condition (Parichy et al., 2009) and can differ even among wild-type strains experiencing the same conditions (see below). Indeed, a size—and implied stage—reported for one strain (eg, AB) may not correspond to the same implied stage for another strain (eg, Tübingen (TU)). By encompassing differences in size–stage relationships, named stages or SSLs allow developmental states to be reported in a common metric.
1.2 PROBLEMS WITH USING SIZE AND AGE AS PROXIES FOR DEVELOPMENTAL STATE
1.2.1 Age is unacceptable as a proxy for development
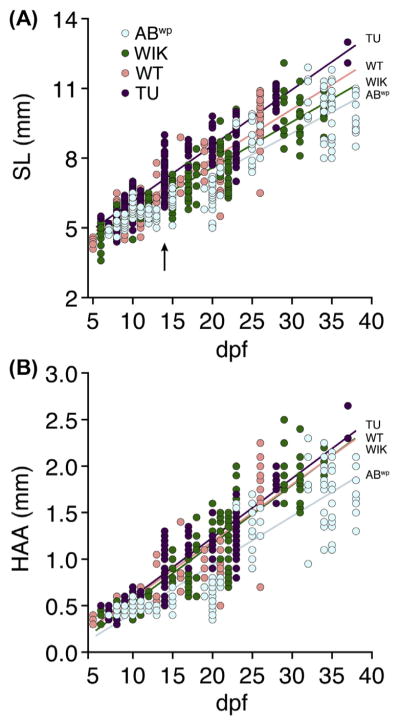
Age, generally reported as days postfertilization (dpf), is a highly unreliable marker of developmental progress. Rates of growth and maturation depend markedly on environmental conditions (Parichy et al., 2009). Further, different genetic backgrounds, even among wild-type laboratory strains, might be assumed to differ in rates of growth and development. Indeed, asynchronies in size and stage arise even within clutches of wild-type fish reared under invariant conditions (Figs. 2 and 3; Kimmel et al., 1995; Parichy et al., 2009): minor environmental, genetic, or stochastic differences in resource utilization and competitive advantage can lead to substantial variation in growth and development, which become increasingly magnified as development proceeds, amplifying heterogeneity within families and differences between families. Thus, age as a developmental indicator becomes increasingly inappropriate at postembryonic stages.
FIGURE 2. Fish growth varies extensively within and between wild-type stocks.
Sizes of individual larvae measured as SL (A) or HAA (B) and plotted by days postfertilization (dpf). Arrow in (A) indicates extensive size variation in TU fish of a single family (see text). Stock-specific growth rates were indicated by significant genetic background × dpf interactions (A: F3,746 = 15.3, P < 0.0001; B: F3,746 = 13.1, P < 0.0001; both dependent variables ln-transformed to normalize variance of residuals). Arrow indicates size variation within a single clutch of TU on a single day (14 dpf). All fish were reared identically in batches of ~20; individuals were measured, staged, and discarded. (See color plate)
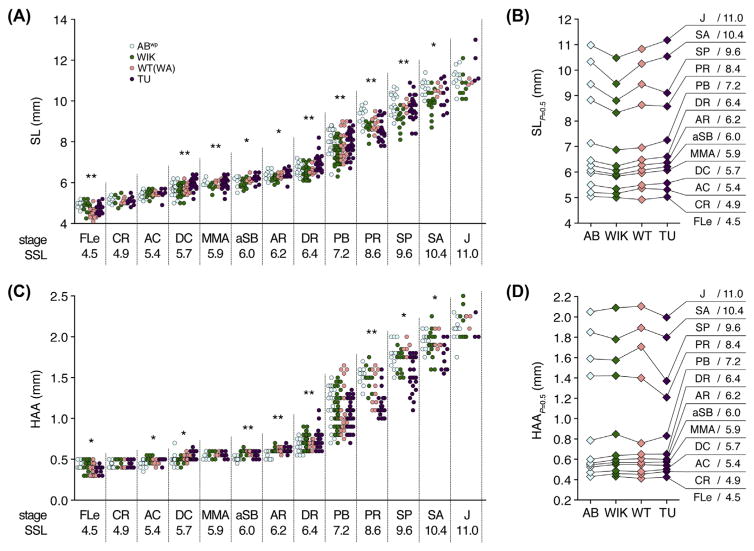
FIGURE 3. Variation in size–stage relationships across wild-type stocks.
(A, C) Stages attained by fish of different SLs (A) or HAAs (C). Named stages and SSLs are indicated on Y-axes. Asterisks within each stage indicate size heterogeneity among stocks (P < 0.05: **, with Bonferroni correction; *, only without Bonferroni correction). (B, C) Sizes at which 50% of individuals are predicted to have transitioned from one stage to the next. Differences in transition probabilities indicate variation in size– stage relationships between strains (B: χ2 = 57.8, d.f. = 30, P < 0.005; D: χ2 = 80.7, d.f. = 30, P < 0.001). Same individuals shown in Fig. 2. (See color plate)
Before staging tools existed, postembryonic stages of development had to be described in very general terms (eg, “larva”) or else in terms of age alone. Now that straightforward and easily diagnosed options are available for postembryonic staging, researchers should not rely on (and peer reviewers are urged to not accept) age as a proxy for development, particularly in reference to postembryonic stages.
