Abstract
We recently reported on the involvement of a RecA-LexA-dependent pathway in the ciprofloxacin-triggered upregulation of fibronectin-binding proteins (FnBPs) by fluoroquinolone-resistant Staphylococcus aureus. The potential additional contribution of the transcription factor sigma B (SigB) to the ciprofloxacin-triggered upregulation of FnBPs was studied in isogenic mutants of fluoroquinolone-resistant strain RA1 (a topoisomerase IV gyrase double mutant of S. aureus NCTC strain 8325), which exhibited widely different levels of SigB activity, as assessed by quantitative reverse transcription-PCR of their respective sigB and SigB-dependent asp23 transcript levels. These mutants were Tn551 insertion sigB strain TE1 and rsbU+ complemented strain TE2, which exhibited a wild-type SigB operon. Levels of FnBP surface display and fibronectin-mediated adhesion were lower in sigB mutant TE1 or higher in the rsbU+-restored strain TE2 compared to their sigB+ but rsbU parent, strain RA1, exhibiting low levels of SigB activity. Steady-state fnbA and fnbB transcripts levels were similar in strains TE1 and RA1 but increased by 4- and 12-fold, respectively, in strain TE2 compared to those in strain RA1. In contrast, fibronectin-mediated adhesion of strains TE1, RA1, and TE2 was similarly enhanced by growth in the presence of one-eighth the MIC of ciprofloxacin, which led to a significantly higher increase in their fnbB transcript levels compared to the increase in their fnbA transcript levels. Increased SigB levels led to a significant reduction in agr RNAIII; in contrast, it led to a slight increase in sarA transcript levels. In conclusion, upregulation of FnBPs by increased SigB levels and ciprofloxacin exposure in fluoroquinolone-resistant S. aureus occurs via independent pathways whose concerted actions may significantly promote bacterial adhesion and colonization.
Staphylococcus aureus is a major pathogen that causes a variety of infections in humans and animals, ranging from minor skin and wound infections to life-threatening diseases (14). This high degree of flexibility in the expression of virulence is due to the highly regulated control and coordinated expression of numerous extracellular and cell wall-associated virulence determinants (1, 10, 13, 17, 32, 42, 46, 50). S. aureus displays a number of surface protein adhesins referred to as microbial surface components that recognize adhesive matrix molecules that promote the binding of several plasma or extracellular matrix host proteins (44). Fibronectin-binding proteins (FnBPs) FnBPA and FnBPB, encoded by the fnbA and the fnbB genes, respectively, play a prominent role in S. aureus attachment and colonization of host tissues or implanted biomaterials (18, 26, 58). FnBPs also promote the endocytic uptake of S. aureus by epithelial and endothelial cell lines and fibroblasts (20, 56).
Several complex regulons, notably, agr and sarA, control expression of cell wall-associated and extracellular virulence determinants in S. aureus in a growth-phase-dependent manner (1, 10, 13, 17, 42). The agr locus encodes a two-component quorum-sensing system that generates two divergent transcripts, RNAII and RNAIII, from two distinct promoters, promoters P2 and P3, respectively. RNAIII controls expression of many virulence genes during the postexponential phase by downregulating a majority of cell wall-associated proteins while upregulating the release of potent exotoxins and proteolytic enzymes (7, 13, 17, 32, 42). In contrast, the sarA locus encodes a single 14.5-kDa DNA-binding protein, but with three upstream promoters driving three overlapping transcripts (10). Like agr, the sarA locus was shown by transcription profiling studies to influence the expression of >100 genes of S. aureus (10, 17, 50). The SarA protein can either directly regulate several target genes (8, 30), e.g., hla (alpha-hemolysin gene), spa (protein A gene), fnbA (61, 63), cna, and sspA, or indirectly influence target gene expression by changing P2 or/and P3 agr promoter activities (11, 39, 63). Several additional sar homologues (sarS, sarR, sarT, sarU, and rot) that are all DNA-binding regulatory elements have recently been revealed by genomic and genetic studies (33, 35, 37, 50, 55). In addition, several of the 16 two-component regulatory systems identified in S. aureus (e.g., arlRS, srrAB, saeRS, lytRS, and yccFG) (16, 19, 22, 43, 45, 64) contribute to the regulation of virulence genes either directly or via the global regulators agr and/or sarA (1, 10, 42, 65).
The alternative transcription factor sigB operon of S. aureus is composed of four open reading frames (rsbU, rsbV, rsbW, and sigB) that show some structural and functional homology to those of Bacillus subtilis (3, 12, 25, 28, 38, 62). The SigB activity is controlled posttranslationally by a multicomponent signal transduction system involving the RsbU, RsbV, and RsbW proteins. The SigB factor, which is normally sequestered by the anti-sigma factor RsbW, may be released from the SigB-RsbW complex by transduction of external signals mediated by RsbU (a phosphatase) that dephosphorylates RsbV, thus permitting its interaction with RsbW. A number of virulence-associated target genes, such as asp23, coa, clfA, sarS, and the P3 promoter of sarA, are reported to be transcriptionally regulated by the level of free SigB (2, 28, 40, 41).
