Abstract
Human Pinx1 protein, associated with shelterin proteins, is widely revealed as a haploinsufficient tumor suppressor. Growing evidence has manifested the deregulation of PinX1 in distinct cancers. Nonetheless, the loss status of PinX1 and its diagnostic, prognostic and clinicopathological significance in Basal-like breast cancer are still unclear. In the present study, the PinX1 expression levels of breast cancer tissues were investigated by qRT-PCR and immunoblotting assays. Then immunohistochemistry (IHC) was performed to detect PinX1 expression on a tissue microarray. The optimal threshold for PinX1 positivity was determined by receiver operating characteristic (ROC) curve analysis. To clarify the probable role of PinX1 in BLBC, the PinX1 knockout and stably over-expressed MDA-MB-231 cell lines were constructed by the CRISPR-Cas9 system and gene transfection. The association of PinX1 expression with cell proliferation, migration and apoptosis of MDA-MB-231 cells were observed by CCK-8 assay, wound healing assay, Transwell assay, flow cytometric analysis and immunoblotting of the cleaved caspase-3 protein level. Our results showed that both PinX1 mRNA and protein expression were downregulated in breast cancer tissues (P<0.05). In IHC analysis, the optimal cut-off parameter for PinX1 positive expression was 62.5% (the AUC was 0.749, P<0.01). PinX1 positivity was 76.9% (10/14) in luminal subtypes, 50% (5/10) in Her2-enriched breast cancer and 27.3% (9/33) in basal-like subtypes. Besides, in 59 invasive ductal breast carcinomas, PinX1 expression was inversely related to histology grade (P<0.05) while it was positively associated with PR status (P<0.05) and ER status (P<0.05). These results indicated that low expression of PinX1 correlated with aggressive clinicopathological significance of breast cancer, especially in the basal-like subtype. Besides, we identified that overexpression of PinX1 inhibited the proliferation rates and migration ability and increased the apoptosis rates of BLBC. Our findings demonstrated that low expression of PinX1 was associated with malignant behaviors in basal-like subtype of breast cancer. PinX1 is likely a feasible biomarker and molecular target of BLBC.
Keywords: PinX1, basal-like breast cancer, malignant behavior, biomarker, molecular target
Introduction
Breast cancer is the the most common malignancy in women worldwide with the highest mortality rate of 12.9 per 100,000 population in both the developed and developing areas (1,2). Based on the expression status of ER, PR, HER2 receptor, p53 and Ki-67, the St. Gallen Consensus listed four broad categories of breast cancer: basal-like breast cancer, the Her2-enriched, luminal A and luminal B subtypes (3). These molecular subtypes markedly influence the clinical treatment strategies, response to therapies, distribution of risk factors and the patients prognosis (4,5).
Triple negative breast cancer (TNBC) refers to the breast cancer that has no expression of ER, PR nor HER2 receptor, which makes up ~15–20% of all the breast cancers with a disproportionate share of mortality (6). As previous studies depicted, a significant part (>90%) of basal-like breast cancers were triple-negative subtypes and 55–81% of TNBCs were BLBCs (7). BLBC is very frequently associated with TNBC. Prognosis of BLBC and TNBC is the poorest among all the subtypes (8). Owing to aggressive clinical courses, higher possibility of distant recurrence and significant shorter survivals (9), TNBC and BLBC are challenging public health concerns. With the advance of biology technologies especially the next generation sequencing, it is obvious that cellular microenvironment, gene mutations and patients innate hallmarks have great influence on disease pathophysiology, treatment sensitivity and recurrence (10). However, the molecular mechanism of breast cancer remains to be elaborated.
Currently, diagnostic and therapeutic strategies for breast cancer are emerging prominently including surgery, neo-adjuvant chemotherapy, endocrine therapy, radiotherapy and molecular targeted therapy (11). Without the molecular targets (ER, PR and Her2) of therapeutic agents for breast cancer, TNBC is relatively sensitive to cytotoxic chemotherapy. Besides, chemotherapy is always combined with neo-adjuvant, adjuvant and metastatic setting (7). However, after the chemotherapy, residual tumor cells mostly survive and provoke recurrent tumor growth (12), which contributes to high recurrence risk and mortality. Yet, there are obstacles and limitations. A targeted therapy relies on a given tumor marker from a host of randomized clinical trails. Besides, it is suitable to the specific population not specific individual patient. Rodler et al (13) revealed that veliparib (ABT-888) combined with cisplatin and vinorelbine possesses antineoplastic activity in TNBC, especially the BRCA mutation-associated TNBC. Growing clinical and biological features are proposed to combine the better predictive carcinoma characteristics and behavior to obtain a more individualized treatment strategy. Concomitantly, new tumor marker development is essential for clinical practice, disease subtyping, predictive diagnosis and prognosis. It is certainly worth screening and validating reliable predictive biomarkers for breast cancer, particularly the TNBC and basal-like subtypes.
Accumulating evidence demonstrates that telomerase plays a pivotal role in maintaining telomere homeostasis and cell immortalization and that telomerase-targeted cancer therapy seems to be effective with less adverse side-effects (14). Pin2/TRF1-interacting telomerase inhibitor 1 (Pinx1) is a haploinsufficient tumor suppressor localized at human chromosome 8p23, the most common deletion region correlated to tumor differentiation and tumorigenicity (15). Tian et al (16) recently showed cervical squamous cell carcinoma patients with PinX1 expression are more sensitive to paclitaxel chemotherapy and PinX1 could be a novel biomarker for CSCC patients who might benefit from paclitaxel. Liu et al (17) reported that PinX1 suppressed the proliferation of bladder urothelial carcinoma cells via inhibiting activity of telomerase. Also, Li et al (18) addressed the function of PinX1 in renal cell carcinomas (RCC) that PinX1 depressed the migration and invasion of RCC. However, there exist few studies in terms of the association between PinX1 expression and its clinical characteristics. Moreover, the concrete molecular mechanism of PinX1 contributing to breast cancer remains poorly understood.
In the present study, we identified that PinX1 was downregulated in breast cancer tissues compared with the adjacent counterparts, especially in the basal-like subtype. Overexpression of PinX1 inhibited the proliferation rates and migration ability, while increased the apoptosis rates of human basal-like breast cancer. Thus, PinX1 is likely a feasible biomarker and a molecular target of basal-like breast cancer.
Materials and methods
Patients and tissue specimens
In the analysis of PinX1 expression by qRT-PCR and western blotting, 26 fresh breast cancer tissues and the paired non-tumor tissue samples were selected from breast cancer patients without preoperative treatment who had undergone surgical intervention during February 2013 to July 2015. The tissue specimens for TMA construction and IHC analysis were previously described (19). All the tissues above were obtained with written informed consent of patients. The Institute Research Medical Ethics Committee of the Nanfang Hospital approved the utilization of these breast cancer samples for research purpose only.
