Abstract
Lipids are essential building blocks synthesized by complex molecular pathways and deposited as lipid droplets (LDs) in cells. LDs are evolutionary conserved organelles found in almost all organisms, from bacteria to mammals. They are composed of a hydrophobic neutral lipid core surrounding by a phospholipid monolayer membrane with various decorating proteins. Degradation of LDs provide metabolic energy for divergent cellular processes such as membrane synthesis and molecular signaling. Lipolysis and autophagy are two main catabolic pathways of LDs, which regulate lipid metabolism and, thereby, closely engaged in many pathological conditons. In this review, we first provide an overview of the current knowledge on the structural properties and the biogenesis of LDs. We further focus on the recent findings of their catabolic mechanism by lipolysis and autophagy as well as their connection ragarding the regulation and function. Moreover, we discuss the relevance of LDs and their catabolism-dependent pathophysiological conditions.
Keywords: Lipid droplets, lipolysis, lipophagy, chaperone-mediated autophagy
Background
Many living organisms store lipids in their cells to produce metabolic energy, in case of insufficient energy sources. Cells preserve lipids by converting them into neutral lipids, such as triacylglycerides (TAG) and sterol esters (SE). These type of lipids are deposited in lipid droplets (LDs) which are also termed as adiposomes, lipid bodies or oil bodies [1]. For a long time, LDs were accepted as inert intracellular vesicles storing neutral lipids in all living organisms. However, recent advances in functional analysis techniques, imaging methods, lipidomics and proteomics technology offer scientists a better understanding of biological properties and functions of LDs. In the last couple of years, many structural and functional proteins were identified and characterized on the surface of LDs, and they were named as perilipins (PLINs) [2]. In addition to their role in energy metabolism, LDs play a role in various cellular events, ranging from protein degradation, sequestration of transcription factors and chromatin components to generation of lipid ligands for certain nuclear receptors, and they serve as fatty acid trafficking nodes [3–5]. Moreover, LDs might be hijacked by various pathogens. Due to these diverse functions, abnormalities of LDs were associated with many pathological conditions [6–8].
The catabolism of LDs into free fatty acids (FAs) is a crucial cellular pathway that is required to generate energy in the form of ATP, and to provide building blocks for biological membrane and hormone synthesis. Lipolysis is a biochemical catabolic pathway that relies on the direct activation of LD-associated lipases, such as adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL) and monoglyceride lipase (MGL) [9, 10]. Together with regulatory protein factors ATGL activator (comperative gene identification-58, CGI-58) and ATGL inhibitor (G0/G1 switch protein 2, G0S2), these lipases constitute the basis for the lipolytic machinery in cells.
Autophagy is one of the two major degradation pathways in cells supporting cell survival by recycling metabolic components under stress conditions (Ubiquitin-proteasome system being the other pathway). It is initiated by sequestering cytosolic organelles or macromolecules in double-membrane vesicles and delivering them to lysosomes for degradation by the lytic enzymes therein. Cellular building blocks are then released back to cytosol and recycled to ensure cellular homeostasis [11, 12]. Basic machanism of autophagy is classified into three different types: Macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy. Macroautophagy is a well-studied pathway, targeting large substrates such as toxic aggregates and degenerated organelles in a selective or non-selective manner [13]. Chaperone-mediated autophagy (CMA) is a selective form of autophagy, targeting specific proteins through the recognition activity of chaperone protein heat shock cognate 70 (Hsc70). Hsc70 delivers the substrates to the lysosomal lumen via lysosomal-associated protein 2a (LAMP2a) [14, 15]. Microautophagy degrades cytoplasmic cargos through a organized invagination of lysosomal membranes and direct engulfment [16].
Recent investigations defined another selective form of macroautophagy termed “lipophagy”. In this autophagy form, LD degradation is controlled by the association of membrane GTPase Rab7 with LDs [17, 18]. Moreover, a significant role of CMA was recently identified in selective degradation of a unique LD-coat protein family called PLINs [19, 20]. Therefore, autophagy pathways play a crucial role in lipid metabolism and the number of publications in this relatively new field is growing exponentially.
In contrast with the traditional view that LDs are simple lipid storage vesicles, because of their dynamic nature and multifunctionality they are today accepted as a distinct intracellular organelles. Recent studies underline physiological importance of these multifunctional organelles, particularly in the context of lipolysis and selective autophagy mechanisms of lipophagy and CMA. Here, we will first summarize biological properties of LDs and then focus on recent findings on molecular regulation of lipolytic and autophagic mechanism. Furthermore, we will discuss the relevance of LDs and related catabolic mechanisms for pathophysiology.
Structure of the lipid droplets
Phospholipid composition
Being different from other membrane-enclosed organelles, LDs have a unique structure with a hydrophobic core of neutral lipids surrounded by a monolayer phospholipid membrane, separating hydrophobic neutral lipids from the aqueous cytoplasmic environment [21].. In the hydrophobic core of LDs, neutral lipids, predominantly TG and SE, are stored at various ratios (Fig. 1). In white adipocytes, TG are primarily stored in LDs as lipid esters, whereas in steroidogenic cells, SE are the main components [22]. Also, depending on the cell type, many other endogenous neutral lipids such as retinyl esters, ether lipids, and free cholesterol are stored in LD cores [23–25]. Interestingly in some cell types, electron microscopy data revealed that membrane-like structures containing ribosomal units were also extending within the core of LDs [26, 27].
Fig. 1.

Basic Morphology of Lipid Droplets
In mammalian cells, the main constituent (up to 60%) of LD monolayer membranes is phosphatidylcholine (PC) that is followed in abundance by phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), sphingomyelin (SM) and lyso forms of PC and PE [28–30]. Phosphatidic acid and free cholesterol are also present in LD surfaces in minor amounts [21, 31]. Although LD formation was believed to take place between the leaflets of endoplasmic reticulum (ER), phospholipid composition of LD membrane differs from that of ER and other organelles [21]. The unique phospholipid membrane composition primarily affects LD synthesis [32], maturation (size) [33, 34] and degradation via lipophagy mechanisms [35, 36]. Changes in LD membrane phospholipid ratios under physiological conditions in various cell types indicate that regulation of phospholipid composition is crucial for homeostasis of this organelle.
Surface protein structure
Besides membrane phospholipid compositions, LD surface proteins are other factors regulating homeostasis and intracellular interactions of LDs. Membrane surface of LD is decorated with several structural and functional proteins [37]. Proteomics studies on LDs in a variety of organisms including bacteria [38–40], plants [41, 42], yeasts [43, 44], insects [45, 46] and mammals [47–49] revealed that over two hundred different proteins are localized to the surface of LDs. Even though LD proteomics studies that were performed in various cell types and organisms resulted in the discovery of different profiles, identified proteins seem to function in common pathways. These proteins were categorized into several distinct functional classes (Table 1).
Table 1.
