Human cytomegalovirus (HCMV) infections are common and lead to lifelong infections. In immunocompetent individuals, primary infections are mostly subclinical or they may be associated with a self-limited mononucleosis-like syndrome. In contrast, infections in immunocompromised hosts (either primary infections, reactivations from latency, or reinfections) are associated with important morbidity and mortality. In patients with AIDS, the introduction of highly active antiretroviral therapy (HAART) has decreased the overall incidence of HCMV disease, mainly retinitis and gastrointestinal infections, by about 80% (50, 105). However, the functional benefit of HAART (i.e., restoration of specific HCMV-specific immune responses) may take up to 3 to 6 months to occur, and some patients do not have access to or do not respond to HAART (3, 40, 67, 104). Thus, HCMV still remains a concern in AIDS patients with CD4 counts <50 to 100 cells/μl.
A specific HCMV syndrome consisting of fever, malaise, arthralgia, and neutropenia may occur in solid-organ transplant (SOT) patients, in particular, those developing a primary HCMV infection (i.e., HCMV-seronegative recipient from a HCMV-seropositive donor [D+/R−]) during the first 3 months posttransplantation. In addition, invasive HCMV disease may involve different organs, such as the lungs, liver, and gastrointestinal tract. In the absence of antiviral intervention, symptomatic HCMV infections occur in approximately 39 to 41% of heart-lung transplant recipients, 9 to 35% of heart transplant recipients, 22 to 29% of liver and pancreas transplant recipients, 8 to 32% of kidney transplant recipients, 50% of kidney-pancreas transplant recipients, and 22% of small-bowel transplant recipients (114), with the highest incidence seen in D+/R− patients. Finally, active HCMV infections have been associated with indirect effects, such as dysfunction or rejection of the transplanted organ, an increased risk for bacterial or fungal opportunistic infections, and accelerated atherosclerosis in heart transplant recipients (109).
Among allogeneic bone marrow transplant (BMT) or hematopoietic stem cell transplant (HSCT) recipients, pneumonia and enteritis are the most common clinical manifestations of HCMV disease. In HCMV-seropositive recipients, active HCMV infections occur in 70 to 80% of patients; and in the absence of antiviral intervention, disease develops in 20 to 35% of those individuals, whereas active infections occur in only 15% of seronegative recipients of marrow from a seropositive donor (102). HCMV pneumonia remains associated with a significant risk of mortality, even when specific antiviral treatment is administered (18, 101). Gastrointestinal disease, alone or in association with pulmonary disease, is the second most common clinical manifestation of HCMV infections in that setting (85).
In addition to AIDS patients and transplant recipients, HCMV infections have been associated with serious complications in other immunocompromised hosts, such as cancer patients, mostly those suffering from hematologic malignancies (48), and children with congenital primary immunodeficiencies (135). Finally, congenital HCMV infections can result in severe sequelae in newborns or during the first years of life (22).
ANTIVIRAL AGENTS FOR TREATMENT OF HCMV INFECTIONS
Three antiviral agents and a prodrug are available for the systemic treatment of HCMV infections, and their mechanisms of action are summarized in Fig. 1. Ganciclovir (GCV; Cytovene, Hoffmann-La Roche) is a deoxyguanosine analogue and in 1988 was the first drug to be approved for the treatment of HCMV. Since then, it has remained the first-line treatment for HCMV infections in immunocompromised patients. Upon entry into HCMV-infected cells, GCV is selectively phosphorylated by a viral protein kinase homologue (the product of the UL97 gene, pUL97). Subsequently, cellular kinases convert GCV monophosphate into GCV triphosphate, which acts as a potent inhibitor of the HCMV DNA polymerase (pol) by competing with dGTP on the enzyme binding site. GCV is also incorporated into the viral DNA, where it slows down and eventually stops chain elongation (7, 15, 122). GCV formulations are available for intravenous (i.v.) or oral administration and as ocular implants for the local treatment of HCMV retinitis. Due to its poor bioavailability (∼6%), efforts were made to develop prodrugs of GCV. Valganciclovir (VGCV; Valcyte; Hoffmann-La Roche) is a new valyl ester formulation of GCV that exhibits about 10 times the bioavailability of GCV following oral administration (107).
FIG. 1.
Mechanisms of action of systemic antivirals approved for treatment of CMV infections. GCV and CDV, once phosphorylated, compete with dNTPs for the binding site on the DNA pol (A) and are incorporated into HCMV DNA (B), thus inhibiting viral DNA replication. FOS directly inhibits viral DNA replication by blocking the pyrophosphate (ppi) binding site (C), thus preventing ppi cleavage from incoming dNTPs and subsequent incorporation of the nucleotide into viral DNA. MP, DP, and TP, monophosphate, diphosphate, and triphosphate, respectively.
The other two compounds approved for systemic treatment of HCMV infections are also potent inhibitors of the viral DNA pol. However, due to their toxicity profiles, they are usually reserved for treatment of patients who have failed or who do not tolerate GCV therapy. Cidofovir (CDV; Vistide; Gilead Sciences) is a nucleotide analogue of cytidine that only requires activation (phosphorylation) by cellular enzymes to exert its antiviral activity (36). Once it is in its diphosphate form, CDV inhibits HCMV DNA pol by a mechanism similar to that of GCV. Foscarnet (FOS; Foscavir; Astra-Zeneca), a pyrophosphate analogue, differs from CDV and GCV both by its mechanism of action and by the fact that it does not require any activation step to exert its antiviral activity. FOS binds to and blocks the pyrophosphate binding site on the viral polymerase, thus preventing incorporation of incoming deoxynucleoside triphosphates (dNTPs) into viral DNA (35). Finally, formivirsen (Vitravene; Novartis) is a 21-nucleotide (nt) antisense with sequence complementary to the HCMV immediate-early-2 mRNA that interferes with HCMV replication at an early stage during the replication cycle (99). Its only indication at present is for the local treatment of HCMV retinitis in AIDS patients.
In addition to the treatment of established HCMV disease, antivirals have also been used to prevent such symptomatic episodes, especially in transplant recipients. The first strategy, defined as universal prophylaxis, consists of administration of an antiviral to all patients during the first 3 months or so after transplantation. The second strategy, referred to as “preemptive therapy,” consists of the use of short courses of antivirals only for high-risk patients on the basis of evidence of active viral replication (e.g., detection of early HCMV antigens, such as the pp65 protein, or sufficient amounts of viral DNA or mRNA) or during intense immunosuppression (e.g., with antithymocyte globulins) (17). Those preventive strategies have shown efficacy in the prevention of classical HCMV disease episodes (i.e., those occurring during the first months following transplantation) in both SOT and HSCT patients (18, 86, 106). However, some studies suggest that prophylaxis may only delay, rather than truly prevent, the onset of HCMV disease in predisposed patients (81, 86, 101, 110, 137).
PHENOTYPIC AND GENOTYPIC ASSAYS FOR EVALUATION OF HCMV DRUG SUSCEPTIBILITY
Two different, albeit complementary, approaches have been developed to assess HCMV drug resistance. In the phenotypic method, the virus is grown in the presence of various concentrations of an antiviral in order to determine the concentration of drug that will inhibit a percentage (more commonly, 50%) of viral growth in cell culture. To date, the plaque reduction assay remains the “gold standard” for phenotypic evaluation of HCMV susceptibility to antiviral agents. In this assay, a standardized viral inoculum is inoculated in different wells. The virus is then allowed to grow for a few days (typically, 7 to 10 days) in the presence of serial drug dilutions before the cells are stained. The number of viral plaques per concentration is first determined. Then, the percentage of viral growth compared to the growth in a control well without antiviral is plotted against the drug concentrations to determine the concentration that will inhibit the growth of 50% of viral plaques (50% inhibitory concentration [IC50]). Even though recent efforts have been made to standardize this assay (77), the interassay and interlaboratory variabilities are still problematic. In addition to the relative subjectivity of this method, there are some differences in the cutoff values that define drug resistance, depending on the laboratory. Similar assays, based on either detection of HCMV DNA by hybridization (39) or quantitative PCR or detection of specific HCMV antigens by enzyme-linked immunosorbent assay (125), flow cytometry (69, 93), immunofluorescence assay (126), or immunoperoxidase assay (51), have also been developed. Among those assays, the results obtained by a commercial DNA-DNA hybridization assay (Hybriwix probe system/cytomegalovirus susceptibility test kit) developed by Diagnostic Hybrids (Athens, Ohio) have shown a good correlation with those obtained by the plaque reduction assay (63, 64, 129). Even if the readout method is more objective, the cutoff values that define resistance are still a matter of debate. Altogether, phenotypic assays are time-consuming and are subject to the selection bias introduced during the growth of mixed viral populations in cell culture (54, 57), and they may lack sensitivity for the detection of low-level resistance or minor resistant subpopulations (34, 54).