Subinhibitory concentrations of antibiotics may downregulate or upregulate specific adhesins (44) or secreted virulence factors (27) of S. aureus, such as FnBPs, collagen-binding protein, or alpha-toxin. In addition, emergence or acquisition of antibiotic resistance determinants by S. aureus, such as methicillin or glycopeptide resistance, can significantly alter expression of global regulators and virulence factors, such as agr (51, 52) and alpha-toxin (52), and can downregulate (54, 59) or upregulate (47) expression of cell wall-associated surface components, such as clumping factor A and FnBPs.
Previous studies performed in our laboratory (5, 6) revealed that subinhibitory concentrations of some antibiotics such as ciprofloxacin could raise the fibronectin-mediated attachment of fluoroquinolone-resistant S. aureus by selectively inducing the fnbB gene. Recently, a RecA-LexA-dependent pathway was shown to mediate the ciprofloxacin-induced fnbB upregulation in S. aureus (4). The aims of the present study were to (i) evaluate a potential additional contribution of SigB to the ciprofloxacin-triggered upregulation of FnBP by using isogenic derivatives of the recently described fluoroquinolone-resistant, grlA gyrA strain RA1 of S. aureus (4), which exhibit widely different levels of SigB activity, and (ii) evaluate the potential contribution of global regulons agr and sarA to either the SigB- or the ciprofloxacin-modulated effects on fnbA or fnbB transcription. These results provide evidence that optimal expression of a stress response factor and triggering of a drug-induced DNA repair system may independently, but in an additive manner, significantly promote FnBP-mediated S. aureus adhesion.
(This study was presented in part at the 13th European Congress for Clinical Microbiology and Infectious Diseases, Glasgow, Scotland.)
MATERIALS AND METHODS
Bacterial strains.
Strain RA1 is a gyrB142 grlA542 gyrAΩ105 quinolone-resistant mutant of NCTC8325 strain ISP794 (4). Strain RA1, like all members of the NCTC8325 family, has an 11-bp deletion in rsbU and yields weakly pigmented colonies on Muller-Hinton agar (MHA).
Strain TE1 (kindly provided by A. L. Cheung) is a Tn551 insertion sigB mutant of RA1 that was constructed by transducing RA1 with a phage lysate of strain ALC1001 carrying the sigB mutation (12, 62). Correct insertion of Tn551 in the sigB locus of strain TE1 was confirmed by PCR assays. Strain TE1 yields completely white colonies on MHA.
Strain TE2 is a derivative of RA1 whose SigB functional activity was restored by transducing RA1 with a phage lysate prepared from strain GP268 (kindly provided by M. Bischoff) rsbU+ rsbV+ rsbW+ sigB+ Tcr (25). Strain TE2 yields strongly pigmented colonies on MHA, and its genotype was verified by a PCR assay.
Identical MICs (32 μg/ml) of ciprofloxacin, determined by a macrodilution method in Mueller-Hinton broth (MHB; Difco, Detroit, Mich.) as described by NCCLS (40a), were recorded for strains RA1, TE1, and TE2.
Assay for bacterial adhesion to fibronectin.
The attachment properties of the S. aureus strains were measured by an adhesion assay with polymethylmethacrylate coverslips coated in vitro with three different concentrations (0.5, 1, and 2 μg/ml) of purified human fibronectin, as described previously (5, 6). Briefly, the strains were grown and metabolically radiolabeled with [3H]thymidine for 5 h without shaking at 37°C in MHB in the presence or absence of one-eighth the MIC (4 μg/ml) of ciprofloxacin. This sub-MIC of ciprofloxacin was previously shown (6) to optimally promote FnBPs without significantly affecting the bacterial growth rate. Thereafter, 107 CFU of washed cultures of radiolabeled bacteria was incubated for 1 h at 37°C with the fibronectin-coated coverslips in human albumin-supplemented phosphate-buffered saline (PBS), as described previously (5). The coverslips were washed, and the amount of radioactivity was determined. Bacterial adhesion data for the different strains whose cell-associated radioactivity and viable counts differed slightly (<20%) were normalized as described previously (57). Relative changes in bacterial adhesion were expressed as the percent increase or decrease in attachment of strains TE1 and TE2 compared to that of strain RA1 grown in ciprofloxacin-free medium or the relative changes in bacterial adhesion between exposure and no exposure to ciprofloxacin for each strain. Each experiment was performed at least three times, and the results were expressed as the mean percent changes ± standard errors of the means (SEMs). The statistical significance of pairwise differences in bacterial adhesion of isogenic strains differing in SigB functional levels or in ciprofloxacin exposure versus no exposure was evaluated by paired t tests of the relative increases or decreases pooled for the three coating concentrations of fibronectin by using P values of <0.05 with a two-tailed significance level (49).
Quantification of FnBPs by flow cytometry.
The FnBP-mediated fibronectin binding displayed by the different strains of S. aureus was monitored by flow cytometry, as described previously (21, 57). The specificity of the flow cytometry data was assessed by parallel analysis of control strains DU5883, a mutant of strain 8325-4 simultaneously defective in expression of both FnBPA and FnBPB, and DU5883(pFNBB4), which overexpresses FnBPB (21, 26, 57).
Relative changes in flow cytometric data were expressed as the percent increase or decrease in fluorescein isothiocyanate (FITC)-labeled fibronectin binding by strains TE1 and TE2 compared to that of strain RA1 grown in ciprofloxacin-free medium or the change in binding between exposure and no exposure to ciprofloxacin for each strain. Each experiment was performed three times, and the results were expressed as mean percent changes ± SEMs. The statistical significance of pairwise differences in FITC-labeled fibronectin binding of isogenic strains differing in SigB functional levels or in ciprofloxacin exposure versus no exposure was evaluated by paired t tests by using P values of <0.05 with a two-tailed significance level (49).