Classification of proteins located on the LD surface
| Protein Group | Short Name | Long Name | References |
|---|---|---|---|
| PAT Family | PLIN | Perilipin | [49, 160] |
| ADRP | Adipose differentiation-related protein | [24, 27, 48, 161–163] | |
| TIP47 | Tail-interacting protein of 47 kD | [48, 160, 163–165] | |
| Lipid & Energy Metabolism | HSL | Hormone sensitive lipase | [49, 160] |
| ATGL (PNPLA2) | Adipose triglyceride lipase | [24, 162, 166] | |
| CGI-49 | CGI-49 protein | [24, 27, 48, 49, 165] | |
| CGI-58 | CGI-58 protein | [24, 27, 49, 160, 162] | |
| Mgll | Monoglyceride lipase | [97, 163] | |
| Tgh | Triacylglycerol hydrolase | [167] | |
| LACS3–4 | Long-chain-fatty-acid–CoA ligase 3–4 | [24, 48, 49, 162, 165] | |
| H105e3 | Sterol-4-carboxylate 3-dehydrogenase | [48, 163–165] | |
| SE | Squalene monooxygenase | [27, 162, 168] | |
| Lss | Lanosterol synthase | [49, 160, 169] | |
| Cpla2 | Cytosolic phospholipase A2 | [24, 169, 170] | |
| Pcyt1a | Phosphocholine cytidylyltransferase A | [169, 171, 172] | |
| Mdh2 | Malate dehydrogenase | [97, 173, 174] | |
| Cyb5r3 | NADH-cytochrome b5 reductase | [24, 49, 162, 173] | |
| Dhrs1 | Dehydrogenase/reductase SDR member 1 | [24, 49, 162, 173] | |
| Dhrs3 | Short-chain dehydrogenase/reductase 3 | [92, 169, 173] | |
| Nsdhl | NAD(P)H steroid dehydrogenase-like | [163, 169, 173] | |
| Acsl1–3 | Long-chain acyl-CoA synthetase 1–3 | [24, 49, 175] | |
| Ldah | LD-associated hydrolase | [173, 176, 177] | |
| Signalling | CHP | Calcium-binding protein p22 | [24, 164, 165, 178] |
| Cav1 | Caveolin1 | [24, 49, 162] | |
| METTL7A | Methyltransferase-like protein 7A | [92, 174, 178] | |
| Membrane trafficking proteins | VIM | Vimentin | [24, 49, 160, 162, 171] |
| ACTB | Actin, cytoplasmic 1/ □-Actin | [24, 27, 163, 165] | |
| Rab10 | Ras-related protein Rab-10 | [24, 27, 48, 162, 165] | |
| Rab 11A | Ras-related protein Rab-11A | [24, 162, 164] | |
| Rab 1a | Ras-related protein Rab-1a | [24, 162, 164, 165] | |
| Rab 1b | Ras-related protein Rab-1b | [48, 164, 165] | |
| Rab 14 | Ras-related protein Rab-14 | [24, 49, 160, 162, 164] | |
| Rab 18 | Ras-related protein Rab-18 | [24, 49, 162, 164, 165] | |
| Rab 5b | Ras-related protein Rab-5b | [24, 162, 165, 166] | |
| Tubulin | Tubulin | [24, 49, 163, 178] | |
| Miscellaneous | Stomatin | Stomatin | [162, 165] |
| HSPA5 | 78 kDa glucose- regulated protein | [24, 48, 49, 160, 162–165] | |
| Hspa1a | Heat shock 70 kDa protein 1A | [24, 49, 161] | |
| FAF2 | FAS-associated factor 2 | [24, 48, 162, 166] | |
| Ancient ubiquitous protein BiP | Ancient ubiquitous protein BiP | [24, 49, 160, 162, 165, 166] | |
| CANX | Calnexin | [49] | |
| HSP 70 | Heat shock protein 70 | [49, 161, 165] | |
| Ribophorin I | Ribophorin I | [49, 160] | |
| ApoB | Apolipoprotein B | [174] | |
| a-synuclein | a-synuclein | [92] | |
| Hepatitis C core protein | Hepatitis C core protein | [175–177] | |
| His2A | Histone 2A | [89] | |
| His2B | Histone 2B | [89] | |
| Alb | Serum albumin | [27, 161, 163] |
In mammalian LDs, predominant proteins are the members of the PAT protein family, an acronym representing perilipin (PLIN), adipocyte differentiation-related protein (ADRP; also called as adipophilin), and TIP47 (tail-interacting protein of 47 kDa) proteins. Members of the PAT family share sequence similarity and the ability to bind intracellular LDs, suggesting that they derived from a common ancestral gene [50]. The most studied member of the PAT family is PLIN, a protein that regulates the access of lipases to neutral lipids in the core of LDs, hence controlling lipid homeostasis. Consistently, elevated basal lipolysis levels were observed in adipocytes obtained from PLIN knockout mice models [51].
LDs also host many other proteins-related to lipid homeostasis. These proteins can be classified according to their functions, including lipid biogenesis (long chain fatty acid CoA ligases, lanosterol synthase, squalene epoxidase), maintainance of intracellular lipid metabolism (long chain fatty acid CoA ligases) and lipid degradation (patatin-like phospholipase domain containing protein 2 (PNPLA2), CGI-58) [52].
In addition to PAT proteins and lipid homeostasis-related proteins in LDs, there are several groups of proteins that are called ‘refugee proteins’ which seem not to be relevant to the known functions of LDs [53]. These proteins are categorized as signaling proteins, membrane trafficking proteins, chaperons and proteins-associated with cellular organelles (Table 1). Signaling-related proteins are classes of proteins that accumulate on the surface of LDs. Major signalling proteins such as, mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K) and Lyn proteins have been shown to localize on LD surfaces [54, 55]. Moreover, caveolins are another group of LD proteins that specifically form a coat by making invaginations and surrounding cellular membranes called caveolae. Caveolae function in endocytosis, signal transduction, cholesterol transport and growth control [33, 56, 57]. Caveolin-1 and caveolin-2 proteins were shown to reside on the surface of LDs. They generate membrane domains where they function as regulators of signaling proteins [58, 59]. Discovery of membrane trafficking proteins on the surface of LDs and the fact that interact with other cellular compartments through intarcellular motility, suggest that LDs are distinct organelles. Membrane trafficking-related proteins consist of five subgroup proteins: Small GTPases governing vesicle formation and motility; motor proteins such as kinesin and myosin that carry LDs on the cytoskeleton; soluble NSF attachment receptor (SNARE) proteins mediate membrane docking and fusion on LDs; vesicular traffic proteins (such as ARFs and COPs) that regulate cargo sorting and vesicle budding; and other membrane trafficking proteins of miscellaneous functions [4].
LD Biogenesis
LD biogenesis is stimulated upon an increase in intracellular free FA levels. In order to prevent lipotoxicity, excessive free fatty acids are converted into neutral lipids and stored in cytosolic LDs. However, recent studies suggested that LDs are not only cytoplasmic but also can be found in the nuclei [60, 61]. Although the morphology of these ‘nuclear LDs’ is similar to their cytoplasmic counterparts, their biogenesis mechanisms remain unexplained. Nuclear LDs are thought to regulate nuclear lipid homeostasis and modulate signaling through lipid molecules [3].
Due to the unique hydrophobic/hydrophilic (amphipathic) structure of LDs, mechanisms of biogenesis of this organelle in the cell attracted much attention and several hypotheses were proposed [62–64]. Being different from most self-replicating organelles, LDs are mainly formed de novo. In addition, LDs could be derived via fission of pre-existing LDs [65]. In prokaryotes, neutral lipid accumulation seems to be initiated at specific lipid domains of plasma membrane and ends in the formation of cytoplasmic LDs [62]. On the other hand, as a eukaryotic LD biogenesis mechanism, there is evidence that LD formation occurs within the leaflets of ER phospholipid bilayer in discrete steps involving neutral lipid synthesis, progressive neutral lipid accumulation in the ER and cytosolic droplet formation. Increased volume of accumulated neutral lipid between the ER bilayer exceeds the solubility limits and LDs are thought to be ‘oiled out’ from the ER bilayer [66]. Consistent with the hypothesis, many electron microscopy studies in various cell types showed that cytosolic LDs are tightly associated with ER [67, 68] (Fig. 2).
Fig. 2.

Neutral Lipid Synthesis, Lipid Droplet (LD) Formation and Growth. a Metabolic pathway of triglyceride (TG) and sterol ester (SE) synthesis, b LD Formation from Endoplasmic Retikulum (ER) by Neutral Lipid Synthesis Enzymes (NLSE). Left panel shows budding model of LDs from ER; middle panel shows bicelle formation model of LDs originating from ER; right panel shows hatching model of LDs from ER
Neutral lipid synthesis
Neutral lipid synthesis is regulated by complex pathways involving lipid metabolism enzymes and structural proteins that are permanently or transiently located to the ER and LDs. The first step of neutral lipid synthesis requires free FA activation. As free FA are chemically inert, their activation step is critical for LD biogenesis pathway. In mammalian cells, acyl-coA synthetase (ACS) enzymes activate long chain FAs to acyl-coA by esterifying with coenzyme A (CoA) (Fig. 2a) [69]. De novo TAG synthesis occurs in a four-step pathway involving glycerol-3-phosphate O-acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), phosphatidic acid phosphatase (PAP) (or lipin), and diacylglycerol acyltransferase (DGAT) enzymes. At the last step of the pathway, FAs, firstly activated to acyl-CoA, are converted to TAGs through DGAT1 and DGAT2 enzymes in mammalian cells. In addition, some LDs store mainly sterol esters (SE) specifically in macrophages, adrenocortical cells, ovarian and testicular interstitial cells. The synthesis of SE is conducted by acyl-CoA cholesterol O-acyltransferases (ACAT1 and ACAT2). The neutral lipid biosynthesis enzymes DGAT1, ACAT1, and ACAT2 were shown to localize on ER domains in mammalian cells [70–72].
