Abstract
Background
Although genome-wide association studies (GWAS) have identified many genetic variants that are strongly associated with lung cancer, these variants have low penetrance and serve as poor predictors of lung cancer in individuals. We sought to increase the predictive value of germline variants by considering their cumulative effects in the context of biologic pathways.
Methods
For individuals in the Environment and Genetics in Lung Cancer Etiology study (1,815 cases/1,971 controls), we computed pathway-level susceptibility effects as the sum of relevant single-nucleotide polymorphism (SNP) variant alleles weighted by their log-additive effects from a separate lung cancer GWAS meta-analysis (7,766 cases/37,482 controls). Logistic regression models based on age, sex, smoking, genetic variants, and principal components of pathway effects and pathway-smoking interactions were trained and optimized in cross-validation, and further tested on an independent dataset (556 cases/830 controls). We assessed prediction performance using area under the receiver operating characteristic curve (AUC).
Results
Compared to typical binomial prediction models which have epidemiologic predictors (AUC = 0.607) in addition to top GWAS variants (AUC = 0.617), our pathway-based smoking-interactive multinomial model significantly improved prediction performance in external validation (AUC = 0.656, P < 0.0001).
Conclusions
Our biologically informed approach demonstrated a larger increase in AUC over non-genetic counterpart models relative to previous approaches that incorporate variants.
Impact
This model is the first of its kind to evaluate lung cancer prediction using subtype-stratified genetic effects organized into pathways and interacted with smoking. We propose pathway-exposure interactions as a potentially powerful new contributor to risk inference.
Keywords: lung cancer, risk prediction, genome-wide association studies, pathway-based analysis, pathway-smoking interactions
INTRODUCTION
Lung cancer (LC) is the most common malignancy worldwide, with 1.3 million new cases per year. It is also the top cause of cancer-related death, responsible for 1.4 million deaths per year (1). Advanced-stage disease is commonly observed at diagnosis; 22% of newly diagnosed LCs have infiltrated regional lymph nodes and 57% have metastasized beyond the lung (2). The National Lung Screening Trial showed that among current smokers between 55 and 74 years of age with a smoking history of at least 30 pack-years, and analogous former smokers who quit for less than 15 years, LC screening by low-dose helical computed tomography (CT) confers a 20% reduction in mortality compared to screening by chest X-ray (3). This finding prompted the U.S. Preventive Services Task Force to recommend annual CT screening for such high-risk individuals (4). However in practice, clinical pursuit of LC usually occurs when patients already present with suggestive symptoms (5). While the 5-year survival of overall LC is 6–15%, that of stage I LC is estimated to be 70%. Therefore better identifying individuals with elevated LC risk that motivates action may enable earlier diagnoses and improve outcomes (6).
The major LC prediction models upon which most subsequent models (7) expand are the Bach model (8), the Spitz model (9), the Liverpool Lung Project (LLP) model (10), and the Tammemagi model (11). Although they differ slightly in population study design, analysis of smokers versus non-smokers, and epidemiologic risk factors under consideration, all of the models agree that ascertaining airborne exposures and history of respiratory illnesses is useful for prediction. In contrast, extended models incorporating variants identified by LC genome-wide association studies (GWAS) have so far demonstrated that germline markers contribute little to risk prediction due to their small effect sizes. For example, adding genome-wide significant single nucleotide polymorphisms (SNPs) rs8034191 and rs402710 increased area under the receiver operating characteristic curve (AUC) of the Bach model by 0.02–0.04 (separate analyses for current, former, and ever smokers). However, training and test sets were drawn from the same study for cross-validation (12). AUC increased by 0.03 with the addition of SNP rs663048 to the LLP model. However, predictions were tested on the same individuals from which the model had been trained (13). The Spitz model with a panel of inflammatory SNPs actually tested predictions on an independent study, but observed a gain of only 0.01 in AUC (14). In a decision tree analysis with 6 SNPs, the 0.008 rise in AUC was not statistically significant (P = 0.056) in external validation (15). Another analysis constructed polygenic risk scores from genome-wide significant SNPs and improved AUC by 0.02 over the baseline non-genetic model in cross-validation (16).
We sought to enhance prediction by modeling genetic risk as the aggregate effects of disease-associated germline variants within biologic pathways. Subtype-specific discriminatory pathways for the two most common LC subtypes, adenocarcinoma (AC) and squamous cell carcinoma (SCC), as well as pathway-smoking interactions, were included to reflect potential histology-dependent and interactive influences as comprehensively as possible. We also derived variant effect sizes, trained our model, and tested our model using three independent datasets that were intentionally not pruned into a consensus set of SNPs in order to evaluate the generalizability of our approach.
MATERIALS AND METHODS
Variant effect sizes from lung cancer GWAS meta-analyses
Fixed effects meta-analysis with inverse variance weighting was conducted using previously reported GWAS of 11,864,235 genotyped and imputed SNPs in 7,766 LC cases (2,424 AC cases and 2,274 SCC cases) and 37,482 controls of European ancestry by the Transdisciplinary Research in Cancer of the Lung (TRICL) consortium (17–25), including stratified analyses for AC and SCC (Table 1). These data are available in the Database of Genotypes and Phenotypes (accession number phs000877.v1.p1).
