Abstract
Tigecycline is an expanded broad-spectrum antibacterial agent that is active against many clinically relevant species of bacterial pathogens, including Klebsiella pneumoniae. The majority of K. pneumoniae isolates are fully susceptible to tigecycline; however, a few strains that have decreased susceptibility have been isolated. One isolate, G340 (for which the tigecycline MIC is 4 μg/ml and which displays a multidrug resistance [MDR] phenotype), was selected for analysis of the mechanism for this decreased susceptibility by use of transposon mutagenesis with IS903φkan. A tigecycline-susceptible mutant of G340, GC7535, was obtained (tigecycline MIC, 0.25 μg/ml). Analysis of the transposon insertion mapped it to ramA, a gene that was previously identified to be involved in MDR in K. pneumoniae. For GC7535, the disruption of ramA led to a 16-fold decrease in the MIC of tigecycline and also a suppression of MDR. Trans-complementation with plasmid-borne ramA restored the original parental phenotype of decreased susceptibility to tigecycline. Northern blot analysis revealed a constitutive overexpression of ramA that correlated with an increased expression of the AcrAB transporter in G340 compared to that in tigecycline-susceptible strains. Laboratory mutants of K. pneumoniae with decreased susceptibility to tigecycline could be selected at a frequency of approximately 4 × 10−8. These results suggest that ramA is associated with decreased tigecycline susceptibility in K. pneumoniae due to its role in the expression of the AcrAB multidrug efflux pump.
Tigecycline is an expanded broad-spectrum antibiotic representing a new class called the glycylcyclines. The glycylcyclines are semisynthetic derivatives of minocycline and have activity against many bacterial pathogens (2, 14, 15). It has been noted that a few species of gram-negative bacteria, including Pseudomonas aeruginosa, Proteus spp., Providencia spp., and Morganella morganii, are intrinsically less susceptible to tigecycline. Previous studies revealed the involvement of multidrug efflux systems such as MexXY and AcrAB in the decreased tigecycline susceptibility of P. aeruginosa and Proteus mirabilis, respectively (3, 22). These pumps belong to the resistance-nodulation-division (RND) family that combines bacterial transporters with a tripartite architecture and broad substrate specificity (9, 12). Due to the broad substrate specificity of RND pumps, their overexpression usually results in the multidrug resistance (MDR) phenotype.
Klebsiella pneumoniae causes infections of wounds, the urinary tract, and the respiratory system. This bacterial species is generally susceptible to tigecycline; however, a few clinical strains with decreased tigecycline susceptibility have been isolated. In this study, one such an isolate, G340, was investigated to determine the mechanism of decreased tigecycline susceptibility in K. pneumoniae.
(These results were reported, in part, previously [M. A. Visalli, S. J. Projan, and P. A. Bradford, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1600, 2002].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Strains G595, G815, and G340 are clinical isolates from various infection types. Plasmid pVJT128 was kindly provided by David Figurski. The strains were propagated at 37°C in Luria-Bertani (LB) broth or agar.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| TOP10 | E. coli cloning strain | Invitrogen |
| INV110 | dam and dcm methylase-deficient E. coli | Invitrogen |
| G340 | Clinical isolate | This study |
| G595 | Clinical isolate | This study |
| G815 | Clinical isolate | This study |
| GC7535 | G340 insertion mutant, IS903φkan inserted in ramA | This study |
| GC7736 | G595 transformed with pCLL3442 | This study |
| GC7737 | GC7535 transformed with pCLL3442 | This study |
| Plasmids | ||
| pCR2.1-TOPO | PCR cloning vector | Invitrogen |
| pUCGm | pUC19 derivative containing gentamicin resistance cassette | 18 |
| pVJT128 | Plasmid carrier vector for IS903φkan transposon | 20 |
| pCLL3441 | pCR2.1-TOPO with cloned ramA gene | This study |
| pCLL3442 | pCLL3441 with cloned gentamicin cassette | This study |
Antibiotic susceptibility testing.