LD Formation from ER
As intracellular FA promote LD formation, synthesized neutral lipids form a ‘lens’ between the leaflets of ER bilayer. Biophysical and in silico predictions suggest that, when TG holding limit of bilayer membranes is reached [23], the lens between ER bilayers is ‘oiled out’ and leads to the formation of nascent LDs. However, it is still an open question how phospholipid monolayer membrane of LDs is derived from ER bilayer membranes. Walther et al. claimed two models about LDs leaving ER membranes (Fig. 2b) [10]. The most widely accepted model states that neutral lipid lens buds out from the ER together with the outer leaflet of the bilayer membrane. This process is thought to be drived by structural LD associated proteins such as PAT proteins in a way that PAT proteins could mediate budding at specific domains of ER bilayer membrane [10]. Their suggested second model states that LDs are excised from both leaflets of the ER membrane bilayer as a bicelle (a lipid monolayer vesicle formed by the fusion of the tips of the outer and inner membranes of the ER with accumulated lipid lens within) [10]. In addition to these models, Fujimoto et al. suggested that LDs are excised from ‘both leaflets of the ER membrane bilayer as a bicelle’ by hatching mechanism (bilayer budding and monolayer membrane formation because of micelle-like isolation of the inner membrane leaflet in the LD lumen) (Fig. 2b) [22]. Although neither of these models rely on solid data, they could hypothetically explain how hydrophobic cores of LDs are surrounded by phospholipid monolayer membranes that are derived from ER membranes and how some ER membrane proteins are targeted to newly formed LDs.
Growth of lipid droplets: the size matters
Once LDs are synthesized, they typically keep growing because of the excessive amount of intracellular FA in cells and reach a final size. The size of LDs varies within a wide range (0.4–100 μm) in different cell types [73]. Even in the same cell, the size of LDs may dramatically differ under changing pathophysiological conditions. Recent studies have shown that subpopulations of LDs may differ in terms of size, morphology, and function within the same cell [74, 75]. Many proteins (e.g. Fsp27, seipin, FITM2 and perilipin1) and lipid factors (e.g. phosphatidylcholine and phosphatidic acid) have been shown to be involved in LD growth mechanisms [73].
One possible mechanism is growth of LDs through fusion of highly mobile smaller LDs [76, 77]. Although there is conspicuous skepticism in the literature, LD fusion rates are high enough to allow LD growth [78], RNAi knockdown of fusion mediator SNARE proteins prominently decreased fusion rates and size of LDs [76]. In addition, mutant form of the yeast homologue of seipin in the mammals regulates phosphatidic acid metabolism and leads to ‘supersized LDs’ by enhancing coalescence of them [31]. Similarly, knockdown of phosphocholine cytidylyltransferase (CCT) enzymes, catalyzing the rate limiting step of phosphatidylcholine synthesis, dramatically induce fusion of LDs, balance surface volume ratio and regulate LD size [79]. Another possible mechanism for growth of droplets is the lateral transfer of synthesized neutral lipids from ER to LDs. If nascent LDs remain attached to ER, as neutral lipid synthesis enzymes on the bilayer membrane of the ER convert FA to neutral lipids, these molecule may be transferred to adjacent LDs [1]. Similar to this mechanism, Gong et al. stated that lipid droplet growth occurs by lipid transfer at LD contact sites between adjacent LDs via Fsp27 (Fat specific protein-27 or cell death-inducing DFF45-like effector C (Cidec)) protein [80]. Ectopic expression of Fsp27 protein promotes formation of unilocular large LDs through stimulation of TG accumulation [80–82]. Alternatively, Fujimoto et al. suggested that neutral lipid synthesis occurs on the surface of LDs [83]. Since phospholipid monolayers of LDs are derived from ER membranes, neutral lipid synthesis enzymes originating from these membranes could be directly transfered to the surface of LDs. Particularly, during lipid loading, DGAT2 enzymes which normally localize to the ER, were shown to localize on the surface of LDs [71, 84]. However, it is ambiguous whether earlier steps of neutral lipid synthesis also take place on the surface of LDs.
Function of LDs other then lipid storage
Interestingly, LDs were involved in various pathological events and innate immunity [85]. LDs were shown to be targeted by various pathogens including viruses, bacteria and parasites. Hepatitis C virus (HCV) was appeared to use the LDs for proliferation and assembly of capsid proteins were shown to take place in the vicinity of host LDs [86]. In addition to HCV, many viruses including GB virus B and dengue virus (DENV) also hijack LDs for viral replication [87, 88]. As LDs may also be related to the pathogenesis of hepatic steatosis caused by viral infections and from this perspective, LDs may become a theurapetic target [22]. Furthermore, LDs may serve in the cytoplasmic sequesteration of proteins such as histones and α-synuclein [37, 52]. In Drosophila embryos, histones are found in high abundance on the surface of LDs and they are rapidly transferred to the nucleus when needed [89]. Recently, histone storage function of LDs was also defined as a defense mechanism against pathogens. Indeed, histones around LDs were shown to possess bactericidal activity [3]. Neuropathological protein aggregates of α-synuclein and amyloid β peptide were shown to be in a close proximity with cytosolic LDs in various cell types [90–92]. In addition, recently Moldavski et al. stated that LDs play important roles as clearing mechanisms of inclusion bodies in cells [93]. Protein sequestration role of LDs may explain the presence of ‘refugee proteins’ that are found associated with LDs, and provide a dynamic regulatory mechanism for protein storage and degradation [53].
The catabolic pathways of LDs
As the LDs reside at the center of cellular lipid and energy homeostasis, their catabolism is strictly under the control of hormones and activation of enzymes. So far, LDs are known to break down mostly by lipolysis, however, recent discoveries that showed a molecular connection between lipolysis and autophagy mechanisms led to the identification of the involvement of selective autophagic forms, lipophagy and CMA, in LD catabolism.
Lipolytic hydrolysis
Under fed conditions, LDs store TAGs mainly in adipose tissues and hydrolysis of ester bonds between long chain FAs and glycerol backbone in TAGs is called “lipolysis”. During the first step of lipolysis, protein kinase A (PKA) phosphorylates one of the PAT protein family member, PLIN1, and leads to its proteasomal degradation [94]. This, results in the release of ATGL activator protein CGI-58, which then initiates TAG breakdown via activated ATGL [95, 96]. ATGL selectively catalyzes the first step of TAG hydrolysis to generate diacylglycerols (DAGs) and free FAs [97]. The second step of lipolysis is depend on the activation of hormone sensitive lipase (HSL) which is a multifunctional enzyme that is capable of hydrolyzing both the first and the second step of lipolysis. HSL hydrolyzes DAGs and produce monoacylglycerol (MAG) and FAs [98]. Within the lipolysis cascade, HSL functions as a rate-limiting enzyme for DAG catabolism [99, 100]. In the last step of lipolysis, MAGs are released into the cytosol and eventually cleaved by MGL to generate glycerol and FA [101] (Fig. 3).
Fig. 3.
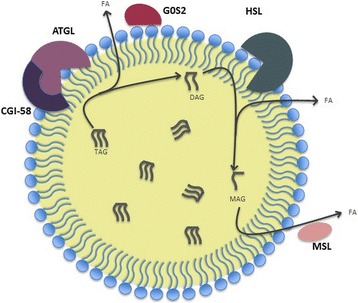
Lipolysis of Lipid Droplets
Under nutrient deprivation, products of lipolysis are secreted from the adipose tissue to the periphery via bloodstream, which are then used for β-oxidation and ATP production. In contrast, in non-adipose tissues, mitochondria or peroxisomes can directly oxidize products of lipolysis through β-oxidation and release acetyl-CoA [102].
Lipophagy
Lipophagy was first described in mouse hepatocytes under starvation [103]. It was shown that LDs were mobilized in order to generate free FAs. Pharmacological or genetic inhibiton of lipophagy resulted in elevated TAG concentrations, increasing concomitantly LD numbers and size. Electron microscopy data confirmed sequesteration of LDs by double-membraned autophagosomes that labeled positive with the autophagy marker LC3 (Microtubule-Associated Proteins 1A and 1B, Light Chain 3). Intriguingly, LC3 as well as PLIN1 and PLIN2 proteins were found in LDs and lysosomes that were isolated from starved livers. In addition, LDs were shown to colocalize with lysosomes, indicating a role for lysosomes in lipid turnover via LDs break down [103]. LDs were shown to be selectively engulfed by a autophagosomes that was followed by the fusion of autophagosomes with lysosomes (autolysosomes) for degradation. Therefore, lipophagy is an LD-selective type of macroautophagy. In line with that, inhibition of the small GTPase dynamin-2 caused the accumulation of LDs in autolysosomes [104] (Fig. 4a).