Cell cultures and plasmids
Breast cancer cell line MDA-MB-231 was obtained from laboratory preservation. It was cultured in Dulbeccos modified Eagles medium (DMEM; Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37°C and 5% CO2. E. coli DH5α (Invitrogen, Carlsbad, CA, USA) was maintained in flasks with LB medium. The plasmid pSpCas9(BB)-2A-GFP (PX458) for gene knockout was aquired from Feng Zhang Lab (#48138; Addgene, Cambridge, MA, USA). pCDNA3.1(+) (Invitrogen) and pEGFP-N1 vectors (Clontech Laboratories, Inc., Mountain View, CA, USA) were used for gene amplification in mammalian cells.
Overexpression and knockout of PinX1 in MDA-MB-231 cells
For PinX1-overexpressed stable cells, the PinX1 ORF was inserted into pEGFP-N1 vector. Then MDA-MB-231 cells were transfected separately with the recombined and empty vectors by Lipofectamine 2000 (Invitrogen). After 24 h, cells were reseeded at 1:4 dilution with a new selective cell culture containing 1300 µg/ml G418 (Sigma-Aldrich, St. Louis, MO, USA) and the selective culture was renewed every 2 days. After 2–3 weeks, the cell clones were obtained by an inverted microscope (Olympus IX71; Olympus, Tokyo, Japan) and grown in maintaining culture with 300 µg/ml of G418 for later experiments. For the knockout of PinX1, three sgRNAs were designed according to the Feng Zhang Lab at http://crispr.mit.edu/ and the sequences were as follows: sgRNA-1, forward, caccgccatcttctctagcatccgc and reverse, aaacgcggatgctagag aagatggc; sgRNA-2, forward, caccgtgatggtagctccgagtccc and reverse, aaacgggactcggagctaccatcac; sgRNA-3, forward, cac cgccgaactgaacacttgccat and reverse, aaacatggcaagtgttcagttc ggc. In addition, sgRNA-3 was selected for its better knockout effects. sgRNA-3 was then cloned into PX458 and MDA-MB-231 cells were transfected with the recombined vector by Lipofectamine 2000. Cells were reseeded into 96-well plates at a density of a single cell/well after 24-h transfection by flow cytometry (BD Biosciences, San Jose, CA, USA). Cell clones were harvested in 2–3 weeks and detected by western blot analysis later to obtain the knockout cells.
RNA extraction and qRT-PCR assays
The PinX1 expression level of 26 pairs of breast malignancy tissues and the normal counterparts were investigated via qRT-PCR. Total RNA of these samples were isolated by RNAiso Plus reagent (Takara Bio, Dalian, China) and dissolved in DEPC-treated water. The RNA concentration and purity were tested by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The denaturing agarose gel electrophoresis was then used to qualify the integrity of total RNA. The qualified RNA were subsquently reverse-transcribed by PrimeScript® RT reagent kit (Takara Bio) depending on the manufacturers instructions. SYBR® Premix Ex Taq™ II (Tli RNase H Plus) kit (Takara Bio) was used for the qRT-PCR on the ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with holding stage of 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 34 sec, followed by the melt curve stage. The forward PinX1 primer was 5-ccagagg agaacgaaaccacg-3 and the reverse was 5-acctgcgtctcagaaa tgtca-3. As an internal reference gene, the forward primer of GAPDH was 5-ctgggctacactgagcacc-3 and the reverse was 5-aagtggtcgttgagggcaatg-3. The relative expression levels of PinX1 was determined by the comparative Ct (ΔΔct) method (20) and the ΔCt was calculated as ΔCt = Ctgene - Ctreference. Each experiment was tested at least in three independent biological repeats.
Immunoblot analysis
The extracted total proteins of breast cancer tissues and the paired normal samples were lysed in the enhanced Radio Immunoprecipitation Assay (RIPA) buffer mixed with 1 mM PMSF (Beyotime Institute of Biotechnology, Haimen, China). Protein concentrations were measured by BCA protein assay kit (Beyotime Institute of Biotechnology). Then 40 µg of each protein specimen was separated by 10% SDS-PAGE. The separated proteins on the gel were transferred onto polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA) by a wet transfer apparatus (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The blots were then blocked by 5% non-fat dry milk dissolved in Tris-buffered saline with Tween-20 (TBST) at room temperature for 1 h. Thereafter, the membranes were, respectively, incubated with primary antibodies diluted by TBST for PinX1 (1:1,000, goat anti-human polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and GAPDH (1:10,000, anti-human, conjugated with HRP; Proteintech Group, Inc., Rosemont, IL, USA) at 4°C overnignt. After washing with TBST for three times, the membranes for PinX1 detection were incubated with rabbit anti-goat IgG conjugated to HRP (1:10,000; Multi Sciences Biotech, Co., Ltd., Hangzhou, China) for 1 h at room temperature. Following 3 washes with TBST, these membranes were visualized by Immobilon Western Chemiluminescent HRP Substrate (Millipore) and exposed to X-ray film. Proteins were quantified by Adobe Photoshop and GAPDH was a loading control. The transfected MDA-MB-231 cells were also lysed in RIPA. The primary antibody for cleaved caspase-3 (1:1000, rabbit anti-human polyclonal; Cell Signaling Technology, Danvers, MA, USA) and the goat anti-rabbit IgG conjugated to HRP were also used in the present study.
Construction of TMA and IHC
In the present study, the construction of TMA in general was referred to standard protocols as previously described (19). The qualified TMA slides were dewaxed by xylene and then rehydrated in distinct gradient of alcohol; 3% hydrogen peroxide was used to compromise the endogenous peroxidase activity of the tissue microarrays for 20 min at room temperature. Thereafter, the slides were immersed in a boiled citrate buffer (50 mM, pH 6.0) for 10 min. After three washes of 0.01 M phosphate-buffered saline (PBS) (pH 7.4). The slides were then blocked by 5% normal goat serum for 30 min, followed with incubation of primary antibody for PinX1 (1:200, goat polyclonal; Santa Cruz Biotechnology) at 4°C overnight and secondary antibody labeled with polymer peroxidase (ZSGB-Bio, Beijing, China). Furthermore, the slides were treated with DAB Horseradish Peroxidase Color Development kit (Beyotime Institute of Biotechnology) followed by hematoxylin counterstaining. In this study, the negative controls were obtained by using PBS instead of anti-PinX1 antibody.
Immunohistochemistry evaluation
Each IHC slide for PinX1 positivity was assessed by staining intensity and the proportion of positive carcinoma cells across the total carcinoma cells semi-quantitatively. Three pathologists who were blinded to the patient clinical data evaluated the slides independently according to the percentage of positive tumor cells. In case of disagreement, the assessments were rescored until consensus was reached. The scores were distributed by 5% increments (0, 5, 10…100%).
