Abstract
A fifth gene cassette containing an aacC gene, aacCA5, was found in an aacCA5-aadA7 cassette array in a class 1 integron isolated from a multiply drug resistant Salmonella enterica serovar Kentucky strain. The AacC-A5 or AAC(3)-Ie acetyltransferase encoded by aacCA5 is related to other AAC(3)-I enzymes and confers resistance to gentamicin.
Acetyltransferases that modify the 3-amino group of aminoglycosides represent one type of enzyme that confers resistance to this important group of antibiotics. The known 3-N-aminoglycoside acetyltransferases [AAC(3) enzymes] are classified into several groups based on phenotypic differences in the specific spectra of aminoglycosides they are able to modify (22). However, they fall into only two clearly distinct groups based on the relationships between the proteins. The four aacC genes in family A, aacC1 [here designated aacCA1 and also sometimes referred to as aac(3)-Ia] (GenBank accession no. X15852) (25), a variant of aacCA1 (97.6% identical) (9, 15) previously named aacC4 (15) and here designated aacCA4, aac(3)-Ib (here designated aacCA2) (L06157) (21), and aac(3)Ic (here designated aacCA3) (AJ511268) (18) belong to the aac(3)-I phenotypic group and are found in gene cassettes. The products of these genes are all small proteins, of 154 to 156 amino acids, that are related to one another (Table 1) and confer resistance to gentamicin, sisomicin, and fortimicin but not to tobramycin, amikacin, or kanamycin. The remaining aacC genes are not found in gene cassettes and encode longer proteins, of 261 to 300 amino acids, that do not appear to be significantly related (generally less than 25% identical) to members of the AAC(3)-I group. This type B protein family currently includes at least 14 distinct members and variants (<2% difference) of some of the members.
TABLE 1.
Relationships between members of the AacC-A or AAC(3)-I protein familya
| Protein | AacC-A1 | AacC-A2 | AacC-A3 | AacC-A4 | AacC-A5 |
|---|---|---|---|---|---|
| AacC-A1 AAC(3)-Ia | — | 71.6 | 59.4 | 95.5 | 51.0 |
| AacC-A2 AAC(3)-Ib | 87.1 | — | 60.1 | 72.1 | 49.0 |
| AacC-A3 AAC(3)-Ic | 74.2 | 77.1 | — | 60.4 | 55.6 |
| AacC-A4 AAC(3)-Id | 98.7 | 86.4 | 74.0 | — | 51.6 |
| AacC-A5 AAC(3)-Ie | 64.1 | 61.4 | 71.9 | 64.1 | — |
Values represent the percent amino acid identities (top right) and similarities (bottom left) between the different AacC-A proteins. Dashes represent the 100% line.
Here, we report the identification of a further aacCA gene cassette that was found in a class 1 integron in a multiply drug resistant Salmonella enterica serovar Kentucky strain.
The isolate.
Salmonella serovar Kentucky SRC73 was isolated in 2001 from spice imported into Australia from India. The strain was serotyped by using standard procedures according to the Kauffman and White scheme (16). Salmonella serovar Kentucky SRC73 was scored as resistant to ampicillin (at 32 μg/ml), gentamicin (2.5 μg/ml), streptomycin (25 μg/ml), spectinomycin (50 μg/ml), sulfathiazole (550 μg/ml), tetracycline (20 μg/ml), and nalidixic acid (50 μg/ml) but susceptible to chloramphenicol (10 μg/ml), trimethoprim (50 μg/ml), kanamycin (10 μg/ml), and ciprofloxacin (2 μg/ml) by using the plate-replicator method as previously described (2, 3). Briefly, antibiotics at the concentrations indicated were in lysed blood Iso Sensitest agar plates (Oxoid, Hampshire, England), and the inoculum was 105 CFU per spot. Plates were incubated overnight at 37°C.
Resistance genes in Salmonella serovar Kentucky SRC73.