In contrast to phenotypic assays, which directly measure the drug susceptibilities of viral isolates, genotypic assays detect the presence of viral mutations known to be associated with drug resistance. Those assays are based on the restriction fragment length polymorphisms (RFLPs) of PCR-amplified DNA fragments and on DNA sequencing of viral genes (UL97 and UL54) that have been involved in HCMV resistance to antivirals. One of the greatest advantages of those assays is that they can be performed directly with clinical specimens (19, 134), thus reducing considerably the time required to obtain results. By omitting the need to grow the virus, such methods also minimize the risks of introducing a selection bias. The limited number of UL97 mutations responsible for GCV resistance has allowed the development of rapid RFLP assays to detect their presence in clinical samples (27, 59). Indeed, approximately 70% of GCV-resistant clinical isolates contain mutations in one of three UL97 codons (codons 460, 594, and 595) (20). Typically, the presence of a given mutation will either obliterate an existing restriction site or create a new one. The difference in RFLP patterns can then be visualized following gel electrophoresis. The major advantages of this assay include its short turnaround time (less than 2 days) and its ability to detect as little as 10 to 20% of a mutant virus in a background of wild-type viruses (27). However, due to reports of new mutations at other codons and by consideration of the possible increased frequency of some mutations associated with lower levels of resistance (possibly as a consequence of the use of oral GCV [34]), the DNA sequence of the entire UL97 region involved in GCV resistance (see below) should be determined for a comprehensive analysis. DNA sequencing is the method of choice for assessment of the presence of mutations associated with drug resistance in the DNA pol gene due to the large number of mutations reported within all conserved regions of this gene (53). Genotypic approaches are fast and sensitive, but interpretation of their results (i.e., discrimination between mutations associated with natural polymorphisms [29, 90] from those related to drug resistance) is not always straightforward. In order to prove that a new mutation is associated with drug resistance, recombinant viruses need to be generated by using either overlapping cosmid and plasmid inserts (37) or marker transfer experiments of the mutated gene in a wild-type virus (4, 5, 30) or a genetically engineered virus (28, 34) prior to testing of this mutant virus in cell-based assays.
CLINICAL SIGNIFICANCE, INCIDENCE, AND RISK FACTORS FOR DRUG-RESISTANT HCMV INFECTIONS
Drug-resistant HCMV strains first emerged as a significant problem in patients with AIDS. Numerous studies have documented the emergence of drug-resistant HCMV strains (detected by phenotypic or genotypic methods) and their correlation with progressive or recurrent HCMV disease (mainly retinitis) during therapy (4, 30, 31, 55, 117, 118, 132, 134). The first study to evaluate the prevalence of GCV resistance in AIDS patients was conducted by evaluating the excretion of GCV-resistant strains in the urine of 31 patients with AIDS treated with i.v. GCV for HCMV retinitis. In that study, no resistant isolates were recovered from patients treated for ≤3 months, whereas 38% of those excreting the virus in their urine after >3 months of GCV treatment, which represented 8% of the entire cohort of patients, were infected with a resistant isolate (43). Since then, larger studies have evaluated the temporal emergence of GCV-resistant strains by either phenotypic (63) or genotypic (20) assays. In all studies, GCV resistance (defined by an IC50 ≥6 to 12 μM) at the initiation of treatment was a rare event (≤2.7% of tested strains). Phenotypic evaluation of isolates from the blood or urine of 95 patients treated with GCV (mostly i.v.) for HCMV retinitis revealed that 7, 12, 27, and 27% of patients excreted a GCV-resistant strain after 3, 6, 9, and 12 months of drug exposure, respectively (63). On the other hand, a more recent study by our group (20) of 148 AIDS patients treated with VGCV for HCMV retinitis has identified the presence of GCV resistance mutations in 2, 7, 9, and 13% of patients after 3, 6, 9, and 12 months of therapy, respectively. The lower incidence of GCV resistance in the latter study, despite the use of more sensitive genotypic methods, might be explained by differences in the study population, notably, improvement in anti-HIV therapy. Due to their less frequent use in the clinic, fewer data on the temporal emergence of FOS- and CDV-resistant HCMV strains in HIV-infected individuals have been reported. One small study evaluated the incidence of phenotypic resistance to FOS and found it to be 9, 26, 37, and 37% after 3, 6, 9, and 12 months of therapy, respectively, by using an IC50 cutoff of 400 μM (64), whereas another one reported lower rates (13, 24, and 37% after 6, 9, and 12 months, respectively) by using an IC50 cutoff of 600 μM (129). The data on CDV resistance (IC50s ≥ 2 to 4 μM) are even more limited, but they seem to indicate a resistance rate similar to those of GCV and FOS (64). Proposed risk factors for the development of HCMV resistance in this patient population include inadequate drug concentrations due to poor penetration into tissue (e.g., the eyes) or poor bioavailability (e.g., oral GCV), a sustained and profound immunosuppression status (CD4 counts <50 cells/μl), frequent discontinuation of treatment due to toxicity, and a high pretherapy HCMV load (41, 102).
HCMV resistance to GCV appears to be an emerging problem in SOT recipients and has been associated with an increased number of asymptomatic and symptomatic viremic episodes, the earlier onset of HCMV disease, graft loss, and an increased risk of death (10). Due to the different strategies used for prevention of HCMV infection and the different immunosuppressive regimens in use at different centers and by consideration of the heterogeneity of the transplant populations, it has been difficult to precisely evaluate the temporal emergence of HCMV resistance in that setting. In lung transplant recipients, the reported incidence of GCV resistance has varied from 3.6 to 9% after median cumulative drug exposures ranging from 79 to 100 days (73, 82, 87). In two of those studies, the incidence of resistance increased to 15.8 to 27% in D+/R− patients (82, 87) and occurred as a late complication, i.e., at a median of 4.4 months after transplantation (82). As opposed to what has been reported in lung transplant recipients, the incidence of GCV resistance in other SOT populations has been much lower in D+/R− patients (81, 87) and very occasional in seropositive (R+) recipients (87). More specifically, Lurain and colleagues (87) studied two cohorts of SOT patients, including heart, liver, and kidney recipients, at two U.S. centers. Phenotypic evaluation for HCMV resistance prompted by either clinical suspicion or positive blood cultures indicated that rates of resistance were generally low (e.g., <0.5%) at one center and varied from 2.2 to 5.6% at another center, depending on the organ transplanted. Another retrospective study by Limaye and colleagues (81) evaluated 240 SOT patients, including 67 D+/R− patients. In their cohort, GCV-resistant HCMV disease developed only in D+/R− patients, with resistance rates of 2.1% for all patients who had received transplants and 7% for D+/R− subjects. In the latter group, HCMV resistance was more frequently seen among kidney-pancreas recipients or pancreas recipients alone (21%) than among kidney (5%) or liver (0%) recipients. Of note, cases of GCV-resistant HCMV infections occurred at a median of 10 months after transplantation, with a median total drug exposure of 194 days (129 days of oral GCV), including two to three treatment courses for HCMV disease per patient. Importantly, GCV-resistant HCMV infections accounted for 20% of cases of HCMV disease that occurred during the first year after transplantation (81).
The first prospective study evaluating the emergence of GCV resistance in SOT recipients was recently reported by our group (21). In that study, molecular methods were used to assess the emergence of UL97 and UL54 mutations associated with GCV resistance in D+/R− patients (175 liver, 120 kidney, 56 heart, 11 kidney-pancreas, and 2 liver-kidney recipients) receiving HCMV prophylaxis with either oral GCV (1 g three times a day) or oral VGCV (900 mg once a day). Among 301 evaluable patients, the incidence of GCV resistance at the end of the prophylactic period (day 100 posttransplantation) was very low in both arms (0 and 1.9% for the VGCV and oral GCV arms, respectively). During the first year following transplantation, GCV resistance-associated mutations were found in 0 and 6.1% of patients at the time of suspected HCMV disease after receiving VGCV and oral GCV prophylaxis, respectively. Of note, however, no lung transplant recipients and a small number of kidney-pancreas recipients were included in the study (21), which might explain at least partly the low level of emergence of GCV resistance in that study. Interestingly, detection of known GCV resistance mutations was not necessarily associated with adverse clinical consequences in the latter study (21). Documented risk factors for the emergence of GCV resistance in SOT patients include the lack of HCMV-specific immunity (as encountered in the D+/R− group) (1, 8), lung or kidney-pancreas transplantation, longer lengths of drug exposure (prophylaxis > preemptive therapy), suboptimal plasma or tissue drug concentrations (as seen with oral GCV), the use of potent immunosuppressive regimens, a high HCMV load, and frequent episodes of HCMV disease (10, 80-82).
Limited data from small-scale studies suggest that the problem of GCV resistance in the BMT-HSCT population might not be as important as what has been observed in SOT recipients and AIDS patients, perhaps because of the more limited exposure to immunosuppression. In a study published by our group (56), molecular methods were used to detect the presence of the most common UL97 mutations associated with GCV resistance in blood samples from HSCT patients selected on the basis of having a positive HCMV PCR result, despite ≥14 days of preemptive i.v. GCV, or a second viremic episode within the first 98 days after transplantation. No UL97 mutations associated with GCV resistance were detected in this cohort of 50 patients (10 of them fulfilled the criteria for genotypic testing presented above) (56). In another study designed to evaluate risk factors and outcomes associated with rising HCMV antigenemia levels during preemptive therapy, Nichols and colleagues (103) prospectively evaluated 119 HSCT patients receiving preemptive GCV or FOS therapy following a positive pp65 antigenemia test result. Among those subjects, 47 (39%) exhibited a significant rise in antigenemia levels, despite antiviral administration, and at least one isolate was available for susceptibility testing from 15 subjects. Only one GCV-resistant isolate was identified in a patient who received GCV therapy for 4 weeks (103). In contrast, Erice et al. (47) reported genotypic or phenotypic evidence of infection with a GCV-resistant HCMV strain in two of five selected patients who had received GCV for a median of 58 days. However, all five patients had also received acyclovir prophylaxis for a median of 47 days, which could have been a predisposition to the selection of a GCV-resistant HCMV strain (95). Of note, the impact of prior acyclovir treatment in selecting for GCV resistance has not been confirmed by another group (42). Even though short courses of GCV therapy appear to be relatively safe in adult BMT patients, the situation might differ in the pediatric setting, as reported by Eckle and colleagues (44). In their study of 42 patients receiving T-cell-depleted transplants from unrelated donors, 3 showed genotypic evidence of GCV resistance, followed by the excretion of a resistant strain after 30 to 93 days of GCV exposure. Of note, in the same study, none of the 37 patients who underwent a similar procedure, but who received their transplant from a mismatched related donor, developed GCV resistance (44). The rapid emergence of GCV resistance was also documented in four of five children with congenital immunodeficiency disorders who underwent T-cell-depleted BMT (135). In those patients, genotypic evidence of GCV resistance was demonstrated after only 7 to 24 days (median, 10 days) of cumulative GCV therapy. Finally, the emergence of GCV-resistant strains has recently been associated with the occurrence of previously uncommon central nervous system HCMV disease and retinitis late after HSCT (57, 133).