Selection of optimal cut-off value
ROC analysis was applied to obtain the PinX1 positivity threshold according to the 0, 1-criterion. The clinicopathological parameters were classified into the following bivariate variables: age (≤50 and >50), clinical stage (I–II and III), pT stage (T1-T2 and T3-T4), pN stage (N0 and N1-N2), histology grade (I and II–III), PR status (positive and negative), ER status (positive and negative), Her2 status (positive and negative) and p53 status (positive and negative). Based on the PinX1 scores and the dichotomized clinicopathological features, the sensitivity and specificity for each test outcome was plotted to generate a ROC curve. The inherent validity of each diagnostic test was measured by the area under the curve (AUC) which combined sensitivity and specificity. As an optimal cut-off value, the PinX1 score was close to the point (0.0 and 0.1) and maximized the sensitivity and specificity. Consequently, tumors were identified as positive with a score above the cut-off threshold while it was negative with an score below the threshold.
CCK-8 cell proliferation assay
The cell viability of MDA-MB-231 cells with different treatments was measured by the Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). A total of 2×103 cells/well were plated into 96-well culture plate and incubated at 37°C for 24, 48 and 72 h, respectively. At each specific time-point, the old medium was replaced by 100 µl fresh medium containing 10 µl CCK-8 solution and cells were incubated for another 2 h. Values of OD450nm were measured by Multiskan FC (Thermo Fisher Scientific).
Cell migration assay
For the wound healing assay, cells with different treatments were plated in 6-well plates at a density of 5×105/well with a monolayer confluence overnight. Then, 10 µl pipette tips were used to make wounds on the single layers. The floated cells were washed away with 1X PBS three times, and cells were incubated at 37°C after adding fresh DMEM without serum. Then images of the wounds at the indicated periods (0, 12, 24 and 36 h) were taken. The 24-well Transwell chambers (BD Biosciences) was used to assess the migration ability of MDA-MB-231 cells. A total of 100 µl 5×104 cells/well was seeded in the upper chamber with serum-free DMEM. In addition, 500 µl DMEM containing 20% FBS was added to the lower chamber. After 24-h incubation, cells were fixed by 100% methanol for 20 min and dyed by 0.1% crystal violate (Amresco, Solon, OH, USA) for 30 min. The migration rate was analyzed after the upper cells were removed.
Apoptotic rate analysis by flow cytometry
MDA-MB-231 cells (2.5×105/well) were plated in 12-well plates overnight before transfected with pCDNA3.1(+) and PinX1-pCDNA3.1(+) by Lipofectamine 2000. After 48-h incubation, cells were harvested for apoptosis analysis by flow cytometry (BD FACSCalibur). For the PinX1 knockout and the wild-type cells, appropriate cells were plated in 12-well plates as well; 40 mM of etoposide was added to cells 24 h later. After 24 h of incubation, cells were harvested and washed by 1X PBS twice. Then cells were resuspended by 500 µl binding buffer, stained by Annexin V-FITC and propidium iodide (PI) according to the Annexin V-FITC apoptosis detection kit (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) and detected by flow cytometry.
Statistical analyses
Statistics shown were analyzed by IBM SPSS Statistics 20.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0. The differences of PinX1 expression between the paired tissues were tested by the Student's paired t-test. The correlation of PinX1 positivity with the clinicopathological parameters was evaluated by the Chi-squared test and the Fishers exact test. With two-tailed tests, P<0.01 or P<0.05 was considered to be statistically significant. The data expressed are mean ± SEM.
Results
The expression of PinX1 mRNA and protein declines substantially in human breast cancer tissues
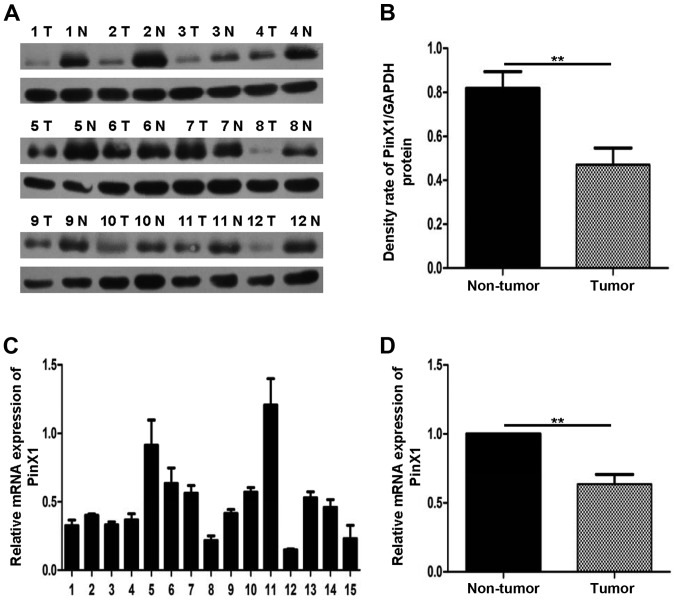
To explore the role of PinX1 in tumorigenesis, we detected the PinX1 mRNA levels of breast cancer and the paired adjacent normal breast tissues. The quantitative real-time PCR (qRT-PCR) assay showed that PinX1 mRNA expression was decreased in 21 out of 26 sample pairs and the fold-changes of the 21 sample pairs were <1. The qRT-PCR result of 15 sample pairs is shown in Fig. 1A. In comparison with the paired adjacent non-tumor breast tissues, the expression of PinX1 in breast cancer tissues was reduced significantly by 0.362 (Fig. 1B; P<0.05).
Figure 1.
PinX1 expression was observed in 26 pairs of breast cancer tissues and the normal counterparts. (A and B) Protein levels of PinX1, as assessed by immunoblot analysis with an anti-PinX1 antibody are downregulated in a majority of cancer tissues compared with the paired normal breast tissues (23/26). Separately, (B) showed significant differences (**P<0.01, two-tailed paired t-test) and (A) presented 12 pairs of the 26 pairs. GAPDH was a loading control; T, tumor; N, non-tumor. (C and D) Quantitative PCR analysis of PinX1 mRNA expression in 26 paired samples. Data are shown relative to those of the internal control gene GAPDH. (D) Significant differences (**P<0.01, two-tailed paired t-test) of PinX1 expression, (C) representing 15 pairs of the total.
Similarly, western blot analysis further confirmed the lower PinX1 expression between 26 paired normal breast tissues and the adjacent counterpart tumor (Fig. 1C, 12 pairs of the total 26 pairs). The densitometry quantification results indicated that PinX1 expression was downregulated in breast cancer tissues and the difference between the two paired groups were of favorably statistical significance (Fig. 1D; P<0.05).
