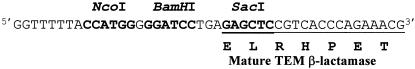Abstract
Escherichia coli, and presumably most other gram-negative bacteria, possesses an efficient protein machinery for recycling its peptidoglycan during cell growth. The major recycled peptidoglycan product is N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid-tetrapeptide. Its uptake from the periplasm into the cytoplasm is carried out via the AmpG protein, an intrinsic membrane protein. In gram-negative bacteria carrying an ampC β-lactamase-inducible gene on their chromosomes, the induction mechanism is directly linked to peptidoglycan recycling. After identification of the different putative hydrophobic segments by computing, the AmpG topology was experimentally determined by using β-lactamase fusion. In the proposed model, AmpG contains 10 transmembrane segments and two large cytoplasmic loops.
The bacterial cell wall contains a cross-linked heteropolymer named peptidoglycan or murein that protects the cell from osmotic lysis and determines its shape. Although this highly cross-linked network forms a rigid and insoluble envelope, the peptidoglycan sacculus is nevertheless continuously remodeled throughout the bacterial cell cycle. As the bacterium grows and divides, extracellular peptidoglycan hydrolases cleave covalent bonds within the peptidoglycan sacculus to allow insertion of new glycan chains during enlargement of the cell and to perform the separation of daughter cells (11, 19). In contrast to Bacillus subtilis, which loses as much as 30% of its peptidoglycan per generation (1), Escherichia coli, and presumably most other gram-negative bacteria, releases only around 5 to 8% of peptidoglycan fragments into the medium per generation (9). This different percentage of released peptidoglycan is the consequence of the presence of a very efficient protein machinery for peptidoglycan recycling in E. coli (8, 23). The major recycled peptidoglycan product is N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid-tetrapeptide (GlucNAc-anhydro-MurNAc-tetrapeptide or anhydromuropeptide) (13, 25). Its uptake, from the periplasm into the cytoplasm, is carried out via the AmpG permease specific for GlucNAc-anhydro-MurNAc or GlucNAc-anhydro-MurNAc-peptide transport (4, 13). In the cytoplasm, the anhydromuropeptide is degraded and the tripeptide reenters the peptidoglycan biosynthesis pathway.
In many gram-negative bacteria, the presence of penicillin or other β-lactam antibiotics outside the cell induces the synthesis of the chromosomal AmpC β-lactamase (20). The inducible ampC β-lactamase gene is transcriptionally controlled by the divergently transcribed regulator gene ampR (15). AmpR is a DNA-binding protein belonging to the LysR superfamily (10), and it has two regulatory properties: (i) in the absence of β-lactam inducer, AmpR is complexed by UDP-MurNAc-pentapeptide and acts as a repressor; (ii) in the presence of a β-lactam antibiotic, the anhydro-MurNAc-oligopeptide (penta-, tetra-, and tripeptides) (5) accumulates in the cytoplasm by concomitant inhibition of membrane d,d-peptidases and the activation of the autolytic system, displaces the UDP-MurNAc-pentapeptide ligand, and converts AmpR into an activator, so that the AmpC β-lactamase is produced (12). The inactivation of ampG by mutation or deletion confers noninducible and microconstitutive β-lactamase phenotypes to the bacterial cell (14, 16). These results clearly demonstrate the links between β-lactamase induction and peptidoglycan recycling. The E. coli ampG gene has been cloned (16) and encodes a 491-amino-acid protein with a molecular mass of 53 kDa. Along the AmpG primary structure, several segments have been identified, and the AmpG protein was predicted to be an integral membrane protein, with 10 transmembrane segments (16).
Our interest in AmpG was stimulated by the fact that this peptidoglycan-specific permease could be used to transport new drugs mimicking the murein recycled compounds into the cytoplasm. These new compounds could directly or indirectly inhibit an enzyme involved in the peptidoglycan biosynthesis. In this way, and to better understand the AmpG transport mechanism, we determined the membrane topology of the AmpG protein by using the β-lactamase fusion procedure described by Broome-Smith et al. (2).