TABLE 1.
Component studies of TRICL lung cancer GWAS meta-analyses
| Cases
|
Controls | |||
|---|---|---|---|---|
| All | ACg | SCCh | ||
| MDACCa | 1150 | 619 | 306 | 1134 |
| UKICRb | 1952 | 465 | 611 | 5200 |
| IARCc | 2533 | 517 | 911 | 3791 |
| Torontod | 331 | 90 | 90 | 499 |
| Germanye | 481 | 186 | 97 | 478 |
| deCODEf | 1319 | 547 | 259 | 26380 |
|
| ||||
| Totals | 7766 | 2424 | 2274 | 37482 |
Individual-level data for training and testing risk prediction models
IARC Lung Cancer Study
representative linkage disequilibrium structure (26) of the TRICL meta-analysis. The IARC study of LC in central Europe recruited subjects from Czech Republic, Hungary, Poland, Romania, Russia, and Slovakia (22). After quality control (QC) defined below, 303,135 SNPs (Illumina HumanHap300) in 1,841 cases and 2,441 controls remain.
Environment and Genetics in Lung Cancer Etiology (EAGLE) Study
prediction training data. The EAGLE study investigated genetic and environmental determinants of LC and smoking persistence in Italians (27). Age and smoking history were ascertained at the time of enrollment, which also coincided with age at LC diagnosis for cases (incident cases). After QC, 501,658 SNPs (Illumina HumanHap550v3.0) in 1,815 cases and 1,971 controls remain.
The Prostate, Lung, Colorectal, and Ovarian Cancer (PLCO) Screening Trial
prediction test data. LC cases were selected from both arms of the screening trial, and were frequency-matched by sex and age in 5-year intervals with controls from the lung and prostate components of PLCO, all European-Americans (28). Age and smoking history were recorded at time of intervention randomization for controls and LC diagnosis for cases. After QC, 502,961 SNPs (Illumina HumanHap550v3.0) in 556 cases and 830 controls remain.
We excluded individuals with more than 10% missing genotypes or any missing age, sex, and smoking pack-years information, SNPs with minor allele frequency less than 5%, SNPs with genotyping rate less than 90%, and SNPs that failed the Hardy-Weinberg test at the 0.0001 significance level (Table 2). Genotype data were then imputed to the 1000 Genomes Project (Phase 3) (29) with haplotype phasing by SHAPEIT (30) using IMPUTE2 v2.3.1 (31). We accepted the best-guess genotypes of imputed SNPs with information measure greater than 0.9. This stringent quality filter was applied because subsequent analyses require hard call genotypes, unlike in GWAS where >0.3–0.4 is accepted to allow non-integer allelic dosages (32). If germline-based LC risk assessment ever becomes a commercial reality with validated medical utility, it is doubtful that relevant genetic markers would be imputed with uncertainty from surrounding markers on a research chip. They would be directly genotyped on a disease-specific chip. We wanted to approximate such a scenario in our training and test datasets.
TABLE 2.
Characteristics of training and test study populations
| EAGLE
|
PLCO
|
||||||
|---|---|---|---|---|---|---|---|
| Cases no. (%) | Controls no. (%) | P a | Cases no. (%) | Controls no. (%) | P a | ||
| Total individuals | 1815 | 1971 | 556 | 830 | |||
| Lung cancer subtype | |||||||
| AC | 753 (41.4) | 267 (48.0) | |||||
| SCC | 466 (25.7) | 122 (22.0) | |||||
| Other | 596 (32.8) | 167 (30.0) | |||||
| Sex | 0.118 | 0.169 | |||||
| Male | 1429 (78.7) | 1509 (76.6) | 341 (61.3) | 477 (57.5) | |||
| Female | 386 (21.3) | 462 (23.4) | 215 (38.7) | 353 (42.5) | |||
| Age | 0.023 | 0.238 | |||||
| ≤ 59 | 395 (21.8) | 502 (25.5) | 106 (19.1) | 177 (21.3) | |||
| 60–64 | 316 (17.4) | 349 (17.7) | 167 (30.0) | 262 (31.6) | |||
| 65–69 | 406 (22.4) | 451 (22.9) | 178 (32.0) | 267 (32.2) | |||
| 70–74 | 399 (22.0) | 400 (20.3) | 105 (18.9) | 124 (14.9) | |||
| ≥ 75 | 299 (16.5) | 269 (13.6) | 0 (0.0) | 0 (0.0) | |||
| Cumulative pack-years smoked | <1×10−10 | <1×10−10 | |||||
| 0 | 138 (7.6) | 633 (32.1) | 51 (9.2) | 75 (9.0) | |||
| 1–15 | 123 (6.8) | 468 (23.7) | 69 (12.4) | 231 (27.8) | |||
| 16–30 | 278 (15.3) | 356 (18.1) | 74 (13.3) | 109 (13.1) | |||
| 31–40 | 306 (16.9) | 194 (9.8) | 106 (19.1) | 168 (20.2) | |||
| 41–50 | 320 (17.6) | 149 (7.6) | 51 (9.2) | 64 (7.7) | |||
| 51–60 | 251 (13.8) | 81 (4.1) | 54 (9.7) | 54 (6.5) | |||
| 61–70 | 120 (6.6) | 29 (1.5) | 46 (8.3) | 42 (5.1) | |||
| 71–80 | 85 (4.7) | 30 (1.5) | 28 (5.0) | 23 (2.8) | |||
| ≥ 81 | 194 (10.7) | 31 (1.6) | 77 (13.8) | 64 (7.7) | |||
| Genetic data | |||||||
| Genotyped SNPs | 501,658 | 502,961 | |||||
| Imputed SNPs | 6,652,756 | 6,535,115 | |||||
Calculated using the χ2 test to compare sex and the t-test to compare age and smoking pack-years.