Whole cell DNA isolated from the Salmonella serovar Kentucky strain by using standard methods (20) was screened by PCR for several known antibiotic resistance genes with primers pairs internal to the genes (Table 2). PCR amplification reactions were carried out with PCR buffer (Roche Molecular Biochemicals, Mannheim, Germany) containing 160 μM of each deoxynucleoside triphosphate, 20 pmol of each primer, approximately 10 to 50 ng of template, and 1 U of Taq DNA polymerase (Roche). Reaction conditions were generally 94 to 96°C for 3 to 5 min; 30 to 40 cycles of 94 to 96°C for 30 s, 53 to 62°C for 30 to 60 s, and 72°C for 30 s to 2 min; and a final incubation at 72°C for 10 to 15 min. A product of the appropriate size for each gene of the strAB gene pair that confers resistance to streptomycin and for the blaTEM ampicillin resistance gene was detected. The tetracycline resistance determinant was tet(A), but spectinomycin resistance was not due to the aadA2 gene. The sul1 gene, which confers resistance to sulfonamides and is found only in association with class 1 integrons, and the sul2 gene were both present.
TABLE 2.
PCR primer pairs
| Primer name | 5′-to-3′ sequence | Location | Product size | Accession no. | Referencea |
|---|---|---|---|---|---|
| L1 | GGCATCCAAGCAGCAAGC | 5′-CS | Variable | M95287.4 | 11 |
| R1 | AAGCAGACTTGACCTGAT | 3′-CS | U12338.2 | 11 | |
| L2 | GACGATGCGTGGAGACC | 5′-CS | 297 | M95287.4 | 19 |
| L3 | CTTGCTGCTTGGATGCC | 5′-CS | M95287.4 | 12 | |
| QS-1 | ATGAAAGGCTGGCTTTTTCTTG | 3′-CS | 722 | U12338.2 | 5 |
| QS-2 | TGAGTGCATAACCACCAGCC | 3′-CS | U12338.2 | 5 | |
| sulI-F | GTGACGGTGTTCGGCATTCT | sul1 | 668 | U12338.2 | 10 |
| sulI-R | TTTACAGGAAGGCCAACGGT | sul1 | U12338.2 | 10 | |
| sulII-F | GGCAGATGTGATCGACCTCG | sul2 | 405 | M28829 | 10 |
| sulII-R | ATGCCGGGATCAAGGACAAG | sul2 | M28829 | 10 | |
| aadA2-L | TGTTGGTTACTGTGGCCG | aadA2 | 538 | X68227 | 14 |
| aadA2-R2 | TGCTTAGCTTCAAGTAAGACG | aadA2 | X68227 | 4 | |
| strA-F | CTTGGTGATAACGGCAATTC | strA | 548 | M95402 | 8 |
| strA-R | CCAATCGCAGATAGAAGGC | strA | M95402 | 8 | |
| strB-F | ATCGTCAAGGGATTGAAACC | strB | 509 | M95402 | 8 |
| strB-R | GGATCGTAGAACATATTGGC | strB | M95402 | 8 | |
| tem-F | TTCTTGAAGACGAAAGGGC | blaTEM | 1,208 | L27758 | 6 |
| tem-R | ACGCTCAGTGGAACGAAAAC | blaTEM | L27768 | 6 | |
| tet(A)-L | GCTACATCCTGCTTGCCTTC | tetA(A) | 210 | X61367 | 14 |
| tet(A)-R | CATAGATCGCCGTGAAGAGG | tetA(A) | X61367 | 14 | |
| tet(B)-L | TTGGTTAGGGGCAAGTTTTG | tetA(B) | 659 | AP000342 | 14 |
| tet(B)-R | GTAATGGGCCAATAACACCG | tetA(B) | AP000342 | 14 | |
| tet(G)-L | CAGCTTTCGGATTCTTACGG | tetA(G) | 844 | S52437 | 14 |
| tet(G)-R | GATTGGTGAGGCTCGTTAGC | tetA(G) | S52437 | 14 |
References for primers are shown.
Gene cassettes in a class 1 integron.