Total RNA extraction.
Cultures of strains RA1, TE1, and TE2 were grown for 5 h in MHB in the absence or presence of 4 μg of ciprofloxacin per ml and were then harvested and immediately processed as described previously (27, 47). Briefly, bacteria were recovered, fixed in acetone-ethanol (1:1), and washed in N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid-sucrose buffer. Samples were treated with ice-cold lysostaphin, and RNA was purified as described previously (47).
Real-time RT-PCR.
mRNA levels were determined by quantitative reverse transcription-PCR (qRT-PCR) with the one-step reverse transcriptase qPCR Master Mix kit (Eurogentec, Seraing, Belgium) and previously described primers and probes (47, 57). The reverse and forward primers and probe specific for the sspA gene were synthesized by Eurogentec (sspAR, 5′-ACTCTTTTAACAAATAAACACGTCGTAGA-3′; sspAF, 5′-GGTTAATTGCAGAAGGGAATGC-3′; sspAP, 5′-AAGCATGAGGATCACCGTGCGTAGC-3′). Conditions for reverse transcription, PCR, detection of fluorescence emission, and normalization of the levels of mRNA of the target genes extracted from the different strains on the basis of their 16S rRNA levels were described previously (47, 57). The statistical significance of strain-specific differences in normalized cycle threshold (CT) values of each transcript was evaluated by paired t tests, and data were considered significant when P was <0.05.
Northern blotting.
Total RNA (16 μg) was separated in a formaldehyde-agarose gel and blotted onto a nylon membrane, as described previously (9, 47). For agr RNAIII detection, a 439-bp RNAIII fragment amplified by PCR with primers RNAIIIa (5′-GTCATTATACGATTTAGTAC- 3′) and RNAIIIb (5′-GGTTATTAAGTTGGGATG-3′) was labeled with digoxigenin-dUTP by using DIG High Prime DNA labeling and detection starter kit II (Roche), as specified by the manufacturer. Blots were prehybridized with DIG Easy Hyb buffer for 1 h at 50°C and then hybridized with the labeled RNAIII probe at 50°C overnight. After the membrane was washed, the hybridized probes were immunodetected with anti-digoxigenin-alkaline phosphatase and then visualized with the chemiluminescence substrate disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3,3.1.13.7]decan]-4-yl) phenyl phosphate (CSPD), as specified by the manufacturer (Roche). Light emission was recorded on X-ray film.
Northern affinity blotting of the sarA transcripts was performed with a 290-bp 32P-labeled sarA probe, as described previously (47).
Measurement of protease activity in culture supernatants.
Proteolytic activity was assayed by determination of the increase in trichloroacetic acid-soluble azopeptides produced upon incubation of the culture supernatants with azocasein (Sigma), as described previously (60).
RESULTS
Modulation of S. aureus adhesion on fibronectin-coated surfaces by SigB levels and ciprofloxacin exposure.
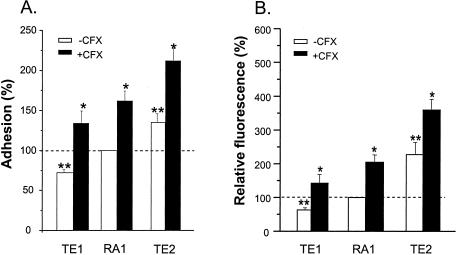
Following growth in ciprofloxacin-free medium, the level of attachment of sigB null mutant strain TE1, averaged over the three fibronectin coating concentrations, was significantly lower (P < 0.01) than that of its rsbU sigB+ parent, strain RA1. In contrast, the average adhesion of the rsbU+ sigB+ strain TE2 (P < 0.01) increased significantly (P < 0.01) compared to that of its parent, strain RA1 (Fig. 1A). Thus, the adhesion values for strains TE1, RA1, and TE2 increased as a function of the predicted increase in their SigB functional activities.
FIG. 1.
Adhesion to fibronectin-coated coverslips (A) or binding of soluble FITC-labeled fibronectin (B) of sigB null strain TE1 and rsbU+-restored strain TE2 grown in either the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of adhesion (A) or fibronectin binding (B) for strain RA1 in ciprofloxacin-free medium. Values represent means + SEMs (error bars) of individual adhesion data accumulated over the three fibronectin coating concentrations in three experiments. *, results significantly different (P < 0.01) for each strain grown in ciprofloxacin-containing MHB from those for strains grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.01) from those for strain RA1 grown in ciprofloxacin-free medium.
To assess whether the stepwise increase in adhesion from strain TE1 to strain RA1 and from strain RA1 to strain TE2 resulted from increased levels of surface display of fibronectin adhesin molecules, FnPB-mediated fibronectin-binding sites were monitored by flow cytometry. Figure 1B shows that strains RA1 and TE2 bound two- and fivefold more FITC-labeled fibronectin (P < 0.01) than TE1, respectively.
Upregulation of bacterial attachment.