Statistical Analyses
The study design overview is presented in Figure 1. We trained logistic regression models on individuals in the EAGLE study to predict LC status for individuals in the PLCO study. In
Yi is the disease status of individual i, having no LC denotes the reference outcome, k an alternative outcome, X a matrix of predictor values, γ a vector of predictor effects, and ε a vector of error terms. Different choices for predictors are described below. In binomial logistic regression, k denotes having LC. For a set of test instances, LC prediction performance was evaluated by AUC applied to the range of risk prediction scores P(Yi = k). In multinomial logistic regression, k denotes having one of several possible LC subtypes: “AC”, “SCC”, or “other lung cancer”. “Other lung cancer” pools together LC subtypes that are neither AC nor SCC, including mixed or unknown histology. Overall LC prediction scores were computed as the linear combination of subtype prediction probabilities: a ⋅ P(AC) + b ⋅ P(SCC) + c ⋅ P(other). Over a grid search on [0.01, 4] in 0.01 increments, optimal a*, b*, c* were identified through 4003 rounds of 5-fold internal cross-validation within the EAGLE study. Then following model training on the entire EAGLE study, LC prediction performance was evaluated by AUC applied to the range of risk prediction scores S = a* ⋅ P(AC) + b* ⋅ P(SCC) + c* ⋅ P(other) for individuals in the PLCO study. Mean, confidence interval, and comparison tests by DeLong’s method (33) of AUC were computed using the R package “pROC”.
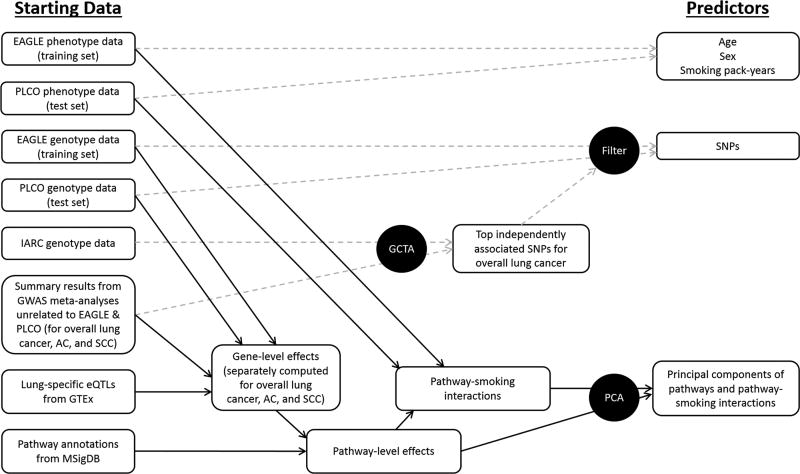
Figure 1.
Schematic overview of study design. Procedures for deriving lung cancer risk predictors from the phenotypes and genotypes of independent datasets are outlined. Dashed arrows refer to methods that have been previously reported or that are similar to those previously reported. Bold arrows refer to new approaches reported herein. AC: adenocarcinoma; EAGLE: Environment and Genetics in Lung Cancer Etiology study; eQTL: expression quantitative loci; GCTA: Genome-wide Complex Trait Analysis; GTEx: Genotype-Tissue Expression project; GWAS: genome-wide association studies; IARC: International Agency for Research on Cancer; PCA: principal component analysis; MSigDB: Molecular Signatures Database; PLCO: Prostate, Lung, Colorectal, and Ovarian cancer screening trial; SCC: squamous cell carcinoma; SNP: single-nucleotide polymorphism.
1. Epidemiologic factors age, sex, and smoking pack-years
Equation 1 was fit to LC status, age, sex, and smoking pack-years of individuals in the EAGLE study, and used to infer LC status of individuals in the PLCO study. In this model and subsequent models, the increases in log odds of having outcome k relative to not having LC with every additional year of age, being female, and every additional pack-year of smoking are β1,k, β2,k and β3,k, respectively.
| (Equation 1) |
2. Epidemiologic factors and top GWAS SNPs
We applied the software Genome-wide Complex Trait Analysis (GCTA) (34) to summary statistics of the TRICL meta-analysis to determine independently associated SNPs through stepwise-selection conditional analysis (26). In the absence of individual-level genotype data for the meta-analysis, GCTA estimated LD structure from the IARC study. Of the 1,343 SNPs deemed to be independently associated (GWAS P < 0.05 and conditional association P < 0.001), 301 SNPs appear in both EAGLE and PLCO (Supplementary Table S1). Using the EAGLE study, Equation 2 was fit to epidemiologic factors plus the top n independently associated SNP(s) ranked by ascending GWAS P-value (1 ≤ n ≤ 301), and was tested on the PLCO study. We reported the highest AUC for each set of 301 binomial and multinomial models. The increase in relative log odds with every additional copy of the variant allele for SNP j is θj,k.