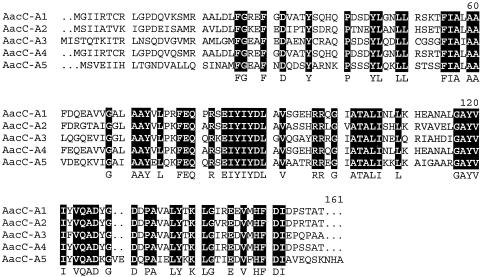
The presence of a class 1 integron was confirmed using primers within the intI1 gene (L2 and L3) and in the 3′-conserved segment (3′-CS) (QS-1 and QS-2) (Table 2). Amplification of the gene cassettes with standard primers in the 5′-CS and 3′-CS (L1 and R1) yielded a product of 1.6 kb, indicating the presence of gene cassettes totaling 1.45 kb. The sequence of this amplicon (GenBank accession no. AY463797) revealed two gene cassettes. The first is 564 bp long and contains an open reading frame with a GTG start codon at positions 22 to 24 relative to the beginning of the cassette that is preceded by a GAGG ribosome binding site at positions 11 to 14. Translation from this GTG gives a protein of 158 amino acids that is related to the known AacC-A [AAC(3)-I] proteins (Table 1), and alignment of the sequences (Fig. 1) revealed 65 completely conserved amino acids. Using the next available number, the gene and cassette were named aacCA5 and the protein AacC-A5 or AAC(3)-Ie.
FIG. 1.
Alignment of AacC-A proteins in the AAC(3)-I family. Amino acids completely conserved in all sequences are shown as white letters on a black background and are indicated by uppercase letters below the sequence. The sequences of AacC-A1 [AAC(3)-Ia], AacC-A2 [AAC(3)-Ib], AacC-A3 [AAC(3)-Ic], and AacC-A4 were obtained from GenBank and have accession nos. U12338, L06157, AJ511268, and AF318077, respectively. AacC-A5 [AAC(3)-Ie] is from this study.
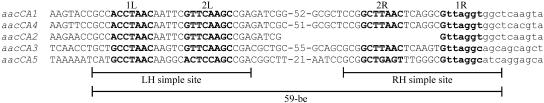
The aacCA5 cassette has a 59-be (59-base element) of 78 bp made up of two simple sites (Fig. 2) and a central region, as is characteristic for 59-be (23). This 59-be is not closely related to those of other cassettes in the aacCA group nor to those of any other known cassettes. The 59-be of the aacCA1 and aacCA4 cassettes, which are closely related, differ at only three positions, as expected if these two cassettes had diverged by accumulation of mutations over their full lengths (17). However, the more distantly related aacCA2 [aac(3)-Ib] cassette (73% identical to aacCA1), for which a complete sequence is not available, appears to have the same 59-be (Fig. 2), as the 34 bp of the aacCA2 59-be sequence present in GenBank accession no. L06157 is identical to the corresponding part of the aacCA1 59-be. In this case, the 59-be may have been acquired recently from the aacCA1 cassette. The 59-be in the aacCA3 [aac(3)-Ic] cassette is most closely related to that of the blaGES/IBC cassette, as noted previously (18). Thus, the aacCA5 and aacCA3 cassettes appear to have separate origins.
FIG. 2.
Alignment of the 59-be of aacCA family gene cassettes. Bases in lowercase letters are derived from the beginning of the cassette. The core sites are in boldface type and are designated 1L, 1R, 2L, and 2R according to reference 23. The extents of the simple sites (LH, left-hand; RH, right-hand) and the 59-be are indicated by bars (bottom). Sequences are from sources given in Fig. 1.
The second cassette in the integron is identical to the aadA7 cassette (13) (GenBank accession no. AF224733), found in an Escherichia coli strain from the ECOR collection that was isolated from a leopard in Washington Zoo in 1973 and in a Shiga toxin-producing E. coli O157:H7 strain (AF234167) (26). In both of these strains, aadA7 is the only cassette in a class 1 integron. The aadA7 gene confers resistance to streptomycin and spectinomycin and accounts for the spectinomycin resistance of SRC73. Since our work was completed, the aacCA5-aadA7 cassette array (AB114632) has been found in Vibrio fluvialis (1), but the aacCA5 gene was incorrectly designated aac(3)-Id. The same cassette array is also found in Vibrio cholerae (AY605683) and Salmonella enterica serovar Newport (AY458224) (7). The aacCA5 cassettes are all identical, but there are minor differences in the aadA7 cassettes (T65G C837Δ in AB114631 and AY605683; T669C A786G in AY458224).