MATERIALS AND METHODS
Prediction of the membrane-spanning segments.
Putative transmembrane α helices in AmpG were identified by a hydrophobic moment plot (7). In this approach, the prediction is achieved by computing the mean hydrophobic moment (<μ>) and the mean hydrophobicity index (<H>) for each possible peptide segments of 11 residues along the AmpG sequence. Following these values, each undecapeptide segment can be predicted to be (i) a monomeric transmembrane segment (T), (ii) a transmembrane segment which could oligomerize (multimeric transmembrane segment or M), or (iii) an amphiphilic segment (S) (6, 7).
Bacterial strains, plasmids, DNA methods, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. For cell selection, kanamycin and chloramphenicol were used at 50 and 30 μg ml−1, respectively. Ampicillin was used at 100 μg ml−1 for PCR-Script SK selection.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli XLI-BlueMRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac (F′proAB lacIqZ ΔM15 Tn10 [Tetr]) | Stratagene |
| E. coli XLI-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac) | Stratagene |
| PCRscript SK | Cloning vector for PCR fragments; Cmr | Stratagene |
| PCRscript SK | Cloning vector for PCR fragments; Ampr | Stratagene |
| pJBS633 | Source of the blaM gene encoding TEM β-lactamase without its signal peptide (mature TEM β-lactamase); Kmr | 2 |
| pGKS171-3 | Source of E. coli ampG gene; Cmr | 14 |
| pYZ4 | pBR322 derivative carrying a modified LacZα peptide under the control of the lacUV5 promoter; Kmr | 27 |
| pCIPblaM | pYZ4 derivative carrying blaM gene under the control of the lacUV5 promoter; for details, see text | This study |
| pCIPFus1 to pCIPFus16 | pCIPblaM derivatives carrying ampG-blaM hybrid; for details, see text | This study |
Standard procedures for DNA manipulations and cell transformation were used (24). Small- and medium-scale preparations of plasmids from E. coli were performed by using a GFX Micro Plasmid Prep kit (Amersham Pharmacia Biotech) and NucleoBond AX (Macherey-Nagel), respectively. PCR fragments were purified by agarose gel electrophoresis. The fragments of interest were recovered from agarose gel by using a GFX-PCR kit (Amersham Pharmacia Biotech). The integrity of the fragment generated by PCR was verified by nucleotide sequencing by the dideoxy chain termination method and with the help of an ALFexpress DNA sequencer (Amersham Pharmacia Biotech).
A 1,130-kb DNA fragment containing the blaM coding region was PCR amplified with the pJBS633 plasmid as template and two primers, blaM_Up (5′TAA GGA TCC TGA GAG CTC CGT CAC CCA GAA ACG3′) and blaM_Down (5′CTA GAA TTC GGT ATC TGC GCT CTG CTG AA3′) (the first codon of the blaM coding region is underlined, and BamHI, SacI, and EcoRI sites are in boldface), designed to introduce flanking BamHI-SacI and EcoRI sites. The amplified fragment was cloned into PCR-Script SK-Cm plasmid. After the amplified sequence was checked by DNA sequencing, the BamHI- and EcoRI-digested fragment was subcloned into the corresponding sites of the pYZ4 plasmid, generating pCIPblaM. In this construct, the blaM gene is not expressed but its 5′ end is flanked by NcoI, BamHI, and SacI unique restriction sites (Fig. 1).
FIG. 1.
Sequence data related to plasmid pCIPblaM. A DNA fragment containing the blaM coding region was PCR amplified with pJBS633 as template and with two primers designed to introduce flanking BamHI-SacI and EcoRI sites. The amplified fragment was cloned into PCR-Script SK-Cm plasmid. After the amplified sequence was checked by DNA sequencing, the BamHI- and EcoRI-digested fragment was subcloned into the corresponding sites of the pYZ4 plasmid, generating pCIPblaM. The blaM gene is not expressed in this construct, but its 5′ end is flanked by NcoI, BamHI, and SacI unique restriction sites, shown in the sequence in boldface type. For more information, see the text.