Tigecycline and minocycline used in this study were from Wyeth Research, Pearl River, N.Y. Tetracycline, acriflavine, ethidium bromide, erythromycin, chloramphenicol, nalidixic acid, novobiocin, trimethoprim, norfloxacin, gentamicin, kanamycin, and IPTG (isopropyl-β-d-thiogalactopyranoside) were obtained from Sigma Chemical Co., St. Louis, Mo. The MICs of various antibacterial agents were determined by standard broth microdilution tests (11). Tests for tigecycline were performed using fresh Mueller-Hinton broth (<12 h old).
DNA techniques.
Standard DNA manipulations such as restriction digestion and molecular cloning were performed as described previously (16). Chemically competent Escherichia coli strains TOP10 and INV110 (Invitrogen, Carlsbad, Calif.) were used for cloning experiments. Transformation was performed as specified by the manufacturer. DNA fragments were gel purified by using a Zymoclean Gel DNA recovery kit (Zymo Research, Orange, Calif.). K. pneumoniae genomic DNA was isolated by using a Puregene tissue kit (Gentra Systems, Inc., Minneapolis, Minn.) and used as a template for PCRs. A 575-bp ramA-containing DNA fragment was amplified by PCR using the primers listed in Table 2. The gel-purified PCR fragment was ligated into the pCR2.1-TOPO vector (Invitrogen). The resulting plasmid, pCLL3441, was modified by cloning a 878-bp XbaI fragment containing a gentamicin resistance cassette from pUCGm into the XbaI site of pCLL3441. The resulting plasmid, pCLL3442, was used in trans-complementation studies. DNA transformations of K. pneumoniae clinical isolates were performed by electroporation with a Gene Pulser II system (Bio-Rad, Hercules, Calif.), using the optimal electroporation settings of 2.5 kV, 25 μF, 200 Ω, and 5 ms.
TABLE 2.
Primers and fluorescent probes used for PCR
| Gene | Product size (bp) | Purpose | Forward primer (5′-3′) | Reverse primer (5′-3′) | Fluorescent probea (5′-3′) |
|---|---|---|---|---|---|
| NAb | NA | Inverse PCR primer 1 | GTTTCCCGTTGAATATGGCTGGG | NA | NA |
| NA | NA | Inverse PCR primer 2 | GCAGTTTCATTTGATGCTCGA | NA | NA |
| ramA | 575 | TOPO cloning | GGATGAACCGTATCAACG | CCATTGAGTATCTGGTGC | NA |
| ramA | 335 | Probe for Northern analysis | GCATATGACGATTTCCGCTCAG | ACTGTGGTTCTCTTTGCG | NA |
| acrA | 557 | Probe for Northern analysis | TTCTGATGCTCTCAGGC | TGACCATTCTGTACCAGC | NA |
| rrsE | 525 | Probe for Northern analysis | GTAGCTAATACCGCATAACGTCGC | GCTACACCTGGAATTCTACC | NA |
| ramA | 66 | RT-PCR | GCATCAACCGCTGCGTATT | CGTTGCAGATGCCATTTCG | ATCGCTCGCCATGCCGGGTAT |
| rrsE | 71 | RT-PCR | TTGACGTTACCCGCAGAAGAA | GCTTGCACCCTCCGTATTACC | TAACTCCGTGCCAGCAGCCG |
Labeled with 6′-FAM at the 5′ end and with TAMRA at the 3′ end.
NA, not applicable.
Transposon mutagenesis.