IHC analysis of PinX1 expression level in breast cancer tissues
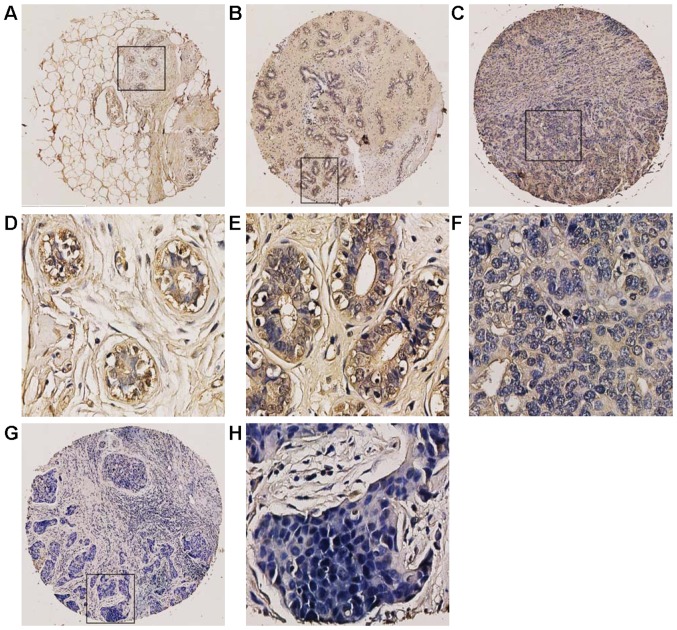
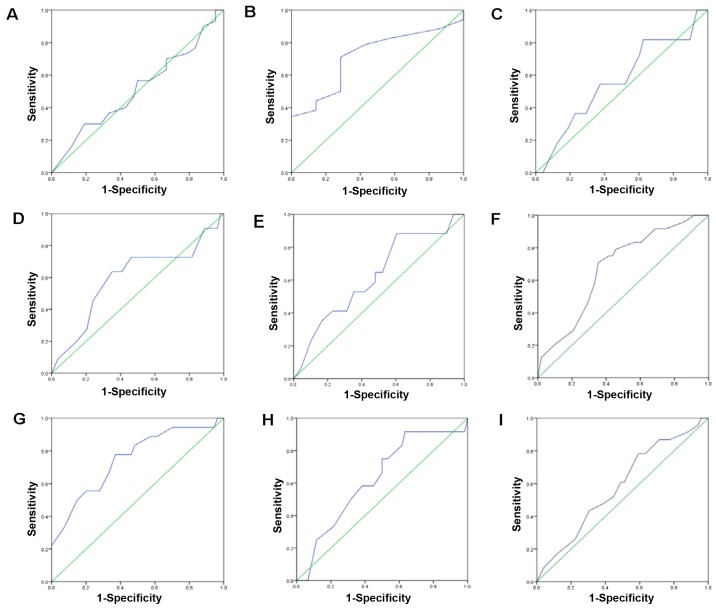
We also used IHC to investigate the PinX1 expression in breast tissues. We found that PinX1 expression varied from breast cancer to normal breast tissues (Fig. 2). The ROC was used to obtain the optimal cut-off value, a threshold for positive and negative PinX1 expression. As is shown in Fig. 3, the ROC curve for PR status is the closest to (0.0 and 0.1) with a maximized sensitivity and specificity. In addition, the optimal cut-off threshold to separate the positive from negative expression of PinX1 was determined as 62.5%. Based on the cut-off value, the association between PinX1 protein expression and the different subtypes of breast cancer was elucidated. The positive expression percentage of PinX1 was 76.9% (10/14) in Luminal A and Luminal B subtypes, 50% (5/10) in Her2-overexpressed and 27.3% (9/33) in basal-like (Table I) subtypes. In the malignant breast carcinomas, PinX1 expression tended to be negative. The results indicated that PinX1 expression was possibly correlated with breast cancer subtypes (P<0.05).
Figure 2.
Immunohistochemical staining of PinX1 was analysed in breast cancer tissues and the normal breast tissues. (A) A cancer adjacent normal breast tissue (case 1) with high expression of PinX1, where almost all tumor cells were positively stained (magnification, ×100). (B) A fibroadenoma tissue (case 3) with >65% of tumor cells positive staining of PinX1 (magnification, ×100). (C) An invasive ductal carcinoma (case 43) exhibited low expression of PinX1, where <45% of tumor cells were positive staining of PinX1 (magnification, ×100). (G) Low expression of PinX1 was obtained in an invasive ductal carcinoma (case 29), where <10% of tumor cells were positive staining of PinX1 (magnification, ×100). Respectively, (D-H) revealed the higher magnification (magnification, ×400) of the specific areas boxed in (A, B, C and G).
Figure 3.
The ROC curve was used to obtain the optimal cut-off value for positive and negative PinX1 expression. The plots of sensitivity and specificity for each clinicopathological parameter was as follows: (A) age at surgery; (B) histology grade; (C) clinical stage; (D) pT status; (E) pN status; (F) ER status; (G) PR status; (H) Her2 status; (I) p53 status.
Table I.
Association of PinX1 expression with subtypes of breast cancer.
| Subtypes | Cases | Negative (%) | Positive (%) | P-valuea |
|---|---|---|---|---|
| Luminal A/B | 13 | 3 (23.1) | 10 (76.9) | 0.008 |
| HER2+ | 10 | 5 (50.0) | 5 (50.0) | |
| Basal like | 33 | 24 (72.7) | 9 (27.3) |
Chi-square test.
Expression of PinX1 associated with clinicopathological parameters of breast cancer
The association between PinX1 expression in 59 invasion ductal breast carcinomas and their clinical pathological characteristics are listed in Table II. The summary demonstrated that decreased PinX1 expression was correlated with the histology grade of breast cancer (P<0.05). Besides, PinX1 expression was positively associated with PR status (P<0.05) and ER status (P<0.05). However, there were no significant relationship between PinX1 expression and other clinicopathological parameters including age at surgery, clinical stage, TNM stage, Her2 status and p53 status (P>0.05).
Table II.
Association of PinX1 expression with clinicopathological parameters in 59 invasive ductal carcinoma patients.
| PinX1 staining | ||||
|---|---|---|---|---|
| Characteristics | Low (%) | High (%) | Total | P-valueb |
| Age at surgery (years)a | ||||
| ≤50 | 19 (61.3) | 12 (38.7) | 31 | 0.746 |
| >50 | 16 (57.1) | 12 (42.9) | 28 | |
| Histology grade | ||||
| I | 0 (0) | 4 (100) | 4 | 0.028c |
| II–III | 34 (61.8) | 21 (38.2) | 55 | |
| Clinical stage | ||||
| I–II | 30 (62.5) | 18 (37.5) | 48 | 0.485 |
| III | 5 (45.5) | 6 (54.5) | 11 | |
| pT status | ||||
| T1-T2 | 28 (59.6) | 19 (40.4) | 47 | 1.000 |
| T3-T4 | 47 (58.3) | 5 (41.7) | 12 | |
| pN status | ||||
| N0 | 27 (64.3) | 15 (35.7) | 42 | 0.222 |
| N1-N2 | 8 (47.1) | 9 (52.9) | 17 | |
| ER status | ||||
| Negative | 29 (72.5) | 11 (27.5) | 40 | 0.003 |
| Positive | 6 (31.6) | 13 (68.4) | 19 | |
| PR status | ||||
| Negative | 32 (66.7) | 16 (33.3) | 48 | 0.040 |
| Positive | 3 (27.3) | 8 (72.7) | 11 | |
| HER2 status | ||||
| Negative | 30 (63.8) | 17 (37.2) | 47 | 0.287 |
| Positive | 5 (41.7) | 7 (58.3) | 12 | |
| p53 status | ||||
| Negative | 10 (55.6) | 8 (45.4) | 18 | 0.696 |
| Positive | 25 (61.0) | 16 (39.0) | 41 | |
Mean age
Chi-square test except c
Fishers exact test.