The various forms of the ampG gene truncated at the 3′ end were amplified by PCR from pGKS171-3 with the oligonucleotides listed in Table 2 as lower primers. The AmpG-UP oligonucleotide (5′-TAACCATGGCCAGTCAATATTTACGTATTTTTCAA-3′), used as the upper primer for the 5′ end of the gene in all amplifications, introduced an NcoI site (shown in boldface type) containing the initiation codon of ampG. The Fus1ampG to Fus14ampG oligonucleotides, used for lower primers, introduced a SacI site at the 3′ end of the amplified fragment. The generated DNA fragments were purified and then cloned with PCR-Script SK-Amp. After its DNA sequence was checked, the recombinant plasmid was cut with both NcoI and SacI enzymes and then inserted between the same sites of the pCIPblaM vector. For fusions 2 to 14 (pCIPFus2 to pCIPFus14), the in-frame insertion resulted in the expression of the truncated form of ampG fused to the mature form of the blaM β-lactamase via short intermediate peptide linkers originating from the creation of SacI site. For fusion 1 (pCIPFus1), the full length of the ampG gene was fused in frame to the blaM gene. In each case, E. coli XL1 Blue MR transformants containing the recombinant plasmids were selected for growth in the presence of kanamycin (50 μg ml−1). The constructs were confirmed by restriction analysis and DNA sequencing of the in-frame fusions after isolation of the plasmids. The ampG-blaM junctions were sequenced with the fluorescent primer BlaMfus, 5′-CY5-TACCGCTGTTGAGATCCAGTTCGAT-3′.
TABLE 2.
Oligonucleotides used for AmpG-TEM hybrid productiona
| Fusion name | Nucleotide position | Fusion position | Sequence |
|---|---|---|---|
| Fus1ampG | C1473 | T491↓ | 5′TTA GAG CTC CGT CAG ATG CGT TTT TCG TAG C3′ |
| Fus2ampG | A1371 | S457↓FLP | 5′ AAG AGC TCA GAG AAG TGC GTC AAA TCC AGC G3′ |
| Fus3ampG | T1260 | A420↓YPA | 5′AAG AGC TCG GTA CGG GAG ATA AAG TTG TCA TTT ACT CG3′ |
| Fus4ampG | G1140 | G380↓WST | 5′AAG AGC TCG CCG TGT GCT TCA ACA AAC CAA CCC3′ |
| Fus5ampG | A1056 | T352↓QFA | 5′TTA GAG CTC AGT AGC GGA AAA TGA CTT A TT ACA TAG CG3′ |
| Fus6ampG | G966 | A322↓VFF | 5′TTA GAG CTC GGC TGC GCCCAT GCT GTA GAG3′ |
| Fus7ampG | G861 | S287↓LFR | 5′TAG AGC TCT GAC AGG CGC TGC ATC AAA ATC3′ |
| Fus8ampG | A774 | G258↓EVG | 5′TTA GAG CTC ACC CGC ATC AAA CCC GAC GCC3′ |
| Fus9ampG | G600 | T200↓IPV | 5′TTA GAG CTC GGT GTC GGT TGG TTC TGG TGC3′ |
| Fus10ampG | G516 | G172↓WQG | 5′TTA GAG CTC GCC CAG CCA TTT ATC TGC CAG3′ |
| Fus11ampG | G516 | T133↓DVL | 5′TTA GAG CTC GGT TTT CCA CGC ATC GAA G3′ |
| Fus12ampG | T318 | Q106↓LRW | 5′TTA GAG CTC TTG GGT GCC TGG TTC GAG AAA3′ |
| Fus13ampG | A222 | P74↓PFF | 5′TTA GAG CTC AGG CGT GTA GCG GTC CAT CAG3′ |
| Fus14ampG | C126 | E42↓NID | 5′TTA GAG CTC GAC CGT CAT CAA GGC CTG3′ |
| Fus15ampG | A459 | R153↓LGM | 5′TTA GAG CTC ACG GTA ACC CAG CAC GCT GAT3′ |
| Fus16ampG | G558 | P186↓CII | 5′TTA GAG CTC GGG GAT CAA CAG TGC CGC CAT3′ |
The SacI restriction sites are shown in boldface type.