The aacCA5 cassette confers resistance to aminoglycosides.
The aacCA5-aadA7 cassette array was amplified by PCR and eluted from a 1% (wt/vol) agarose gel using an Amicon Bioseparations Ultrafree-DNA kit (Millipore Corp., Bedford, Massachusetts), and ligated into pCR-Script (PCR-Script Amp cloning kit; Stratagene, La Jolla, California) using the manufacturer's protocols. The cloned fragment was recovered by transformation with selection on LB agar containing ampicillin (50 μg/ml) and gentamicin (8 μg/ml). Susceptibilities to gentamicin, tobramycin, amikacin, netilmicin, and kanamycin for E. coli strain 294 (24) containing either pCR-Script or pCR-Script with the cassette array were determined by using the CDS method for antibiotic disks (Oxoid, Basingstoke, Hampshire, United Kingdom). The cloned fragment conferred resistance to gentamicin (5- versus 11-mm zone size), but not to tobramycin, amikacin, netilmicin, or kanamycin, consistent with the presence of an AAC(3)-I-type aminoglycoside acetyltransferase. Resistance to sisomicin has also been demonstrated (1). The cassette array also conferred resistance to streptomycin and spectinomycin, consistent with the presence of the aadA7 cassette. SRC73 was resistant to the same set of aminoglycosides, indicating that the aacCA5 and aadA7 cassettes are sufficient to account for the observed aminoglycoside resistance.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to GenBank under accession no. AY463797.
Acknowledgments
R.S.L. was supported in part by a grant from the University of Wollongong, and S.R.P. was supported in part by grant no. 192108 from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Amavisit, P., P. F. Markham, D. Lightfoot, K. G. Whithear, and G. F. Browning. 2001. Molecular epidemiology of Salmonella Heidelberg in an equine hospital. Vet. Microbiol. 80:85-98. [DOI] [PubMed] [Google Scholar]
- 3.Bettelheim, K. A., M. A. Hornitzky, S. P. Djordjevic, and A. Kuzevski. 2003. Antibiotic resistance among verocytotoxigenic Escherichia coli (VTEC) and non-VTEC isolated from domestic animals and humans. J. Med. Microbiol. 52:155-162. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briñas, L., M. Zarazaga, Y. Sáenz, F. Ruiz-Larrea, and C. Torres. 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46:3156-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebreyes, W. A., and C. Altier. 2002. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M.-F. I. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leverstein-van Hall, M. A., A. Paauw, A. T. A. Box, H. E. M. Blok, J. Verhoef, and A. C. Fluit. 2002. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lévesque, C., and P. H. Roy. 1993. PCR analysis of integrons., p. 590-594. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. Mayo Foundation, Rochester, Minn.
- 12.Maguire, A. J., D. F. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng, L. K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., G. F. Weldhagen, C. De Champs, and P. Nordmann. 2002. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum β-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 49:561-565. [DOI] [PubMed] [Google Scholar]
- 16.Popoff, M. Y., and L. Le Minor. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Institute Pasteur, Paris, France.
- 17.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 18.Riccio, M. L., J.-D. Docquier, E. Dell'Amico, F. Luzzaro, G. Amicosante, and G. M. Rossolini. 2003. Novel 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ic, from a Pseudomonas aeruginosa integron. Antimicrob. Agents Chemother. 47:1746-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallen, B., A. Rajoharison, S. Desvarenne, and C. Mabilat. 1995. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb. Drug Resist. 1:195-202. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 24.Talmadge, K., and W. Gilbert. 1980. Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene 12:235-241. [DOI] [PubMed] [Google Scholar]
- 25.Wohlleben, W., W. Arnold, L. Bissonnette, A. Pelletier, A. Tanguay, P. H. Roy, G. C. Gamboa, G. F. Barry, E. Aubert, J. Davies, and S. A. Kagan. 1989. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC(3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217:202-208. [DOI] [PubMed] [Google Scholar]
- 26.Zhao, S., D. G. White, B. Ge, S. Ayers, S. Friedman, L. English, D. Wagner, S. Gaines, and J. Meng. 2001. Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl. Envron. Microbiol. 67:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]