1.2.2 Size alone is a poor proxy for development, but can add valuable information to stage
The size of a fish can be measured in a number of dimensions, including SL and myotome Height at the Anterior margin of the Anal fin (HAA). SL is correlated with developmental progress and explains significantly more developmental variance than age alone (Fuiman, Poling, & Higgs, 1998; Parichy et al., 2009). Yet, size and stage are far from perfectly correlated, even among wild-type zebrafish, and differences in growth and development rate due to environmental, genetic, or stochastic differences can significantly shift the sizes at which milestones are reached (see below). Further, many mutations and experimental manipulations disrupt the relationships between size and stage.
Although size is not generally a reliable indicator for postembryonic developmental stage, it can nevertheless be useful when referencing specific strains, as for mutants or transgenic lines in which size–stage relationships are known to be disrupted (eg, McMenamin et al., 2014; McMenamin, Minchin, Gordon, Rawls, & Parichy, 2013 and see below). Particularly when coupled with staging assignments, SL can add valuable information about strain-specific size–stage relationships (see discussion on composite staging, below).
1.3 VARIATION IN GROWTH AND DEVELOPMENTAL TIMING
Although embryonic and postembryonic normal tables focused on single wild-type strains, one might anticipate that differences in growth and development exist between strains, particularly given the tremendous genetic diversity harbored by zebrafish and the stark differences even between laboratory strains (Howe et al., 2013; Parichy, 2015; Patowary et al., 2013). Indeed, the SSL-staging convention was based in part on the assumption that strain variation likely exists (Parichy et al., 2009). We therefore sought to assess the potential for differences in size–stage relationships by examining fish of four wild-type genetic backgrounds, ABwp (an isolate of AB maintained by inbreeding since 2000), TU, WIK (Wild India Kolkata), and WT(WA) (Wild-type, WIK x AB; generated by crossing WIK and ABwp, and used for prior analyses).
As observed previously, sizes (and thus, growth rates) varied considerably even among individuals of the same genetic background examined on the same day (eg, arrow in Fig. 2A), illustrating the imprecision associated with age as an indicator of phenotype. These analyses also confirmed differences in growth rates among wild-type stocks (Fig. 2). For example, TU fish grew significantly faster than ABwp overall, as measured by SL and HAA.
Each genetic background also exhibited a somewhat different relationship between size and developmental state (Fig. 3). Although mean SLs associated with developmental milestones were close to archetypal SSLs, there were significant heterogeneities across strains in size–stage relationships. For example, WIK fish reached developmental milestones at SLs that were typically smaller than other stocks (Fig. 3B), whereas TU fish reached some developmental milestones at HAAs that were smaller than other stocks (Fig. 3D).
These data represent only limited sampling (three to four families per genetic background) and thus do not conclusively define strain-specific differences. Nevertheless, these heterogeneities—even among these fish reared under identical conditions—demonstrate the importance of using proper staging methods to report developmental progress, rather than relying on age or size as proxies. Two examples from these data serve to further illustrate this point (Fig. 4): (1) A wild-type fish reported to be “20 dpf” could range anywhere from 5.1 to 9.8 mm SL, with corresponding stages ranging between anal fin condensation (AC) and onset of posterior squamation (SP), encompassing much of what is considered to be “metamorphosis” (Parichy et al., 2009), (2) A wild-type fish reported to be “6.3 mm SL” could likewise imply stages ranging from dorsal fin condensation (DC) to pelvic fin bud (PB), again encompassing a tremendous diversity in morphological, physiological, and transcriptomic states. For the majority of research contexts, then, authors are urged to use either named stages or SSLs for reporting developmental progress.
FIGURE 4. Developmental ambiguities associated with age and size.
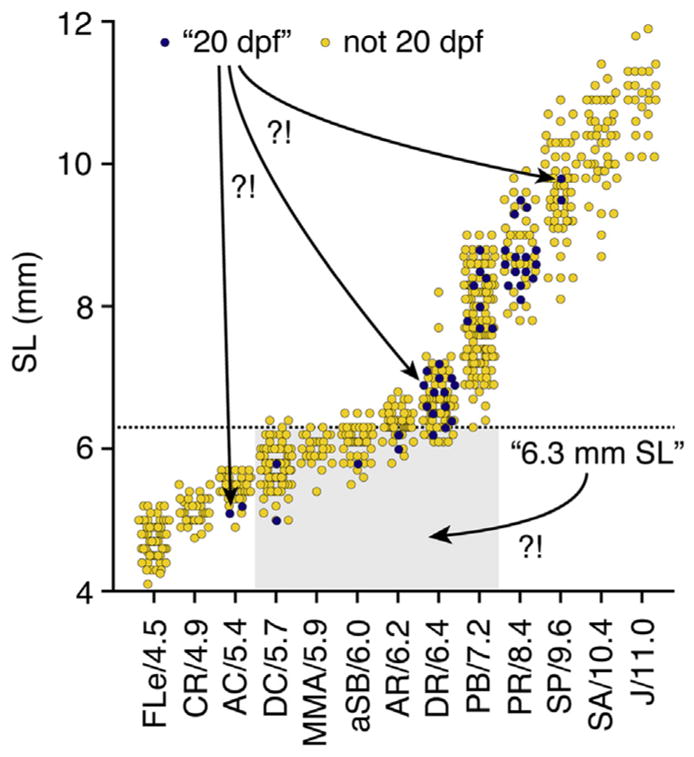
Data from Figs. 2 and 3 replotted to illustrate ambiguities (“?!”) in developmental stages when only a given day (20 dpf) or a given absolute size (6.3 mm) is reported.