| (Equation 2) |
3. Epidemiologic factors, top GWAS SNPs, and SNP-smoking interactions
Corresponding smoking interactions of the top n independently associated SNPs were added to the previous model. The increases in relative log odds with every additional pack-year of smoking and copy of the variant allele for SNP j are and θj,k + ξj,k ⋅ packyearsi, respectively.
| (Equation 3) |
4. Epidemiologic factors and biologic pathways
For nominally significant SNPs in the TRICL meta-analysis (association P < 0.05) that may affect a gene through either residing within the gene or influencing the gene’s expression in lung (eQTL FDR < 0.05 in the Genotype-Tissue Expression project) (35), we determined the number of variant alleles possessed by individuals in the EAGLE and PLCO studies. Although some SNPs are available in one study but not the other, the following construction of pathway predictors does not depend on identical genotype data between the two studies. Gene-level susceptibility effects were first computed for every individual in both studies as variant allele count (0, 1, or 2) multiplied by additive effect size (log odds ratio) of each gene’s LC-associated mapped SNP from the meta-analysis. We then derived pathway-level susceptibility effects for all individuals with respect to every curated pathway in the Molecular Signatures Database (36) as the sum of effects for genes relevant to that pathway. Using a randomly sampled 80% of the EAGLE study, we identified pathways that exhibit significant differences (t-test P < 0.05) between LC cases and controls. Principal components (PCs) of pathway z-scores were separately derived for the internal training and testing data (remaining 20% of the EAGLE study) using the R package “FactoMineR”. A binomial logistic regression model was then fit to epidemiologic factors plus the top p PCs of the 80% EAGLE training set, and tested on the 20% EAGLE test set. This process was repeated 30×100 times for positive integers p ≤ 30 and across one hundred 80/20 re-samplings of EAGLE to optimize AUC. Using the optimal p*, we trained Equation 4 on the entire EAGLE study to infer LC status in the PLCO study. The increase in relative log odds with every unit increment of PC x is ϕx,k.
| (Equation 4) |
5. Epidemiologic factors, biologic pathways, and pathway-smoking interactions
We repeated the previous analysis, except PCs were derived for both pathways and pathway-smoking interactions (Supplementary Table S2). Pathway-smoking interactions were constructed as the product of pathway effects and corresponding cumulative smoking pack-years.
6. Epidemiologic factors, subtype-specific biologic pathways, and subtype-specific pathway-smoking interactions
We repeated the previous analysis, except with pathways and pathway-smoking interactions from stratified GWAS meta-analyses for AC and SCC (Supplementary Table S3–S4), in addition to those for overall LC. With respect to the top p overall LC PCs, top q AC PCs, and top r SCC PCs, 303×100 rounds of internal cross-validation for positive integers p, q, r ≤ 30 across one hundred 80/20 re-samplings of EAGLE were performed to optimize AUC. Solutions p*, q*, r* were then used to train Equation 5 on the entire EAGLE study for prediction in the PLCO study. The increases in relative log odds with every unit increment of PCs x, yand z for overall LC, AC, and SCC are ϕx,k, ψy,k, and ωz,k, respectively.
| (Equation 5) |
Classification accuracy
We computed net reclassification improvement (NRI) (37) for the pathway-based models compared to the SNP-based binomial model as reference, using the R package “PredictABEL” (Table 3). The decision score cutoff for each model was chosen as the receiver operating characteristic (ROC) curve point with minimum Euclidean distance to coordinate (0, 1). This cutoff was also used to compute Cohen’s kappa (38) between predicted and observed outcomes (no cancer or lung cancer); it is a measure of concordance beyond what would be expected by chance alone, ranging from 0 (concordance due to chance) to 1 (perfect concordance). Likewise for the multinomial models, a predicted disease subtype (no cancer, AC, SCC, or other lung cancer) for each individual was chosen as the one with highest probability and Cohen’s kappa was computed.
TABLE 3.
