Abstract
Background:
The atypical antipsychotic clozapine is effective in treatment-resistant schizophrenia; however, the success or failure of clozapine therapy is substantially affected by the variables that impact the clozapine blood concentration. Thus, elucidating the inter-individual differences in clozapine pharmacokinetics can facilitate the personalized therapy.
Methods:
Since a potential role in clozapine metabolism is assigned to CYP1A2, CYP2C19, CYP2D6 and CYP3A enzymes, the association between the patients’ CYP status (CYP genotypes, CYP expression) and clozapine clearance was evaluated in 92 psychiatric patients.
Results:
The patients’ CYP2C19 or CYP2D6 genotypes and CYP1A2 expression seemed to have no effect on clozapine serum concentration, whereas CYP3A4 expression significantly influenced the normalized clozapine concentration (185.53±56.53 in low expressers vs 78.05±29.57 or 66.52±0.25 (ng/mL)/(mg/kg) in normal or high expressers, P<.0001), in particular that the patients expressed CYP1A2 at a relatively low level. The functional CYP3A5*1 allele seemed to influence clozapine concentrations in those patients who expressed CYP3A4 at low levels. The dose requirement for the therapeutic concentration of clozapine was substantially lower in low CYP3A4 expresser patients than in normal/high expressers (2.18±0.64 vs 4.98±1.40 mg/kg, P<.0001). Furthermore, significantly higher plasma concentration ratios of norclozapine/clozapine and clozapine N-oxide/clozapine were observed in the patients displaying normal/high CYP3A4 expression than in the low expressers.
Conclusion:
Prospective assaying of CYP3A-status (CYP3A4 expression, CYP3A5 genotype) may better identify the patients with higher risk of inefficiency or adverse reactions and may facilitate the improvement of personalized clozapine therapy; however, further clinical studies are required to prove the benefit of CYP3A testing for patients under clozapine therapy.
Keywords: personalized clozapine therapy, CYP3A4 expression, CYP3A5 genotype, CYP1A2 expression
Significance Statement
Inter-individual variability of cytochrome P450 enzymes (CYP1A2, CYP3A4, CYP2C19, CYP2D6) has been attributed to substantial differences in clozapine plasma concentrations and in drug response. The patients’ CYP2C19 and CYP2D6 genotypes appeared to have no effect on steady-state clozapine concentrations, even as CYP1A2 expression influenced neither the clozapine levels nor the norclozapine/clozapine ratio. The relative importance of these enzymes in clozapine clearance seemed to be lower, whereas the patients’ CYP3A-status (CYP3A4 expression and CYP3A5 genotype) was found to potentially influence normalized clozapine concentration and dose requirement, in particular that the patients expressed CYP1A2 at a relatively low level. For the optimal plasma concentrations (200–600 ng/mL), CYP3A-status guided clozapine dosing was proposed (4.98 mg/kg for the normal/high CYP3A4 expressers and CYP3A5*1 carriers, whereas 2.18 mg/kg for low CYP3A4 expressers). Personalized medication taking patients’ CYP3A status into account may facilitate the improvement of individual clozapine therapy, leading to the dosage optimization for a more effective therapy.
Introduction
Clozapine, the prototype of second-generation antipsychotics, is efficacious against both positive and negative symptoms of schizophrenia and potently reduces aggression and the risk of suicide. It is proved to be effective for therapy of treatment-resistant patients (Lally et al., 2016). The CATIE study found that switching to clozapine after therapeutic failure was more effective than to any other newer antipsychotic (McEvoy et al., 2006), and the SOHO program confirmed the superiority of clozapine over other antipsychotics (Attard et al., 2012). However, there are some adverse effects; for example, weight gain, increased fasting glucose concentration, seizures, and sedation are more common with clozapine than with other atypical antipsychotics (Asenjo-Lobos et al., 2010). Monitoring of hematological parameters is obligatory because of the risk for agranulocytosis (Ng et al., 2014). The plasma concentration of clozapine rather than the dose is in close association with most side effects. The incidence of adverse events is more frequent in patients with plasma concentrations >750 ng/mL (Ulrich et al., 2003). At the same dose, considerable variability in clozapine concentration has been observed, which is attributed to the substantial inter-individual differences in clozapine metabolism (Couchman et al., 2010); therefore, therapeutic drug monitoring is highly recommended (Hiemke et al., 2011).