Upregulation of bacterial attachment by the different strains by growth in the presence of one-eighth the MIC of ciprofloxacin (4 μg/ml) was not significantly influenced by modulation of their SigB functional levels. The average ciprofloxacin-promoted increase in the level of adhesion over the three fibronectin coating concentrations was significant (P < 0.01) and equivalent for strains TE1, RA1, and TE2 (Fig. 1). Flow cytometry (Fig. 1B) also showed a similar approximately twofold increase in the level of FnBP surface display in each strain grown in the presence of one-eighth the MIC of ciprofloxacin.
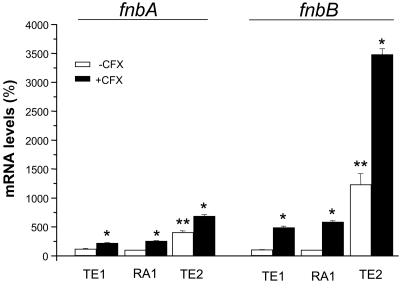
Upregulation of fnbA and fnbB transcripts by SigB levels and ciprofloxacin exposure.
The steady-state fnbA and fnbB mRNA levels of strains RA1, TE1, and TE2 were also influenced by their respective functional SigB levels and growth conditions in the presence versus absence of ciprofloxacin. After growth in ciprofloxacin-free medium, fnbA mRNA levels were equivalent in strains RA1 and TE1 but were significantly increased fourfold in strain TE2 compared to those in strain RA1 (Fig. 2). Under similar conditions, fnbB mRNA levels were also equivalent in strains RA1 and TE1 but were sharply (P < 0.01) increased 12-fold in strain TE2 compared to those in strain RA1 (Fig. 2). Upregulation of both fnb transcripts in ciprofloxacin-free medium occurred not only in 5-h cultures but also during early-exponential-phase growth. Control experiments demonstrated that fnbA and fnbB mRNA levels significantly (P < 0.01) increased by 6- and 18-fold, respectively, in TE2 cells compared to the levels in RA1 cells grown for 2 h.
FIG. 2.
Steady-state levels of the fnbA (left) and fnbB (right) transcripts of strains TE1 and TE2 grown in the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of those of strain RA1 grown in ciprofloxacin-free medium. mRNA levels were determined by real-time RT-PCR and were normalized on the basis of their 16S rRNA levels. Values represent the means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.01) for each strain grown in ciprofloxacin-containing MHB compared to those for the strain grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.01) from those for strain RA1 grown in ciprofloxacin-free medium.
As expected from bacterial adhesion and flow cytometry data (Fig. 1), the relative increases in fnbA and fnbB transcript levels triggered in the different strains by growth in the presence of 4 μg of ciprofloxacin per ml were not correlated with their respective SigB functional levels. The ciprofloxacin-mediated increases in fnbB transcript levels in strains TE1, RA1, and TE2 were 5.0-, 5.8-, and 2.8-fold, respectively, being significantly (P < 0.01) greater than the 2.5-, 1.9-, and 1.7-fold increases in fnbA transcript levels, respectively, recorded in these strains (Fig. 2).
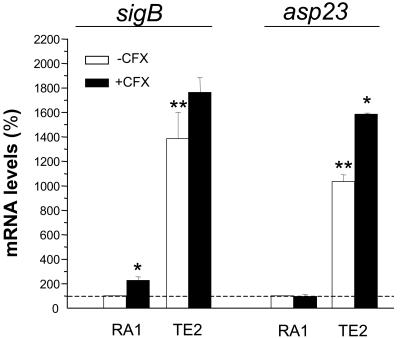
Assessment by qRT-PCR of SigB functional levels in the different strains.
The sigB transcript levels were strongly (13.8-fold) elevated in the rsbU+-restored strain TE2 compared to those in its rsbU parent, strain RA1 (Fig. 3). A similar trend was observed in the transcript levels of the asp23 gene, frequently used as a marker for SigB functional activity, which showed a 10.3-fold increase in strain TE2 compared to that in strain RA1. A striking finding was the 100-fold decrease in the asp23 transcript levels recorded in strain TE1 compared to those in RA1, which were too small to be visualized in Fig. 3. These data confirm the strong reduction of SigB functional levels in the sigB null mutant compared to those in its sigB+ but rsbU parent.
FIG. 3.
Steady-state mRNA levels of the sigB (left) and asp23 (right) genes of strain TE2 grown in the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of those of strain RA1 grown in ciprofloxacin-free medium. mRNA levels were determined by real-time RT-PCR and were normalized on the basis of their 16S rRNA levels. Values are the means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.05) for each strain grown in ciprofloxacin-containing MHB compared to those for the strain grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.01) from those for strain RA1 grown in ciprofloxacin-free medium. The asp23 transcripts levels of strain TE1 were too small to be visualized (see text).
Ciprofloxacin exposure led to a significant (P < 0.05) increase in the sigB transcript levels of strain RA1 but not those of strain TE2. On the other hand, ciprofloxacin exposure led to significantly (P < 0.05) increased asp23 levels in strains TE1 (data not shown) and TE2 but not in strain RA1 (Fig. 3).
Influences of SigB levels and ciprofloxacin exposure on agr transcript levels.