Transposon mutagenesis with IS903φkan was performed essentially as described previously (20). Briefly, the transposon carrier plasmid pVJT128 was electroporated into G340, and transformants were selected on LB plates containing 200 μg of chloramphenicol/ml. Seven individual colonies were selected, inoculated into LB broth containing 1 mM IPTG and 200 μg of chloramphenicol/ml, and then propagated overnight with shaking to induce transposition. Clones with transposon insertions were selected by plating aliquots of overnight culture onto LB plates containing 50 μg of kanamycin/ml. Tigecycline-susceptible transposon mutants were isolated by replica plating with selection for colonies that grew on LB plates containing 50 μg of kanamycin/ml but not on LB plates containing 2 μg of tigecycline/ml. The carrier plasmid was cured by serial passage in chloramphenicol-free medium. Transposon insertions were mapped by an inverse PCR as described previously (21), using outward-facing primers (Table 2) (20). The products of inverse PCR were cloned into the pCR2.1-TOPO vector, and the nucleotide sequence was determined with an ABI 3700 automated sequencer (Applied Biosystems, Foster City, Calif.) using universal sequencing primers. The site of transposon insertion was determined by submitting sequence batches to the NCBI BLAST database (http://www.ncbi.nlm.nih.gov/BLAST).
Mutation frequency.
The frequency of spontaneous mutations leading to decreased tigecycline susceptibility in two tigecycline-sensitive clinical isolates, G595 and G815, was determined essentially as described previously for the estimation of the frequency of MDR (4). Cells were grown overnight in LB broth and inoculated in triplicate on LB plates containing 4 μg of tigecycline/ml using an inoculum size of 109 CFU per plate. After overnight incubation, the colonies were counted and the mutation frequency was calculated using the method of the median (8).
Northern blot hybridization.
DNA fragments containing ramA, acrA, and rrsE were amplified by PCR (Table 2). 32P-labeled probes were generated by random prime labeling of gel-purified PCR fragments by using a High Prime kit (Roche Diagnostics, Mannheim, Germany) with Redivue 5′-[α-32P]dCTP (Amersham, Piscataway, N.J.) as the source of the 32P isotope. Total RNA was isolated from mid-log-phase bacterial cultures by using an RNAeasy kit (QIAGEN, Valencia, Calif.). RNA samples were separated on a 1.2% agarose-0.66 M formaldehyde gel and transferred to a Hybond-N+ nylon membrane (Amersham). The membrane was hybridized with labeled probes, washed, and exposed to BioMax MS X-ray film (Kodak, Rochester, N.Y.) according to standard molecular procedures (1). X-ray images were scanned on a ScanJet ADF scanner (Hewlett-Packard, Palo Alto, Calif.), and densitometry analysis was performed by using Quantity One 4.1.1 software (Bio-Rad).
RT-PCR.
Oligonucleotide primers and probes used for real-time (RT)-PCR (Table 2) were designed with Primer Express Software version 2.0 (Applied Biosystems) and purchased from QIAGEN. The probes were labeled by the manufacturer with the reporter dye 6-carboxyfluorescein (6′-FAM) at the 5′ end and with the quencher dye 6-carboxytetramethylrodamine (TAMRA) at the 3′ end. DNase-treated RNA template was prepared from mid-log-phase bacterial cultures by using an RNAeasy kit (QIAGEN). RT-PCR was performed by using a Taqman One-Step RT-PCR master mix reagents kit (Applied Biosystems) on an ABI Prism 7900HT sequence detection system (Applied Biosystems). A typical RT-PCR sample (20 μl) contained 5 μl of a serial dilution of RNA template (range, 0.04 ng/ml to 400 μg/ml), 4.38 μl of nuclease-free water (Ambion, Austin, Tex.), 10 μl of AmpliTaq Gold DNA polymerase mix (2×), 0.5 μl of RT enzyme mix (40×), 0.04 μl of 100 μM solutions of both forward and reverse gene-specific primers, and 0.04 μl of a 100 μM solution of gene-specific probe. Each sample was run in duplicate. The critical threshold cycle (CT) numbers were determined by the detection system software. Relative quantification of target (ramA) gene expression was performed by normalization to an endogenous reference (rrsE) as recommended by the manufacturer. Briefly, the amount of target is given as 2−ΔΔT, where ΔΔT is the difference between the CT values for a target and a reference.
Western blot analysis.