Fig. 4.

Lipid Droplet Degradation by Autophagy Mechanisms. a Lipophagy b Chaperone Mediated Autophagy (CMA)-dependent degradation
Substrate labeling is a general mechanism used by cellular degradation machineries. So, how does lipophagic machinery preferentially recognize LDs as substrates? Proteomic analyses of LDs from different organisms showed that membrane GTPase Rab7 is associated with LDs and suggested a universal role of Rab7 in the regulation of LD docking and degradation [105]. Involvement of GTPases in endosome-autophagosome interactions [106] is made Rab7 a strong candidate for selection of LDs as lipophagic substrate. Activation of lipophagy in starved hepatocellular cells lead to significant increase in Rab7-LD association while Rab7 mutants, defective in binding to Rab7-interacting lysosomal protein (RILP) failed to promote lipophagy [18]. Furthermore, starvation-activated Rab7 was shown to lead to the recruitment of multivesicular bodies to the LD-autophagosome complex and induced the formation of amphisomes through fusion of autophagosomes and endosomes [18]. In this study, microscopy data revealed an interaction between LDs, autophagosomes and lysosomes, which is consistent with the concept that LDs do not fuse directly with lysosomes [103]. In addition to hepatocytes, the essential role of Rab7 for autolysosome-mediated LD degradation during hormone-stimulated lipophagy was also shown in adipocyte cells. In these cells, LD-associated PLIN1 protein inhibited lipophagy by blocking Rab7 binding to the LD surface [18]. Consequently, Rab7 was identified as a protein regulating LD docking via its effector protein RILP and FYVE and coiled-coil domains containing protein 1 (FYCO1) that directly connects the cargo to LC3 proteins on autophagosome membranes [18].
In adipocytes, the membrane curvature protein Bif-1 was found as a novel regulator protein for lipophagy [107]. Induction of lipophagy has been shown to promote Bif-1-dependent degradation of LD-associated PLIN1, and deficiency of this protein led to decreased TAG hydrolysis, indicating that Bif-1 is essential for lipophagy-dependent PLIN1 degradation, and thereby LD break down [107]. Note that, membrane curvature varies according to LD size, however, whether Bif-1 affects LD targeting by lipophagy through its effects on LD size, is still unknown.
Although the role of lipophagy in LD degradation was first described in hepatocytes [103, 108], several recent studies shown activation of this mechanism in various cell types, such as neurons [109, 110], glial cells [109], foam cells [111], enterocytes [112], T cells [113], fibroblasts, adipocytes [114] and prostate carcinoma cells [115]. Moreover, lipophagy was demonstrated in yeast [116], C. elegans [117] and some fungus species [118]. Indeed, mechanisms that trigger lipophagy may vary in each cell type, and it could be context-dependent. It is likely that cells adapted lipophagy mechanisms in order to better deal with extreme conditons, such as lipotoxicity, nutrient deprivation etc. However, the core components that are essential for lipophagy seem to be conserved in most cell types.
Chaperone-mediated autophagy (CMA)
More recently, involvement of another selective type of autophagy, CMA, in degradation of LD-associated PLINs proteins, PLIN2 and PLIN3 was described [19]. In this context, activation of CMA was shown to induce PLIN2 and PLIN3 turnover, which caused recruitment of ATGL and lipophagic protein ATG, leading to the degradation of LDs. Consistantly, inhibiton of PLINs degradation in CMA-deficient model systems, resulted in both reduced ATGL recruitment, and lipophagy activation gave rise to LD accumulation. Therefore, functional CMA was shown to be essential for the removal of LD-associated proteins, which in turn led to the accumulation of lipases and lipophagy proteins on specific regions of LDs [19] (Fig. 4b).
Cross-talk between lipolysis and lipophagy/CMA mechanisms
Even though basic molecular mechanisms of lipolysis, lipophagy and CMA are different, recent investigations indicate that these independent pathways actually cross-talk. For example, ATG2A, an essential autophagy protein was also found on LDs, and genetic knock-down of ATG2A and ATG2B resulted in defective lipophagy as well as LD accumulation [119]. A recent study with starved mouse embryonic fibroblasts (MEF) demonstrated that FAs derived from LDs end-up in mitochondria which is required only for ATGL-dependent lipolysis activation not for lipophagy [120]. In the same study, size and number of LDs increased as result of autophagic degradation of other cellular membrane components, confirming that overall lipid content that recycled via autophagy, contributed to the growth of LDs. As mentioned before, Bif-1 regulating curvature of various membranes including autophagic membranes, was identified as a protein involved in lipophagy regulation in adipose tissues [107]. PLIN2 and PLIN3 are CMA substrates, and they contribute to the cross-talk between lipolysis and CMA. Promotion of ATGL and lipophagy proteins by CMA activation was shown to trigger not only lipophagy but also lipolysis, indicating that PLIN2 and PLIN3 undergo CMA-mediated degradation before the initiation of lipolysis [19]. Moreover, lipolysis has been shown to depend on PLIN2 phosphorylation by AMPK, which occured after the interaction of PLIN2 with CMA chaperone protein Hsc70 [20]. A complex cross-talk between lipolysis and lipophagy has been described in brown adipose tissue, in which both central nervous system (CNS) and local cytosolic lipases were involved. Here, the autophagosome marker LC3 recruited ATGL and HSL to LDs, supporting coordinated regulation of lipolysis by both CNS and peripheral protein-protein interactions [121].
On the other hand, the existance of a reverse relationship between lipolysis and lipophagy has also been shown in which lipolysis-dependent LDs breakdown regulated autophagosome formation. LDs was shown in close proximity of autophagosomes with transient ‘kiss and run’ interactions contributing to autophagosome biogenesis [122]. In this process, patatin-like phospholipase domain containing protein-5 (PNPLA5), a lipase localized on the surface of LDs, was shown to be needed for optimal initiation of autophagosome formation by generating DAG, a building block for phospholipid constituents of the double-membrane structure [122]. Consistent with this, additional evidence has been provided by a yeast study, demonstrating that deletion of TAG and SE synthesis enzymes resulted in the inhibition lipophagy [123]. In a very recent yeast study, absence of LDs caused morphological changes in the ER due to defective FA synthesis, compromised autophagosome biogenesis [124]. Therefore, active lipolysis on the LD surface or LD-ER contact sites is required for the de novo formation of autophagosomes [122–124].
Pathophysiological relevance of lipolysis, lipophagy and CMA
As LDs play central role in the regulation of intracellular lipid and energy metabolism, abnormalities of LD-related mechanisms, including lipolysis, lipophagy and CMA are involved in many pathological and physiological conditions. For example, neutral lipid storage disease (NLSD), atherosclerosis, obesity etc. [8, 125].
Neutral lipid storage disease (NLSD)
Neutral Lipid Storage Disease (NLSD) is a heterogenous group of rare autosomal recessive disorder characterized by abnormal accumulation of LDs in multiple tissues and lymphocytes. Mutations in one of the two genes coding for ATGL or CGI-58 (proteins located on the surface of LDs) are associated with the NLSD [126, 127]. Both ATGL and CGI-58 coordinately function in the lipolysis of LDs; however, mutations in each gene result in different symptoms. Mutations in CGI-58 (a coactivator of ATGL), are associated with NLSD with ichthyosis known as Chanarin-Dorfmann syndrome (CDS) characterized by defective permeability barrier of the skin [128–130]; whereas, ATGL mutations lead to severe NLSD with cardiac myopathy (NLDSM) [131]. These symptomatic differences may be related to the different functions of CGI-58 independent from ATGL. For example, CGI-58 was shown to have acyltransferase activity in phosphatidic acid biosynthesis and play an ATGL-independent role in the phospholipid metabolism [132]. It was suggested that, apart from its lipolytic activity, CGI-58 may facilitate the utilization of hydrolysis products of TGs as phospholipids, and maintain TG homeostasis [132].