Performance of lung cancer prediction models
| Logistic regression model | AUC (95% C.I., P) a | NRI (95% C.I., P) b | Cohen’s κ (95% C.I.) |
|---|---|---|---|
| 1) Epidemiologic: age, sex, pack-years (PY) | |||
| Binomial | 0.607 (0.577–0.637) | 0.172 (0.120–0.224) | |
| Multinomial | 0.608 (0.577–0.638) | 0.081 (0.037–0.125) | |
| 2) Epidemiologic + top independently associated SNPs c | |||
| Binomial | 0.617 (0.587–0.647, ref.) | (ref.) | 0.194 (0.141–0.246) |
| Multinomial | 0.617 (0.587–0.647, 0.618) | 3.6% (0.9–6.4, 0.0104) | 0.095 (0.047–0.144) |
| 3) Epidemiologic + top independently associated SNPs c and their interactions with PY | |||
| Binomial | 0.619 (0.589–0.650, 0.598) | 2.9% (0.7–5.1, 0.0097) | 0.195 (0.143–0.247) |
| Multinomial | 0.620 (0.590–0.650, 0.586) | 2.2% (0.1–4.3, 0.0438) | 0.101 (0.052–0.150) |
| 4) Epidemiologic + PCs of top overall pathways | |||
| Binomial | 0.621 (0.591–0.651, 0.568) | 5.3% (1.8–8.8, 0.0027) | 0.200 (0.146–0.253) |
| Multinomial | 0.621 (0.591–0.651, 0.582) | 5.0% (1.7–8.4, 0.0033) | 0.105 (0.058–0.152) |
| 5) Epidemiologic + PCs of top overall pathways and their interactions with PY | |||
| Binomial | 0.630 (0.600–0.660, 3.28 × 10−2) | 4.9% (2.5–7.4, 0.0002) | 0.207 (0.154–0.261) |
| Multinomial | 0.631 (0.600–0.661, 2.94 × 10−2) | 5.0% (2.4–7.6, 0.0002) | 0.125 (0.077–0.173) |
| 6) Epidemiologic + PCs of top subtype-specific pathways and their interactions with PY | |||
| Binomial | 0.651 (0.621–0.681, 8.78 × 10−4) | 8.9% (5.7–12.1, <0.0001) | 0.226 (0.171–0.281) |
| Multinomial | 0.656 (0.626–0.685, 6.11 × 10−5) | 11.7% (7.3–16.0, <0.0001) | 0.152 (0.108–0.195) |
Changes in AUC were computed relative to the #2 binomial model as reference.
Net reclassification improvement values (%) were computed relative to the #2 binomial model as reference. Classification was determined based on choosing a risk prediction score cutoff for each model that corresponds to the ROC curve point with minimum Euclidean distance to coordinate (0, 1).
Three hundred and one independently associated SNPs were identified by stepwise-selection conditional analysis of the TRICL overall lung cancer GWAS meta-analysis. Logistic regression models that include the top n SNPs were tested, for 1 ≤ n ≤ 301. Optimal n yielding the highest AUC were 30 and 39 for models 2 and 3, respectively.
RESULTS
In order to assess differences in the distribution of demographic variables between cases and controls in the EAGLE and PLCO studies, we used the χ2 test to compare categorical variables and t-test to compare continuous variables (Table 2). Cases were much more likely to have longer smoking histories. LC exhibits modest association with age in the EAGLE study, but not in the PLCO study.
Logistic regression models were trained on the EAGLE study and tested on the PLCO study. A baseline binomial model with only age, sex, and smoking pack-years as predictors attained an AUC of 0.607 (#1 in Table 3). Consistent with previous findings (7), adding top SNPs implicated in overall LC GWAS hardly increased AUC, by around 0.01 (#2 in Table 3). Diminishing returns came from further adding smoking interactions with these SNPs (#3 in Table 3). To our knowledge, existing studies of genetic interactions with smoking in LC tend to be discovery in nature and have not evaluated risk prediction (14, 39–41). Hence comparable AUC gains are unavailable. While incorporating top PCs of discriminatory pathway-level effects from overall LC GWAS negligibly boosted AUC as well (#4 in Table 3), significant improvements were achieved by including pathway-smoking interactions along with pathways derived from AC and SCC subtype-specific GWAS (#s 5–6 in Table 3).
AUC is usually favored in the early phase of prediction modeling because setting decision cutoffs is not required. However, a common critique by clinical researchers is that a specific cutoff must be selected in order to assess patient impact (42). We chose the LC prediction score cutoff for NRI that equally values sensitivity and specificity. Compared to the SNP-based binomial model, our best model correctly net re-classified 11.7% of individuals in the PLCO study. Sensitivity improved from 56% to 66% and specificity improved from 61% to 63%. A recent external validation study of several popular non-genetic LC prediction models, also treating true positives and false positives as equal tradeoffs, yielded sensitivities all less than 66% despite having incorporated many more epidemiologic risk factors, such as asbestos exposure, COPD, hay fever, and family history of LC (43). Our model’s specificity was not as high though. These findings suggest that genetic considerations may be more important for identifying cases, while epidemiologic considerations may be more important for identifying controls. Genetics-informed LC prediction models have reported AUCs, but not sensitivities and specificities at discrete ROC curve cutoffs against which to further compare (7, 12–16).