MOLECULAR MECHANISMS OF HCMV RESISTANCE TO CURRENT ANTIVIRAL AGENTS
As anticipated from its mode of action (Fig. 1), HCMV resistance to GCV can be the result of alterations in two different viral gene products, namely, pUL97 and the viral DNA pol (pUL54). The UL97 protein is responsible for GCV monophosphorylation in HCMV-infected cells (83, 122) and is thus involved in HCMV resistance only to GCV. In fact, this mechanism of resistance (i.e., decreased phosphorylation of GCV) was recognized in laboratory-derived mutants (13, 119) and clinical resistant HCMV strains (12) even before pUL97 was identified as the protein responsible for such activation. Since then, studies of recombinant HCMV UL97 mutants (88) and recombinant vaccinia viruses expressing wild-type or mutated forms of UL97 (2, 62) have confirmed this mechanism of resistance. Several genotypic studies have identified UL97 mutations in over 90% of GCV-resistant HCMV clinical isolates (34, 49, 65, 66, 116), implying that impaired drug phosphorylation is the most important mechanism of GCV resistance in HCMV.
Even though the precise role of pUL97 in the HCMV replication cycle has still not been clearly elucidated, major advances in our understanding of its biological function have been recently made. The assumption that pUL97 has an important role in the viral replication cycle was confirmed by showing that the replication capacities of recombinant viruses with a UL97 deletion were severely impaired (108). Whether or not mutations associated with GCV resistance have an influence on viral fitness in the absence of selective pressure remains controversial. Data from one study reported that single amino acid substitutions associated with GCV resistance could result in a loss of viral fitness ranging from 3.5 to 13% (45), whereas another group (33, 34) found no significant loss of replication when residues 595 to 603 or residues 591 to 607 were deleted. Nevertheless, clinical strains with UL97 mutations appear to be fully pathogenic (79, 113, 117). Other insights into the biology of pUL97 came from biochemical and sequence analyses. Even though the protein is able to phosphorylate GCV, acyclovir, and pencyclovir (two other nucleoside analogues mainly used to treat herpes simplex virus [HSV] infections) (123, 139), the natural nucleosides (dA, dC, dT, and dG) are not phosphorylated by pUL97 (94, 97). Indeed, pUL97 shows no homology with known nucleoside kinases but, rather, shares sequence homology with protein tyrosine kinases and bacterial phosphotransferases (25, 58). Those homologies have allowed the recognition of conserved regions within the gene (Fig. 2). Finally, pUL97 has been shown to be a structural component of the virion with autophosphorylation properties (130), and independent studies have suggested a role for the protein in DNA replication, probably through phosphorylation of the DNA pol processivity factor (pUL44) (91, 131), as well as in DNA encapsidation and/or nuclear egress (72, 131).
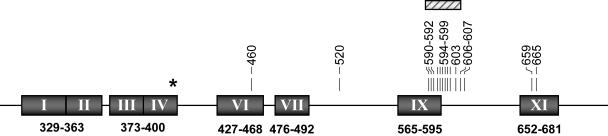
FIG. 2.
HCMV UL97 mutations conferring GCV resistance. HCMV UL97 conserved regions are represented by shaded boxes. The numbers under the boxes indicate the positions (codon numbers) of these conserved regions (58). Vertical bars indicate the presence of amino acid substitutions, while the hatched box indicates a region (codons 590 to 607) in which diverse codon deletions (from 1 to 17 codons) have been reported. Other mutations potentially associated with GCV resistance have been described at codons 466 and 521, but no susceptibility testing results were available (20, 44). *, the position of the UL97 mutation (codon 397) involved in maribavir (1263W94) resistance (14).
To date, UL97 mutations associated with GCV resistance that result in either amino acid substitutions or short (1- to 17-amino-acid) in-frame deletions have been found at codons 460 and 520 or in a region defined by codons 590 to 607 (Fig. 2), which are thought to be involved in ATP binding and substrate recognition, respectively. Only a few amino acid substitutions potentially associated with GCV resistance have been reported at other residues. Two of those are located at the 3′ extremity of the gene (Fig. 2), and the other two (V466M [20] and P521L [44]) are located close to codons known to be involved in GCV resistance (codons 460 and 520). However, the last two mutations were identified in clinical samples with no corresponding isolates, DNA pol mutations conferring GCV resistance were also present in one case (P521L), and their role in GCV resistance has not been confirmed by marker transfer experiments. Readers are referred to a recent review by our group (53) for an exhaustive list of UL97 mutations associated with GCV resistance. Nevertheless, some important aspects related to those mutations are worth mentioning. The cumulative results obtained from three recent studies that have documented the emergence of UL97 mutations in clinical isolates (66, 87) or in blood samples (20) from 61 AIDS and SOT patients are in general agreement with the proposed frequency of UL97 mutations, based on characterization of 76 independent UL97 mutants gathered in a single laboratory over years (34). Those data suggest that mutations A594V (30 to 34.5%), L595S (20 to 24%), M460V (11.5 to 14.5%), and H520Q (5 to 11.5%) represent the most frequent UL97 mutations present in GCV-resistant mutants. Other frequent UL97 mutations associated with resistance include C592G and C603W. On the basis of marker transfer experiments, mutation M460V (7-fold) (27), mutation C603W (8-fold) (30), deletion of codons 595 to 603 (8.4-fold) (33), mutation H520Q (10-fold) (59), mutation L595S (4.9- to 11.5-fold) (27, 34), mutation A594V (10.7-fold) (27), mutation C607Y (12.5-fold) (6), and deletion of codon 595 (13.3-fold) (4) appear to be associated with the highest rate of increase in GCV resistance over that of the parental strain, whereas mutations C592G, A594T, and E596G and deletion of codon 600 seem to confer only a modest decrease in susceptibility (34). Interestingly, analysis of the GCV-phosphorylating activity of mutated UL97 genes expressed in a recombinant vaccinia virus expression system would have predicted that mutations H520Q and M460V confer the highest decrease in GCV susceptibility (2).
The second viral protein involved in HCMV resistance to all currently approved systemic antivirals is the viral DNA pol (pUL54). The catalytic domain of the HCMV DNA pol, like the polymerases of other herpesviruses, is composed of eight highly conserved regions (regions I to VII and δ region C) that partially overlap with three conserved Exo motifs (motifs I to III) (Fig. 3) (9, 16, 78, 115, 138). On the basis of homology with other α-like DNA polymerases, it has been proposed that the Exo I to III motifs constitute the 3′ to 5′ exonuclease site of the enzyme (9, 16, 115). However, enzymatic analyses of purified mutated HCMV DNA pol proteins (L501F and K513N) suggest that the N-terminal portion of δ region C would also participate in this specific enzyme activity (38, 68). On the other hand, conserved regions II to V (Fig. 3) were proposed to be involved in the polymerization activity of DNA pol in general (16, 78), as in that of HCMV (136). However, more recent enzymatic studies of a purified HCMV DNA pol mutated in the N-terminal portion of δ region C (K513N) (38) and of purified HSV DNA pol proteins mutated in the Exo II and III motifs and in δ region C (61, 74) show that the two enzymatic functions (i.e., polymerization and 3′ to 5′ exonuclease activities) may not behave independently in herpesviruses and may indeed overlap (Fig. 3). By considering those two enzymatic activities while keeping in mind that GCV triphosphate and CDV diphosphate both act as alternate substrates for the DNA pol enzyme, three possible mechanisms of resistance have been postulated for those two antivirals: (i) a decreased affinity of the enzyme for the inhibitor, (ii) a decreased selective incorporation of the inhibitor into the elongating DNA chain, or (iii) an enhanced selective excision from the DNA chain of the incorporated inhibitors (37). In the case of FOS, which is not incorporated into the elongating DNA, only the first mechanism of resistance (i.e., decreased affinity of the enzyme) would apply.
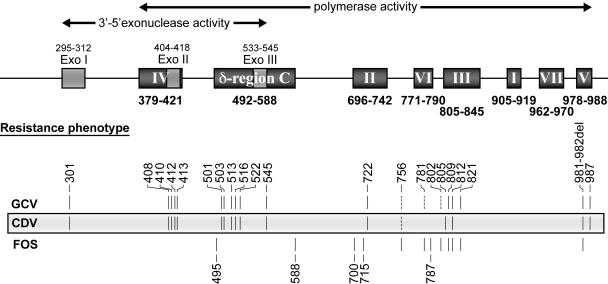
FIG. 3.