1.4 CUSTOMIZING STAGING FOR PARTICULAR APPLICATIONS
For most purposes, it is appropriate to examine developmental milestones with reference to the postembryonic normal table and to report developmental progress either as named stage or SSL, depending on how finely development needs to be parsed (named stages will be coarser owing to the relative paucity of discrete, externally visible indicators of developmental progress). Indeed, named stages and SSLs even apply reasonably well to several other species of Danio, despite differences in absolute size relative to zebrafish. If size–stage relationships differ greatly, however, it may be useful to report a composite stage, consisting of the named stage and the actual size at which the stage has been achieved (eg, “DR:6.3 mm SL”; for further discussion see Parichy et al., 2009).
Finally, a particular complication arises when reporting on mutants, treatments, or species in which the sequence of development is itself so altered that individual developmental milestones are no longer informative about overall phenotype. For example, zebrafish lacking thyroid hormone inflate their anterior swim bladder lobe much later, and out of sequence, relative to other milestones attained by wild-type siblings (McMenamin et al., 2014). Under such circumstances, it may be preferable to report a strain-specific SL in millimeters, along with enough strain-specific descriptive information for the SL “shorthand” to be interpretable in other contexts (eg, different rearing conditions in another lab).
1.5 EFFICIENT STAGING
The easiest way to evaluate developmental progress is under a stereomicroscope, using transmitted light, reflected light, or both to reveal specific features. Fish are anesthetized (eg, using MS222), then lifted from the anesthetic using either a spoon or a transfer pipet (depending on the fish’s size), and placed with very little water on a slide or clear dish. A flat ruler, preferably with 0.5 mm markings, or stage and ocular micrometers, can be used to measure SL, which can provide a starting point for identifying the normal table-defined named stage or SSL. Once an investigator is proficient at staging, multiple fish can be examined at the same time or in rapid succession. Note, however, that larvae can be sensitive to anesthetics, particularly between anal fin ray appearance (AR) and PB+ (following pelvic fin bud appearance; 6.2–7.6 SSL), and care should be taken if recovery from anesthesia is necessary.
1.6 SAMPLE COLLECTION
There is always variation in size and stage even within low density, growth-optimized wild-type clutches (eg, Figs. 2 and 3), so that often only a subset of individuals will be at a particular stage at any given time. Thus, if a large number of fish are needed at a specific stage, it is advisable to either (1) rear large numbers of fish and collect individuals when a maximum number are at the desired stage or (2) collect fish piecemeal as they reach the requisite stage, immediately analyzing or storing them for later use (eg, in paraformaldehyde, RNAlater®, or other media as appropriate).
Importantly, sizes and stages within a clutch should show a roughly normal distribution. Large amounts of variability, highly skewed, or bimodal distributions of sizes can indicate suboptimal growth conditions and a need to further optimize rearing practices. The rearing protocols described in the following section can help maximize developmental uniformity and facilitate collection of large samples.
2. PART II: REARING FISH FOR USE AT POSTEMBRYONIC STAGES
Although zebrafish develop relatively rapidly, researchers interested in postembryonic processes must wait for days or weeks for their stages of interest to be reached. Particularly when working at these postembryonic stages, it is critical to achieve rates of growth and development that are both rapid and as uniform as possible. In this section we present some general strategies we use for achieving relatively rapid rates of growth and development (additional information on our procedures can be found through http://faculty.washington.edu/dparichy/).
2.1 GENERAL RECOMMENDATIONS
2.1.1 Food guidelines
Frequent feedings with high-quality food are essential to maximize growth rates and developmental uniformity within clutches. Our fish are fed three times daily (including weekends and holidays) with four different types of food: live marine rotifers, live Artemia, a fine-grained juvenile food suspended in water, and a coarse-grained adult flake food. Fish are fed in the morning, no more than 2 h after lights-on, at midday, and in the evening ~2 h before lights-off. Many tanks receive multiple types of food (up to three types), so a clear labeling system is essential. We use differently colored adhesive plastic tabs (eg, Post-it® brand) to indicate both types and quantities of food for each tank, and these are updated twice weekly to account for changing needs as fish develop. To reduce direct competition and thereby “level the playing field” between slightly larger and smaller fish, we feed the largest type of food first, allowing the largest individuals to satiate themselves before providing smaller types of food more appropriate to slightly smaller individuals in a tank.
Since every facility will produce different densities of rotifers and Artemia, and since the amount of each food needed will vary depending on the number and size of fish in a tank and the system setup (flow-through rate, tank size, etc.), we do not specify food quantities here. In general, fish should be fed with only as much flake food or Artemia as they will consume within ~15 min, though we feed rotifers more liberally and try to balance fish–rotifer encounter rates with rotifer quality (see below).
