Abstract
The erm gene product confers clindamycin resistance on Staphylococcus aureus. We report a clindamycin clinical failure where resistance developed on therapy in a D-test-positive strain. D tests of 91 clindamycin-susceptible, erythromycin-resistant S. aureus isolates showed that 68% of methicillin-susceptible and 12.3% of methicillin-resistant S. aureus strains were D-test positive.
Clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus (clindamycin-erythromycin discordant) may develop clindamycin resistance (7, 8). The erm gene product is a ribosome methylase whose expression is normally minimal. Erythromycin induces the production of this methylase, which is why these strains are erythromycin resistant, but mutations in the promoter region of erm allow production of methylase without an inducer (18, 19). These mutants are stably erythromycin and clindamycin resistant. Since erythromycin resistance can occur with other mechanisms (e.g., efflux pumps and enzymatic modification) (15), the D-test identifies inducible resistance that might presage mutational clindamycin constitutive resistance. The D-test is performed by placing clindamycin and erythromycin disks at an edge-to-edge distance of 15 to 20 mm and looking for flattening of the clindamycin zone nearest the erythromycin disk (2). A positive D-test suggests the presence of an erm gene that could result in constitutive clindamycin resistance and clinical failure.
There are few published clinical failures of clindamycin with emergence of resistance (1, 3, 9, 12, 16). However, there are also reports of successful use of clindamycin in treating patients with D-test-positive isolates (3, 9). We report a clinical failure with documented emergence of resistance. In order to avoid poor clinical outcomes but retain the usefulness of clindamycin, it would be helpful to know the prevalence of inducible resistance in clindamycin-erythromycin discordant bacteria. This prevalence varies by geographic location, patient age, bacterial species, and bacterial susceptibility profile (4, 5, 10, 11, 13, 14, 16). For example, a pediatric population in Houston, Tex., with methicillin-resistant S. aureus (MRSA) had a D-test positivity rate of 2.2% (9) compared to a similar population of children with MRSA in Chicago, Ill., whose D-test positivity rate was 94% (3). We determined the prevalence of D-test positivity in clindamycin-erythromycin discordant S. aureus isolates in our institution in early 2004. We tested isolates from infected body sites where clindamycin might be considered for therapy.
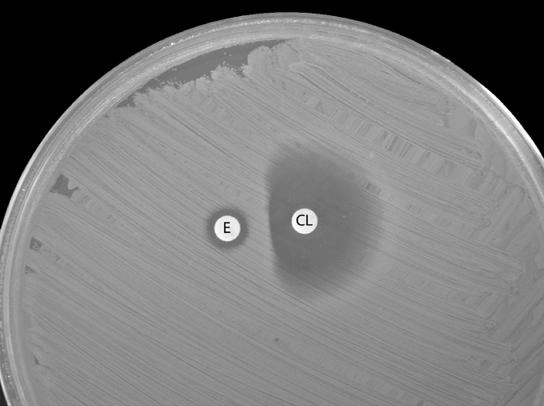
The patient was a 44-year-old man with fever, chills, and increased low back pain 10 days after a lumbar discectomy. At admission he had a temperature of 102.4°F and complained of 3 days of erythema, bloody drainage, and pain along his incision. He had a wound exploration, and the tissue Gram stain showed white blood cells and gram-positive cocci. Fluid and blood cultures grew clindamycin-erythromycin discordant methicillin-susceptible S. aureus (MSSA), which was treated with 2 weeks of oxacillin followed by 2 weeks of oral clindamycin. Ten days after completing clindamycin therapy, he had severe back pain, chills, a temperature of 101.6°F, and purulent drainage from his wound. A magnetic resonance image showed new evidence of lumbar osteomyelitis, discitis, and enhancement of paravertebral musculature with a small fluid collection. On vancomycin, he had surgical debridement. Bone, blood, and wound cultures all grew MSSA with the same antibiogram as that of his prior isolate except for new clindamycin resistance. He successfully completed therapy with intravenous (oxacillin) and oral (combinations of minocycline, trimethoprim-sulfamethoxazole, and rifampin) antibiotics for a total of 5 months. The D-test on the original fluid isolate was positive (Fig. 1); however, this result was not available at the time of his original therapy. The original S. aureus wound isolate and the blood and bone isolates collected after completion of primary therapy were indistinguishable by ribotyping with EcoRI restriction enzyme (Christiana Care Health Services, Wilmington, Del.).
FIG. 1.
D-test positivity on the original wound culture isolate from the case patient. E, erythromycin; CL, clindamycin.
We collected all 168 S. aureus isolates from wounds, abscesses, bone, cerebrospinal fluid, pleural fluid, ascitic fluid, and joint fluid from 1 January to 28 February 2004 at Temple University Hospital (Philadelphia, Pa.). The strains (adult, pediatric, inpatient, and outpatient) were stored in glycerol at −74°C until batch testing in April 2004. Antibiotic susceptibilities were determined by the Vitek-1 test (bioMerieux, Hazelwood, Mo.); erythromycin-intermediate strains were considered resistant. The D-test was performed on clindamycin-erythromycin discordant strains by preparing an 0.5 McFarland suspension of bacteria in tryptic soy broth (2) inoculated on Mueller-Hinton plates incubated at 35°C for 18 to 24 h with erythromycin (15 μg) and clindamycin (2 μg) disks placed 17 mm apart (edge to edge). Controls with known D-test positivity and negativity were also tested. Results were photographed and visually inspected by two investigators.