HCMV DNA polymerase (UL54) mutations conferring resistance to antivirals. HCMV DNA pol conserved regions are represented by shaded boxes. Numbers under and over the boxes indicate the positions (codon numbers) of these conserved regions (9, 16, 78, 115, 138). Vertical bars indicate the positions of mutations that have been associated with drug resistance. Other mutations (at codons 304, 393, 406, 521, 691, 695, 737, 751, 830, 834, 841, 961, and 972) potentially associated with drug resistance have been reported, but the presence in the same isolate of other known UL54 resistance mutations or the lack of phenotypic evaluation have precluded the ability to make an association with drug resistance (23, 26, 44, 49, 75, 112, 116). Note that resistant strains containing reported mutations were not necessarily tested for all antivirals listed. Dashed lines indicate that independent marker transfer experiments (codons 781 and 802) or substitutions by different amino acids (codon 756) have resulted in discrepant results.
The first evidence that DNA pol alterations could be involved in drug resistance came from analyses of laboratory-derived mutants (89, 121) as well as from a plaque-purified clinical isolate (124). The laboratory-derived mutants showed both GCV phosphorylation defects and decreased susceptibility to GCV as well as to nucleotide analogues (CDV and adefovir) that do not require prior virus-dependent activation. Sequence analyses indeed revealed alterations in three distinct conserved regions of the mutant DNA pol genes. When recombinant viruses were generated by transfer of those mutated pol genes into the genome of the reference strain AD169, GCV phosphorylation levels were found to return to normal but the recombinant viruses remained resistant (to a lower level) to GCV as well as to the other DNA pol inhibitors previously mentioned (89, 121). The plaque-purified clinical isolate exhibited GCV resistance in the presence of wild-type levels of GCV phosphorylation. The mutant was also resistant to CDV, adefovir, and FOS, which strongly suggested that the target of all those antivirals (the DNA pol) was the site of the alteration (124). From the mutations reported thus far, some conclusions on the regions involved in drug resistance can be drawn. In general, mutations located within the Exo I motif, region IV (including Exo II), the N-terminal extremity of δ region C (including Exo III), and region V are mostly associated with resistance to GCV and CDV. On the other hand, mutations located within the C-terminal extremity of δ region C and within or next to conserved regions II and VI seem to be mostly involved in FOS resistance. Finally, mutations reported within or next to conserved region III appear to be associated with various drug resistance or hypersusceptibility phenotypes (Fig. 3). It would therefore be tempting to define binding sites for dNTPs and pyrophosphates on the basis of the distribution of those HCMV mutations. However, the distribution of mutations affecting susceptibilities to adefovir and lobucavir (two other nucleotide or nucleoside analogues) (37) as well as the drug phenotypes associated with mutations in the same conserved regions of HSV and varicella-zoster virus DNA pol genes (53) preclude such conclusions from being made. This probably reflects the complex folding of the protein, which brings together distant conserved regions to form the specific binding sites, and definitive conclusions should await protein crystallization. Readers are referred to Fig. 3 and to a recent review (53) for an extensive list of DNA pol mutations associated with various drug resistance profiles. Among the most frequent DNA pol mutations associated with drug resistance are V715M, V781I, and L802M, which confer resistance to FOS, and F412C, L501I/F, and P522S, which confer resistance to GCV and CDV. Mutation A809V, which confers resistance to GCV and FOS, has also been reported with some frequency. Importantly, some mutations (E756K and V812L and the deletion of codons 981 and 982) have been associated with resistance to all three antivirals (28, 38). With regard to the levels of resistance, mutations L501I and K513N and deletion of codons 981 to 982 have been associated with a 6- to 8-fold decrease in GCV susceptibility (28, 37, 38) and mutations F412C/V, K513N, and A987G have been associated with a 10- to 18-fold decrease in CDV susceptibility (30, 37, 38), whereas mutations D588N, V715M, E756K, L802M, and T821I seem to confer 5.5- to 21-fold increases in resistance to FOS (5, 28, 30, 37, 98). A few UL54 mutations have been studied in marker transfer experiments for their effects on viral fitness. Among those, mutations T700A and V715 M (conserved region II) (5), K513N (δ region C) (38), and D301N (Exo I motif) (28) were shown to significantly reduce the yield of progeny virus in cell culture supernatants, whereas some others (D413E, T503I, L516R, and E756K/D) were associated with only a modest attenuation of viral replication (28). In the case of HCMV DNA pol mutants selected during GCV therapy, it should be noted that UL97 mutations have generally been shown to emerge first and to confer a low level of resistance (IC50 < 30 μM), whereas the subsequent emergence of UL54 mutations usually leads to a high level of drug resistance (IC50 > 30 μM) (49, 65, 116).
CONCLUSIONS AND PERSPECTIVES
When and how to monitor for HCMV resistance.
HCMV resistance to antivirals should be primarily suspected in the context of patients who have been exposed to an antiviral for substantial periods of time (typically, after >3 to 4 months of treatment in AIDS patients and after long-term prophylaxis in transplant recipients), especially if some risk factors are present (i.e., D+/R− SOT, lung or kidney-pancreas transplant, and AIDS with CD4 counts <50 cells/μl). In addition, clinical resistance is more likely if active viral replication (high or increasing levels of DNAemia, antigenemia, or viremia) persists, despite treatment with the maximum i.v. doses of the antivirals (80, 102). On the other hand, rising antigenemia levels during the first 2 weeks of antiviral therapy in HSCT recipients have not been associated with antiviral resistance but, rather, have been associated with host and other transplant-related factors (52, 103). Whenever antiviral resistance is suspected, phenotypic and/or genotypic investigation for resistance should be undertaken when these tests are available. As discussed above, genotypic methods are fast and more convenient and provide useful information for selection of an alternative treatment. However, identification of mutations of unknown significance remains problematic, and for that reason, phenotypic assays may still be necessary. The choice of the sample to be analyzed may also have some importance. Some studies have reported that there is a good correlation between the genotypes of strains detected in the eyes and the blood (93.5%) (60) and between isolates detected in the blood and urine (87.5%) (65) of AIDS patients with HCMV retinitis. However, there have been at least some reports of resistant HCMV strains restricted to specific body compartments (44, 84). This suggests that resistance assessment solely on the basis of the findings from blood or urine isolates may be suboptimal in some cases.
Management of infections caused by resistant HCMV strains.
Resistance should be suspected when stable or rising viral loads (especially DNAemia levels) or the persistence of clinical symptoms is observed 1 week or more after the receipt of appropriate full-dose i.v. antiviral therapy. In this context, clinical decisions on disease management should be based on genotypic analysis of UL97 and UL54 genes (when available), the immune status of the patients (e.g., patients with poor immune status, such as high-risk D+/R− recipients and lung transplant recipients), and disease severity (i.e., sight- or life-threatening conditions) (41, 80). Despite its limitation, as mentioned above, genotypic testing for resistance is more practical and rapid (results are available in 72 to 96 h) than phenotypic assays. Thus, ideally, rescue therapy should be based on the results of the genotypic assays. In centers where genotypic testing is unavailable or is performed infrequently, initial management should avoid the use of drugs with similar pathways of resistance. For instance, patients failing GCV should be given FOS in the absence of any sequencing data due to possible, albeit rare, UL54 mutations that usually confer resistance to both GCV and CDV. On the other hand, if UL97 and UL54 sequencing data are available and indicate that only UL97 mutations are present, then CDV therapy can be attempted. Other empirical options for patients failing GCV therapy could consist of treatment reinduction of the patient with higher-than-normal doses of GCV (up to 10 mg/kg of body weight i.v. twice a day) or the use of combination therapy with reduced GCV and FOS doses (100, 120), although these strategies are associated with significant toxicity and can be clinically risky in patients with life- or sight-threatening diseases. Whenever possible, improvement of the patient's immune status (i.e., reduction of the immunosuppressive regimen in transplant recipients or aggressive antiretroviral therapy in AIDS patients) should also be considered. The HCMV load should be carefully monitored (once weekly) while the patient is receiving therapy, and treatment should be continued for at least 1 week after the viral load becomes undetectable.
Investigational compounds.
In addition to efforts that are being made to develop an oral formulation of CDV (11), new compounds with anti-HCMV activity are being developed. These drugs belong to different classes that include benzimidazole derivatives, 4-sulfonamide-substituted naphthalene derivatives, benzathidazine-modified acyclonucleosides, tricyclic inhibitors, indolocarbazoles, and an experimental immunosuppressive agent (46, 96). Among the promising candidates, maribavir (1263W94; GlaxoSmithKline) is an l-ribofuranosyl derivative of BDCRB (2-bromo-5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole; a benzimidazole derivative) that has been shown to have good bioavailability and toxicity profiles in humans (71, 128) and that has been associated with a potent inhibitory effect on HCMV replication in vitro (14, 71) and in vivo (76). Maribavir is thought to prevent the exit of nucleocapsids from the nucleus (nuclear egress) and DNA replication by direct inhibition of pUL97 (Fig. 2) (14, 72), and mutations conferring resistance to this drug have been mapped to this gene (14) as well as the UL27 gene (32, 70). Other promising compounds include another derivative of the prototype compound BDCRB, the d-ribopyranosyl derivative GW275175X (GlaxoSmithKline), as well as the nonnucleosidic 4-sulfonamide-substituted naphthalene derivative tomeglovir (BAY-384766; Bayer). Both compounds showed good inhibitory activities against HCMV replication (92, 111, 127) and seem to interfere with cleavage of concatameric DNA molecules and encapsidation, which involve pUL89 and pUL56, respectively (24, 127).