Then analyses of the positive staining of PinX1 were stratified by different breast cancer subtypes in the breast cancer tissues. Among all the 33 basal-like/triple-negative breast cancer cases, we found that the mean positive staining was 81.1% and the mean negative staining was 28.1%. Significantly, PinX1 was downregulated in basal-like/triple-negative breast cancer (P<0.01), which indicated that PinX1 may be correlated with the development and progression of basal-like breast cancer.
PinX1 attenuates proliferation in a human basal-like breast cancer cell line
To clarify the probable role of PinX1 in basal-like breast cancer, we constructed the MDA-MB-231 cell lines that stably overexpressed PinX1 (PinX1-pEGFP-N1) and the negative control (pEGFP-N1). We adopted CRISPR-Cas9 system (21) to knock out the PinX1 gene of MDA-MB-231 cells. PinX1 expression levels were detected by immunoblot analysis (Fig. 4A and B). Then Cell Counting kit-8 (CCK- 8) assay was employed to investigate the impact of PinX1 on cell proliferation rates. The results showed that overexpression of PinX1 significantly inhibited the proliferation of MDA-MB-231 cells while PinX1 knockout promoted cell proliferation (Fig. 4C and D).
Figure 4.
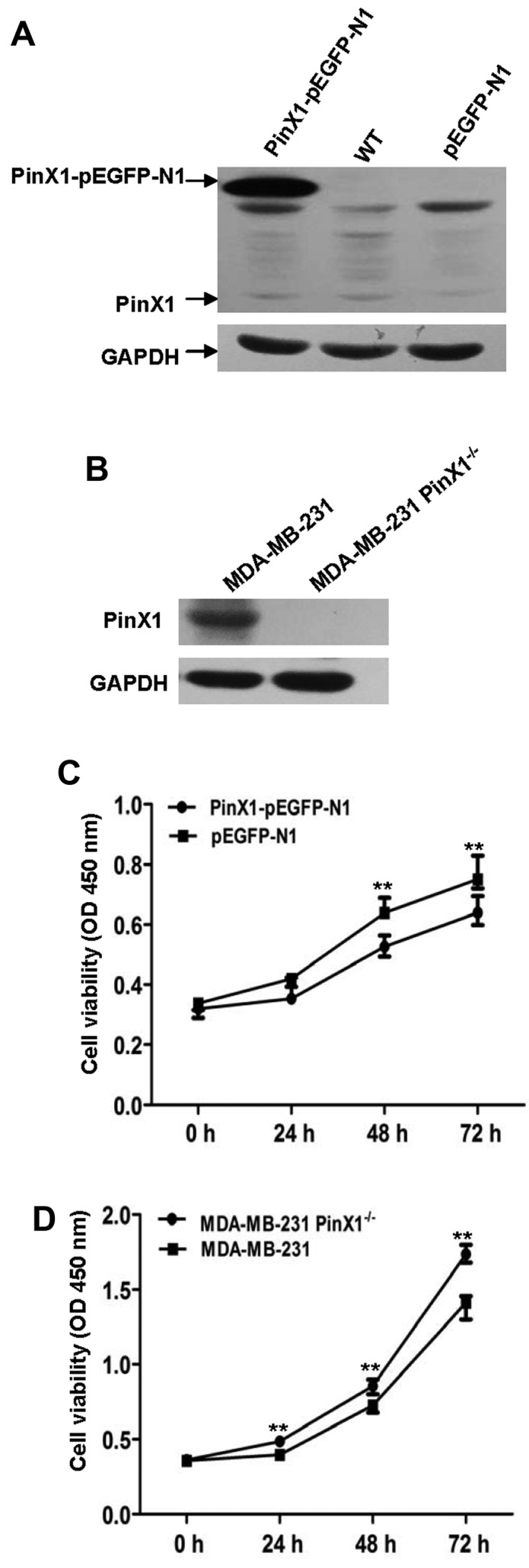
PinX1 inhibits proliferation in a human basal-like breast cancer cell line. (A) Immunoblot analysis of PinX1 expression levels in the PinX1-overexpressed group (PinX1-pEGFP-N1), wild-type MDA-MB-231 (WT) and negative control (pEGFP-N1). GAPDH is a loading control. (B) Immunoblot analysis of PinX1 expression levels in the PinX1-knockout group (PinX1−/−) and the wild-type control (MDA-MB-231). (C and D) The Cell Counting kit-8 (CCK-8) assay was applied to detect the proliferation rates at specific periods (0, 24, 48 and 72 h). Overexpression of PinX1 inhibits the proliferation of MDA-MB-231 while PinX1 knockout promotes its proliferation. **P<0.01.
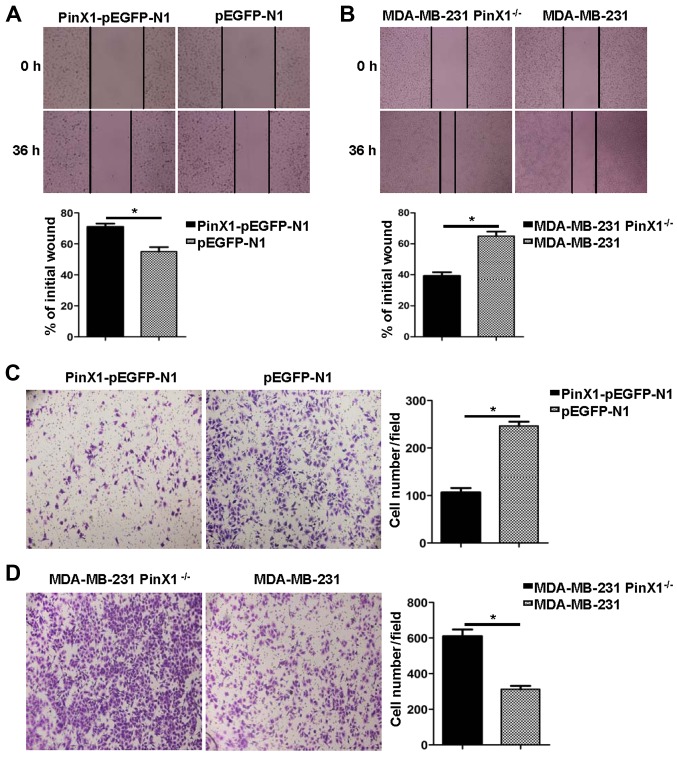
PinX1 suppresses the migration ability of MDA-MB-231 cells
Subsequently, the wound healing assay was used to assess the migration ability of MDA-MB-231 cells that received distinct treatments. As was shown in Fig. 5A and B, the migration rate was inhibited when the PinX1 was overexpressed. However, it was significantly reversed when the PinX1 was knocked out. In the Transwell assay, different experimental groups of cells were fixed and stained by crystal violet after 24-h incubation. Then, the number of cells penetrating the membrane was analysed by a digital light microscope. Results suggested that the cell number/field was drastically increased in the PinX1 knockout group (Fig. 5D). On the contrary, the cell migration ability was markedly impaired when PinX1 was upregulated (Fig. 5C).