Clozapine is extensively metabolized forming two major metabolites, the pharmacologically active norclozapine and the inactive clozapine N-oxide as well as minor hydroxylated metabolites (Dain et al., 1997). Norclozapine contributes to the unique antipsychotic profile of clozapine with low risk for extrapyramidal side effects and improvement of cognitive function; however, it has some attributes distinct from those of the parent compound (norclozapine: D2/D3 partial agonist, clozapine: D2/D3 inverse agonist/antagonist) (Lameh et al., 2007). Various cytochrome P450 (CYP) enzymes (CYP1A2, CYP2C19, CYP2D6, CYP3A4) have been demonstrated to be involved in clozapine metabolism; however, the clinical relevance of these enzymes has to be clarified. CYP3A4 seems to catalyze N-oxidation of clozapine, whereas the formation of norclozapine appears to be more complex (Linnet and Olesen, 1997). CYP1A2 and CYP3A4 were assumed to be the major enzymes responsible for the N-demethylation pathway (Fang et al., 1998), and several clinical reports provided indirect evidence for their participation in clozapine metabolism. Concomitant treatment with fluvoxamine, the potent inhibitor of several CYPs (CYP1A2, CYP2C19, CYP3A4), significantly decreased the plasma norclozapine/clozapine ratios and patients’ clozapine dose requirement (Lu et al., 2000; Jones et al., 2007). Cigarette smoking, inducing CYP1A2 expression, seems to increase clozapine clearance; however, CYP1A2 genetic polymorphisms do not have significant clinical effect (Weide et al., 2003). In vitro studies have demonstrated that at high clozapine concentrations, CYP3A4 plays a primary role in N-demethylation (Olesen and Linnet, 2001; Eiermann et al., 1997, Fang et al., 1998). The relative importance of CYP2C19 in clozapine N-demethylation was demonstrated in vitro (Linnet and Olesen, 1997; Fang et al., 1998), and a minor role of CYP2D6 in metabolic pathways other than N-demethylation was assumed (Fischer et al., 1992; Olesen and Linnet, 2001). A possible contribution of CYP2C19 to in vivo clozapine metabolism was suggested by Jaquenoud-Sirot et al. (2009); however, in vivo investigations by Dahl et al. (1994) and Ghassabian et al. (2010) failed to confirm the correlation between the CYP2C19 or CYP2D6 phenotypes and the serum clozapine concentrations or the norclozapine/clozapine ratio.
CYP activities can display more than 100-fold inter-individual variability (Temesvári et al., 2012), which is partly attributed to genetic factors. Genetic polymorphisms of CYP enzymes can explain some differences in clozapine metabolism; however, nongenetic factors (hormones, diseases, age, medication, smoking) can modify CYP activities, resulting in transient poor (or extensive) metabolism. The genotype determines the potential for the expression of functional or nonfunctional CYP enzyme, whereas nongenetic factors give rise to altered phenotypes. Homozygous wild genotype, predicted to be translated to CYP enzyme with normal function, may be switched into poor (or extensive) metabolism due to phenoconversion (Shah and Smith, 2015). Patients’ clozapine-metabolizing capacity can be estimated by the evaluation of CYP genotypes and by CYP expression. We previously described a complex diagnostic system (CYPtest) that determines drug metabolizing capacity by CYP genotyping and by the current CYP expression in leukocytes. CYP3A4 and CYP1A2 mRNA levels in leukocytes were proven to inform about the hepatic CYP3A4 and CYP1A2 activities (Temesvári et al., 2012). Information on patients’ CYP status can refine the personalized clozapine therapy, facilitating the appropriate dosage, and can predict the risk of outlying from the therapeutic concentration range.
The goals of the present study were to estimate the association between the patients’ drug-metabolizing capacity (CYP expressions and CYP genotypes) and their clozapine therapy (dose, clozapine levels), and to analyze the influence of CYP-status on the patients’ dose requirements, which can contribute to the improvement of patients’ personalized medication.
METHODS
Patients
Ninety-two inpatients at the Department of Psychiatry and Psychotherapy, Semmelweis University (Budapest, Hungary) were enrolled. The patients on stable clozapine therapy for at least 2 weeks were included in the study. Written informed consent was obtained from all participants. The study was approved by the Hungarian Committee of Science and Ethics. Patients’ demographic data and medication was recorded (Table 1). Of 92 patients, 18 patients were prescribed clozapine as monotherapy, whereas the majority of the patients received concomitant medications (haloperidol, risperidone, aripiprazole, or amisulpride) or anticonvulsant drugs (clonazepam, valproic acid, or lamotrigine) as adjunctive therapy. Nearly one-half of the patients (n=41) received beta-adrenergic blockers (propranolol or metoprolol). The clozapine therapy was applied according to the conventional clinical protocol, initiated at low dosage (12.5–25 mg/d), and subsequently titrated in increments of 12.5 to 50 mg/d until the optimal response was achieved.
Table 1.