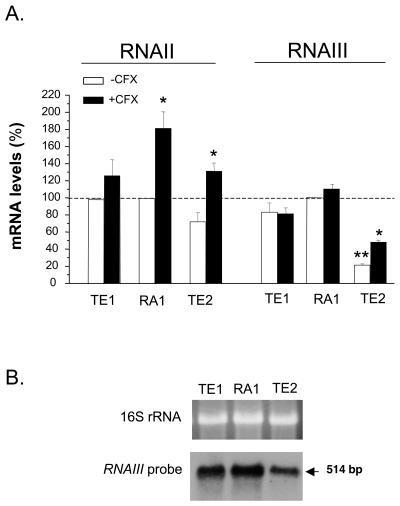
To detect any potential change in the activity of the global regulator agr that might contribute to either the SigB-mediated or the ciprofloxacin-triggered upregulation of fnb genes, we assayed the agr RNAII and RNAIII levels in strains TE1, RA1, and TE2. While the RNAII levels of strains grown in ciprofloxacin-free medium were not significantly different (Fig. 4A), RNAIII levels were similar in strains TE1 and RA1 but showed a significant 79% decline in strain TE2 compared to those in the other strains, as confirmed by Northern blotting analysis (Fig. 4B).
FIG. 4.
(A) Steady-state levels of RNAII and RNAIII of strains TE1 and TE2 grown in the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of those of strain RA1 grown in ciprofloxacin-free medium. mRNA levels were determined by real-time RT-PCR and were normalized on the basis of their 16S rRNA levels. Values represent the means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.05) for each strain grown in ciprofloxacin-containing MHB compared to those for the strain grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.05) from those for strain RA1 grown in ciprofloxacin-free medium. (B) Northern blot analysis of RNAIII in strains RA1, TE1, and TE2.
Ciprofloxacin exposure led to a significant (P < 0.05) but slight (less than twofold) increase in RNAII levels for strains RA1 and TE2 but not strain TE1. On the other hand, ciprofloxacin exposure led to significantly (P < 0.01) increased RNAIII levels only in strain TE2 and not in strains TE1 and RA1.
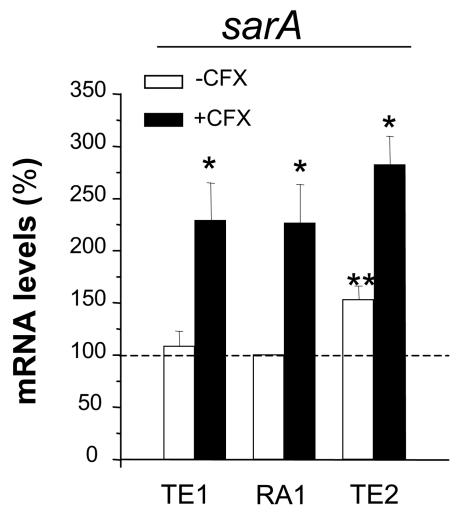
Impacts of SigB levels and ciprofloxacin exposure on sarA transcript levels.
We next evaluated whether the global regulator sarA could contribute to either the SigB-mediated or the ciprofloxacin-triggered upregulation of fnb genes. While overall the steady-state sarA mRNA levels assayed by qRT-PCR were equivalent in strains TE1 and RA1, they showed a slight (53%) but significant (P < 0.05) increase in the rsbU+-restored strain TE2 compared to those in strain RA1 (Fig. 5A). Northern blotting (data not shown) confirmed a stepwise increase in the levels of the SigB-dependent promoter P3-driven transcript from strain TE1 to RA1 and from strain RA1 to TE2, as expected from previous studies (2, 3, 15, 24, 34).
FIG. 5.
Steady-state levels of the sarA transcripts of strains TE1 and TE2 grown in the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of those of strain RA1 grown in ciprofloxacin-free medium. mRNA levels were determined by real-time RT-PCR and were normalized on the basis of their 16S rRNA levels. Values represent the means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.05) for each strain grown in ciprofloxacin-containing MHB compared to those for the strain grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.05) from those for strain RA1 grown in ciprofloxacin-free medium.
Ciprofloxacin exposure led to a significant (P < 0.05) approximately twofold increase in sarA transcript levels in all three strains, strains TE1, RA1, and TE2 (Fig. 5A).
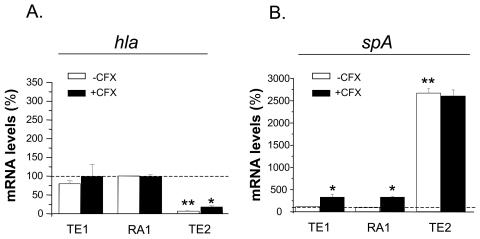
Impacts of SigB levels and ciprofloxacin exposure on hla and spa transcript levels.
Since previous reports demonstrated the strong impacts of the agr, sarA, and sigB regulons on the expression of hemolysin and protein A (1, 2, 10, 17, 42), we assayed the transcript levels of the target genes hla and spa in the SigB-modulated strains grown in the absence or the presence of ciprofloxacin. Striking differences in the hla and the spa transcript levels of strain TE2 compared to those of strains RA1 and TE1 were found. In the rsbU+-restored strain, hla transcript levels were <10% of the levels recorded in RA1 and TE1, the levels of which in the last two strains were nearly equivalent (Fig. 6A). These strain-specific differences in hla mRNA levels likely accounted for the strongly reduced hemolytic zones produced on sheep blood agar by strain TE2 compared to those produced by strains RA1 and TE1 (data not shown). An inverse situation was found for spa transcript levels, which were increased by >25-fold in strain TE2 compared to the low levels recorded in strains RA1 and TE2, which were nearly equivalent in the last two strains (Fig. 6B).
FIG. 6.