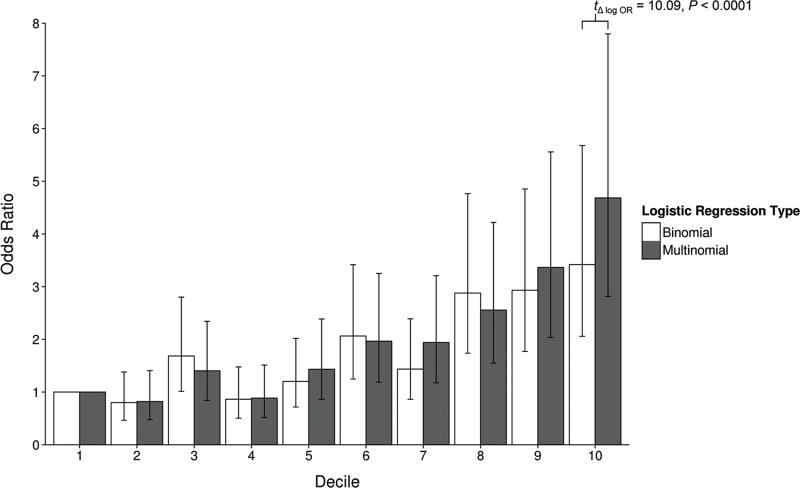
Multinomial logistic regressions assigned LC subtypes to individuals in the PLCO study. Although Cohen’s kappa of the most comprehensive model is nearly double that of the non-genetic model, classification accuracies across multinomial models are low in absolute terms. As expected, all of the binomial models exhibit higher kappas, mainly due to the difference in difficulty of predicting 2 outcomes versus 4 outcomes. Overall LC scores generated by merging LC subtype probabilities from the multinomial models also achieved AUCs similar to those of counterpart binomial models. However, the value of the multinomial approach becomes apparent when examining ordered LC prediction scores. With individuals in the bottom decile of scores from the best multinomial model (#6 in Table 3) as reference, those in the upper deciles have odds ratios of developing LC that follow a more evenly graded progression with a wider spread, compared to predicted risk deciles from the partner binomial model. In particular, the difference in stratification at the extremes is highly significant (Figure 2). Based on the multinomial model, odds of developing LC for PLCO subjects in the top predicted risk decile is 4.68 times the odds in the bottom decile. The binomial model was not as proficient in identifying the highest and lowest risk individuals, producing a corresponding odds ratio of 3.42.
Figure 2.
Odds ratios by risk prediction score deciles. Individuals in the PLCO study were grouped into deciles based on their risk prediction scores from the #6 binomial and multinomial models in Table 3. The odds of having lung cancer for each decile were compared to the odds for the lowest decile. Bars denote the 95% confidence interval for these odds ratios.
Beyond these deciles, constructing clinically meaningful risk categories is outside the scope of this study. Nevertheless, we still identified individuals at high risk for LC in a way that could at least be compared with the U.S. Preventive Services Task Force criteria: adults aged 55–80 years with at least 30 pack-years of smoking history, including former smokers who quit for less than 15 years (4). Of the 1,386 individuals in the PLCO test set, 777 meet this criteria and 362 (47%) actually developed LC. On the other hand, the 777 highest scoring individuals in PLCO from the #6 multinomial model consist of 435 (56%) LC cases, yielding a positive predictive value increase of 9%.
DISCUSSION
We put forward a method of improving LC risk prediction that is innovative in several ways. Since LC is a heterogeneous disease, we used SNP effect sizes from overall LC and subtype-stratified GWAS. While selection of SNP predictors has traditionally involved filtering SNPs by GWAS P-value or suspected functions (12–16), we aggregated relevant SNP effects into pathway-level effects. Many studies have already warned about the shortcomings of conducting mainly biostatistical polygenic analyses of GWAS for complex disease prediction (44), even if a larger than expected portion of “missing heritability” has been extrapolated to be hidden rather than actually missing (34). Our biologic integration also included constructing pathway-smoking interactions, the first use of pathway-exposure interactions in germline genetic prediction.
SNP-gene correspondences were established based on intragenic or lung eQTL status, a refinement of the usual proximity approach. As an exercise, we re-computed pathway effects by mapping SNPs to genes within 10 kilobases (kb) and discovered that the differences between LC cases and controls became less pronounced (data not shown). This suggests previous pathway-based analyses of GWAS that mapped SNPs to genes based solely on chromosomal position may have introduced noise. Not all genetic variations within 10 kb of a gene influence its expression. And some variants farther than 10 kb away can also influence expression. Pathway values derived using the position method did not distinguish cases from controls as strikingly because the sums of gene-level effects may have included genes that were improperly linked to disease-associated SNPs from GWAS.
Furthermore, simultaneous consideration of LC subtypes through multinomial logistic regression identified individuals with the highest and lowest LC risk better than binomial regression. This finding is in line with a study that demonstrated jointly predicting risk for schizophrenia, bipolar disorder, and depression using SNP data is more effective than making separate predictions (45). It is believed that genetic correlation among several diseases implies a variant affecting risk for one disease will tend to be informative of risk for correlated diseases as well. However, kappa values from joint disease prediction are not impressive. Similar to GWAS of most complex diseases, many associated genetic risk loci exist for LC, and their effect sizes have been measured to be quite small and with noise (44). Therefore prediction accuracy at the single disease (subtype) level is similarly poor. Nevertheless, merging subtype risks following multinomial analysis likely enhanced the signal-to-noise ratio of genetic effects (46) and has contributed to the more clinically important endeavor of better assessing overall LC risk. After all, motivation for LC screening does not depend on probable cancer subtype.