REFERENCES
- 1.Baldanti, F., D. Lilleri, G. Campanini, G. Comolli, A. L. Ridolfo, S. Rusconi, and G. Gerna. 2004. Human cytomegalovirus double resistance in a donor-positive/recipient-negative lung transplant patient with an impaired CD4-mediated specific immune response. J. Antimicrob. Chemother. 53:536-539. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti, F., D. Michel, L. Simoncini, M. Heuschmid, A. Zimmermann, R. Minisini, P. Schaarschmidt, T. Schmid, G. Gerna, and T. Mertens. 2002. Mutations in the UL97 ORF of ganciclovir-resistant clinical cytomegalovirus isolates differentially affect GCV phosphorylation as determined in a recombinant vaccinia virus system. Antivir. Res. 54:59-67. [DOI] [PubMed] [Google Scholar]
- 3.Baldanti, F., S. Paolucci, A. Parisi, L. Meroni, and G. Gerna. 2002. Emergence of multiple drug-resistant human cytomegalovirus variants in 2 patients with human immunodeficiency virus infection unresponsive to highly active antiretroviral therapy. Clin. Infect. Dis. 34:1146-1149. [DOI] [PubMed] [Google Scholar]
- 4.Baldanti, F., E. Silini, A. Sarasini, C. L. Talarico, S. C. Stanat, K. K. Biron, M. Furione, F. Bono, G. Palu, and G. Gerna. 1995. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J. Virol. 69:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldanti, F., M. R. Underwood, S. C. Stanat, K. K. Biron, S. Chou, A. Sarasini, E. Silini, and G. Gerna. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldanti, F., M. R. Underwood, C. L. Talarico, L. Simoncini, A. Sarasini, K. K. Biron, and G. Gerna. 1998. The Cys607→Tyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients. Antimicrob. Agents Chemother. 42:444-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balfour, H. H., Jr. 1999. Antiviral drugs. N. Engl. J. Med. 340:1255-1268. [DOI] [PubMed] [Google Scholar]
- 8.Benz, C., G. Holz, D. Michel, S. Awerkiew, V. Dries, D. Stippel, T. Goeser, and D. H. Busch. 2003. Viral escape and T-cell immunity during ganciclovir treatment of cytomegalovirus infection: case report of a pancreatico-renal transplant recipient. Transplantation 75:724-727. [DOI] [PubMed] [Google Scholar]
- 9.Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. [DOI] [PubMed] [Google Scholar]
- 10.Bhorade, S. M., N. S. Lurain, A. Jordan, J. Leischner, J. Villanueva, R. Durazo, S. Creech, W. T. Vigneswaran, and E. R. Garrity. 2002. Emergence of ganciclovir-resistant cytomegalovirus in lung transplant recipients. J. Heart Lung Transplant. 21:1274-1282. [DOI] [PubMed] [Google Scholar]
- 11.Bidanset, D. J., J. R. Beadle, W. B. Wan, K. Y. Hostetler, and E. R. Kern. 2004. Oral activity of ether lipid ester prodrugs of cidofovir against experimental human cytomegalovirus infection. J. Infect. Dis. 190:499-503. [DOI] [PubMed] [Google Scholar]
- 12.Biron, K. K. 1991. Ganciclovir-resistant human cytomegalovirus clinical isolates; resistance mechanisms and in vitro susceptibility to antiviral agents. Transplant. Proc. 23:162-167. [PubMed] [Google Scholar]
- 13.Biron, K. K., J. A. Fyfe, S. C. Stanat, L. K. Leslie, J. B. Sorrell, C. U. Lambe, and D. M. Coen. 1986. A human cytomegalovirus mutant resistant to the nucleoside analog 9-([2-hydroxy-1-(hydroxymethyl)ethoxy]methyl)guanine (BW B759U) induces reduced levels of BW B759U triphosphate. Proc. Natl. Acad. Sci. USA 83:8769-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biron, K. K., S. C. Stanat, J. B. Sorrell, J. A. Fyfe, P. M. Keller, C. U. Lambe, and D. J. Nelson. 1985. Metabolic activation of the nucleoside analog 9-([2-hydroxy-1-(hydroxymethyl)ethoxy]methyl)guanine in human diploid fibroblasts infected with human cytomegalovirus. Proc. Natl. Acad. Sci. USA 82:2473-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco, L., A. Bernad, M. A. Blasco, and M. Salas. 1991. A general structure for DNA-dependent DNA polymerases. Gene 100:27-38. [DOI] [PubMed] [Google Scholar]
- 17.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeckh, M., W. Leisenring, S. R. Riddell, R. A. Bowden, M.-L. Huang, D. Myerson, T. Stevens-Ayers, M. E. D. Flowers, T. Cunningham, and L. Corey. 2003. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood 101:407-414. [DOI] [PubMed] [Google Scholar]
- 19.Boivin, G., S. Chou, M. R. Quirk, A. Erice, and M. C. Jordan. 1996. Detection of ganciclovir resistance mutations and quantitation of cytomegalovirus (CMV) DNA in leukocytes of patients with fatal disseminated CMV disease. J. Infect. Dis. 173:523-528. [DOI] [PubMed] [Google Scholar]
- 20.Boivin, G., C. Gilbert, A. Gaudreau, I. Greenfield, R. Sudlow, and N. A. Roberts. 2001. Rate of emergence of cytomegalovirus (CMV) mutations in leukocytes of patients with acquired immunodeficiency syndrome who are receiving valganciclovir as induction and maintenance therapy for CMV retinitis. J. Infect. Dis. 184:1598-1602. [DOI] [PubMed] [Google Scholar]
- 21.Boivin, G., N. Goyette, C. Gilbert, N. Roberts, K. Macey, C. Paya, M. D. Pescovitz, A. Humar, E. Dominguez, K. Washburn, E. Blumberg, B. Alexander, R. Freeman, N. Heaton, and E. Covington. 2004. Absence of cytomegalovirus-resistance mutations after valganciclovir prophylaxis, in a prospective multicenter study of solid-organ transplant recipients. J. Infect. Dis. 189:1615-1618. [DOI] [PubMed] [Google Scholar]
- 22.Boppana, S. B., R. F. Pass, W. J. Britt, S. Stagno, and C. A. Alford. 1992. Symptomatic congenital cytomegalovirus infection: neonatal morbidity and mortality. Pediatr. Infect. Dis. J. 11:93-99. [DOI] [PubMed] [Google Scholar]
- 23.Bowen, E. F., J. M. Cherrington, P. D. Lamy, P. D. Griffiths, M. A. Johnson, and V. C. Emery. 1999. Quantitative changes in cytomegalovirus DNAemia and genetic analysis of the UL97 and UL54 genes in AIDS patients receiving cidofovir following ganciclovir therapy. J. Med. Virol. 58:402-407. [PubMed] [Google Scholar]
- 24.Buerger, I., J. Reefschlaeger, W. Bender, P. Eckenberg, A. Popp, O. Weber, S. Graeper, H. D. Klenk, H. Ruebsamen-Waigmann, and S. Hallenberger. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chee, M. S., G. L. Lawrence, and B. G. Barrell. 1989. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 70:1151-1160. [DOI] [PubMed] [Google Scholar]
- 26.Cherrington, J. M., M. D. Fuller, P. D. Lamy, R. Miner, J. P. Lalezari, S. Nuessle, and W. L. Drew. 1998. In vitro antiviral susceptibilities of isolates from cytomegalovirus retinitis patients receiving first- or second-line cidofovir therapy: relationship to clinical outcome. J. Infect. Dis. 178:1821-1825. [DOI] [PubMed] [Google Scholar]
- 27.Chou, S., A. Erice, M. C. Jordan, G. M. Vercellotti, K. R. Michels, C. L. Talarico, S. C. Stanat, and K. K. Biron. 1995. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J. Infect. Dis. 171:576-583. [DOI] [PubMed] [Google Scholar]
- 28.Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 29.Chou, S., N. S. Lurain, A. Weinberg, G. Y. Cai, P. L. Sharma, and C. S. Crumpacker. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou, S., G. Marousek, S. Guentzel, S. E. Follansbee, M. E. Poscher, J. P. Lalezari, R. C. Miner, and W. L. Drew. 1997. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 176:786-789. [DOI] [PubMed] [Google Scholar]
- 31.Chou, S., G. Marousek, D. M. Parenti, S. M. Gordon, A. G. Lavoy, J. G. Ross, R. C. Miner, and W. L. Drew. 1998. Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J. Infect. Dis. 178:526-530. [DOI] [PubMed] [Google Scholar]
- 32.Chou, S., G. I. Marousek, A. E. Senters, M. G. Davis, and K. K. Biron. 2004. Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir. J. Virol. 78:7124-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou, S., and C. L. Meichsner. 2000. A nine-codon deletion mutation in the cytomegalovirus UL97 phosphotransferase gene confers resistance to ganciclovir. Antimicrob. Agents Chemother. 44:183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou, S., R. H. Waldemer, A. E. Senters, K. S. Michels, G. W. Kemble, R. C. Miner, and W. L. Drew. 2002. Cytomegalovirus UL97 phosphotransferase mutations that affect susceptibility to ganciclovir. J. Infect. Dis. 185:162-169. [DOI] [PubMed] [Google Scholar]
- 35.Chrisp, P., and S. P. Clissold. 1991. Foscarnet. A review of its antiviral activity, pharmacokinetic properties and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs 41:104-129. [DOI] [PubMed] [Google Scholar]