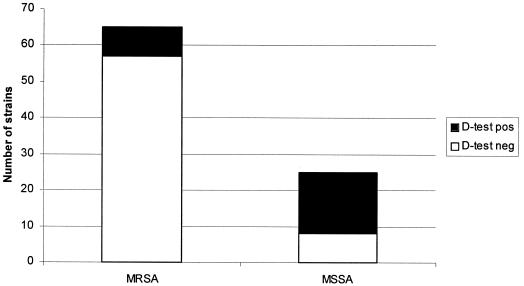
Ninety-one (54.2%) of 168 isolates were clindamycin-erythromycin discordant. There was 100% concordance of the D-test results between the investigators. The majority (72.2%) of the discordant strains were MRSA; however, only 8 (12.3%) of 65 strains were D-test positive (Fig. 2). Twenty-five (27.5%) of the 91 discordant strains were MSSA, but 17 (68%) of the 25 strains were D-test positive. Overall, 25 (27.5%) of the 91 discordant strains were D-test positive.
FIG. 2.
Summary of all clindamycin-erythromycin discordant isolates.
The mechanisms of clindamycin resistance have been studied for years, and the D-test was developed to identify potential clindamycin resistance so that possible ineffective therapy is not started when conventional tests show clindamycin MICs within the susceptible range (≤0.5 μg/ml). Molecular markers for the erm gene are available, but they are costly and inconvenient for everyday use (8, 15, 17). The D-test is easy to perform and interpret, reproducible, and inexpensive but still not universally used. There is a good correlation of standard D-tests with a modification applied to purity plates done at the time of specimen preparation for the Vitek-2 test (6).
Clindamycin is frequently used to treat skin and bone infections because of its tolerability, cost, oral form, and good tissue penetration. With this high prevalence of D-test positivity, why are clindamycin clinical failures seldom reported? It may take time for a mutant strain to develop, and the immune system may have already controlled the infection. New agents active against gram-positive bacteria have been developed, so clindamycin may now be used less often. Finally, although a D-test-positive isolate may mutate on therapy, the mutation rate in clinical infections is unknown and may be rare.
We studied the prevalence of D-test positivity in isolates that might be treated with clindamycin. We documented a clindamycin failure when resistance developed in a D-test-positive MSSA isolate. S. aureus clindamycin susceptibility reporting should be suppressed when erythromycin resistance is found (unless a D-test is negative). If D-testing is delayed until resistance testing is complete, the results may not be available for maximal clinical utility. Alternatively, the clinician may have to request it specifically.
It may be risky to use clindamycin when erythromycin testing shows a resistant or intermediate phenotype. The risk might currently be lower with MRSA, but this disparity may not continue or not be true elsewhere. Routine D-testing might allow clinicians to retain confidence in clindamycin when erythromycin resistance is present.
REFERENCES
- 1.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 2.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank, A. I., J. F. Marcinak, P. D. Mangat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton-Miller, J. M. T., and S. Shah. 2000. Patterns of phenotypic resistance to the macrolide-lincosamide-ketolide-streptogramin group of antibiotics in staphylococci. J. Antimicrob. Chemother. 46:941-949. [DOI] [PubMed] [Google Scholar]
- 5.Jennssen, W. D., S. Thakker-Varia, D. T. Dubin, and M. P. Weinstein. 1987. Prevalence of macrolides-lincosamides-streptogramin B resistance and erm gene classes among clinical strains of staphylococci and streptococci. Antimicrob. Agents Chemother. 31:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen, J. H., S. A. Crawford, M. L. McElmeel, and K. R. Fiebelkorn. 2004. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J. Clin. Microbiol. 42:1800-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 9.Martines-Aquilar, G., W. A. Hammerman, E. O. Mason, Jr., and S. L. Kaplan. 2003. Clindamycin treatment of invasive infections caused by community-acquired, methicillin-resistant and methicillin-susceptible Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 22:593-598. [DOI] [PubMed] [Google Scholar]
- 10.Nicola, F. G., L. K. McDougal, J. W. Biddle, and F. C. Tenover. 1998. Characterization of erythromycin-resistant isolates of Staphylococcus aureus recovered in the United States from 1958 through 1969. Antimicrob. Agents Chemother. 42:3024-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panagea, S., J. D. Perry, and F. K. Gould. 1999. Should clindamycin be used as treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob. Chemother. 44:577-582. [DOI] [PubMed] [Google Scholar]
- 12.Rao, G. G. 2000. Should clindamycin be used in treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob. Chemother. 45:715-716. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez, M. L., K. K. Flint, and R. N. Jones. 1993. Occurrence of macrolide-lincosamide-streptogramin resistance among staphylococcal clinical isolates at a university medical center. Diagn. Microbiol. Infect. Dis. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 14.Sattler, C. A., E. O. Mason, Jr., and S. L. Kaplan. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr. Infect. Dis. J. 21:910-916. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz, F.-J., J. Petridou, A. C. Fluit, U. Hadding, G. Peters, and C. von Eiff. 2000. Distribution of macrolide-resistant genes in Staphylococcus aureus blood-culture isolates from fifteen German university hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 19:385-387. [DOI] [PubMed] [Google Scholar]
- 16.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 17.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisblum, B., and V. Demohn. 1969. Erythromycin-inducible resistance in Staphylococcus aureus: survey of antibiotic classes involved. J. Bacteriol. 98:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]