Topological analysis and ampicillin resistance of cells expressing β-lactamase fusion proteins.
To probe the presence of the β-lactamase moiety outside or inside the cell, the method of Broome-Smith et al. (2) was used. In this method, a single cell producing a periplasmic β-lactamase fusion can survive and form a colony on an agar plate containing 10 μg of ampicillin ml−1, 50 μg of kanamycin ml−1, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), whereas a cell producing a cytoplasmic β-lactamase fusion is ampicillin sensitive (single cell screening). On the contrary, when a clone producing a cytoplasmic β-lactamase moiety is plated at high density (i.e., “toothpicked”), it does grow (patch screening). Indeed, in the latter case, ampicillin-induced lysis releases β-lactamase into the medium, where it hydrolyses the ampicillin and thus enables the remaining cells to survive. In patch screening, colony growth is independent of the location, cytosolic or periplasmic, of the β-lactamase moiety and indicates the production of functional AmpG-β-lactamase hybrid.
The ampicillin resistance of individual cells of E. coli XL1 Blue MR harboring the pCIPFusx plasmids was determined by plating appropriate dilutions of exponential phase cultures onto Luria-Bertani (LB) agar plates containing 0, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, and 200 μg of ampicillin ml−1. The MIC of ampicillin was estimated as the lowest inhibitory concentration that inhibits cell growth.
Cellular compartments and β-lactamase activity.
The periplasmic, cytoplasmic, and membrane fractions of the cells were obtained by lysozyme treatment as described by Lindström et al. (17). β-Lactamase activity was determined by using nitrocefin as substrate (21).
RESULTS AND DISCUSSION
Prediction of transmembrane segments and construction and analysis of ampG-x-blaM fusions.
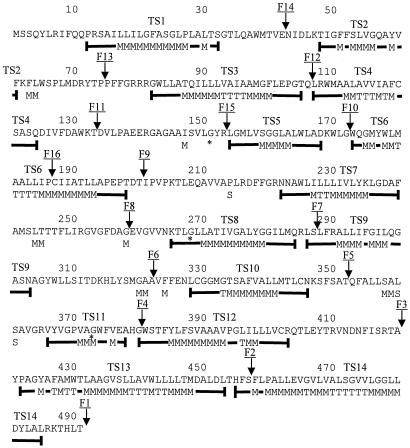
A hydrophobic moment plot analysis of the AmpG polypeptide chain by the method of Eisenberg et al. (7) highlighted 14 hydrophobic segments (TS1 to TS14), which might be 14 putative transmembrane segments (for details, see Fig. 2 and Materials and Methods).
FIG. 2.
Prediction of E. coli AmpG transmembrane segments. Eisenberg prediction symbols T, M, and S represent monomeric transmembrane segments, multimeric transmembrane segments, and amphiphilic α helices, respectively. These symbols are placed at the positions corresponding to the centers of the sliding spans of 11 residues used to perform the prediction. The segments corresponding to the thick lines under the amino acid sequence and including the Eisenberg symbols represent the length of the predicted transmembrane segment by the following rule: three consecutive M or T symbols are required for a nucleus of transmembrane segment. TSx represents hydrophobic segments deduced from the Eisenberg analysis. Arrows indicate the positions of AmpG-Bla M fusions (for details, see Table 2 and the text). The Gly-Asp substitutions in the three characterized AmpG mutants yielding a noninducible β-lactamase phenotype are marked by asterisks (for details, see the text and reference 19).
To determine the membrane topology of AmpG deduced from the hydrophobic properties of the polypeptide chain (see above), 14 directed fusions, ampG-x-blaM, were created between a truncated ampG gene generated by PCR and the blaM gene encoding a leaderless TEM β-lactamase. To obtain ampG-x-blaM hybrids, the pCIPblaM expression plasmid (a derivative of pYZ4) (27) was constructed. It allows an in-frame blaM cloning of a PCR-amplified ampG-x fragment containing NcoI and SacI restriction sites at its 5′ and 3′ ends, respectively (for details, see Materials and Methods).