- 36.Cihlar, T., and M. S. Chen. 1996. Identification of enzymes catalyzing two-step phosphorylation of cidofovir and the effect of cytomegalovirus infection on their activities in host cells. Mol. Pharmacol. 50:1502-1510. [PubMed] [Google Scholar]
- 37.Cihlar, T., M. Fuller, and J. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cihlar, T., M. D. Fuller, A. S. Mulato, and J. M. Cherrington. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382-393. [DOI] [PubMed] [Google Scholar]
- 39.Dankner, W. M., D. Scholl, S. C. Stanat, M. Martin, R. L. Sonke, and S. A. Spector. 1990. Rapid antiviral DNA-DNA hybridization assay for human cytomegalovirus. J. Virol. Methods 28:293-298. [DOI] [PubMed] [Google Scholar]
- 40.Deayton, J., A. Mocroft, P. Wilson, V. C. Emery, M. A. Johnson, and P. D. Griffiths. 1999. Loss of cytomegalovirus (CMV) viraemia following highly active antiretroviral therapy in the absence of specific anti-CMV therapy. AIDS 13:1203-1206. [DOI] [PubMed] [Google Scholar]
- 41.Drew, W. 2003. Cytomegalovirus disease in the highly active antiretroviral therapy era. Curr. Infect. Dis. Rep. 5:257-265. [DOI] [PubMed] [Google Scholar]
- 42.Drew, W. L., R. Anderson, W. Lang, R. C. Miner, G. Davis, and J. Lalezari. 1995. Failure of high-dose oral acyclovir to suppress CMV viruria or induce ganciclovir-resistant CMV in HIV antibody positive patients. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:289-291. [DOI] [PubMed] [Google Scholar]
- 43.Drew, W. L., R. C. Miner, D. F. Busch, S. E. Follansbee, J. Gullett, S. G. Mehalko, S. M. Gordon, W. F. Owen, Jr., T. R. Matthews, and W. C. Buhles. 1991. Prevalence of resistance in patients receiving ganciclovir for serious cytomegalovirus infection. J. Infect. Dis. 163:716-719. [DOI] [PubMed] [Google Scholar]
- 44.Eckle, T., L. Prix, G. Jahn, T. Klingebiel, R. Handgretinger, B. Selle, and K. Hamprecht. 2000. Drug-resistant human cytomegalovirus infection in children after allogeneic stem cell transplantation may have different clinical outcomes. Blood 96:3286-3289. [PubMed] [Google Scholar]
- 45.Emery, V. C., A. V. Cope, E. F. Bowen, D. Gor, and P. D. Griffiths. 1999. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 190:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emery, V. C., and A. F. Hassan-Walker. 2002. Focus on new drugs in development against human cytomegalovirus. Drugs 62:1853-1858. [DOI] [PubMed] [Google Scholar]
- 47.Erice, A., N. Borrell, W. Li, W. J. Miller, and H. H. Balfour, Jr. 1998. Ganciclovir susceptibilities and analysis of UL97 region in cytomegalovirus (CMV) isolates from bone marrow recipients with CMV disease after antiviral prophylaxis. J. Infect. Dis. 178:531-534. [DOI] [PubMed] [Google Scholar]
- 48.Erice, A., S. Chou, K. K. Biron, S. C. Stanat, H. H. Balfour, Jr., and M. C. Jordan. 1989. Progressive disease due to ganciclovir-resistant cytomegalovirus in immunocompromised patients. N. Engl. J. Med. 320:289-293. [DOI] [PubMed] [Google Scholar]
- 49.Erice, A., C. Gil-Roda, J. L. Perez, H. H. Balfour, K. J. Sannerud, M. N. Hanson, G. Boivin, and S. Chou. 1997. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J. Infect. Dis. 175:1087-1092. [DOI] [PubMed] [Google Scholar]
- 50.Forrest, D. M., E. Seminari, R. S. Hogg, B. Yip, J. Raboud, L. Lawson, P. Phillips, M. T. Schechter, M. V. O'Shaughnessy, and J. S. Montaner. 1998. The incidence and spectrum of AIDS-defining illnesses in persons treated with antiretroviral drugs. Clin. Infect. Dis. 27:1379-1385. [DOI] [PubMed] [Google Scholar]
- 51.Gerna, G., F. Baldanti, M. Zavattoni, A. Sarasini, E. Percivalle, and M. G. Revello. 1992. Monitoring of ganciclovir sensitivity of multiple human cytomegalovirus strains coinfecting blood of an AIDS patient by an immediate-early antigen plaque assay. Antivir. Res. 19:333-345. [DOI] [PubMed] [Google Scholar]
- 52.Gerna, G., A. Sarasini, D. Lilleri, E. Percivalle, M. Torsellini, F. Baldanti, and M. G. Revello. 2003. In vitro model for the study of the dissociation of increasing antigenemia and decreasing DNAemia and viremia during treatment of human cytomegalovirus infection with ganciclovir in transplant recipients. J. Infect. Dis. 188:1639-1647. [DOI] [PubMed] [Google Scholar]
- 53.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Update 5:88-114. [DOI] [PubMed] [Google Scholar]
- 54.Gilbert, C., and G. Boivin. 2003. Discordant phenotypes and genotypes of cytomegalovirus (CMV) in patients with AIDS and relapsing CMV retinitis. AIDS 17:337-341. [DOI] [PubMed] [Google Scholar]
- 55.Gilbert, C., J. Handfield, E. Toma, R. Lalonde, M. G. Bergeron, and G. Boivin. 1998. Emergence and prevalence of cytomegalovirus UL97 mutations associated with ganciclovir resistance in AIDS patients. AIDS 12:125-129. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert, C., J. Roy, R. Bélanger, R. Delage, C. Béliveau, C. Demers, and G. Boivin. 2001. Lack of emergence of cytomegalovirus UL97 mutations conferring ganciclovir (GCV) resistance following preemptive GCV therapy in allogeneic stem cell transplant recipients. Antimicrob. Agents Chemother. 45:3669-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamprecht, K., T. Eckle, L. Prix, C. Faul, H. Einsele, and G. Jahn. 2003. Ganciclovir-resistant cytomegalovirus disease after allogeneic stem cell transplantation: pitfalls of phenotypic diagnosis by in vitro selection of an UL97 mutant strain. J. Infect. Dis. 187:139-143. [DOI] [PubMed] [Google Scholar]
- 58.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 59.Hanson, M. N., L. C. Preheim, S. Chou, C. L. Talarico, K. K. Biron, and A. Erice. 1995. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob. Agents Chemother. 39:1204-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu, H., D. A. Jabs, M. S. Forman, B. K. Martin, J. P. Dunn, D. V. Weinberg, and J. L. Davis. 2002. Comparison of cytomegalovirus (CMV) UL97 gene sequences in the blood and vitreous of patients with acquired immunodeficiency syndrome and CMV retinitis. J. Infect. Dis. 185:861-867. [DOI] [PubMed] [Google Scholar]
- 61.Hwang, Y. T., B. Y. Liu, D. M. Coen, and C. B. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ijichi, O., D. Michel, T. Mertens, K. Miyata, and Y. Eizuru. 2002. GCV resistance due to the mutation A594P in the cytomegalovirus protein UL97 is partially reconstituted by a second mutation at D605E. Antivir. Res. 53:135-142. [DOI] [PubMed] [Google Scholar]
- 63.Jabs, D. A., C. Enger, J. P. Dunn, and M. Forman. 1998. Cytomegalovirus retinitis and viral resistance: ganciclovir resistance. J. Infect. Dis. 177:770-773. [DOI] [PubMed] [Google Scholar]
- 64.Jabs, D. A., C. Enger, M. Forman, and J. P. Dunn. 1998. Incidence of foscarnet resistance and cidofovir resistance in patients treated for cytomegalovirus retinitis. Antimicrob. Agents Chemother. 42:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jabs, D. A., B. K. Martin, M. S. Forman, J. P. Dunn, J. L. Davis, D. V. Weinberg, K. K. Biron, and F. Baldanti. 2001. Mutations conferring ganciclovir resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 183:333-337. [DOI] [PubMed] [Google Scholar]
- 66.Jabs, D. A., B. K. Martin, M. S. Forman, J. P. Dunn, J. L. Davis, D. V. Weinberg, K. K. Biron, F. Baldanti, and H. Hu. 2001. Longitudinal observations on mutations conferring ganciclovir resistance in patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis: The Cytomegalovirus and Viral Resistance Study Group Report Number 8. Am. J. Ophthalmol. 132:700-710. [DOI] [PubMed] [Google Scholar]
- 67.Jacobson, M. A., H. Stanley, C. Holtzer, T. P. Margolis, and E. T. Cunningham. 2000. Natural history and outcome of new AIDS-related cytomegalovirus retinitis diagnosed in the era of highly active antiretroviral therapy. Clin. Infect. Dis. 30:231-233. [DOI] [PubMed] [Google Scholar]
- 68.Kariya, M., S. Mori, and Y. Eizuru. 2000. Comparison of human cytomegalovirus DNA polymerase activity for ganciclovir-resistant and -sensitive clinical strains. Antivir. Res. 45:115-122. [DOI] [PubMed] [Google Scholar]
- 69.Kesson, A., F. Zeng, A. Cunningham, and W. Rawlinson. 1998. The use of flow cytometry to detect antiviral resistance in human cytomegalovirus. J. Virol. Methods 71:177-186. [DOI] [PubMed] [Google Scholar]