2.1.2 Rotifer rearing and preparation
While a fish lab focused on embryonic stages can likely thrive without using rotifers, a lab requiring large numbers of postembryonic fish will find rotifers valuable for promoting growth and developmental uniformity. Typically, rotifers are marine Brachionus plicatilis (Reed Mariculture) that have been acclimated to lower salinities, other varieties of marine and freshwater rotifers can be used as well (Aoyama et al., 2015; Best, Adatto, Cockington, James, & Lawrence, 2010; Lawrence, James, & Mobley, 2015; Lawrence, Sanders, & Henry, 2012). Many rotifer-culturing systems are available, ranging from simple aerated buckets to sophisticated recirculating systems appropriate for a large, high-throughput fish facility. We employ two recirculating systems (aquatic habitats) for redundancy. In these, we maintain 60 L cultures of ~2 × 108 rotifers (~3300 rotifers/mL) in rotifer water (see Section 3), from which we harvest ~6 L (~2 × 107 rotifers) daily per system. To feed the rotifers, cultures are dosed by peristaltic pump with preserved algae (eg, Nanno 3600 Instant Algae, Reed Mariculture, Campbell CA), and to maintain constant pH and prevent ammonia toxicity (obviating a biological filter), they are simultaneously dosed with a proportionate amount of ChlorAm-X/Bicarb (see Section 3). By examining samples under a microscope, rotifer populations are monitored for health (color, swimming speed, presence of eggs) and counted in morning and evening to estimate population numbers. Both food and harvest levels are modulated based on these absolute numbers, as well as population growth rates [r = ln(N − N0)/t, where N and N0 are current and starting population sizes and t is time elapsed] calculated over short-term (since harvest) or long-term (over several days). To preserve overall viability of the culture, a smaller harvest would be dictated by an unexpectedly small population size or growth rate since last harvest, or a net negative growth rate over several days. We have found that quantitative population assessments, and corresponding adjustments in food and harvest, are critical to maximizing rotifer production and culture stability.
Provided directly to fish, we find that algae-fed rotifers increase fish growth as compared to other diets. However, it is even more beneficial to supplement rotifers with Rotimac (see Section 3) immediately before introducing to the fish. We supplement the rotifer harvest with Rotimac an hour before they are fed to the fish (since rotifers are fed to fish three times per day, the harvested rotifers are supplemented with Rotimac three times per day). The harvest is then concentrated and excess Rotimac removed by gentle straining through a 23 μm screen, and rotifers are resuspended in an appropriately small volume of rotifer water from which they can be pipetted into fish tanks. In order to minimize ammonia levels in static water fish tanks, several milliliters of 10% ChlorAm-X/Bicarb (see Section 3) are added to the strained rotifer harvest before feeding to the fish.
2.1.3 Supplementation with brine shrimp
Although rotifers and flake feeds (see below) are the major types of food provided to growing larvae, some stocks, particularly if genetically growth-compromised, are supplemented with newly hatched brine shrimp, Artemia. Such supplementation is limited to tanks on flowing water as feeding Artemia to fish in static water is not recommended unless quantities are monitored very carefully and the water is changed frequently. Although Artemia cannot be substituted for rotifers or other very small foods (eg, Paramecia) at the very early stages of larval fish development; the commercial availability of Artemia cysts that can be stored for long periods and the ease of hatching Artemia without the maintenance required for maintaining rotifer in high-density rotifer cultures continue to make them a useful food supply. It also seems likely that mixed diets combining rotifers, Artemia, and other foods have benefits over diets comprising just single food types (Lawrence et al., 2015).
2.2 FISH-REARING PROTOCOLS
2.2.1 Embryo rearing
The embryonic period lasts from fertilization until hatching. Embryos are sorted for fertilization within a few hours and then placed at low densities into 100 × 15 mm Petri dishes filled to ~3 mm with 10% Hanks solution (Westerfield, 2000). To inhibit bacterial and fungal growth, a single drop (~50 μL) of stock methylene blue (0.005% weight/volume) is added to each dish. Since dead embryos can foul the water and can kill or inhibit growth of the entire clutch, dishes are sorted daily to remove any individuals that are dead, or failing to thrive. We typically rear only 20–40 embryos per Petri dish, so that embryos can be transferred directly to tanks at an appropriate density for long-term rearing.
2.2.2 Larva rearing: static water
The larval period lasts from hatching through the juvenile stage (11 SSL). Once larvae have inflated swim bladders (pSB:3.5, ~4 dpf), they are transferred from their dish into a tank containing ~4 cm larval water (see Section 3). The relatively low water level maximizes surface area for gas exchange and allows for high encounter rates between larvae and a given number of rotifers. Since high salinity maximizes the survival of rotifers and inhibits bacterial growth, larval water contains more salt than regular fish system water. For routine stock maintenance, we aim for no more than 20–30 larvae per 2.8 L tank; even lower densities can support faster growth rates, though larvae are sometimes reared at higher density as well, if they are to be harvested prior to adulthood.