Steady-state levels of the hla and spa transcripts of strains TE1 and TE2 grown in the absence or the presence of 4 μg of ciprofloxacin (CFX) per ml (one-eighth the MIC) expressed as the percentages of those of strain RA1 grown in ciprofloxacin-free medium. mRNA levels were determined by real-time RT-PCR and were normalized on the basis of their 16S rRNA levels. Values represent the means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.01) for each strain grown in ciprofloxacin-containing MHB compared to those for the strain grown in ciprofloxacin-free MHB; **, results significantly different (P < 0.05) from those for strain RA1 grown in ciprofloxacin-free medium.
While ciprofloxacin exposure led to a significant increase in the hla transcript levels of strain TE2 but not those of strains RA1 and TE1, an inverse situation was seen with spa transcripts, whose levels were elevated in strains RA1 and TE1 but not strain TE2.
Expression of extracellular proteases in the isogenic SigB-modulated strains.
To evaluate putative strain-dependent differences in the levels of production and the extracellular release of proteases, which might account for the differential remodeling of surface-expressed FnBPs, we assayed the levels of the V8 protease gene transcripts (sspA) in strains RA1, TE1, and TE2 by qRT-PCR and extracellular proteolytic activity by the colorimetric detection of azopeptides in the culture supernatants incubated with azocasein (60). While the sspA transcript levels in 5-h cultures of strains RA1 and TE1 were equivalent, those of strain TE2 were slightly reduced (<50%) compared to those of strain RA1, thus confirming repression of this serine protease in the SigB-restored strain TE2 compared to its expression in the nonrestored isogenic derivatives (30). However, phenotypic detection of proteolytic activity in supernatants of 5-h cultures revealed only background levels of azopeptides. The levels released from 5-h cultures of the isogenic SigB-modulated strains, which represented <10% of those released from overnight (18-h) cultures of the same strains, did not allow assessment of strain-specific differences in the levels of production of extracellular proteases. In contrast, phenotypic detection of proteolytic activity of the SigB-modulated strains grown for 18 h revealed a ca. 75% reduction in extracellular proteolytic activity for strain TE2 compared to those for strains RA1 and TE1, in agreement with previous observations (30). Taken together, these data suggest that extracellular protease release from 5-h cultures of the SigB-modulated strains probably plays a minor role, if any, on FnBP surface display. However, differential remodeling of surface-expressed FnBPs is likely to play a more important role in cultures at more advanced stages of growth, as expected from previous studies (28, 30, 31).
DISCUSSION
Growing evidence suggests that expression and surface display of FnBPs in S. aureus are regulated by a complex network of global regulators, transcription factors, and stress response pathways (2-7, 29, 31, 53, 54, 57, 59, 61, 63). Besides the previously reported growth phase- and quorum sensing-controlled effects of agr and sarA regulons on transcription of fnb genes and/or FnBP surface display (7, 53, 61, 63), a variety of environmental and/or stressful conditions may also alter expression of fibronectin adhesins. These diverse situations include switching to small-colony variant phenotypes (57), acquisition of methicillin resistance (48, 54, 59), emergence of teicoplanin resistance (47), and exposure of fluoroquinolone-resistant S. aureus strains to subinhibitory levels of ciprofloxacin (4-6). Except for the indirect impact of the methicillin resistance element, which does not affect fnb transcription but which is believed to interfere with FnBP surface display via production of the pls surface protein (29, 54), all other conditions mentioned above were shown to regulate fnb transcription (4-6, 57). The upregulation of FnBP expression by specific environmental and stressful stimuli, including fluoroquinolone exposure, may play a significant role in promoting S. aureus attachment to and colonization of host tissues or implanted biomaterials (58). In addition, the higher levels of surface display of FnBPs involved in fibronectin-mediated bridging with the host receptor integrin α5β1 may also increase their endocytic uptake by nonprofessional phagocytes (20, 56), as demonstrated with hemB mutants of S. aureus displaying small-colony variant phenotypes (57).
The production of isogenic derivatives of fluoroquinolone-resistant grlA gyrA double-mutant strain RA1 of S. aureus, which displayed widely different levels of SigB activity, allowed exploration of the potential interaction of the SigB-mediated and ciprofloxacin-triggered pathways. Combination of transcriptional and phenotypic assays provided indirect, although consistent, evidence that the SigB-mediated and the ciprofloxacin-triggered responses involve separate regulatory networks whose characterization is still incomplete. We recently reported (4) on the contribution of a RecA-LexA pathway on the induction of fibronectin binding via selective upregulation of the fnbB gene, which did not require any functional agr or sarA activities. This study extends those previous findings by showing that the SigB functional activity does not interfere with the ciprofloxacin-triggered transcriptional and phenotypic responses. While the strong induction of the fnbB gene by a sub-MIC of ciprofloxacin was confirmed in the SigB-modulated derivatives of strain RA1, the fnbA gene was also induced, although to a much lower extent. The ciprofloxacin-triggered fnbA induction may possibly result from the longer exposure of each strain with ciprofloxacin in this study compared to the shorter 20-min exposure in the previous study (4), which failed to significantly induce fnbA. Further studies are required to elucidate the molecular basis of the differential ciprofloxacin-triggered fnbA versus fnbB upregulation.