Fusion sites (Fig. 2) were chosen to be located in the middle of the putative cytoplasmic or periplasmic loops (F2 to F14) (Fig. 2) connecting hydrophobic segments deduced by hydrophobic moment plot analysis or at the C-terminal end of AmpG (F1). E. coli XL1 Blue MR harboring pCIPFusx-derivative plasmids (pCIPFus1 to pCIPFus14, Table 1) and carrying ampG-x-blaM hybrids was selected for its ability to grow on LB agar plates containing 50 μg of kanamycin ml−1. The expression of the ampG-x-blaM hybrids in these recombinant E. coli cells was highlighted by their ability to grow when patched onto LB agar containing 10 μg of ampicillin ml−1, 50 μg of kanamycin ml−1, and 1 mM IPTG. Indeed, under these conditions, ampicillin resistance was independent of the localization (cytosolic or periplasmic) of the β-lactamase moiety (2). All of the fusions constructed conferred ampicillin resistance in patch screening, thereby clearly indicating that they all expressed ampG-x-blaM fusions.
To assess the cytoplasmic or periplasmic location of the β-lactamase moiety, the MIC of ampicillin for single cells (single cell screening) was determined by plating appropriate dilutions of exponential cell cultures on LB agar plates containing 10 μg of ampicillin ml−1, 50 μg of kanamycin ml−1, and 1 mM IPTG. Under these conditions, the failure to confer ampicillin resistance indicates that the BlaM moieties of the fusions are cytoplasmic. In contrast, the BlaM moieties of the fusion proteins that conferred ampicillin resistance are expected to be translocated across the bacterial cytoplasmic membrane and exposed in the periplasm. This was the case for AmpG-x-BlaM fusions at amino acids S457, A322, G258, Q106, and E42, which allowed growth on plates containing an ampicillin concentration of ≥40 μg ml−1 (Table 2). The fusions at amino acids T491, A420, G380, T352, S287, T200, G172, T133, and P74 did not confer ampicillin resistance and were considered to have a cytoplasmic β-lactamase moiety (Table 2). In these cases, the ratio of the cytoplasmic to periplasmic β-lactamase activity ranged from 5/1 to 50/1 and the periplasmic activity was indeed much too small to confer sufficient ampicillin resistance. In addition, part of the so-called periplasmic activity might be due to a small proportion of the cell lysis during the separation of the various cellular compartments.
Topology of AmpG.
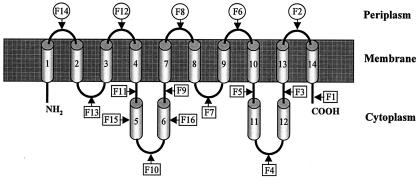
The AmpG topological model derived from ampicillin resistance data contains 10 transmembrane hydrophobic segments: TS1, TS2, TS3, TS4, TS7, TS8, TS9, TS10, TS13, and TS14 (Fig. 3) which delimit six cytoplasmic hydrophilic domains, including the N- and C-terminal ends, and five periplasmic hydrophilic domains. In this model, the putative transmembrane segments TS5, TS6, TS11and TS12 are not included in the membrane and are located in the cytoplasm. Except for BlaM fusions F5 (T352) and F10 (G172), the BlaM fusions were in agreement with the transmembrane segments predicted by Eisenberg analysis (see above). The presence of prolyl residues in the middle of the hydrophobic segments TS6 and TS12 (Pro186 and Pro394, Fig. 2) suggests the existence of a truncated α helix or of a turn in these hydrophobic segments. By inserting a β turn in these hydrophobic segments, another possibility for the polypeptide chain to cross the membrane would be expected if it adopts a β-stranded structure. In this case, a hydrophobic segment would give rise to two hydrophobic β strands. To probe this hypothesis, we have included the transmembrane segments TS5 and TS11 in this study, although they do not contain a prolyl residue in their sequence, because the four hydrophobic segments, TS5, TS6, TS11, and TS12, would generate eight hydrophobic β strands which could be associated in the membrane to form a β-barrel structure. To rule out the hypothesis of the presence of β-stranded segments, two additional blaM fusions were generated at the positions of the putative β turns in TS5 and TS6 (fusions F15 and F16, Table 2). The resulting fusions were ampicillin sensitive when they were grown as single cells, indicating that the fusion points (R153 and P186) were located in the cytoplasm. No additional fusions have been performed with TS11 and TS12, because they share similar hydrophobic properties with TS5 and TS6. In conclusion, the final model for AmpG topology is that derived from ampicillin resistance data (Fig. 3).