Figure 5.
PinX1 suppresses the migration ability of MDA-MB-231 cells in vitro. (A) The migration ability of the PinX1-overexpressed and the control groups were tested by wound healing assay in a 36-h recovery time (top, magnification, ×20). Bottom, statistical analysis of the migration rates, presented relative to the initial wound area. (B) The migration ability of the PinX1 knockout and the wild-type groups was tested by wound healing assay in a 36-h recovery time (top, magnifications, ×10). Bottom, statistical analysis of the migration rates, presented relative to the initial wound area. (C and D) Transwell assay was also used to measure the migration ability of the different experimental groups (left, magnifications, ×10). Right, statistical analyses of the migrated cell number per field. At least three independent experiments were performed (*P<0.05, two-tailed unpaired t-test) and the data expressed are mean ± SEM.
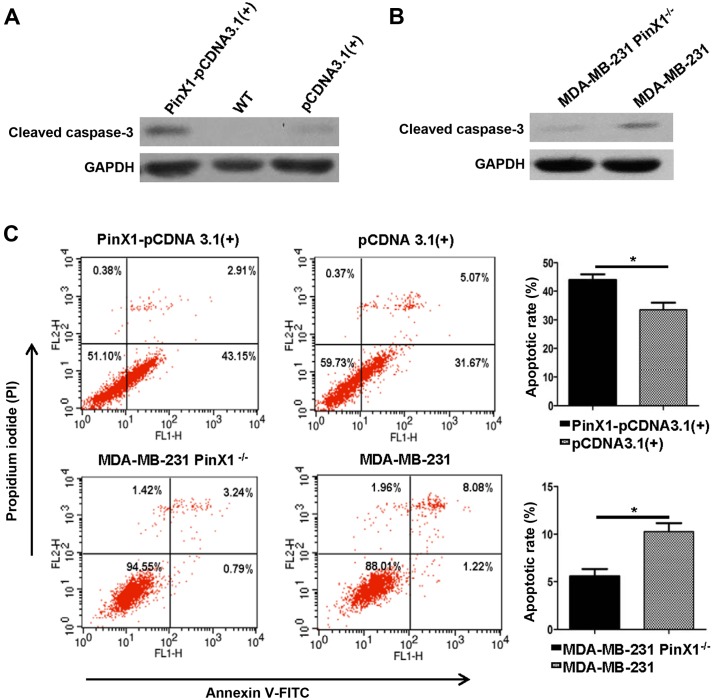
Overexpression of PinX1 provoked apoptosis of MDA-MB-231 in vitro
Previous reports indicated that PinX1 could regulate the telomerase activity and induce apoptosis of nasopharyngeal carcinoma cells (22). Whether PinX1 expression levels were correlated with apoptosis of breast cancer is still unclear. Here we detected the expression levels of cleaved caspase-3, a marker for cell apoptosis by western blot analysis. After transfected with PinX1-pCDNA3.1(+) and pCDNA3.1(+) for 48 h, MDA-MB-231 cells were harvested for later analysis. The PinX1 knockout cells were exposed to etoposide (40 mM) for 24 h. We observed a higher expression level of cleaved caspase-3 in the PinX1 overexpressed group [PinX1-pCDNA3.1(+)] compared with the negative control [pCDNA3.1(+)] (Fig. 6A). There was a higher expression of cleaved caspase-3 in wild-type MDA-MB-231 than the PinX1 knockout cells (Fig. 6B). Consistently, the flow cytometric analysis revealed that overexpression of PinX1 significantly augmented the number of apoptotic cells in MDA-MB-231 and PinX1 knockout markedly decreased the proportion of apoptotic cells (Fig. 6C). These findings indicated that PinX1 may play an essential role in breast cancer cell apoptosis.
Figure 6.
Overexpression of PinX1 induces apoptosis of MDA-MB-231 cells in vitro. (A and B) Western blot analysis was used to measure the expression levels of cleaved caspase-3 in different experimental groups. GAPDH was a loading control. (C) Flow cytometric analysis (left) of the apoptotic rate of the PinX1-overexpressed group with its negative control and the PinX1 knockout group with its counterpart. Quantitative analyses of the apoptotic rate on the right. At least three independent experiments were preformed (*P<0.05, two-tailed unpaired t-test) and the data expressed are mean ± SEM.
Discussion
Breast cancer is the most etiologically heterogeneous malignancy in women encompassing distinct clinical, morphological and molecular entities (23). Unlike the other subtypes of breast cancer, TNBC is highly aggressive. Patients diagnosed of TNBC are younger. Although TNBC is chemosensitive, a higher risk of relapse and an increased tendency to metastasize to visceral organs make TNBC a challenge for current oncologists (24–26). However, recent clinical therapy for TNBC remains limited. Though chemotherapy has shown significant results in clinical practice, previous research revealed that TNBC was not subjected to hormone therapy or Her2-directed agents (i.e. trastuzumab) with the lack of hormone receptors or Her2 receptors (27,28). Besides, Wu et al (29) reported that neo-adjuvant docetaxel plus epirubicin chemotherapy increased pathological complete remission (pCR) rate of TNBC patients with significantly worse survival. During the last decades, with the widespread use of high-throughput technologies, precision medicine contributes to specific molecular drivers unique to specific individual for targeted therapies. The occurrence of various therapeutic targets (i.e. BRCA1, PARP-1 and EGFR) benefit TNBC patients markedly (30). Nonetheless, as a cancer marker, overexpressed Ki-67 fails to prognosticate DFS or OS in basal-like breast cancer (31). Tumor-targeted biomarkers of good prognostic significance, analytic performance, and clinical utility are urgent to be identified and validated to prompt patient survival and life quality.