In contrast to the ciprofloxacin-triggered responses, those promoted by genetic modulation of SigB functional activity appear to be more complex at both the transcriptional and the phenotypic levels, as supported by a recent microarray-based analysis of the S. aureus SigB regulon (2). Under our experimental conditions, similar transcript levels were recorded for either fnbA or fnbB when those of sigB null strain TE1 were compared with those of its rsbU parent, strain RA1. In contrast, there was a sharp, although disproportionate, increase in fnbB transcript levels compared with fnbA transcript levels in rsbU+-restored strain TE2 compared to those in its rsbU parent, strain RA1. The selective increase in fnbB transcript levels over fnbA transcript levels in the rsbU+-restored strain TE2 compared to those in its rsbU parent, strain RA1, is an original observation whose molecular basis is unknown at present. The fnbA and fnbB transcriptional dose-response data contrasted with the smoother increase in the levels of FnBP surface display from strain TE1 to TE2 via RA1, as recorded by bacterial adhesion and flow cytometry assays. The molecular basis of these contrasting data is not understood, and its elucidation will require improved understanding of the SigB-controlled pathway and its interactions with other regulatory networks controlling expression of fibronectin adhesins. Since expression of extracellular proteases was shown to be downregulated by high SigB and sarA functional levels (30) and upregulated by agr (42), we assayed in our set of SigB-modulated strains the levels of extracellular proteases that could potentially alter the half-lives of surface-exposed FnBPs (31, 36). The levels of proteolytic activity recorded in supernatants from 5-h cultures were too low to allow assessment of strain-specific differences. While qRT-PCR data indicated a decreased level of expression of the V8 protease gene sspA in the SigB-restored strain TE2 compared to that in the nonrestored isogenic derivatives, the extracellular protease release seems to be too marginal to play a major role in 5-h cultures. Previous reports (28, 30, 31) have shown that production of proteases mainly occurs during the late exponential and postexponential phases of growth. Thus, our initial hypothesis that strain-dependent differences in the production and extracellular release of proteases may account for the lack of correlation between fnb transcription and fibronectin adhesion was not supported by our experimental data. This discrepancy between fnb transcript levels and FnBP surface display may possibly result from differences in fnbA and/or fnbB mRNA decay between strain TE2 and strain RA1 or TE2 or from the presence of saturating amounts of cell wall-anchored FnBP molecules that may be displayed on bacterial cell surfaces.
The potential contribution of the major global regulators agr and sarA to increased levels of both fnb transcripts in the rsbU+-restored strain TE2 compared to those in strains RA1 and TE1 was also evaluated. It should be emphasized that transcript levels from global regulators and their putative target genes from 5-h cultures grown without shaking cannot be directly compared with those from cultures grown with rotatory shaking, whose growth rates and final biomasses are much higher. In contrast to agr RNAII levels, which were equivalent in all three strains, RNAIII levels were reduced by less than 1 order of magnitude in strain TE2 compared to the levels in strains RA1 and TE1, thus confirming the previously reported downregulation of the agr response regulator in strains displaying fully functional levels of SigB compared to its expression in their SigB-defective derivatives (3, 28). While the decreased RNAIII levels in strain TE2 compared to those in strains RA1 and TE1 may explain, at least in part (17), the changes in hla and spa transcript levels recorded for strain TE2 compared to those recorded for strains RA1 and TE1, the molecular details of the RNAIII-mediated downregulation of each fnb gene are still unknown. Since no selective effect of RNAIII on fnbB transcript levels compared to the effect on fnbA transcript levels has yet been reported, it is likely that other global regulatory systems or transcription factors may play a significant role in this complex regulatory process (1, 10, 42).
The impact of SigB functional levels on the activities of the sarA regulon is controversial. While some studies indicate a SigB-promoted upregulation of sarA transcription (2, 3, 23) and translation (24), other studies indicate no change in SarA protein levels (28) or even a SigB-promoted downregulation of sarA (12). These conflicting observations may potentially arise from either technical variables or significant differences in the genetic backgrounds of the strains examined. In our study, overall sarA transcript levels determined by real-time RT-PCR were increased by less than twofold in rsbU+-restored strain TE2 compared to the levels in strains RA1 and TE1. Nevertheless, it is unlikely that increased sarA transcript levels may account for increased fnbB mRNA levels, in particular, because it was established previously (61, 63) that sarA upregulates fnbA transcription but not fnbB transcription.
In conclusion, the results presented here provide evidence that optimal expression of a stress response factor and triggering of a drug-induced DNA repair system may independently, but in an additive manner, lead to an impressive >30-fold increase in fnbB transcript levels and promote S. aureus attachment to fibronectin. Ongoing studies performed in our laboratory also provide preliminary evidence that increased levels of FnBP expression in the SigB-restored strain TE2 grown in the presence of ciprofloxacin can upregulate FnBP uptake by nonprofessional phagocytes (A. Renzoni, unpublished data). Further studies of SigB-regulated pathways by using combined approaches of transcription profiling, targeted mutagenesis, and functional assays are required to better understand how multiresistant clinical isolates may benefit from the highly flexible regulation of S. aureus colonization and virulence.
Acknowledgments
This study was supported in part by research grants 3200-63710.00 and 3200B0-103951 (to P.V.) and research grant 632-57950.99 (to J.S.) from the Swiss National Science Foundation. D. Li acknowledges the support from the International Society of Infectious Diseases Fellowship Program.
We thank M. Bento and E. Huggler for technical assistance and A. L. Cheung and M. Bischoff for providing strains TE1 and GP268, respectively.