Obesity
Obesity is a common health problem, which is characterized by excessive lipid storage in various cell types such as adipocytes, muscle cells and hepatocytes. Different types and degrees of obesity were shown to be directly related with the lipophagic activity of cells and the size of their lipid storages [133]. Additionally, lipophagic activity was found to be upregulated in the adipose tissue of obese people, and especially in those who have particularly high intra-abdominal fat accumulation. In these people, visceral lipid distribution or hypertrophic adipocytes could also be seen concomitantly with insulin resistance [134]. Under these conditons, upregulation of lipophagy might protect cells against lipotoxicity by clearing excessive intracellular lipids, and decrease obesity-dependent mortaility in patients [134]. In fact, the amplitude of lipophagic activation may depend on the metabolic properties of individuals. For example, in patients diagnosed with type-2 diabetes which is characterized by insulin resistance, inhibition of the main energy regulator pathway, mTOR, was described as the responsible mechanism for lipophagic upregulation [135]. Yet, enhanced LD formation can sometimes lead to excessive release of FAs from LDs, and lipophagy may favor intracellular lipotoxicity under these conditons [135].
Accumulation of LDs in tissues in high amounts may cause chronic inflammation which is identified as one of the hallmarks of obesity-related metabolic disorders. Indeed, enlarged LDs in the adipose tissue may lead to cellular remodeling, and finally lead to macrophage-driven chronic inflammation [136]. Activation of macrophages is a energy-dependent mechanism and relies on FA oxidation, suggesting a possible involvement of lipophagy in this mechanism. Accordingly, lipophagy was shown to increase with obesity in adipose tissue macrophages; however, their activation was not affected, suggesting that lipophagy only regulates elavated intracellular lipid level in the adipose tissue [114]. Liu et al. have shown decreased level of lipophagy with obesity in hepatic macrophages, and inhibiton of lipophagy was shown to favor the immune responses by promoting proinflammatory macrophage activation [137]. Very recently, lipophagy was found as an essential catabolic mechanism in adipose tissue macrophages, and it increased with obesity. But intriguingly, genetic or pharmacological inhibition of lipophagy did not change lipid balance, indicating existence of another pathway critical to lysosome TG hydrolysis [138]. All these data indicate that the role of lipophagy in macrophage-driven inflammation may depend on metabolic conditions of obesity or inflammatory stimulus.
Fatty liver diseases
The important role of lipophagy regarding LD breakdown in hepatocytes suggests that impaired lipophagy may contribute to the development of steatotic liver diseases such as non-alcoholic and alcoholic steatohepatitis [133]. These diseases are characterized by increased level of lipid storage in LDs, and they progressively lead to the development of chronic liver injury and its complications, such as fibrosis and hepatocellular cancer [139, 140].
Promotion of lipophagy by resveratrol treatment was shown to attenuate methionine choline-induced non-alcoholic steatohepatitis [141]. Consistently, in a recent study, lipophagy has been found to play an important protective role in methionine choline-induced non-alcoholic steatohepatitis [142]. Lipophagy has also an important protective role in acute alcohol-induced hepatic steatosis and injury [143]. Likewise, impairment of both non-selective macroautophagy and selective CMA promoted oxidant-induced liver injury, indicating that oxidative stress resistance and liver injury is connected to autophagic activity [144]. Another report also support the protective role of lipophagy against chronic alcohol-induced hepatotoxicity caused by oxidative stress [145].
Liver fibrosis may be the first step of more serious cirrhotic liver conditions. Hepatic stellate cells (HSCs) activation for the development of this condition, and lipophagy plays a role as well [146, 147]. HSCs are quiescent cells having large lipid stores that are metabolized during activation. Increased lipophagy was shown in HSCs upon fibrotic stimuli and HSC-specific inhibiton of lipophagy blocked their activation, leading to improved liver fibrosis in mice and in human tissues [147]. On the other hand, controversy exist over the effect of lipophagy modulation in this disease. For example, pharmacological induction of lipophagy was shown to be a promissing treatment for liver fibrosis in alpha-1 antitrypsin deficiency [148]. Even though HSCs activation is a common phenomenon for fibrogenesis among tissues, whether lipophagy mediates fibrosis in other organs is still unknown.
Today, it is well known that after many years of chronic non-alcoholic or alcoholic steatotic liver diseases or hepatitis virus infection hepatocellular carcinoma (HCC) might develop. Hepatitis B virus (HBV) and hepatitis C virus (HCV) replication was shown to be induced by autophagy activation both in vitro and in vivo studies [149, 150]. A number of reviews have described the role of non-selective and/or selective autophagy in HCC, and accumulating data indicate changes of autophagic responses in this disease [133, 151]. However, there are discrepancies among studies that were published so far, and development of autophagy-based novel therapeutic strategies for HCC require further studies.
Cholesterol ester storage disease (CESD)
Cholesterol ester storage disease (CESD) is an autosomal recessive genetic disease caused by the mutation of the LIPA gene encoding lysosomal acid lipases (LAL). Deficient LAL activity lead to insufficient lipolysis and eventually intracellular accumulation of cholesterol ester and TGs [152]. The most severe form of CESD is identified as Wolman Disease that mostly presents in infancy and mainly result in infant fatality. Affected cells are characterized by the presence of lysosomes that are filled with excessive LDs and cholesterol clumps [153]. The involvement of lipophagy was shown in a study analyzed LD mobilization in macrophages [111]. LDs are the major organelles for cholesterol storage and the study demonstrated that cholesterol efflux from macrophage foam cells was regulated by lipophagy, which rely on LAL function. In line with these results, genetic knock-down of lipophagy in macrophages or knockout in the whole mice resulted in insufficient clearance of cholesterol in cells and tissues [111].
Atherosclerosis
Accumulation of cholesterol in macrophage foam cells also contribute to another common disease, atherosclerosis. The main cause of the disease is the accumulation of excessive cholesterol in arterial walls eventually leading to heart failure, stroke and cardiac dysfunction. Excessive cholesterol is esterified by ACAT enzymes and stored as cholesterolester (CE) in LDs of macrophages. Studies revealed that LD-associated proteins ACAT1 and ABCA1 regulating cholesterol esterification play crucial roles in atherosclerosis. In hyperlipidemic mice, deletion of ACAT1 results in both impairment of ABCA1 dependent cholesterol efflux and elevated atherosclerosis [154]. Moreover, among the PAT protein family of LDs, adipose differentiation-related protein (ADFP) was strongly associated with foam cell formation and atherosclerosis [155, 156].
The importance of lipophagic activation in controlling intracellular LDs level and the recent finding that lipophagy contribute LD mobilization in macrophages provides a new point of view in the pathogenesis of atherosclerosis. In addition to the dysfunction of above mentioned LD-associated proteins, it is likely that defective lipophagy may compromise the chronic exposure to excessive circulating lipids of the artery wall macrophages and may result in their remodelling into “foam cells”. However, the significance of insufficient lipophagy underlying foam cell formation or progression is still not known. On the other hand, in a recent study on the tumor-supressor gene programmed cell death 4 (PDCD4), it was demonstrated that the protein inhibited autophagy in macrophages, and Pdcd4 knockout mice displayed increased atherosclerosis, indicating a link between autophagy and atherogenesis [157].
Lipodystrophies
Apart from excessive accumulation of lipids, deficiency of LDs in cells may lead to lipodystrophies. Particularly, defects in the synthesis of neutral lipids lead to deficiencies in LD formation and lead to the formation of abnormal or degenerative white adipose tissue. Genetic causes of lipodystrophies include various genes encoding proteins related to LD synthesis, storage and regulation such as acylglycerol-phosphate acyltransferase (AGPAT2), seipin (BCSL2) and caveolin (CAV1). In addition, lamin A/C (LMNA), peroxisome proliferator-activated receptor-γ (PPARG), Akt2/protein kinase B (AKT2), and endoprotease Face-1 (ZMPSTE24) were identified as defective genes in partial lipodystrophies [158]. Insufficient LD biosynthesis in adipose tissue often leads to massive hepatic steatosis with subsequent metabolic abnormalities, including insulin resistance, diabetes, and hypertension [159].
Conclusion
In recent years, except the neutral lipid storage function of LDs, understanding of their various metabolic roles and intracellular interactions with many compartments makes this field as an attractive research area. Novel findings in selective types of autophagy, lipophagy and CMA, also contributed to a better of understanding of intracellular catabolic pathways regulating LDs. More importantly, signals that madiate the crosstalk between lipolysis and lipophagy lead to the consideration of these mechanisms as promising therapeutic targets. However there are a large number of questions that still need to be answered. Particularly, the pathophysiological relevance of LDs and their catabolic mechanisms must be further examined in order to discover potential therapies for common metabolic diseases such as obesity or atherosclerosis.
Acknowledgements
Not applicable.