- 70.Komazin, G., R. G. Ptak, B. T. Emmer, L. B. Townsend, and J. C. Drach. 2003. Resistance of human cytomegalovirus to the benzimidazole l-ribonucleoside maribavir maps to UL27. J. Virol. 77:11499-11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koszalka, G. W., N. W. Johnson, S. S. Good, L. Boyd, S. C. Chamberlain, L. B. Townsend, J. C. Drach, and K. K. Biron. 2002. Preclinical and toxicology studies of 1263W94, a potent and selective inhibitor of human cytomegalovirus replication. Antimicrob. Agents Chemother. 46:2373-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruger, R. M., W. D. Shannon, M. Q. Arens, J. P. Lynch, G. A. Storch, and E. P. Trulock. 1999. The impact of ganciclovir-resistant cytomegalovirus infection after lung transplantation. Transplantation 68:1272-1279. [DOI] [PubMed] [Google Scholar]
- 74.Kuhn, F. J., and C. W. Knopf. 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′-exonuclease domain. J. Biol. Chem. 271:29245-29254. [DOI] [PubMed] [Google Scholar]
- 75.Kuo, I. C., Y. Imai, C. Shum, D. F. Martin, B. D. Kuppermann, and T. P. Margolis. 2003. Genotypic analysis of cytomegalovirus retinitis poorly responsive to intravenous ganciclovir but responsive to the ganciclovir implant. Am. J. Ophthalmol. 135:20-25. [DOI] [PubMed] [Google Scholar]
- 76.Lalezari, J. P., J. A. Aberg, L. H. Wang, M. B. Wire, R. Miner, W. Snowden, C. L. Talarico, S. Shaw, M. A. Jacobson, and W. L. Drew. 2002. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 46:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Landry, M. L., S. Stanat, K. Biron, D. Brambilla, W. Britt, J. Jokela, S. Chou, W. L. Drew, A. Erice, B. Gilliam, N. Lurain, J. Manischewitz, R. Miner, M. Nokta, P. Reichelderfer, S. Spector, A. Weinberg, B. Yen-Lieberman, and C. Crumpacker. 2000. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob. Agents Chemother. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Larder, B. A., S. D. Kemp, and G. Darby. 1987. Related functional domains in virus DNA polymerases. EMBO J. 6:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li, W., F. Anwar, J. Jesurrun, and A. Erice. 1999. Cytomegalovirus UL97 and glycoprotein B (gB) sequences in tissues from immunocompromised patients with ganciclovir-resistant virus infection. Scand. J. Infect. Dis. 31:549-553. [DOI] [PubMed] [Google Scholar]
- 80.Limaye, A. P. 2002. Ganciclovir-resistant cytomegalovirus in organ transplant recipients. Clin. Infect. Dis. 35:866-872. [DOI] [PubMed] [Google Scholar]
- 81.Limaye, A. P., L. Corey, D. M. Koelle, C. L. Davis, and M. Boeckh. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645-649. [DOI] [PubMed] [Google Scholar]
- 82.Limaye, A. P., G. Raghu, D. M. Koelle, J. Ferrenberg, M. L. Huang, and M. Boeckh. 2002. High incidence of ganciclovir-resistant cytomegalovirus infection among lung transplant recipients receiving preemptive therapy. J. Infect. Dis. 185:20-27. [DOI] [PubMed] [Google Scholar]
- 83.Littler, E., A. D. Stuart, and M. S. Chee. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160-162. [DOI] [PubMed] [Google Scholar]
- 84.Liu, W., B. D. Kuppermann, D. F. Martin, R. A. Wolitz, and T. P. Margolis. 1998. Mutations in the cytomegalovirus UL97 gene associated with ganciclovir-resistant retinitis. J. Infect. Dis. 177:1176-1181. [DOI] [PubMed] [Google Scholar]
- 85.Ljungman, P. 1996. Cytomegalovirus infections in transplant patients. Scand. J. Infect. Dis. Suppl. 100:59-63. [PubMed] [Google Scholar]
- 86.Lowance, D., H. H. Neumayer, C. M. Legendre, J. P. Squifflet, J. Kovarik, P. J. Brennan, D. Norman, R. Mendez, M. R. Keating, G. L. Coggon, A. Crisp, and I. C. Lee. 1999. Valacyclovir for the prevention of cytomegalovirus disease after renal transplantation. N. Engl. J. Med. 340:1462-1470. [DOI] [PubMed] [Google Scholar]
- 87.Lurain, N. S., S. M. Bhorade, K. J. Pursell, R. K. Avery, V. V. Yeldandi, C. M. Isada, E. S. Robert, D. J. Kohn, M. Q. Arens, E. R. Garrity, A. J. Taege, M. G. Mullen, K. M. Todd, J. W. Bremer, and B. Yen-Lieberman. 2002. Analysis and characterization of antiviral drug-resistant cytomegalovirus isolates from solid organ transplant recipients. J. Infect. Dis. 186:760-768. [DOI] [PubMed] [Google Scholar]
- 88.Lurain, N. S., L. E. Spafford, and K. D. Thompson. 1994. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J. Virol. 68:4427-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lurain, N. S., K. D. Thompson, E. W. Holmes, and G. S. Read. 1992. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J. Virol. 66:7146-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lurain, N. S., A. Weinberg, C. S. Crumpacker, and S. Chou. 2001. Sequencing of cytomegalovirus UL97 gene for genotypic antiviral resistance testing. Antimicrob. Agents Chemother. 45:2775-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311:60-71. [DOI] [PubMed] [Google Scholar]
- 92.McSharry, J. J., A. McDonough, B. Olson, S. Hallenberger, J. Reefschlaeger, W. Bender, and G. L. Drusano. 2001. Susceptibilities of human cytomegalovirus clinical isolates to BAY38-4766, BAY43-9695, and ganciclovir. Antimicrob. Agents Chemother. 45:2925-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McSharry, J. M., N. S. Lurain, G. L. Drusano, A. Landay, J. Manischewitz, M. Nokta, M. O'Gorman, H. M. Shapiro, A. Weinberg, P. Reichelderfer, and C. Crumpacker. 1998. Flow cytometric determination of ganciclovir susceptibilities of human cytomegalovirus clinical isolates. J. Clin. Microbiol. 36:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Metzger, C., D. Michel, K. Schneider, A. Luske, H. J. Schlicht, and T. Mertens. 1994. Human cytomegalovirus UL97 kinase confers ganciclovir susceptibility to recombinant vaccinia virus. J. Virol. 68:8423-8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Michel, D., S. Hohn, T. Haller, D. Jun, and T. Mertens. 2001. Aciclovir selects for ganciclovir-cross-resistance of human cytomegalovirus in vitro that is only in part explained by known mutations in the UL97 protein. J. Med. Virol. 65:70-76. [PubMed] [Google Scholar]
- 96.Michel, D., and T. Mertens. 2004. The UL97 protein kinase of human cytomegalovirus and homologues in other herpesviruses: impact on virus and host. Biochim. Biophys. Acta 1697:169-180. [DOI] [PubMed] [Google Scholar]
- 97.Michel, D., I. Pavic, A. Zimmermann, E. Haupt, K. Wunderlich, M. Heuschmid, and T. Mertens. 1996. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J. Virol. 70:6340-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mousavi-Jazi, M., L. Schloss, W. L. Drew, A. Linde, R. C. Miner, J. Harmenberg, B. Wahren, and M. Brytting. 2001. Variations in the cytomegalovirus DNA polymerase and phosphotransferase genes in relation to foscarnet and ganciclovir sensitivity. J. Clin. Virol. 23:1-15. [DOI] [PubMed] [Google Scholar]
- 99.Mulamba, G. B., A. Hu, R. F. Azad, K. P. Anderson, and D. M. Coen. 1998. Human cytomegalovirus mutant with sequence-dependent resistance to the phosphorothioate oligonucleotide fomivirsen (ISIS 2922). Antimicrob. Agents Chemother. 42:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mylonakis, E., W. M. Kallas, and J. A. Fishman. 2002. Combination antiviral therapy for ganciclovir-resistant cytomegalovirus infection in solid-organ transplant recipients. Clin. Infect. Dis. 34:1337-1341. [DOI] [PubMed] [Google Scholar]
- 101.Nguyen, Q., R. Champlin, S. Giralt, K. Rolston, I. Raad, K. Jacobson, C. Ippoliti, D. Hecht, J. Tarrand, M. Luna, and E. Whimbey. 1999. Late cytomegalovirus pneumonia in adult allogeneic blood and marrow transplant recipients. Clin. Infect. Dis. 28:618-623. [DOI] [PubMed] [Google Scholar]
- 102.Nichols, W., and M. Boeckh. 2001. Cytomegalovirus infections. Curr. Treat. Options Infect. Dis. 3:78-91. [Google Scholar]
- 103.Nichols, W. G., L. Corey, T. Gooley, W. L. Drew, R. Miner, M. Huang, C. Davis, and M. Boeckh. 2001. Rising pp65 antigenemia during preemptive anticytomegalovirus therapy after allogeneic hematopoietic stem cell transplantation: risk factors, correlation with DNA load, and outcomes. Blood 97:867-874. [DOI] [PubMed] [Google Scholar]
- 104.O'Sullivan, C. E., W. L. Drew, D. J. McMullen, R. Miner, J. Y. Lee, R. A. Kaslow, J. G. Lazar, and M. S. Saag. 1999. Decrease of cytomegalovirus replication in human immunodeficiency virus infected-patients after treatment with highly active antiretroviral therapy. J. Infect. Dis. 180:847-849. [DOI] [PubMed] [Google Scholar]