Bacterial cells were grown in LB broth to an A600 of 0.7 to 1.0. Approximately 1 μg of the total cell lysate proteins was loaded onto a 10% Tris-HCl gel (Bio-Rad) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (7). The proteins were transferred from the polyacrylamide gel to a nitrocellulose filter (Bio-Rad) by use of a Multiphor II system (Amersham). The membrane was blocked for 1 h at 4°C with shaking in TBST (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20) supplemented with 5% nonfat dry milk (Bio-Rad). The membrane was probed with an anti-AcrA polyclonal antibody (1:10,000; gift from H. Zgurskaya, University of Oklahoma, Norman) at room temperature while being shaken. The membrane was washed in TBST buffer three times at room temperature and probed for 1 h with antirabbit antibodies conjugated to horseradish peroxidase (1:50,000; Pierce, Rockford, Ill.). The blots were developed using SuperSignal West Pico solutions (Pierce) and exposed to X-ray film (BioMax MR; Kodak). X-ray images were scanned on a ScanJet ADF scanner (Hewlett-Packard), and densitometry analysis was performed by using Quantity One 4.1.1 software (Bio-Rad).
RESULTS
Estimation of mutation frequency.
The MIC of tigecycline for G340, a clinical strain of K. pneumoniae, was 4 μg/ml, which represents a 16-fold increase over the MIC for the tigecycline-susceptible isolate G595 (Table 3). In addition, G340 displayed the MDR phenotype in that the MICs of a number of other antibiotics for this strain were also elevated (Table 3). The discovery of such a strain prompted an investigation to determine a frequency of spontaneous mutations that results in decreased tigecycline susceptibility in K. pneumoniae. Tigecycline-susceptible clinical isolates G595 and G815 were plated on plates containing tigecycline at a concentration 16 times the MIC for each strain, after which tigecycline-resistant colonies emerged at a frequency of 3.5 × 10−8 for G595 and 4.4 × 10−8 for G815. This result is consistent with the mutation frequency of 2.2 × 10−8 reported previously for the emergence of MDR in K. pneumoniae strain ECL8 (4).
TABLE 3.
Antibiotic susceptibility
| Strain | RamA overexpression | MIC (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGCa | MIN | TET | ACR | EtBr | ERY | CHL | NA | NOV | TRM | NOR | ||
| G595 | − | 0.25 | 2 | 1 | 128 | 1,024 | 128 | 4 | 4 | 128 | 0.5 | 0.125 |
| GC7736 | + | 8 | 64 | 32 | 512 | 2,048 | 256 | 256 | >64 | >512 | 16 | 2 |
| G340 | + | 4 | 32 | 16 | 256 | 2,048 | 256 | 128 | 64 | >512 | 2 | 1 |
| GC7535 | − | 0.25 | 1 | 1 | 128 | 512 | 128 | 256 | 4 | 128 | 0.25 | 0.06 |
| GC7737 | + | 16 | >128 | 64 | 512 | 2,048 | 256 | >512 | >64 | >512 | 16 | 4 |
Abbreviations: TGC, tigecycline; MIN, minocycline; TET, tetracycline; ACR, acriflavine; EtBr, ethidium bromide; ERY, erythromycin; CHL, chloramphenicol; NA, nalidixic acid; NOV, novobiocin; TRM, trimethoprim; NOR, norfloxacin.
Transposon mutagenesis and mapping.
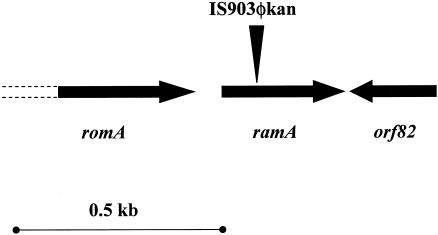
To elucidate the mechanism of the decreased tigecycline susceptibility in K. pneumoniae, strain G340 was subjected to transposon mutagenesis using IS903φkan, which is carried on a plasmid and therefore can be maintained in the host prior to the induction of transposition (20). Mutagenesis of G340 with IS903φkan resulted in the selection of a tigecycline-susceptible transposon insertion mutant, GC7535. The site of IS903φkan insertion in GC7535 was mapped by inverse PCR to a 102-bp position within the ramA gene (Fig. 1). This gene is a part of a previously characterized genetic locus that includes romA and orf82 (Fig. 1), as well as a putative cation transporter (not shown) (4).