Funding
This work is supported by Hacettepe University Scientific Research Projects Coordination Unit (Project number 014 D04 101,019–546). G.O is supported by The Scientific and Technological Research Council of Turkey (TUBITAK)- BIDEB 2211, as scholarship student for her PhD study.
Availability of data and materials
Not applicable.
Authors’ contributions
All authors made contribution in writing the manuscript. All authors read and approved the final manuscript.
Author’s information
D.G. is a recipient of the Turkish Academy of Sciences (TUBA) GEBIP Award, EMBO Strategical Development and Installation Grant (EMBO-SDIG), IKU Prof. Dr. Onder Oztunali Science Award and TGC Sedat Simavi Award. D.G and O.K. are recipient of 2015 Elginkan Foundation Technology Research Award.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ACS
Acyl-coA synthetase
- ADFP
Adipose differentiation-related protein
- ADRP
Adipocyte differentiation-related protein
- AGPAT
1-acylglycerol-3-phosphate O-acyltransferase
- AKT2
Akt2/protein kinase B
- ATGL
Adipose triglyceride lipase
- BCSL2
Seipin
- CAV1
Caveolin
- CCT
Phosphocholine cytidylyltransferase
- CESD
Cholesterol ester storage disease
- CGI-58
Comperative gene identification-58
- Cidec
Cell death-inducing DFF45-like effector C
- CMA
Chaperone mediated autophagy
- DAG
Diacylglycerols
- DENV
Dengue virus
- DGAT
diacylglycerol acyltransferase
- ER
Endoplasmic reticulum
- Fsp27
Fat specific protein-27
- FYCO1
FYVE and coiled-coil domains containing protein 1
- GPAT
Glycerol-3-phosphate O-acyltransferase
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- Hsc70
Heat shock cognate 70
- HSL
Hormone sensitive lipase
- LAL
Lysosomal acid lipases
- LAMP2a
Lysosomal-associated protein 2a
- LC3
Microtubule-Associated Proteins 1A and 1B, Light Chain 3
- LD
Lipid droplet
- LMNA
Lamin A/C
- MAG
Monoacylglycerol
- MAPK
Mitogen-activated protein kinase
- MEF
Mouse embryonic fibroblasts
- MGL
Monoglyceride lipase
- NLDS
Neutral lipid storage disease
- NLDSM
NLSD with cardiac myopathy
- PAP
phosphatidic acid phosphatase
- PC
Phosphatidylcholine
- PDCD4
Programmed cell death 4
- PE
Phosphatidylethanolamine
- PI
Phosphatidylinositol
- PI3K
Phosphatidylinositol 3-kinase
- PKA
Protein kinase A
- PLIN
Perilipin
- PNPLA2
Patatin-like phospholipase domain containing protein 2
- PPARG
peroxisome proliferator-activated receptor-γ
- PS
Phosphatidylserine
- RILP
Rab7-interacting lysosomal protein
- SE
Sterol ester
- SM
Sphingomyelin
- SNARE
Soluble NSF attachment receptor
- TAG
Triacylglyceride
- TIP47
Tail-interacting protein of 47 kDa
- ZMPSTE24
Endoprotease Face-1
Contributor Information
Gizem Onal, Email: gizemkaragedikli@gmail.com.
Ozlem Kutlu, Email: ozlemkutlu@sabanciuniv.edu.
Devrim Gozuacik, Email: dgozuacik@sabanciuniv.edu.
Serap Dokmeci Emre, Email: semre@hacettepe.edu.tr.
References
- 1.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie E, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 3.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–R481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herker E, Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol Metab. 2011;22:241–248. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan Q-W, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krahmer N, Farese RV, Jr, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross DA, Silver DL. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Crit Rev Biochem Mol Biol. 2014;49:304–326. doi: 10.3109/10409238.2014.931337. [DOI] [PubMed] [Google Scholar]
- 9.Ducharme NA, Bickel PE. Minireview: lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149:942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- 10.Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Oral O, Akkoc Y, Bayraktar O, Gozuacik D. Physiological and pathological significance of the molecular cross-talk between autophagy and apoptosis. Histol Histopathol. 2016;31:479–498. doi: 10.14670/HH-11-714. [DOI] [PubMed] [Google Scholar]
- 13.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 15.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–26. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lizaso A, Tan K-T, Lee Y-H. β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9:1228–1243. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaushik S, Cuervo AM. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 2016;12:432–438. doi: 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilfling F, Haas JT, Walther TC, Farese RV., Jr Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartz R, Li W-H, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Blaner WS, O'Byrne SM, Wongsiriroj N, Kluwe J, D'Ambrosio DM, Jiang H, Schwabe RF, Hillman EM, Piantedosi R, Libien J. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta Mol Cell Biol Lipids. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3 [DOI] [PMC free article] [PubMed]
- 27.Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chitraju C, Trotzmuller M, Hartler J, Wolinski H, Thallinger GG, Lass A, Zechner R, Zimmermann R, Kofeler HC, Spener F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J Lipid Res. 2012;53:2141–2152. doi: 10.1194/jlr.M028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochim Biophys Acta. 1831;2013:589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 30.McIntosh AL, Storey SM, Atshaves BP. Intracellular lipid droplets contain dynamic pools of sphingomyelin: ADRP binds phospholipids with high affinity. Lipids. 2010;45:465–477. doi: 10.1007/s11745-010-3424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, Huang X, Parton RG, Wenk MR, Walther TC, Yang H. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanghellini J, Wodlei F, von Grünberg H. Phospholipid demixing and the birth of a lipid droplet. J Theor Biol. 2010;264:952–961. doi: 10.1016/j.jtbi.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Blouin CM, Le Lay S, Eberl A, Köfeler HC, Guerrera IC, Klein C, Le Liepvre X, Lasnier F, Bourron O, Gautier J-F. Lipid droplet analysis in caveolin-deficient adipocytes: alterations in surface phospholipid composition and maturation defects. J Lipid Res. 2010;51:945–956. doi: 10.1194/jlr.M001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. 2012;287:2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalscheuer R, Wältermann M, Alvarez H, Steinbüchel A. Preparative isolation of lipid inclusions from Rhodococcus opacus and Rhodococcus ruber and identification of granule-associated proteins. Arch Microbiol. 2001;177:20–28. doi: 10.1007/s00203-001-0355-5. [DOI] [PubMed] [Google Scholar]
- 39.Ding Y, Yang L, Zhang S, Wang Y, Du Y, Pu J, Peng G, Chen Y, Zhang H, Yu J. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res. 2012;53:399–411. doi: 10.1194/jlr.M021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low KL, Shui G, Natter K, Yeo WK, Kohlwein SD, Dick T, Rao SP, Wenk MR. Lipid droplet-associated proteins are involved in the biosynthesis and hydrolysis of triacylglycerol in Mycobacterium bovis bacillus Calmette-Guerin. J Biol Chem. 2010;285:21662–21670. doi: 10.1074/jbc.M110.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jolivet P, Boulard C, Bellamy A, Larré C, Barre M, Rogniaux H, d'Andréa S, Chardot T, Nesi N. Protein composition of oil bodies from mature Brassica napus seeds. Proteomics. 2009;9:3268–3284. doi: 10.1002/pmic.200800449. [DOI] [PubMed] [Google Scholar]
- 42.Jolivet P, Roux E, d’Andrea S, Davanture M, Negroni L, Zivy M, Chardot T. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2004;42:501–509. doi: 10.1016/j.plaphy.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grillitsch K, Connerth M, Köfeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta Mol Cell Biol Lipids. 1811;2011:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beller M, Riedel D, Jänsch L, Dieterich G, Wehland J, Jäckle H, Kühnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Soulages JL, Firdaus SJ, Hartson S, Chen X, Howard AD, Arrese EL. Developmental changes in the protein composition of Manduca sexta lipid droplets. Insect Biochem Mol Biol. 2012;42:305–320. doi: 10.1016/j.ibmb.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouchoux J, Beilstein F, Pauquai T, Guerrera IC, Chateau D, Ly N, Alqub M, Klein C, Chambaz J, Rousset M. The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol Cell. 2011;103:499–517. doi: 10.1042/BC20110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 49.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 50.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta Mol Cell Biol Lipids. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tansey J, Sztalryd C, Gruia-Gray J, Roush D, Zee J, Gavrilova O, Reitman M, Deng C-X, Li C, Kimmel A. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Yu W, Cassara J, Weller PF. Phosphatidylinositide 3-kinase localizes to cytoplasmic lipid bodies in human polymorphonuclear leukocytes and other myeloid-derived cells. Blood. 2000;95:1078–1085. [PubMed] [Google Scholar]