Incorporating subtype-specific pathways and pathway-smoking interactions increased AUC of overall LC prediction by 0.05 and 0.04 over the baseline non-genetic model and the baseline model plus top independently associated SNPs, respectively, in external validation. Primarily limited by available data, our best model achieved an AUC of 0.656. While this AUC is far from being qualified to sway individual clinical decisions, we emphasize the significance of our gain, rather than the absolute value of AUC. The current, most thorough non-genetic models that assess many other predictors in addition to age, sex, and smoking pack-years, such as history of respiratory illnesses, occupational exposures, and level of education, have achieved AUC of 0.797 in external validation (47). This performance can be expected to advance another 0.03–0.05 upon inclusion of pathway-based and interactive predictors because pathway effects are largely uncorrelated with epidemiologic risk factors (Supplementary Tables S2–S4). The projected attenuation is attributed to potential overlapping genetic effects on disease and other epidemiologic factors not ascertained here. Such a gain would bring us ever closer the goal of accurately identifying high-risk individuals for early LC screening.
A drawback of this study is difficulty interpreting the biologic impacts of genetic factors on risk prediction. From the training data, we derived pathways and pathway-smoking interactions that discern LC cases from healthy controls (Supplementary Tables S2–S4). Of note, top pathways for AC and SCC are mostly different, and some pathway-smoking interactions are more significant than their corresponding non-interacted pathways. Prominent examples include mechanisms of Bcl11b (a zinc finger protein) in AC and regulation of Smad2/3/4 (downstream transducers of signaling by transforming growth factor β) in SCC. However, the precise risk-influencing effects of pathways and their constituent genes are masked by PC regression. Coefficient estimates cannot be ascribed to any single factor from the original data. This masking through orthogonalization somewhat absolves the concern that evaluating so many interactive terms would inevitably lead to false positive biologic inferences. The rankings presented in Supplementary Tables S2–S4 may still merit new experimental pursuits though; actions of Bcl11b and Smad4 have been implicated in tumorigenesis of colon AC (48) and head and neck SCC (49), respectively. In addition, transforming to PCs has allowed removal of multicollinearity among many related pathways, an inherent feature of modern pathway databases, without discarding pathways altogether since the subtle distinctions may be important.
Another weakness of this study, along with all existing LC prediction efforts, is poor ability to model risk for never-smokers. They lack the top risk factor for LC and their pathway-smoking interactions are all zero. Adding pathways did not significantly augment AUC among PLCO never-smokers compared to the baseline non-genetic model (data not shown). With additional information and the adaptability of our pathway-exposure interactions, however, this inadequacy has the potential to change in future studies. Other exposures besides smoking, such as second-hand smoke, pollution, asbestos, various dusts, and diet, can increase risk for LC as well (50). Modeling interactions between a variety of exposure risk factors and genetic pathway effects, especially pathways for AC as it is the major tumor subtype among never-smokers (51), may reveal new insights.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Transdisciplinary Research in Cancer of the Lung consortium. This research was supported by the National Institutes of Health (P30CA023108, U19CA148127, R01CA149462, and P20GM103534 to CI Amos), and the National Science Foundation Graduate-K12 Fellowship in collaboration with Kimball Union Academy (DGE-0947790 to DC Qian).
Footnotes
Disclosure of Potential Conflicts of Interest
None declared
References
- 1.Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SEER Stat Fact Sheets. Lung and Bronchus Cancer. Bethesda, MD: National Cancer Institute; 2015. [Google Scholar]
- 3.The National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphrey L, Deffebach M, Pappas M, Baumann C, Artis K, Priest Mitchell J, Zakher B, Fu R, Slatore C. Evidence Synthesis No. 105. Rockville, MD: Agency for Healthcare Research and Quality; 2013. Screening for Lung Cancer: Systematic Review to Update the U.S. Preventive Services Task Force Recommendation. [PubMed] [Google Scholar]
- 5.Chen X, Gorlov IP, Ying J, Merriman KW, Kimmel M, Lu C, et al. Initial medical attention on patients with early-stage non-small cell lung cancer. PloS One. 2012;7:e32644. doi: 10.1371/journal.pone.0032644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field JK, Chen Y, Marcus MW, McRonald FE, Raji OY, Duffy SW. The contribution of risk prediction models to early detection of lung cancer. J Surgical Oncol. 2013;108:304–311. doi: 10.1002/jso.23384. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Oldani MJ, Zhao X, Huang X, Qian D. A review of cancer risk prediction models with genetic variants. Cancer Inform. 2014;13:19–28. doi: 10.4137/CIN.S13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach PB, Kattan MW, Thornquist MD, Kris MG, Tate RC, Barnett MJ, et al. Variations in lung cancer risk among smokers. J Natl Cancer Inst. 2003;95:470–478. doi: 10.1093/jnci/95.6.470. [DOI] [PubMed] [Google Scholar]