- 105.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 106.Paya, C. V., J. A. Wilson, M. J. Espy, I. G. Sia, M. J. Debernardi, T. F. Smith, R. Patel, G. Jenkins, W. S. Harmsen, D. J. Vanness, and R. H. Wiesner. 2002. Preemptive use of oral ganciclovir to prevent cytomegalovirus infection in liver transplant patients: a randomized, placebo-controlled trial. J. Infect. Dis. 185:854-860. [DOI] [PubMed] [Google Scholar]
- 107.Pescovitz, M. D., J. Rabkin, R. M. Merion, C. V. Paya, J. Pirsch, R. B. Freeman, J. O'Grady, C. Robinson, Z. To, K. Wren, L. Banken, W. Buhles, and F. Brown. 2000. Valganciclovir results in improved oral absorption of ganciclovir in liver transplant recipients. Antimicrob. Agents Chemother. 44:2811-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Razonable, R. R., and C. V. Paya. 2002. β-Herpesviruses in transplantation. Rev. Med. Microbiol. 13:163-176. [Google Scholar]
- 110.Razonable, R. R., A. Rivero, A. Rodriguez, J. Wilson, J. Daniels, G. Jenkins, T. Larson, W. C. Hellinger, J. R. Spivey, and C. V. Paya. 2001. Allograft rejection predicts the occurrence of late-onset cytomegalovirus (CMV) disease among CMV-mismatched solid organ transplant patients receiving prophylaxis with oral ganciclovir. J. Infect. Dis. 184:1461-1464. [DOI] [PubMed] [Google Scholar]
- 111.Reefschlaeger, J., W. Bender, S. Hallenberger, O. Weber, P. Eckenberg, S. Goldmann, M. Haerter, I. Buerger, J. Trappe, J. A. Herrington, D. Haebich, and H. Ruebsamen-Waigmann. 2001. Novel non-nucleoside inhibitors of cytomegaloviruses (BAY 38-4766): in vitro and in vivo antiviral activity and mechanism of action. J. Antimicrob. Chemother. 48:757-767. [DOI] [PubMed] [Google Scholar]
- 112.Scott, G. M., M. A. Isaacs, F. Zeng, A. M. Kesson, and W. D. Rawlinson. 2004. Cytomegalovirus antiviral resistance associated with treatment induced UL97 (protein kinase) and UL54 (DNA polymerase) mutations. J. Med. Virol. 74:85-93. [DOI] [PubMed] [Google Scholar]
- 113.Seo, S. K., A. Regan, T. Cihlar, D. C. Lin, F. Boulad, D. George, V. K. Prasad, T. E. Kiehn, and B. Polsky. 2001. Cytomegalovirus ventriculoencephalitis in a bone marrow transplant recipient receiving antiviral maintenance: clinical and molecular evidence of drug resistance. Clin. Infect. Dis. 33:e105-e108. [DOI] [PubMed] [Google Scholar]
- 114.Sia, I. G., and R. Patel. 2000. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin. Microbiol. Rev. 13:83-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Simon, M., L. Giot, and G. Faye. 1991. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 10:2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith, I. L., J. M. Cherrington, R. E. Jiles, M. D. Fuller, W. R. Freeman, and S. A. Spector. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69-77. [DOI] [PubMed] [Google Scholar]
- 117.Smith, I. L., M. Shinkai, W. R. Freeman, and S. A. Spector. 1996. Polyradiculopathy associated with ganciclovir-resistant cytomegalovirus in an AIDS patient: phenotypic and genotypic characterization of sequential virus isolates. J. Infect. Dis. 173:1481-1484. [DOI] [PubMed] [Google Scholar]
- 118.Smith, I. L., I. Taskintuna, F. M. Rahhal, H. C. Powell, E. Ai, A. J. Mueller, S. A. Spector, and W. R. Freeman. 1998. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch. Ophthalmol. 116:178-185. [DOI] [PubMed] [Google Scholar]
- 119.Stanat, S. C., J. E. Reardon, A. Erice, M. C. Jordan, W. L. Drew, and K. K. Biron. 1991. Ganciclovir-resistant cytomegalovirus clinical isolates: mode of resistance to ganciclovir. Antimicrob. Agents Chemother. 35:2191-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Studies of Ocular Complications of AIDS Research Group and AIDS Clinical Trials Group. 1996. Combination foscarnet and ganciclovir therapy vs monotherapy for the treatment of relapsed cytomegalovirus retinitis in patients with AIDS. Arch. Ophthalmol. 114:23-33. [DOI] [PubMed] [Google Scholar]
- 121.Sullivan, V., K. K. Biron, C. Talarico, S. C. Stanat, M. Davis, L. M. Pozzi, and D. M. Coen. 1993. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob. Agents Chemother. 37:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162-164. [DOI] [PubMed] [Google Scholar]
- 123.Talarico, C. L., T. C. Burnette, W. H. Miller, S. L. Smith, M. G. Davis, S. C. Stanat, T. I. Ng, Z. He, D. M. Coen, B. Roizman, and K. K. Biron. 1999. Acyclovir is phosphorylated by the human cytomegalovirus UL97 protein. Antimicrob. Agents Chemother. 43:1941-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tatarowicz, W. A., N. S. Lurain, and K. D. Thompson. 1992. A ganciclovir-resistant clinical isolate of human cytomegalovirus exhibiting cross-resistance to other DNA polymerase inhibitors. J. Infect. Dis. 166:904-907. [DOI] [PubMed] [Google Scholar]
- 125.Tatarowicz, W. A., N. S. Lurain, and K. D. Thompson. 1991. In situ ELISA for the evaluation of antiviral compounds effective against human cytomegalovirus. J. Virol. Methods 35:207-215. [DOI] [PubMed] [Google Scholar]
- 126.Telenti, A., and T. F. Smith. 1989. Screening with a shell vial assay for antiviral activity against cytomegalovirus. Diagn. Microbiol. Infect. Dis. 12:5-8. [DOI] [PubMed] [Google Scholar]
- 127.Underwood, M. R., R. G. Ferris, D. W. Selleseth, M. G. Davis, J. C. Drach, L. B. Townsend, K. K. Biron, and F. L. Boyd. 2004. Mechanism of action of the ribopyranoside benzimidazole GW275175X against human cytomegalovirus. Antimicrob. Agents Chemother. 48:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang, L. H., R. W. Peck, Y. Yin, J. Allanson, R. Wiggs, and M. B. Wire. 2003. Phase I safety and pharmacokinetic trials of 1263W94, a novel oral anti-human cytomegalovirus agent, in healthy and human immunodeficiency virus-infected subjects. Antimicrob. Agents Chemother. 47:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weinberg, A., D. A. Jabs, S. Chou, B. K. Martin, N. S. Lurain, M. S. Forman, and C. Crumpacker. 2003. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 187:777-784. [DOI] [PubMed] [Google Scholar]
- 130.Wolf, D., A. Honigman, J. Lazarovits, E. Tavor, and A. Panet. 1998. Characterization of the human cytomegalovirus UL97 gene product as a virion-associated protein kinase. Arch. Virol. 143:1223-1232. [DOI] [PubMed] [Google Scholar]
- 131.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wolf, D. G., D. J. Lee, and S. A. Spector. 1995. Detection of human cytomegalovirus mutations associated with ganciclovir resistance in cerebrospinal fluid of AIDS patients with central nervous system disease. Antimicrob. Agents Chemother. 39:2552-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wolf, D. G., N. S. Lurain, T. Zuckerman, R. Hoffman, J. Satinger, A. Honigman, N. Saleh, E. S. Robert, J. M. Rowe, and Z. Kra-Oz. 2003. Emergence of late cytomegalovirus central nervous system disease in hematopoietic stem cell transplant recipients. Blood 101:463-465. [DOI] [PubMed] [Google Scholar]
- 134.Wolf, D. G., I. L. Smith, D. J. Lee, W. R. Freeman, M. Flores Aguilar, and S. A. Spector. 1995. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J. Clin. Investig. 95:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wolf, D. G., I. Yaniv, A. Honigman, I. Kassis, T. Schonfeld, and S. Ashkenazi. 1998. Early emergence of ganciclovir-resistant human cytomegalovirus strains in children with primary combined immunodeficiency. J. Infect. Dis. 178:535-538. [DOI] [PubMed] [Google Scholar]
- 136.Ye, L. B., and E. S. Huang. 1993. In vitro expression of the human cytomegalovirus DNA polymerase gene: effects of sequence alterations on enzyme activity. J. Virol. 67:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zaia, J. A., G. M. Gallezhawkins, B. R. Tegtmeier, A. Terveer, X. L. Li, J. C. Niland, and S. J. Forman. 1997. Late cytomegalovirus disease in marrow transplantation is predicted by virus load in plasma. J. Infect. Dis. 176:782-785. [DOI] [PubMed] [Google Scholar]
- 138.Zhang, J., D. W. Chung, C. K. Tan, K. M. Downey, E. W. Davie, and A. G. So. 1991. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry 30:11742-11750. [DOI] [PubMed] [Google Scholar]
- 139.Zimmermann, A., D. Michel, I. Pavic, W. Hampl, A. Luske, J. Neyts, E. De Clercq, and T. Mertens. 1997. Phosphorylation of aciclovir, ganciclovir, penciclovir and S2242 by the cytomegalovirus UL97 protein: a quantitative analysis using recombinant vaccinia viruses. Antivir. Res. 36:35-42. [DOI] [PubMed] [Google Scholar]