The phenomenon that telomerase activity in cancer is highly elevated is known for several decades (32). Increasing research supports the potential of telomerase as a clinical therapeutic target. In general, telomere and telomerase influence the tumor formation in different manner (33). Telomere shortening and eventual uncapping are signals for cell to become senescent and stop division via p53 and Rb pathways while rare cells may escape from death and compromise the p53 and Rb pathways with a telomere maintenance mechanism, typically in the way of prompted telomerase expression. What is worse, these survived cells secrete tumor-stimulating factors abetting cancers derived from neighboring cells (34). As a shelterin-related factor, PinX1 binds directly with both TRF1 and telomerase. Originally, PinX1 was found to be an intrinsic telomerase inhibitor and a conceivable tumor suppressor. However, PinX1 expression patterns are contradictory between different types of carcinomas. It was reported that PinX1 was negatively expressed in colorectal, prostate and gastric carcinomas while it was overexpressed in oesophageal squamous cell carcinomas (19,35–37), demonstrating that abnormalities and effects of PinX1 in tumorigenesis are complex and probably tumor-type-specific. In the present study, we explored potential significance of PinX1 in breast malignancy. We investigated the PinX1 expression levels in 26 pairs of breast cancer samples and the adjacent normal counterparts by western blotting and qRT-PCR. We found that PinX1 was downregulated in breast cancer tissues compared with the paired normal breast tissue samples (P<0.05), which was in consistent with a previous study.
IHC remains an indispensable research tool in routine tumor clinicopathology, which is widely used to study diagnostic and prognostic characteristics of neoplasms (38). To further confirm the function of PinX1 in breast cancer, IHC for PinX1 was performed in 75 breast cancer specimens. We applied a specific scoring system to evaluate PinX1 immunoreactivity, depending on the percentage of PinX1 positive cells. To make a reliable and reproducible assessment without predetermination and arbitrariness, three pathologists scored separately until they reached a consensus outcome. Subsequently, ROC curve analysis helped to obtain the optimal cut-off value for PinX1 positivity according to distinct clinicopathological parameters. Our results suggested that reduced PinX1 expression was correlated with advanced histology grade. Besides, negative PinX1 expression was consistent with the negative status of ER and PR. Moreover, stratification by four subtypes of breast cancer was performed. The data indicated that PinX1 expression levels were correlated with the subtypes of breast cancer. In addition, PinX1 was significantly downregulated in basal-like breast cancer. Consequently, we hypothesized that low expression of PinX1 probably play a critical role in the development and progression of basal-like/triple-negative breast cancer.
To validate our hypotheses in basal-like breast cancer, we constructed knocked out and stably overexpressed PinX1 in the MDA-MB-231 basal-like breast cancer cell line. Then influence of PinX1 on cell proliferation rates were detected by CCK-8 assay. We found that overexpression of PinX1 decreased the proliferation rate of MDA-MB-231 cells, while knockout of PinX1 expression level markedly reversed the effects. Besides, we performed wound healing assay and trans- well assay to further explore whether PinX1 was associated with the migration of basal-like breast cancer. Consistently, high expression level of PinX1 suppressed the number of migratory cells. It has been demonstrated that overexpression of PinX1 induced esophageal epithelial cell apoptosis by downregulating telomerase activity (39). To further confirm the relationship between PinX1 and breast cancer apoptosis, we performed immunoblot analysis to assess the cleaved caspase-3 (a marker for apoptosis) expression levels. Notably, the expression level of cleaved caspase-3 was increased with the higher expression of PinX1. The cleaved caspase-3 was downregulated when PinX1 was knocked out. The flow cytometric analysis of apoptosis supported the conclusion that PinX1 promoted also basal-like cell apoptosis.
We confirmed the significant role of PinX1 as a breast cancer suppressor. PinX1 was deregulated in breast cancer tissues while it was upregulated in the normal samples. Among distinguishing subtypes of breast cancer tissues, PinX1 expression was reduced in the basal-like subtype compared with other subtypes. Moreover, overexpression of PinX1 inhibited the proliferation rates and migration ability and increased the apoptosis rates of a human basal-like cancer. Our data clarified the importance of PinX1 in basal-like breast cancer and could have a fundamental impact on the development of basal-like breast cancer. PinX1 may be a potential therapic target of basal-like breast cancer. However, the underlying mechanisms of PinX1 contributing to basal-like breast cancer remain to be further studied.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (81372154, 81672588) and the Science and Technology Planning Project of Guangdong Province, China (2013B021800146).
Glossary
Abbreviations
- ER
estrogen receptor
- PR
progesterone receptor
- HER2 receptor
human epidermal growth factor receptor-2 receptor
- BLBC
basal-like breast cancer
- TNBC
triple-negative breast cancer
- BRCA
breast cancer susceptibility gene
- Pinx1
Pin2/TRF1-interacting telomerase inhibitor 1
- qRT-PCR
quantitative real-time polymerase chain reaction
- TMA
tissue microarray
- AUC
the area under the curve
- CCK-8
Cell Counting kit-8
- pCR
pathological complete remission
- EGFR
epidermal growth factor receptor
- PARP-1
poly(ADP-ribose) polymerase 1
- OS
overall survival
- DFS
disease-free survival
- TRF1
telomeric repeat binding factor 1
References
- 1.Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: linking molecular mechanisms to disease prognosis. Brief Bioinform. 2015;16:461–474. doi: 10.1093/bib/bbu020. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ, Albain KS, André F, Bergh J, et al. Panel members: Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallejos CS, Gómez HL, Cruz WR, Pinto JA, Dyer RR, Velarde R, Suazo JF, Neciosup SP, León M, de la Cruz MA, et al. Breast cancer classification according to immunohistochemistry markers: Subtypes and association with clinicopathologic variables in a Peruvian hospital database. Clin Breast Cancer. 2010;10:294–300. doi: 10.3816/CBC.2010.n.038. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez-Fort MK, Case EC, Rosen AC, Cerci FB, Wu S, Lacouture ME. Rash to the mTOR inhibitor everolimus: Systematic review and meta-analysis. Am J Clin Oncol. 2014;37:266–271. doi: 10.1097/COC.0b013e318277d62f. [DOI] [PubMed] [Google Scholar]
- 6.Gelmon K, Dent R, Mackey JR, Laing K, McLeod D, Verma S. Targeting triple-negative breast cancer: Optimising therapeutic outcomes. Ann Oncol. 2012;23:2223–2234. doi: 10.1093/annonc/mds067. [DOI] [PubMed] [Google Scholar]
- 7.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2010;15(Suppl 5):39–48. doi: 10.1634/theoncologist.2010-S5-39. [DOI] [PubMed] [Google Scholar]