- 55.Yu W, Bozza PT, Tzizik DM, Gray JP, Cassara J, Dvorak AM, Weller PF. Co-compartmentalization of MAP kinases and cytosolic phospholipase A2 at cytoplasmic arachidonate-rich lipid bodies. Am J Pathol. 1998;152:759. [PMC free article] [PubMed] [Google Scholar]
- 56.Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, Okamoto T, Lisanti MP. Caveolins, liquid-ordered domains, and signal transduction. Mo Cell Biol. 1999;19:7289–7304. doi: 10.1128/MCB.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurzchalia TV, Partan RG. Membrane microdomains and caveolae. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/S0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 58.Fujimoto T, Kogo H, Ishiguro K, Tauchi K, Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J Cell Biol. 2001;152:1079–1086. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Meer G. Caveolin, cholesterol, and lipid droplets? J Cell Biol. 2001;152:F29–F34. doi: 10.1083/jcb.152.5.F29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uzbekov R, Roingeard P. Nuclear lipid droplets identified by electron microscopy of serial sections. BMC Res Notes. 2013;6:386. doi: 10.1186/1756-0500-6-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Layerenza J, González P, de Bravo MG, Polo M, Sisti M, Ves-Losada A. Nuclear lipid droplets: a novel nuclear domain. Biochim Biophys Acta Mol Cell Biol Lipids. 1831;2013:327–340. doi: 10.1016/j.bbalip.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, Von Landenberg P. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- 63.Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- 64.Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta Rev Biomembr. 2000;1469:101–120. doi: 10.1016/S0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]
- 65.Long AP, Manneschmidt AK, VerBrugge B, Dortch MR, Minkin SC, Prater KE, Biggerstaff JP, Dunlap JR, Dalhaimer P. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic. 2012;13:705–714. doi: 10.1111/j.1600-0854.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- 66.Ohsaki Y, Suzuki M, Fujimoto T. Open questions in lipid droplet biology. Chem Biol. 2014;21:86–96. doi: 10.1016/j.chembiol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 69.Ellis JM, Frahm JL, Li LO, Coleman RA. Acyl-coenzyme A synthetases in metabolic control. Curr Opin Lipidol. 2010;21:212. doi: 10.1097/MOL.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khelef N, Buton X, Beatini N, Wang H, Meiner V, Chang T-Y, Farese RV, Maxfield FR, Tabas I. Immunolocalization of Acyl-Coenzyme A: CholesterolO-Acyltransferase in Macrophages. J Biol Chem. 1998;273:11218–11224. doi: 10.1074/jbc.273.18.11218. [DOI] [PubMed] [Google Scholar]
- 71.Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV., Jr The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, Rapoport TA. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep. 2014;6:44–55. doi: 10.1016/j.celrep.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang H, Galea A, Sytnyk V, Crossley M. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol. 2012;24:509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 74.Zhang S, Wang Y, Cui L, Deng Y, Xu S, Yu J, et al. Morphologically and Functionally Distinct Lipid Droplet Subpopulations. Sci Rep. 2016;6 [DOI] [PMC free article] [PubMed]
- 75.Thiam AR, Beller M. The why, when and how of lipid droplet diversity. J Cell Sci. 2017. Doi:10.1242/jcs.192021. [DOI] [PubMed]
- 76.Boström P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Fernandez-Rodriguez J, Ericson J, Nilsson T, Borén J. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 77.Boström P, Rutberg M, Ericsson J, Holmdahl P, Andersson L, Frohman MA, Borén J, Olofsson S-O. Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler Thromb Vasc Biol. 2005;25:1945–1951. doi: 10.1161/01.ATV.0000179676.41064.d4. [DOI] [PubMed] [Google Scholar]
- 78.Digel M, Ehehalt R, Fullekrug J. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 2010;584:2168–2175. doi: 10.1016/j.febslet.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 79.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chakladar A, Czech MP. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 82.Keller P, Petrie JT, De Rose P, Gerin I, Wright WS, Chiang S-H, Nielsen AR, Fischer CP, Pedersen BK, MacDougald OA. Fat-specific protein 27 regulates storage of triacylglycerol. J Biol Chem. 2008;283:14355–14365. doi: 10.1074/jbc.M708323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujimoto Y, Itabe H, Kinoshita T, Homma KJ, Onoduka J, Mori M, Yamaguchi S, Makita M, Higashi Y, Yamashita A. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- 84.Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem. 2011;286:21330–21339. doi: 10.1074/jbc.M110.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roingeard P, Melo RC. Lipid droplet hijacking by intracellular pathogens. Cell Microbiol. 2016;19 doi: 10.1111/cmi.12688. [DOI] [PubMed] [Google Scholar]
- 86.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 87.Hourioux C, Patient R, Morin A, Blanchard E, Moreau A, Trassard S, Giraudeau B, Roingeard P. The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut. 2007;56:1302–1308. doi: 10.1136/gut.2006.108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samsa MM, Mondotte JA, Iglesias NG, Assunção-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 90.Gómez-Ramos P, Asunción MM. Ultrastructural localization of intraneuronal Abeta-peptide in Alzheimer disease brains. J Alzheimers Dis. 2007;11:53–59. doi: 10.3233/JAD-2007-11109. [DOI] [PubMed] [Google Scholar]
- 91.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cole NB, Murphy DD, Grider T, Rueter S, Brasaemle D, Nussbaum RL. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein alpha-synuclein. J Biol Chem. 2002;277:6344–6352. doi: 10.1074/jbc.M108414200. [DOI] [PubMed] [Google Scholar]
- 93.Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M. Lipid Droplets Are Essential for Efficient Clearance of Cytosolic Inclusion Bodies. Dev Cell. 2015;33:603–610. doi: 10.1016/j.devcel.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 94.Souza SC, Muliro KV, Liscum L, Lien P, Yamamoto MT, Schaffer JE, Dallal GE, Wang X, Kraemer FB, Obin M. Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J Biol Chem. 2002;277:8267–8272. doi: 10.1074/jbc.M108329200. [DOI] [PubMed] [Google Scholar]
- 95.Gruber A, Cornaciu I, Lass A, Schweiger M, Poeschl M, Eder C, Kumari M, Schoiswohl G, Wolinski H, Kohlwein SD. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem. 2010;285:12289–12298. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zechner R, Zimmermann R, Eichmann TO. Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 98.Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, Tornqvist H, Zechner R, Zimmermann R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 99.J-i O, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujimoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 101.Taschler U, Radner FP, Heier C, Schreiber R, Schweiger M, Schoiswohl G, Preiss-Landl K, Jaeger D, Reiter B, Koefeler HC. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J Biol Chem. 2011;286:17467–17477. doi: 10.1074/jbc.M110.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D’Andrea S. Lipid droplet mobilization: The different ways to loosen the purse strings. Biochimie. 2016;120:17–27. doi:10.1016/j.biochi.2015.07.010. [DOI] [PubMed]
- 103.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, McNiven MA. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol. 2013;203:315–326. doi: 10.1083/jcb.201306140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 106.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y, Takahashi Y, Desai N, Zhang J, Serfass JM, Shi Y-G, et al. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci Rep. 2016;6 [DOI] [PMC free article] [PubMed]
- 108.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, De Vries R, Arias E, Harris S, Sulzer D. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khaldoun SA, Emond-Boisjoly MA, Chateau D, Carriere V, Lacasa M, Rousset M, Demignot S, Morel E. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol Biol Cell. 2014;25:118–132. doi: 10.1091/mbc.E13-06-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaini RR, Hu C-AA. Synergistic killing effect of chloroquine and androgen deprivation in LNCaP cells. Biochem Biophys Res Commun. 2012;425:150–156. doi: 10.1016/j.bbrc.2012.07.054. [DOI] [PubMed] [Google Scholar]