FIG. 3.
Proposed topological model of E. coli AmpG protein. Arrows indicate the positions of AmpG-BlaM fusions that have been constructed (for details, see Table 2 and Fig. 2). Digits represent hydrophobic segments deduced from the Eisenberg analysis.
The transmembrane topology deduced from the TEM β-lactamase fusions shows that the AmpG N and C termini are exposed in the cytoplasm and that the number of transmembrane segments must be an even number. F3, F4, F9, and F10 fusions exclude the TS5, TS6, TS11, and TS12 segments as transmembrane α helices. These results are in contradiction with those obtained by the Eisenberg analysis for the prediction of the transmembrane segments. Before we discuss the reasons that these four hydrophobic segments are not embedded in the cytoplasmic membrane, we note that it was unexpected that these four segments could be grouped by pairs according to their length and their hydrophobicity. Indeed, TS5 and TS11 contain 14 and 13 residues, respectively, whereas TS6 and TS12 contain 26 and 24 residues, respectively. For the first pair, the length of the α helices is probably too short to cross the cytoplasmic membrane if we accept that a minimum length of 18 residues is required to do so. In our prediction, TS2 also contains 14 residues. However, the hydrophobicity of TS2 is higher than that of TS5 and of TS11. The two hydrophobic segments for the second pair, TS6 and TS12, contain a central prolyl residue. Though prolyl residues are frequently found in transmembrane segments, in the case of AmpG these prolyl residues could act as α-helix breakers and give rise to α helices too short to span the membrane. On the other hand, if TS5 and TS11 are not in the membrane and if TS6 is a transmembrane segment, TS11 and points of fusion F4 and F5 would be exposed in the periplasm, which contradicts results of the TEM-β-lactamase fusion experiments. Finally, in the topological model proposed in Fig. 3, the positive-inside rule of von Heijne (26) is respected for the orientation of transmembrane α helices (data not shown). Out of the 10 transmembrane segments previously predicted by Lindquist et al. (16), 8 are in agreement with our topological model and correspond to TS1, TS3, TS4, TS6, TS7, TS8, TS10, TS12, TS13, and TS14. The two remaining transmembrane segments predicted by these authors were TS6 and TS12 and have been eliminated in our topological model and replaced by the TS2 and TS9 transmembrane segments. The same authors also reported three AmpG mutants yielding a noninducible phenotype. The mutations are identical Gly-Asp substitutions located at positions 151, 268, and 373, respectively (Fig. 2). Interestingly, the G151D and G373D mutations are located in two large cytoplasmic domains of AmpG and probably highlight the importance of these cytoplasmic domains in the AmpG transport mechanism. The last mutation, G268D, is located at the beginning of the transmembrane segment TS8 and probably destabilizes the transmembrane scaffolding.
A recent study of the AmpG permease substrate specificity and mechanism of transport shows that carbonyl cyanide m-chlorophenylhydrazone (CCCP) prevents the uptake of GlucNAc-anhydro-MurNAc or GlucNAc-anhydro-MurNAc-tetrapeptide by AmpG (4). This inhibition by CCCP suggests that the AmpG permease is a single component permease dependent on the proton motive force as demonstrated for some members of the sugar transport system belonging to the oligosaccharide-H+ symporter family (22). With respect to the transport mechanism of AmpG, the two large cytoplasmic loops containing TS5, TS6, TS11, and TS12 may be involved in a scissors-type mechanism similar to that envisioned for lipid flipping by the E. coli MsbA ABC transporter (3) or in a swinging rearrangement as proposed for the E. coli BtucD ABC transporter (18).