- 9.De Laurentiis M, Cianniello D, Caputo R, Stanzione B, Arpino G, Cinieri S, Lorusso V, De Placido S. Treatment of triple negative breast cancer (TNBC): Current options and future perspectives. Cancer Treat Rev. 2010;36(Suppl 3):S80–S86. doi: 10.1016/S0305-7372(10)70025-6. [DOI] [PubMed] [Google Scholar]
- 10.Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Esposito A. Highlights from the 14th St Gallen International Breast Cancer Conference 2015 in Vienna: Dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer. E Cancer Med Sci. 2015;9:518. doi: 10.3332/ecancer.2015.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson ER, Li S, Kennedy M, Payne S, Kilibarda K, Groth J, Bowie M, Parilla-Castellar E, De Ridder G, Marcom PK, et al. Chemotherapy enriches for an invasive triple-negative breast tumor cell subpopulation expressing a precursor form of N-cadherin on the cell surface. Oncotarget. 2016;7:84030–84042. doi: 10.18632/oncotarget.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodler ET, Kurland BF, Griffin M, Gralow JR, Porter P, Yeh RF, Gadi VK, Guenthoer J, Beumer JH, Korde L, et al. Phase I study of veliparib (ABT-888) combined with cisplatin and vinorelbine in advanced triple-negative breast cancer and/or BRCA mutation-associated breast cancer. Clin Cancer Res. 2016;22:2855–2864. doi: 10.1158/1078-0432.CCR-15-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Tergaonkar V. Noncanonical functions of telomerase: Implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74:1639–1644. doi: 10.1158/0008-5472.CAN-13-3568. [DOI] [PubMed] [Google Scholar]
- 15.Zhou XZ, Huang P, Shi R, Lee TH, Lu G, Zhang Z, Bronson R, Lu KP. The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. J Clin Invest. 2011;121:1266–1282. doi: 10.1172/JCI43452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian XP, Qian D, He LR, Huang H, Mai SJ, Li CP, Huang XX, Cai MY, Liao YJ, Kung HF, et al. The telomere/telomerase binding factor PinX1 regulates paclitaxel sensitivity depending on spindle assembly checkpoint in human cervical squamous cell carcinomas. Cancer Lett. 2014;353:104–114. doi: 10.1016/j.canlet.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Liu JY, Qian D, He LR, Li YH, Liao YJ, Mai SJ, Tian XP, Liu YH, Zhang JX, Kung HF, et al. PinX1 suppresses bladder urothelial carcinoma cell proliferation via the inhibition of telomerase activity and p16/cyclin D1 pathway. Mol Cancer. 2013;12:148. doi: 10.1186/1476-4598-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li HL, Han L, Chen HR, Meng F, Liu QH, Pan ZQ, Bai J, Zheng JN. PinX1 serves as a potential prognostic indicator for clear cell renal cell carcinoma and inhibits its invasion and metastasis by suppressing MMP-2 via NF-κB-dependent transcription. Oncotarget. 2015;6:21406–21420. doi: 10.18632/oncotarget.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi R, Zhao Z, Zhou H, Wei M, Ma WL, Zhou JY, Tan WL. Reduced expression of PinX1 correlates to progressive features in patients with prostate cancer. Cancer Cell Int. 2014;14:46. doi: 10.1186/1475-2867-14-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai XF, Shen CX, Wen Z, Qian YH, Yu CS, Wang JQ, Zhong PN, Wang HL. PinX1 regulation of telomerase activity and apoptosis in nasopharyngeal carcinoma cells. J Exp Clin Cancer Res. 2012;31:12. doi: 10.1186/1756-9966-31-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toss A, Cristofanilli M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015;17:60. doi: 10.1186/s13058-015-0560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris PG, Murphy CG, Mallam D, Accordino M, Patil S, Howard J, Omuro A, Beal K, Seidman AD, Hudis CA, et al. Limited overall survival in patients with brain metastases from triple negative breast cancer. Breast J. 2012;18:345–350. doi: 10.1111/j.1524-4741.2012.01246.x. [DOI] [PubMed] [Google Scholar]
- 25.Ogata H, Kikuchi Y, Natori K, Shiraga N, Kobayashi M, Magoshi S, Saito F, Osaku T, Kanazawa S, Kubota Y, et al. Liver metastasis of a triple-negative breast cancer and complete remission for 5 years after treatment with combined bevacizumab/paclitaxel/carboplatin: Case Report and Review of the Literature. Medicine (Baltimore) 2015;94:e1756. doi: 10.1097/MD.0000000000001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartholomeusz C, Xie X, Pitner MK, Kondo K, Dadbin A, Lee J, Saso H, Smith PD, Dalby KN, Ueno NT. MEK inhibitor selumetinib (AZD6244; ARRY-142886) prevents lung metastasis in a triple-negative breast xancer xenograft model. Mol Cancer Ther. 2015;14:2773–2781. doi: 10.1158/1535-7163.MCT-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burnett JP, Korkaya H, Ouzounova MD, Jiang H, Conley SJ, Newman BW, Sun L, Connarn JN, Chen CS, Zhang N, et al. Trastuzumab resistance induces EMT to transform HER2+ PTEN to a triple negative breast cancer that requires unique treatment options. Sci Rep. 2015;5:15821. doi: 10.1038/srep15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal SK, Childs BH, Pegram M. Triple negative breast cancer: Unmet medical needs. Breast Cancer Res Treat. 2011;125:627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Li S, Jia W, Su F. Response and prognosis of taxanes and anthracyclines neoadjuvant chemotherapy in patients with triple-negative breast cancer. J Cancer Res Clin Oncol. 2011;137:1505–1510. doi: 10.1007/s00432-011-1029-6. [DOI] [PubMed] [Google Scholar]
- 30.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, et al. TBCRC 001: Randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang JE, Wecsler JS, Press MF, Tripathy D. Molecular markers for breast cancer diagnosis, prognosis and targeted therapy. J Surg Oncol. 2015;111:81–90. doi: 10.1002/jso.23732. [DOI] [PubMed] [Google Scholar]
- 32.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 33.Johnson FB. PinX1 the tail on the chromosome. J Clin Invest. 2011;121:1242–1244. doi: 10.1172/JCI57024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng W, Jiao N, Li N, Wan X, Luo S, Zhang Y. Decreased expression of PinX1 protein predicts poor prognosis of colorectal cancer patients receiving 5-FU adjuvant chemotherapy. Biomed Pharmacother. 2015;73:1–5. doi: 10.1016/j.biopha.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Qian D, Zhang B, He LR, Cai MY, Mai SJ, Liao YJ, Liu YH, Lin MC, Bian XW, Zeng YX, et al. The telomere/telomerase binding factor PinX1 is a new target to improve the radiotherapy effect of oesophageal squamous cell carcinomas. J Pathol. 2013;229:765–774. doi: 10.1002/path.4163. [DOI] [PubMed] [Google Scholar]
- 37.Wang HB, Wang XW, Zhou G, Wang WQ, Sun YG, Yang SM, Fang DC. PinX1 inhibits telomerase activity in gastric cancer cells through Mad1/c-Myc pathway. J Gastrointest Surg. 2010;14:1227–1234. doi: 10.1007/s11605-010-1253-4. [DOI] [PubMed] [Google Scholar]
- 38.Swanson PE. Immunohistochemistry as a surrogate for molecular testing: A review. Appl Immunohistochem Mol Morphol. 2015;23:81–96. doi: 10.1097/PAI.0000000000000181. [DOI] [PubMed] [Google Scholar]
- 39.Zuo J, Wang DH, Zhang YJ, Liu L, Liu FL, Liu W. Expression and mechanism of PinX1 and telomerase activity in the carcinogenesis of esophageal epithelial cells. Oncol Rep. 2013;30:1823–1831. doi: 10.3892/or.2013.2649. [DOI] [PubMed] [Google Scholar]