FIG. 1.
Map of the chromosomal region around the IS903φkan insertion site in GC7535. The genetic organization of the ramA locus of K. pneumoniae was described previously (4). Open reading frames are shown by arrows; the unsequenced portions of romA are shown by dashed lines. The site of the IS903φkan insertion in ramA is shown by a vertical triangle.
Susceptibility testing and trans-complementation studies.
The transposon insertion in the ramA gene of G340 resulted in suppression of the MDR phenotype, as shown by the decrease in the MICs of minocycline, tetracycline, ethidium bromide, nalidixic acid, novobiocin, trimethoprim, and norfloxacin (Table 3). In addition, the MIC of tigecycline decreased substantially (16-fold; from 4 to 0.25 μg/ml) (Table 3). This result indicated that the ramA locus was linked to the decreased tigecycline susceptibility in G340.
To assess whether inactivation of the ramA gene is the primary factor responsible for the observed phenotype of a transposon insertion mutant, a typical trans-complementation experiment was performed. pCLL3442, a multicopy plasmid that carries full-length ramA, was introduced into transposon mutant GC7535. In the resulting strain, GC7737, the MDR phenotype was restored in that the MICs were either equal to or higher than those observed for the original parental strain, G340 (Table 3). Such an increase over the original MICs might be due to the multicopy nature of the carrier plasmid as well as to the elevated level of constitutive ramA transcription from the plasmid-encoded promoter.
Analysis of ramA and acrA expression.
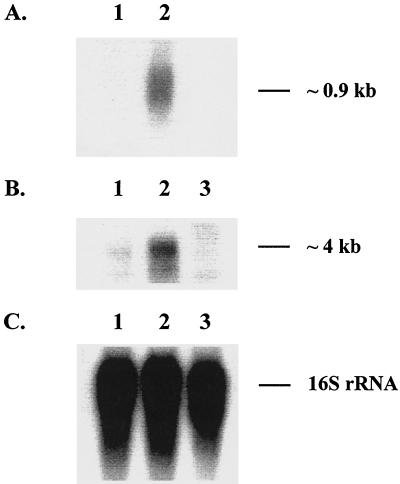
The ramA-encoded protein, RamA, is related to the DNA-binding proteins MarA and SoxS from the AraC family of transcriptional activators (4). Recent studies showed that transcription of ramA was increased in fluoroquinolone-resistant clinical isolates of K. pneumoniae (17). To test whether a similar scenario existed for the decreased tigecycline susceptibility, Northern blot analysis of ramA expression was performed. As shown in Fig. 2, the amount of ramA transcript was significantly larger in G340 than in the tigecycline-susceptible strain G595. According to densitometry analysis, there was an approximate 12-fold increase in ramA expression in G340 over that of G595. An additional quantitative analysis of ramA expression was performed by RT-PCR. By use of this technique, the level of ramA expression was determined to be about 40-fold higher in G340 than in G595 (Table 4). These results indicated that decreased tigecycline susceptibility was linked to ramA overexpression in K. pneumoniae. In addition, the MICs of the various substrates were elevated for GC7736, which contains pCLL3442 expressing ramA (Table 3).
FIG. 2.
Northern blot analysis of ramA and acrA expression. Total bacterial RNA was isolated from mid-log-phase cultures of G595 (lane 1), G340 (lane 2), and GC7535 (lane 3) and hybridized with 32P-labeled DNA probes specific to either ramA (A), acrA (B), or rrsE (C). Hybridization with rrsE that encodes 16S rRNA served as an RNA loading control.
TABLE 4.
RT-PCR analysis of ramA expression
| Gene | Strain | CTa | Relative expression level versus rrsEb | Overexpression ratioc |
|---|---|---|---|---|
| ramA | G340 | 26.5 | 2.09e − 3 | 36.7 |
| G595 | 31.3 | 5.69e − 5 | ||
| rrsE | G340 | 17.6 | ||
| G595 | 17.2 |
CT values represent averages for duplicate samples with a final RNA concentration of 1 ng/ml.