Larvae in static water are fed with Rotimac-supplemented rotifers (with ChlorAm-X/Bicarb) dispersed in drops across the tank three times a day from a pipet. It is important to provide enough rotifers that young larvae encounter them easily. However, rotifers are most nutritious shortly after they, themselves, have been fortified with Rotimac; it is therefore also important to provide few enough rotifers that Rotimac does not have time to clear from the rotifers’ systems before being eaten by larvae. Careful examination of fish (to see if they are eating and growing) and their tanks (to assess left-overs and water quality) are critical to getting the balance right. During the static-water phase, tanks need to be carefully monitored to ensure that water neither becomes turbid (indicating bacterial bloom) nor evaporates, concentrating salts and waste products; if these are problems, fish should be transferred to fresh larval water.
2.2.3 Larva rearing: flowing water
When the larvae have reached ~6–8 SSL (depending on screen size), they can be transferred to flowing system water, with a low rate of water turnover and with mesh screens to prevent larvae from being flushed into the system. Larvae on flowing water are fed with both large quantities of rotifers (milliliters rather than drops) and either newly hatched Artemia or juvenile food (see Section 3) three times a day. To facilitate rapid feeding, juvenile food is suspended in fishroom water and dispensed from a 3 mL plastic pipet in 0.5 mL increments. Since the food particles tend to settle to the bottom, feeders take care to swirl the suspension regularly.
2.2.4 Juvenile and adult rearing
Once juvenile fish are sufficiently large, larval screens are removed and fish are transitioned to a flake food diet. Juveniles are initially fed a mix of both juvenile food and adult flake food (see Section 3) three times per day. Once fish reach adulthood (A:26), they can be fed with adult food alone. Many adults are fed only twice daily (morning and evening), though adults being frequently bred are given a supplementary midday feeding to support gamete production. Diets of fish being bred frequently are sometimes also supplemented with live Artemia three times per day. Careful monitoring and adjustment of food levels prevents overfeeding and therefore the accumulation of detritus and blooms of harmful fungi or bacteria, and this in turn minimizes the need to clean tanks. Indeed, we find that frequent tank changing is stressful on fish and personnel alike; we find that fish in tanks well “conditioned” with algae and presumably beneficial bacteria and mucous from the fish themselves are most healthy and productive (as might be expected given their natural habitats (Engeszer, Patterson, Rao, & Parichy, 2007; Parichy, 2015)).
2.3 WORKING WITH SLOW-GROWING OR DEVELOPMENTALLY COMPROMISED FISH
Certain mutations cause developmental or growth defects arising at postembryonic stages; indeed many of these mutants are of interest for their postembryonic defects. The general recommendations laid out above (high food, low density) are especially critical when working with growth-compromised strains, which may perish under suboptimal conditions.
If working with backcrosses or intercrosses of heterozygotes, wild-type individuals can often outcompete their slow-growing siblings, so it is important to sort frequently and separate the smallest from the largest individuals. Depending on the relative rates of growth and the nature of the mutant phenotypes, sorting can be performed weekly, either with a plastic pipet directly from the tanks, in Petri dishes by eye or under a microscope, or using size-specific fish graders. Once identified, mutants may need to be kept under different rearing conditions than their wild-type counterparts, likely requiring longer periods of an exclusively rotifer diet and static water. When fish are kept in static larval water for prolonged periods, tanks are carefully monitored for bacterial blooms and waste buildup, and are changed whenever the water appears fouled.
3. RECIPES
For recipes not listed here see Westerfield (2000).
-
Larval water (for rearing young larvae in static tanks)
0.44% (w/v) Instant Ocean (Instant Ocean, Blacksburg, VA) in RO water
(168 g Instant Ocean per 10 gallons RO water)
-
Rotifer water (for maintaining cultures of low saline-acclimated marine rotifers)
1.48% (w/v) Instant Ocean in RO water
(2.80 kg Instant Ocean per 50 gallons RO water)
-
Rotimac
8.3% (w/v) Rotimac powder (Aquafauna, Hawthorne, CA) in RO water
(75 g Rotimac powder in 900 mL RO water—homogenize thoroughly in a blender, aliquot into 50 mL tubes, and store frozen)
-
ChlorAm-X/Bicarb
10% (w/v) ChlorAm-X powder (Reed Mariculture, Campbell, CA)
5% NaHCO3 in RO water
-
Juvenile food
200 g Larval AP 100 (100–150 μM size) (Zeigler, Gardners, PA)
50 g spirulina powder (Aquafauna Bio-marine, Inc.)
1 kg ArteMac #2 (100–200 μM size) (Aquafauna, Hawthorne, CA)
(mix well, store frozen in sealed ziplock bags feed in suspension)
-
Adult flake food
200 g Larval AP 100 (250–450 μM size) (Zeigler, Gardners, PA)
50 g spirulina powder (Aquafauna Bio-marine, Inc.)
1 kg ArteMac #2 (300–500 μM size) (Aquafauna, Hawthorne, CA)
(mix well, store frozen in sealed ziplock bags)
Acknowledgments
Thanks to Parichy lab members for help rearing the fish. Supported by NIH R01 GM111233 and NIH R03 HD074787 to DMP and K99 GM105874 to SKM.