TABLE 3.
AmpG-TEM hybrid characterization for antibiotic single cell resistance and determination of AmpG topologya
| Characteristic | Fusion
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F15 | F16 | |
| Amino acid position | T491 | S457 | A420 | G380 | T352 | A322 | S287 | G258 | T200 | G172 | T133 | Q106 | P74 | E42 | R153 | P186 |
| KmR | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| Kmr + Ampr + IPTG | S | R | S | S | S | R | S | R | S | S | S | R | S | R | S | S |
| MIC (μg/ml) | 10 | 60 | 10 | 10 | 10 | 50 | 10 | 90 | 10 | 10 | 10 | 40 | 10 | >200 | 10 | 10 |
| β-lactamase moiety location | In | Out | In | In | In | Out | In | Out | In | In | In | Out | In | Out | In | In |
All AmpG-TEM fusions grew on LB agar plates supplemented with kanamycin, ampicillin, and IPTG when they were replicated in patch. This result indicates that the recombinant E. coli cells express the ampG-x-blaM in-frame fusion (for details, see text). The final concentration of IPTG was 1 mM. Kmr, kanamycin resistant (50 μg/ml); Ampr, ampicillin resistant (10 μg/ml); R, resistant; S, sensitive; MIC, ampicillin MIC for E. coli recombinant cells induced by IPTG; In, β-lactamase domain exposed in the cytoplasm; Out, β-lactamase domain exposed in the periplasm.
Acknowledgments
We thank Jean-Marie Frère for insightful discussion and critical reading of the manuscript.
This work was supported by the Belgian Program on Interuniversity Poles of Attraction initiated by the Federal Office for Scientific, Technical and Cultural Affairs (PAI no. P5/33), the Fond National de la Recherche Scientifique (FNRS; crédit aux chercheurs no. 1.5201.02 and FRFC no. 2.4530.03) and the European Commission (Targeted Research Project COBRA). B.J. is a FNRS associate researcher. A.C. and M.D. were supported by Coopération Technique Belge (CTB-BCT Brussels) and the Ministère de la Région Wallonne (DGTRE; contract βA4, no. 114830), respectively. R.B. is FNRS Research Director. This work was supported by the Ministère de la Région Wallonne (Contract Protmem no. 14540).
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis and turnover, p. 381-410. In A. L Sonenshein, J. A. Hoch and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 2.Broome-Smith, J. K., M. Tadayyon, and Y. Zhang. 1990. Beta-lactamase as a probe of membrane protein assembly and protein export. Mol. Microbiol. 4:1637-1644. [DOI] [PubMed] [Google Scholar]
- 3.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberg, D. 1984. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 53:595-623. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 8.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162:391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höltje, J. V., and E. I. Tuomanen. 1991. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J. Gen. Microbiol. 137:441-454. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, C., J. M. Frere, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC β-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg, F., L. Westman, and S. Normark. 1985. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc. Natl. Acad. Sci. USA 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindquist, S., K. Weston-Hafer, H. Schmidt, C. Pul, G. Korfmann, J. Erickson, C. Sanders, H. H. Martin, and S. Normark. 1993. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol. Microbiol. 9:703-715. [DOI] [PubMed] [Google Scholar]
- 17.Lindström, E. B., H. G. Boman, and B. B. Steele. 1970. Resistance of Escherichia coli to penicillins. VI. Purification and characterization of the chromosomally mediated penicillinase present in ampA-containing strains. J. Bacteriol. 101:218-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 19.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Normak, S. 1994. Mechanism of chromosomal beta-lactamase induction in Gram-negative bacteria, p. 485-503. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands.
- 21.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pao, S. S., I. T. Paulsen, and M. H. J. Saier. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Templin, M. F., A. Ursinus, and J. V. Höltje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y. B., and J. K. Broome-Smith. 1990. Correct insertion of a simple eukaryotic plasma-membrane protein into the cytoplasmic membrane of Escherichia coli. Gene 96:51-57. [DOI] [PubMed] [Google Scholar]