- 116.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lapierre LR, Gelino S, Meléndez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nguyen LN, Bormann J, Le GTT, Stärkel C, Olsson S, Nosanchuk JD, Giese H, Schäfer W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet Biol. 2011;48:217–224. doi: 10.1016/j.fgb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 119.Velikkakath AKG, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, Singh R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Velázquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet–mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reue K. A thematic review series: lipid droplet storage and metabolism: from yeast to man. J Lipid Res. 2011;52:1865–1868. doi: 10.1194/jlr.E020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fischer J, Lefèvre C, Morava E, Mussini J-M, Laforêt P, Negre-Salvayre A, Lathrop M, Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 127.Lefèvre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret J-L, Weissenbach J. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Emre S, Ünver N, Evans SE, Yüzbaşıoğlu A, Gürakan F, Gümrük F, Karaduman A. Molecular analysis of Chanarin-Dorfman syndrome (CDS) patients: Identification of novel mutations in the ABHD5 gene. Eur J Med Genet. 2010;53:141–144. doi: 10.1016/j.ejmg.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 129.Nur BG, Gencpinar P, Yuzbasıoglu A, Emre SD, Mihci E. Chanarin-Dorfman syndrome: Genotype-Phenotype Correlation. Eur J Med Genet. 2015;58:238–242. doi: 10.1016/j.ejmg.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 130.Waheed N, Cheema HA, Suleman H, Mushtaq I, Fayyaz Z. Chanarin-Dorfman Syndrome. J Coll Physicians Surg Pak. 2016;26:787. [PubMed] [Google Scholar]
- 131.Schweiger M, Lass A, Zimmermann R, Eichmann TO. Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–EE96. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 132.Ghosh AK, Ramakrishnan G, Chandramohan C, Rajasekharan R. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J Biol Chem. 2008;283:24525–24533. doi: 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology. 2016;150:328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR, Greenberg AS, Elazar Z, Bashan N, Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 135.Ost A, Svensson K, Ruishalme I, Brannmark C, Franck N, Krook H, Sandstrom P, Kjolhede P, Stralfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med. 2010;16:235–246. doi: 10.2119/molmed.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hubler MJ, Kennedy AJ. Role of lipids in the metabolism and activation of immune cells. J Nutr Biochem. 2016;34:1–7. doi: 10.1016/j.jnutbio.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grijalva A, Xu X, Ferrante AW. Autophagy is dispensable for macrophage mediated lipid homeostasis in adipose tissue. Diabetes. 2016. 2016;65(4):967–80. doi:10.2337/db15-1219. [DOI] [PMC free article] [PubMed]
- 139.Czaja MJ. Function of autophagy in nonalcoholic fatty liver disease. Dig Dis Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dolganiuc A, Thomes PG, Ding WX, Lemasters JJ, Donohue TM. Autophagy in Alcohol-Induced Liver Diseases. Alcohol Clin Exp Res. 2012;36:1301–1308. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ji G, Wang Y, Deng Y, Li X, Jiang Z. Resveratrol ameliorates hepatic steatosis and inflammation in methionine/choline-deficient diet-induced steatohepatitis through regulating autophagy. Lipids Health Dis. 2015;14:1. doi: 10.1186/s12944-015-0139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen R, Wang Q, Song S, Liu F, He B, Gao X. Protective role of autophagy in methionine–choline deficient diet-induced advanced nonalcoholic steatohepatitis in mice. Eur J Pharmacol. 2016;770:126–133. doi: 10.1016/j.ejphar.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 143.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lu Y, Cederbaum AI. Autophagy Protects against CYP2E1/Chronic Ethanol-Induced Hepatotoxicity. Biomol Ther. 2015;5:2659–2674. doi: 10.3390/biom5042659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thoen LF, Guimarães EL, Dollé L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 147.Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 149.Rautou P-E, Cazals-Hatem D, Feldmann G, Mansouri A, Grodet A, Barge S, Martinot-Peignoux M, Duces A, Bièche I, Lebrec D. Changes in autophagic response in patients with chronic hepatitis C virus infection. Am J Pathol. 2011;178:2708–2715. doi: 10.1016/j.ajpath.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taguwa S, Kambara H, Fujita N, Noda T, Yoshimori T, Koike K, Moriishi K, Matsuura Y. Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol. 2011;85:13185–13194. doi: 10.1128/JVI.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dash S, Chava S, Chandra PK, Aydin Y, Balart LA, Wu T. Autophagy in hepatocellular carcinomas: From pathophysiology to therapeutic response. Hepat Med. 2016;8:9. doi: 10.2147/HMER.S63700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reynolds T. Cholesteryl ester storage disease: a rare and possibly treatable cause of premature vascular disease and cirrhosis. J Clin Pathol. 2013:doi:10.1136/jclinpath-2012-201302. [DOI] [PubMed]
- 153.Assmann G, Fredrickson D. Acid lipase deficiency: Wolman's disease and cholesteryl ester storage disease. In: Stanbury JB et al., editors. Metabolic basis of inherited disease. New York: McGraw Hill; 1983.
- 154.Su YR, Dove DE, Major AS, Hasty AH, Boone B, Linton MF, Fazio S. Reduced ABCA1-Mediated Cholesterol Efflux and Accelerated Atherosclerosis in Apolipoprotein E–Deficient Mice Lacking Macrophage-Derived ACAT1. Circulation. 2005;111:2373–2381. doi: 10.1161/01.CIR.0000164236.19860.13. [DOI] [PubMed] [Google Scholar]
- 155.Larigauderie G, Furman C, Jaye M, Lasselin C, Copin C, Fruchart J-C, Castro G, Rouis M. Adipophilin enhances lipid accumulation and prevents lipid efflux from THP-1 macrophages potential role in atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:504–510. doi: 10.1161/01.ATV.0000115638.27381.97. [DOI] [PubMed] [Google Scholar]
- 156.Paul A, Chang BH-J, Li L, Yechoor VK, Chan L. Deficiency of adipose differentiation-related protein impairs foam cell formation and protects against atherosclerosis. Circ Res. 2008;102:1492–1501. doi: 10.1161/CIRCRESAHA.107.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang T, Zhang L, Hu J, Duan Y, Zhang M, Lin J, Man W, Pan X, Jiang Z, Zhang G. Mst1 participates in the atherosclerosis progression through macrophage autophagy inhibition and macrophage apoptosis enhancement. J Mol Cell Cardiol. 2016;98:108–116. doi: 10.1016/j.yjmcc.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 158.Garg A, Agarwal AK. Lipodystrophies: disorders of adipose tissue biology. Biochim Biophys Acta. 2009;1791:507–513. doi: 10.1016/j.bbalip.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hegele RA. Phenomics, lipodystrophy, and the metabolic syndrome. Trends Cardiovasc Med. 2004;14:133–137. doi: 10.1016/j.tcm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 160.Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- 161.III JRY, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 162.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 163.Turró S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernández MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 164.Wan H-C, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- 166.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta Mol Cell Res. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 167.Wang H, Wei E, Quiroga AD, Sun X, Touret N, Lehner R. Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol Biol Cell. 2010;21:1991–2000. doi: 10.1091/mbc.E09-05-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Khor VK, Ahrends R, Lin Y, Shen W-J, Adams CM, Roseman AN, Cortez Y, Teruel MN, Azhar S, Kraemer FB. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One. 2014;9:e105047. doi: 10.1371/journal.pone.0105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jäckle H, Beller M. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Sci. 2013;126:2198–2212. doi: 10.1242/jcs.120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Goo Y-H, Son S-H, Kreienberg PB, Paul A. Novel Lipid Droplet–Associated Serine Hydrolase Regulates Macrophage Cholesterol MobilizationSignificance. Arterioscler Thromb Vasc Biol. 2014;34:386–396. doi: 10.1161/ATVBAHA.113.302448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Heid H, Rickelt S, Zimbelmann R, Winter S, Schumacher H, Dörflinger Y, Kuhn C, Franke WW. On the formation of lipid droplets in human adipocytes: the organization of the perilipin–vimentin cortex. PLoS One. 2014;9:e90386. doi: 10.1371/journal.pone.0090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005:163–74. [DOI] [PubMed]
- 173.Pol A, Martin S, Fernández MA, Ingelmo-Torres M, Ferguson C, Enrich C, Parton RG. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. 2005;16:2091–2105. doi: 10.1091/mbc.E04-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- 176.Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem. 2002;277:4261–4270. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 177.Alonzi T, Agrati C, Costabile B, Cicchini C, Amicone L, Cavallari C, Della Rocca C, Folgori A, Fipaldini C, Poccia F. Steatosis and intrahepatic lymphocyte recruitment in hepatitis C virus transgenic mice. J Gen Virol. 2004;85:1509–1520. doi: 10.1099/vir.0.19724-0. [DOI] [PubMed] [Google Scholar]
- 178.Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang H, Zhang S, Peng G, Yang F. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein aI. J Proteome Res. 2011;10:4757–4768. doi: 10.1021/pr200553c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