- 9.Spitz MR, Hong WK, Amos CI, Wu X, Schabath MB, Dong Q, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst. 2007;99:715–726. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 10.Cassidy A, Myles JP, van Tongeren M, Page RD, Liloglou T, Duffy SW, et al. The LLP risk model: an individual risk prediction model for lung cancer. Br J Cancer. 2008;98:270–276. doi: 10.1038/sj.bjc.6604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tammemagi CM, Pinsky PF, Caporaso NE, Kvale PA, Hocking WG, Church TR, et al. Lung cancer risk prediction: Prostate, Lung, Colorectal And Ovarian Cancer Screening Trial models and validation. J Natl Cancer Inst. 2011;103:1058–1068. doi: 10.1093/jnci/djr173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoggart C, Brennan P, Tjonneland A, Vogel U, Overvad K, Ostergaard JN, et al. A risk model for lung cancer incidence. Cancer Prev Res. 2012;5:834–846. doi: 10.1158/1940-6207.CAPR-11-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raji OY, Agbaje OF, Duffy SW, Cassidy A, Field JK. Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: the Liverpool Lung Project. Cancer Prev Res. 2010;3:664–669. doi: 10.1158/1940-6207.CAPR-09-0141. [DOI] [PubMed] [Google Scholar]
- 14.Spitz MR, Amos CI, Land S, Wu X, Dong Q, Wenzlaff AS, et al. Role of selected genetic variants in lung cancer risk in African Americans. J Thorac Oncol. 2013;8:391–397. doi: 10.1097/JTO.0b013e318283da29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissfeld JL, Lin Y, Lin HM, Kurland BF, Wilson DO, Fuhrman CR, et al. Lung cancer risk prediction using common SNPs located in GWAS-identified susceptibility regions. J Thorac Oncol. 2015;10:1538–1545. doi: 10.1097/JTO.0000000000000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Yang L, Zhao X, Wang J, Qian J, Chen H, et al. Prediction of lung cancer risk in a Chinese population using a multifactorial genetic model. BMC Med Genet. 2012;13:118. doi: 10.1186/1471-2350-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen T, Matakidou A, Houlston R GELCAPS Consortium. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS) BMC Cancer. 2008;8:244. doi: 10.1186/1471-2407-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 22.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 23.Scelo G, Constantinescu V, Csiki I, Zaridze D, Szeszenia-Dabrowska N, Rudnai P, et al. Occupational exposure to vinyl chloride, acrylonitrile and styrene and lung cancer risk (Europe) Cancer Causes Control. 2004;15:445–452. doi: 10.1023/B:CACO.0000036444.11655.be. [DOI] [PubMed] [Google Scholar]
- 24.Sauter W, Rosenberger A, Beckmann L, Kropp S, Mittelstrass K, Timofeeva M, et al. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. [DOI] [PubMed] [Google Scholar]
- 25.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ANthropometric Traits (GIANT) Consortium, DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–675. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes RB, Sigurdson A, Moore L, Peters U, Huang WY, Pinsky P, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005;592:147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Delaneau O, Marchini J The 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 31.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 34.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pencina MJ, D’Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 39.VanderWeele TJ, Asomaning K, Tchetgen Tchetgen EJ, Han Y, Spitz MR, Shete S, et al. Genetic variants on 15q25.1, smoking, and lung cancer: an assessment of mediation and interaction. Am J Epidemiol. 2012;175:1013–1020. doi: 10.1093/aje/kwr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudd MF, Webb EL, Matakidou A, Sellick GS, Williams RD, Bridle H, et al. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006;16:693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xun X, Wang H, Yang H, Wang H, Wang B, Kang L, et al. CLPTM1L genetic polymorphisms and interaction with smoking and alcohol drinking in lung cancer risk: a case-control study in the Han population from northwest China. Medicine. 2014;93:e289. doi: 10.1097/MD.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leening MJ, Vedder MM, Witteman JC, Pencina MJ, Steyerberg EW. Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician’s guide. Ann Intern Med. 2014;160:122–131. doi: 10.7326/M13-1522. [DOI] [PubMed] [Google Scholar]
- 43.Li K, Husing A, Sookthai D, Bergmann M, Boeing H, Becker N, et al. Selecting High-Risk Individuals for Lung Cancer Screening: A Prospective Evaluation of Existing Risk Models and Eligibility Criteria in the German EPIC Cohort. Cancer Prev Res. 2015;8:777–785. doi: 10.1158/1940-6207.CAPR-14-0424. [DOI] [PubMed] [Google Scholar]
- 44.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier R, Moser G, Chen GB, Ripke S, Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Coryell W, et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet. 2015;96:283–294. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo G, Zhao F, Wang Y, Zhang Y, Du L, Su G. Comparison of single-trait and multiple-trait genomic prediction models. BMC Genet. 2014;15:30. doi: 10.1186/1471-2156-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tammemagi MC, Katki HA, Hocking WG, Church TR, Caporaso N, Kvale PA, et al. Selection criteria for lung-cancer screening. N Engl J Med. 2013;368:728–736. doi: 10.1056/NEJMoa1211776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakamaki A, Katsuragi Y, Otsuka K, Tomita M, Obata M, Iwasaki T, et al. Bcl11b SWI/SNF-complex subunit modulates intestinal adenoma and regeneration after gamma-irradiation through Wnt/beta-catenin pathway. Carcinogenesis. 2015;36:622–631. doi: 10.1093/carcin/bgv044. [DOI] [PubMed] [Google Scholar]
- 49.Korc M. Smad4: gatekeeper gene in head and neck squamous cell carcinoma. J Clin Invest. 2009;119:3208–3211. doi: 10.1172/JCI41230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samet JM, Avila-Tang E, Boffetta P, Hannan LM, Olivo-Marston S, Thun MJ, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–299. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.