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisognano, C., W. L. Kelley, T. Estoppey, P. Francois, J. Schrenzel, D. Li, D. P. Lew, D. C. Hooper, A. L. Cheung, and P. Vaudaux. 2004. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 5.Bisognano, C., P. Vaudaux, D. P. Lew, E. Y. W. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisognano, C., P. Vaudaux, P. Rohner, D. P. Lew, and D. C. Hooper. 2000. Induction of fibronectin-binding proteins and increased adhesion of quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 44:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. [DOI] [PubMed] [Google Scholar]
- 9.Brown, T., and K. Mackey. 1997. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.1-4.9.16. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 10.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung, A. L., S. J. Projan, and H. Gresham. 2002. The genomic aspect of virulence, sepsis, and resistance to killing mechanisms in Staphylococcus aureus. Curr. Infect. Dis. Rep. 4:400-410. [DOI] [PubMed] [Google Scholar]
- 14.Crossley, K. B., and G. L. Archer. 1997. The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 15.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, B., P. Vaudaux, M. Magnin, Y. El Mestikawy, R. A. Proctor, D. P. Lew, and H. Vasey. 1996. Novel animal model for studying the molecular mechanisms of bacterial adhesion to bone-implanted metallic devices: role of fibronectin in Staphylococcus aureus adhesion. J. Orthop. Res. 14:914-920. [DOI] [PubMed] [Google Scholar]
- 19.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 20.Fowler, T., E. R. Wann, D. Joh, S. A. Johansson, T. J. Foster, and M. Höök. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79:672-679. [DOI] [PubMed] [Google Scholar]
- 21.Francois, P., J. Schrenzel, C. Stoerman-Chopard, H. Favre, M. Herrmann, T. J. Foster, D. P. Lew, and P. Vaudaux. 2000. Identification of plasma proteins adsorbed on hemodialysis tubing that promote Staphylococcus aureus adhesion. J. Lab. Clin. Med. 135:32-42. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 24.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene, C., D. McDevitt, P. François, P. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 27.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huesca, M., R. Peralta, D. N. Sauder, A. E. Simor, and M. J. McGavin. 2002. Adhesion and virulence properties of epidemic Canadian methicillin-resistant Staphylococcus aureus strain 1: identification of novel adhesion functions associated with plasmin-sensitive surface protein. J. Infect. Dis. 185:1285-1296. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor SarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 33.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGavin, M. J., C. Zahradka, K. Rice, and J. E. Scott. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 40.Nair, S. P., M. Bischoff, M. M. Senn, and B. Berger-Bächi. 2003. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 71:4167-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. NCCLS, Wayne, Pa. Infect. Immun. 71:4167-4170.12819110 [Google Scholar]
- 41.Nicholas, R. O., T. Li, D. McDevitt, A. Marra, S. Sucoloski, P. L. Demarsh, and D. R. Gentry. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 43.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 44.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 45.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 47.Renzoni, A., P. Francois, D. Li, W. L. Kelley, D. P. Lew, P. Vaudaux, and J. Schrenzel. 2004. Modulation of fibronectin adhesins and other virulence factors in a teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice, K., M. Huesca, D. Vaz, and M. J. McGavin. 2001. Variance in fibronectin binding and fnb locus polymorphisms in Staphylococcus aureus: identification of antigenic variation in a fibronectin binding protein adhesin of the epidemic CMRSA-1 strain of methicillin-resistant S. aureus. Infect. Immun. 69:3791-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosner, B. 1990. Analysis of variance, p. 474-526. In M. R. Payne, S. Hankinson, and S. London (ed.), Fundamentals of biostatistics. PWS-KENT Publishing Company, Belmont, Calif.
- 50.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakoulas, G., G. M. Eliopoulos, R. C. J. Moellering, R. P. Novick, L. Venkataraman, C. Wennersten, P. C. Degirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 52.Sakoulas, G., G. M. Eliopoulos, R. C. J. Moellering, C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saravia-Otten, P., H. P. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savolainen, K., L. Paulin, B. Westerlund-Wikström, T. J. Foster, T. K. Korhonen, and P. Kuusela. 2001. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 69:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of α-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha, B., P. P. Francois, O. Nüsse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell. Microbiol. 1:101-118. [DOI] [PubMed] [Google Scholar]
- 57.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaudaux, P., P. Francois, D. P. Lew, and F. A. Waldvogel. 2000. Host factors predisposing to and influencing therapy of foreign body infections, p. 1-26. In F. A. Waldvogel and A. L. Bisno (ed.), Infections associated with indwelling medical devices. American Society for Microbiology, Washington, D.C.
- 59.Vaudaux, P., V. Monzillo, P. Francois, D. P. Lew, T. J. Foster, and B. Berger-Bächi. 1998. Introduction of the mec element (methicillin resistance) into Staphylococcus aureus alters in vitro functional activities of fibrinogen and fibronectin adhesins. Antimicrob. Agents Chemother. 42:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Windle, H. J., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. Van Wamel, and A. L. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 62.Wu, S. W., H. De Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong, Y. Q., A. S. Bayer, M. R. Yeaman, W. Van Wamel, A. C. Manna, and A. L. Cheung. 2004. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect. Immun. 72:1832-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yarwood, J. M., and P. M. Schlievert. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 112:1620-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]