Relative expression is calculated as 2−ΔΔT. ΔΔT = CT(target) − CT(reference), where the target is ramA and the reference is rrsE.
The overexpression ratio is the ratio between the relative expression level of the target gene in G340 versus that in G595.
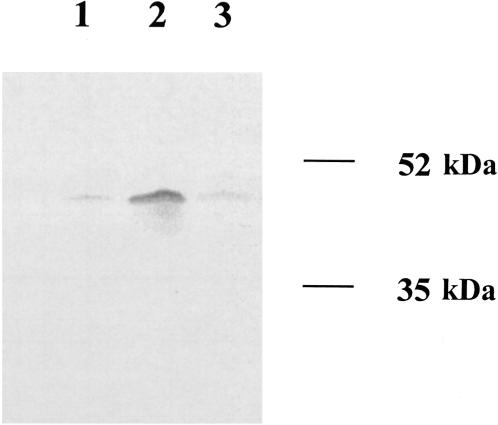
It was previously reported that ramA overexpression was observed in conjunction with an elevated expression of the multidrug efflux pump AcrAB in K. pneumoniae, suggesting that RamA might be involved in the regulation of AcrAB production (17). To investigate this hypothesis, Northern blot analysis of acrA expression was performed with G595, G340, and the ramA knockout mutant GC7535. As shown in Fig. 2, the level of acrA transcription in G340 exceeded that seen in either G595 or GC7535. Since acrA is located upstream of acrB and both genes share a common promoter (5), the size of the acrA transcript (∼4 kb) was consistent with the size of a bicistronic acrAB mRNA, implying that both of the components of the AcrAB pump were overexpressed in G340. Densitometry analysis showed that the level of acrA expression was about 3.4-fold higher in G340 than in either G595 or GC7535. The amount of AcrA protein was elevated in G340, as revealed by Western blot analysis of cell lysates (Fig. 3). Densitometry analysis showed about a fourfold excess of AcrA in G340 than in G595 or GC7535. These results suggested that RamA was indeed a part of the regulatory pathway that governs the production of AcrAB in K. pneumoniae.
FIG. 3.
Western blot analysis of AcrA expression. Total cell lysate proteins of G595 (lane 1), G340 (lane 2), and GC7535 (lane 3) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with polyclonal anti-AcrA antibodies (gift from H. Zgurskaya).
DISCUSSION
Following selection with tigecycline in the laboratory, spontaneous mutants of K. pneumoniae with decreased tigecycline susceptibility emerged at a frequency of about 10−8. Such an order of magnitude suggests that less-susceptible colonies can arise as a result of single-step mutations. Despite this fact, the majority of K. pneumoniae clinical isolates are susceptible to tigecycline at typical MICs of 0.25 to 1 μg/ml. To date, only a few isolates that result in tigecycline MICs exceeding 1 μg/ml have been collected.
This study demonstrated that one of the formerly described MDR determinants, ramA, is involved in reduced tigecycline susceptibility in K. pneumoniae. The ramA-encoded protein, RamA, belongs to the family of transcriptional activators that also includes MarA, SoxS, and Rob, which are known for their role in promoting antibiotic resistance due to the up-regulation of the multidrug efflux pump AcrAB-TolC (10, 13, 19, 23). As shown recently, the AcrAB efflux system is associated with reduced susceptibility to tigecycline in P. mirabilis (22). The present study revealed that the increased tigecycline MIC for K. pneumoniae strain G340 correlated with the constitutive overexpression of ramA, suggesting that RamA might act as an activator of expression of Acr transporter. A similar hypothesis was proposed recently when elevated levels of ramA transcription coincided with increased expression of AcrAB in K. pneumoniae strains resistant to fluoroquinolones (17). In contrast, expression levels of known Acr regulators such as marA and soxS were unchanged (17). The results presented here further support the role of RamA in the regulation of the Acr pump by linking the overexpression of ramA to up-regulation of AcrAB and the inactivation of ramA to the down-regulation of AcrAB expression. Similar to fluoroquinolone-resistant isolates, no increase in marA expression was detected (data not shown), indicating that induction of AcrAB expression by RamA is not mediated through overexpression of MarA. Due to the broad substrate specificity of the Acr pump, these data might explain the increased efflux of tetracycline and chloramphenicol that was previously observed in strains that overexpressed ramA and provide a mechanism for the previously established role of ramA as a determinant of MDR in K. pneumoniae (4).