References
- Ablain J, Zon LI. Of fish and men: using zebrafish to fight human diseases. Trends in Cell Biology. 2013;23(12):584–586. doi: 10.1016/j.tcb.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Müller T, … Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139(2):366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Akiva A, Malkinson G, Masic A, Kerschnitzki M, Bennet M, Fratzl P, … Yaniv K. On the pathway of mineral deposition in larval zebrafish caudal fin bone. Bone. 2015;75:192–200. doi: 10.1016/j.bone.2015.02.020. [DOI] [PubMed] [Google Scholar]
- Ali S, Champagne DL, Spaink HP, Richardson MK. Zebrafish embryos and larvae: a new generation of disease models and drug screens. Birth Defects Research Part C: Embryo Today: Reviews. 2011;93(2):115–133. doi: 10.1002/bdrc.20206. [DOI] [PubMed] [Google Scholar]
- Aoyama Y, Moriya N, Tanaka S, Taniguchi T, Hosokawa H, Maegawa S. A novel method for rearing zebrafish by using freshwater rotifers (Brachionus calyciflorus) Zebrafish. 2015;12(4):288–295. doi: 10.1089/zeb.2014.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka Y, Terai S, Sakaida I, Nishina H. The expanding role of fish models in understanding non-alcoholic fatty liver disease. Disease Models Mechanisms. 2013;6(4):905–914. doi: 10.1242/dmm.011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin PJ, Goizet C, Raldúa D. Zebrafish models of human motor neuron diseases: advantages and limitations. Progress in Neurobiology. 2014;118:36–58. doi: 10.1016/j.pneurobio.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Best J, Adatto I, Cockington J, James A, Lawrence C. A novel method for rearing first-feeding larval zebrafish: polyculture with Type L saltwater rotifers (Brachionus plicatilis) Zebrafish. 2010;7(3):289–295. doi: 10.1089/zeb.2010.0667. [DOI] [PubMed] [Google Scholar]
- Bird NC, Mabee PM. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Developmental Dynamics. 2003;228(3):337–357. doi: 10.1002/dvdy.10387. [DOI] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Post-embryonic nerve-associated precursors to adult pigment cells: genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genetics. 2011;7(5):e1002044. doi: 10.1371/journal.pgen.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. The ISME Journal. 2015 doi: 10.1038/ismej.2015.142. http://dx.doi.org/10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed]
- Cass AN, Servetnick MD, McCune AR. Expression of a lung developmental cassette in the adult and developing zebrafish swim bladder. Evolution Development. 2013;15(2):119–132. doi: 10.1111/ede.12022. [DOI] [PubMed] [Google Scholar]
- Coffin AB, Brignull H, Raible DW, Rubel EW. The lateral line system. Springer; 2014. Hearing loss, protection, and regeneration in the larval zebrafish lateral line; pp. 313–347. [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132(5):1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- Cubbage CC, Mabee PM. Development of the cranium and paired fins in the zebra-fish, Danio rerio (Ostariophysi, Cyprinidae) Journal of Morphology. 1996;229(2):121–160. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- De Vrieze E, van Kessel M, Peters H, Spanings F, Flik G, Metz J. Prednisolone induces osteoporosis-like phenotype in regenerating zebrafish scales. Osteoporosis International. 2014;25(2):567–578. doi: 10.1007/s00198-013-2441-3. [DOI] [PubMed] [Google Scholar]
- Drummond IA. Kidney development and disease in the zebrafish. Journal of the American Society of Nephrology. 2005;16(2):299–304. doi: 10.1681/ASN.2004090754. [DOI] [PubMed] [Google Scholar]
- Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, … Kimmel CB. FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Developmental Biology. 2013;13(1):23. doi: 10.1186/1471-213X-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizondo MR, Arduini BL, Paulsen J, MacDonald EL, Sabel JL, Henion PD, … Parichy DM. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Current Biology. 2005;15(7):667–671. doi: 10.1016/j.cub.2005.02.050. [DOI] [PubMed] [Google Scholar]
- Elizondo MR, Budi EH, Parichy DM. Trpm7 regulation of in vivo cation homeostasis and kidney function involves stanniocalcin 1 and fgf23. Endocrinology. 2010;151(12):5700. doi: 10.1210/en.2010-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4(1):21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- de Esch C, Slieker R, Wolterbeek A, Woutersen R, de Groot D. Zebrafish as potential model for developmental neurotoxicity testing: a mini review. Neurotoxicology and Teratology. 2012;34(6):545–553. doi: 10.1016/j.ntt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Fero K, Yokogawa T, Burgess HA. Zebrafish models in neurobehavioral research. Springer; 2011. The behavioral repertoire of larval zebra-fish; pp. 249–291. [Google Scholar]
- Flynn EJ, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) Journal of Lipid Research. 2009;50(8):1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuiman LA, Poling KR, Higgs DM. Quantifying developmental progress for comparative studies of larval fishes. Copeia. 1998;1998(3):602–611. [Google Scholar]
- Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends in Genetics. 2013;29(11):611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2(5):559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A, Wada H, Dambly-Chaudière C. Flow sensing in air and water. Springer; 2014. Patterning the posterior lateral line in Teleosts: evolution of development; pp. 295–318. [Google Scholar]
- Godwin J. The promise of perfect adult tissue repair and regeneration in mammals: learning from regenerative amphibians and fish. Bioessays. 2014;36(9):861–871. doi: 10.1002/bies.201300144. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, … Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DW, Oh ES, Park SH, Chang YT, Kim CH, Choi SY, Williams DR. A novel zebrafish human tumor xenograft model validated for anti-cancer drug screening. Molecular BioSystems. 2012;8(7):1930–1939. doi: 10.1039/c2mb05501e. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends in Pharmacological Sciences. 2014;35(2):63–75. doi: 10.1016/j.tips.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, DeLaurier A, Ullmann B, Dowd J, McFadden M. Modes of developmental outgrowth and shaping of a craniofacial bone in zebrafish. PLoS One. 2010;5(3):e9475. doi: 10.1371/journal.pone.0009475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizil C, Kaslin J, Kroehne V, Brand M. Adult neurogenesis and brain regeneration in zebrafish. Developmental Neurobiology. 2012;72(3):429–461. doi: 10.1002/dneu.20918. [DOI] [PubMed] [Google Scholar]
- Kondo S, Watanabe M. Black, yellow, or silver: which one leads skin pattern formation? Pigment Cell Melanoma Research. 2015;28(1):2–4. doi: 10.1111/pcmr.12328. [DOI] [PubMed] [Google Scholar]
- Laizé V, Gavaia PJ, Cancela ML. Fish: a suitable system to model human bone disorders and discover drugs with osteogenic or osteotoxic activities. Drug Discovery Today: Disease Models. 2014;13:29–37. [Google Scholar]
- Lawrence C, James A, Mobley S. Successful replacement of Artemia salina nauplii with marine rotifers (Brachionus plicatilis) in the diet of preadult zebrafish (Danio rerio) Zebrafish. 2015;12(5):366–371. doi: 10.1089/zeb.2015.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C, Sanders E, Henry E. Methods for culturing saltwater rotifers (Brachionus plicatilis) for rearing larval zebrafish. Zebrafish. 2012;9(3):140–146. doi: 10.1089/zeb.2012.0771. [DOI] [PubMed] [Google Scholar]
- Lee J, Freeman JL. Zebrafish as a model for investigating developmental lead (Pb) neurotoxicity as a risk factor in adult neurodegenerative disease: a mini-review. Neurotoxicology. 2014;43:57–64. doi: 10.1016/j.neuro.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Löhr H, Hammerschmidt M. Zebrafish in endocrine systems: recent advances and implications for human disease. Annual Review of Physiology. 2011;73:183–211. doi: 10.1146/annurev-physiol-012110-142320. [DOI] [PubMed] [Google Scholar]
- Longo S, Riccio M, McCune AR. Homology of lungs and gas bladders: insights from arterial vasculature. Journal of Morphology. 2013;274(6):687–703. doi: 10.1002/jmor.20128. [DOI] [PubMed] [Google Scholar]
- Marjoram L, Bagnat M. Infection, inflammation and healing in zebrafish: intestinal inflammation. Current Pathobiology Reports. 2015;3(2):147–153. doi: 10.1007/s40139-015-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone G, Wilson KS, Mullins JJ, Tucker CS, Denvir MA. Temporal cohesion of the structural, functional and molecular characteristics of the developing zebrafish heart. Differentiation. 2015;89(5):117–127. doi: 10.1016/j.diff.2015.05.001. [DOI] [PubMed] [Google Scholar]
- McMenamin SK, Bain EJ, McCann AE, Patterson LB, Eom DS, Waller ZP, … Parichy DM. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345(6202):1358–1361. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMenamin SK, Minchin JE, Gordon TN, Rawls JF, Parichy DM. Dwarfism and increased adiposity in the gh1 mutant zebrafish vizzini. Endocrinology. 2013;154(4):1476–1487. doi: 10.1210/en.2012-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin J, Rawls JF. In vivo analysis of white adipose tissue in zebrafish. Methods in Cell Biology. 2011;105:63–86. doi: 10.1016/B978-0-12-381320-6.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Yang E, Neelkantan N, Mikhaylova A, Arnold R, Poudel MK, … Kalueff AV. Developing ‘integrative’ zebrafish models of behavioral and metabolic disorders. Behavioural Brain Research. 2013;256:172–187. doi: 10.1016/j.bbr.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Norton W, Bally-Cuif L. Adult zebrafish as a model organism for behavioural genetics. BMC Neuroscience. 2010;11(1):90. doi: 10.1186/1471-2202-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton WH, Stumpenhorst K, Faus-Kessler T, Folchert A, Rohner N, Harris MP, … Bally-Cuif L. Modulation of fgfr1a signaling in zebrafish reveals a genetic basis for the aggression–boldness syndrome. The Journal of Neuroscience. 2011;31(39):13796–13807. doi: 10.1523/JNEUROSCI.2892-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM. Advancing biology through a deeper understanding of zebrafish ecology and evolution. Elife. 2015:4. doi: 10.7554/eLife.05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Developmental Dynamics. 2009;238(12):2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Spiewak JE. Origins of adult pigmentation: diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell Melanoma Research. 2015;28(1):31–50. doi: 10.1111/pcmr.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualetti S, Banfi G, Mariotti M. The zebrafish scale as model to study the bone mineralization process. Journal of Molecular Histology. 2012;43(5):589–595. doi: 10.1007/s10735-012-9425-z. [DOI] [PubMed] [Google Scholar]