The study presented here leaves several questions open. First, the exact mechanism of ramA overexpression in K. pneumoniae is unknown, and it is possible that RamA production is inducible. Because K. pneumoniae is generally susceptible to tigecycline, it is unlikely that tigecycline serves as an inducer of either RamA or AcrAB production in this organism. It is possible that RamA plays some physiological role and perhaps is produced in response to certain environmental stimuli. In addition to K. pneumoniae, the close homologs of RamA are identified in Enterobacter cloacae (6), Enterobacter aerogenes (GenBank accession number AJ404625) and Salmonella enterica (24), whereas other bacteria, e.g., E. coli, lack a RamA equivalent, suggesting that RamA might function in certain species-specific environmental niches. It is also possible that overexpression of ramA results in the overproduction of MDR pumps other than AcrAB. Further experiments, e.g., involving disruption of the acrAB locus in GC7535 and G340, are required to establish whether overexpression of acrAB is the only factor contributing to decreased tigecycline susceptibility in K. pneumoniae.
Acknowledgments
We thank David Figurski for providing plasmid pVJT128 and Helen Zgurskaya for a kind gift of anti-AcrA antibodies.
REFERENCES
- 1.Ausubel, F. M., et al. (ed.). 1987. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 2.Boucher, H. W., C. B. Wennersten, and G. M. Eliopoulos. 2000. In vitro activities of the glycylcycline GAR-936 against gram-positive bacteria. Antimicrob. Agents Chemother. 44:2225-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean, C. R., M. A. Visalli, S. J. Projan, P. E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George, A. M., R. M. Hall, and H. W. Stokes. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909-1920. [DOI] [PubMed] [Google Scholar]
- 5.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu, T., M. Ohta, N. Kido, Y. Arakawa, H. Ito, T. Mizuno, and N. Kato. 1990. Molecular characterization of an Enterobacter cloacae gene (romA) which pleiotropically inhibits the expression of Escherichia coli outer membrane proteins. J. Bacteriol. 172:4082-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Lea, D. E., and C. A. Coulsen. 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 9.Lomovskaya, O., and W. J. Watkins. 2001. Efflux pumps: their role in antibacterial drug discovery. Curr. Med. Chem. 8:1699-1711. [DOI] [PubMed] [Google Scholar]
- 10.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5 and information supplement M100-S10. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 13.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen, P. J., P. A. Bradford, W. J. Weiss, T. M. Murphy, P. E. Sum, and S. J. Projan. 2002. In vitro and in vivo activities of tigecycline (GAR-936), daptomycin, and comparative antimicrobial agents against glycopeptide-intermediate Staphylococcus aureus and other resistant gram-positive pathogens. Antimicrob. Agents Chemother. 46:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Schneiders, T., S. G. Amyes, and S. B. Levy. 2003. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 19.Tanaka, T., T. Horii, K. Shibayama, K. Sato, S. Ohsuka, Y. Arakawa, K. Yamaki, K. Takagi, and M. Ohta. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697-702. [DOI] [PubMed] [Google Scholar]
- 20.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visalli, M. A., E. Murphy, S. J. Projan, and P. A. Bradford. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob. Agents Chemother. 47:665-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yassien, M. A., H. E. Ewis, C. D. Lu, and A. T. Abdelal. 2002. Molecular cloning and characterization of the Salmonella enterica serovar Paratyphi B rma gene, which confers multiple drug resistance in Escherichia coli. Antimicrob. Agents Chemother. 46:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]