- Patowary A, Purkanti R, Singh M, Chauhan R, Singh AR, Swarnkar M, … Sivasubbu S. A sequence-based variation map of zebrafish. Zebrafish. 2013;10(1):15–20. doi: 10.1089/zeb.2012.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Disease Models Mechanisms. 2014;7(7):739–743. doi: 10.1242/dmm.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Current Opinion in Chemical Biology. 2015;24:58–70. doi: 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R, Slanchev K, Kraus C, Knyphausen P, Eming S, Hammerschmidt M. Adult zebrafish as a model system for cutaneous wound-healing research. Journal of Investigative Dermatology. 2013;133(6):1655–1665. doi: 10.1038/jid.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. The ISME Journal. 2011;5(10):1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. Journal of Clinical Investigation. 2012;122(7):2337. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Gut P. Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cellular and Molecular Life Sciences. 2015:1–12. doi: 10.1007/s00018-014-1816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Haass C. Genomic editing opens new avenues for zebrafish as a model for neurodegeneration. Journal of Neurochemistry. 2013;127(4):461–470. doi: 10.1111/jnc.12460. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Strahle U, Scholpp S. Neurogenesis in zebrafish-from embryo to adult. Neural Development. 2013;8(3):8104–8108. doi: 10.1186/1749-8104-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Stemple DL, Barroso I. The emerging use of zebrafish to model metabolic disease. Disease Models Mechanisms. 2013;6(5):1080–1088. doi: 10.1242/dmm.011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JT, Fishman MC. From zebrafish to human: modular medical models. Annual Review of Genomics and Human Genetics. 2002;3(1):311–340. doi: 10.1146/annurev.genom.3.031402.131506. [DOI] [PubMed] [Google Scholar]
- Singh AP, Nüsslein-Volhard C. Zebrafish stripes as a model for vertebrate colour pattern formation. Current Biology. 2015;25(2):R81–R92. doi: 10.1016/j.cub.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Singleman C, Holtzman NG. Analysis of postembryonic heart development and maturation in the zebrafish, Danio rerio. Developmental Dynamics. 2012;241(12):1993–2004. doi: 10.1002/dvdy.23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJM. The composition of the zebrafish intestinal microbial community varies across development. The ISME Journal. 2015 doi: 10.1038/ismej.2015.140. http://dx.doi.org/10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed]
- Stewart AM, Braubach O, Spitsbergen J, Gerlai R, Kalueff AV. Zebrafish models for translational neuroscience research: from tank to bedside. Trends in Neurosciences. 2014;37(5):264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teck Ho Lee R, Thiery JP, Carney TJ. Dermal fin rays and scales derive from mesoderm, not neural crest. Current Biology. 2013;23(9):R336–R337. doi: 10.1016/j.cub.2013.02.055. [DOI] [PubMed] [Google Scholar]
- Thomas ED, Cruz IA, Hailey DW, Raible DW. There and back again: development and regeneration of the zebrafish lateral line system. Wiley Interdisciplinary Reviews: Developmental Biology. 2015;4(1):1–16. doi: 10.1002/wdev.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Kawakami K. Size control during organogenesis: development of the lateral line organs in zebrafish. Development, Growth Differentiation. 2015;57(2):169–178. doi: 10.1111/dgd.12196. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mechanisms of Development. 2005;122(2):157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wehner D, Weidinger G. Signaling networks organizing regenerative growth of the zebrafish fin. Trends in Genetics. 2015;31(6):336–343. doi: 10.1016/j.tig.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 4. Eugene, Oregon: University of Oregon Press; 2000. [Google Scholar]
- White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nature Reviews Cancer. 2013;13(9):624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tomkovich S, Jobin C. Could a swimming creature inform us on intestinal diseases? Lessons from zebrafish. Inflammatory Bowel Diseases. 2014;20(5):956–966. doi: 10.1097/01.MIB.0000442923.85569.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Huang J, Ye J. A fresh look at zebrafish from the perspective of cancer research. Journal of Experimental and Clinical Cancer Research. 2015;34(1):1–9. doi: 10.1186/s13046-015-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang Z, Collins JE, Andrews RM, Stemple D, Gong Z. Comparative transcriptome analyses indicate molecular homology of zebrafish swim bladder and mammalian lung. PLoS One. 2011;6(8):e24019. doi: 10.1371/journal.pone.0024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupanc GKH. Adult neurogenesis in teleost fish. In: Seki T, Sawamoto K, Parent JM, Alvarez-Buylla A, editors. Neurogenesis in the adult brain I: Neurobiology. Springer; 2011. p. 137. [Google Scholar]