Patients’ Demographic and Clinical Characteristics
| n | % | |
|---|---|---|
| Patients | 92 | |
| Sex, male/female | 38/54 | 41.3/58.7 |
| Diagnosis | ||
| Schizophrenia | 73 | 79.4 |
| Schizoaffective disorder | 15 | 16.3 |
| Other | 4 | 4.3 |
| Current smokers | 35 | 38.0 |
| Age (y), median (min-max) | 42 (17–74) | |
| Clozapine daily dose (mg), median (min-max) | 200 (50–700) | |
| Bodyweight (kg), median (min-max) | 76 (51–129) | |
| Serum levels, median (min-max) | ||
| Clozapine (ng/mL) | 232 (23–1400) | |
| Norclozapine (ng/mL) | 156 (16–894) | |
| Clozapine N-oxide (ng/mL) | 90 (0–383) | |
| CYP genotype | ||
| CY2C19* | ||
| *1/*1 | 35 | 38.0 |
| *1/mut | 23 | 25 |
| mut/mut | 2 | 2.2 |
| mut/*17 | 6 | 6.5 |
| *1/*17 | 20 | 21.7 |
| *17/*17 | 6 | 6.5 |
| CYP2D6** | ||
| *1/*1 | 36 | 39.2 |
| *1/mut | 37 | 40.2 |
| mut/mut | 14 | 15.2 |
| *1/*1xN | 5 | 5.4 |
| CYP3A4 | ||
| *1/*1 | 74 | 80.4 |
| *1/*1B | 5 | 5.4 |
| *1/*22 | 13 | 14.2 |
| CYP3A5 | ||
| *1/*3 | 10 | 10.9 |
| *3/*3 | 82 | 89.1 |
| CYP expression | ||
| CYP1A2 | ||
| low expressers | 65 | 70.7 |
| normal expressers | 27 | 29.3 |
| CYP3A4 | ||
| low expressers | 27 | 29.3 |
| normal expressers | 62 | 67.4 |
| high expressers | 3 | 3.3 |
*CYP2C19 mut: *2, *3 and *4; **CYP2D6 mut: *3, *4, *5, *6, *10 and *41.
Assaying CYP status
Patients’ CYP status was determined by CYP2C19, CYP2D6, CYP3A4, and CYP3A5 genotyping and by analyzing CYP1A2 and CYP3A4 expression in leukocytes. Genomic DNA and leukocytes were isolated from the peripheral blood samples as previously described by Temesvári et al. (2012). Hydrolysis single-nucleotide polymorphism analysis for CYP2C19*2, CYP2C19*3, CYP2C19*4, CYP2C19*17, CYP2D6*3, CYP2D6*4, CYP2D6*6, CYP2D6*10, CYP2D6*41, CYP3A4*1B, CYP3A4*22, and CYP3A5*3 was performed using TaqMan probes (BioSearch Technologies, Novato, CA). CYP2D6*5 (gene deletion) and CYP2D6 duplication were analyzed by TaqMan Copy Number Assay (ThermoFisher Scientific, Waltham, MA). Since CYP1A2 expression was found to be poorly associated with the presence of CYP1A2 SNPs (Ferrari et al., 2012), the patients’ CYP1A2 activity was estimated by CYP1A2 mRNA levels in leukocytes, and not by CYP1A2-genotype.
For assaying CYP expressions, RNA was isolated from leukocytes, reverse transcribed into single-stranded cDNA using the Maxima First Strand cDNA Synthesis Kit (ThermoFisher Scientific), and then real-time PCR was performed using KAPA Fast Probes Mastermix (KAPA Biosystems, Cape Town, South Africa) and TaqMan probes. The quantities of CYP1A2 and CYP3A4 mRNAs relative to that of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were determined. GAPDH expression is constant in all cells and independent of experimental conditions; therefore, its expression was set to 1 and CYP mRNA levels were normalized by GAPDH expression. Three categories of CYP expressions were applied to describe low, normal, and high expressers. The cut-off values for CYP mRNA levels in leukocytes have been previously established on the basis of the cut-off values for the hepatic CYP enzyme activities (CYP1A2: phenacetin O-dealkylation; CYP3A4: nifedipine oxidation or midazolam 1’- and 4-hydroxylation). The cut-off values for CYP1A2 (10–5 and 5*10–4) and CYP3A4 (10–6 and 10–4) allowed a distinction between low, normal, and high expressers (Temesvári et al., 2012).
Blood Concentrations of Clozapine and Its Metabolites
The blood samples were taken 12 hours after the evening dose of clozapine. The steady-state concentrations of clozapine, norclozapine, and clozapine N-oxide were determined by LC-MS/MS. Chromatographic separation was performed using an Inertsil ODS-4 (75×2.1 mm, 3 µm) column (GL Sciences Inc., Tokyo, Japan) and mobile phases of acetonitrile and 0.1% formic acid in gradient running mode. The samples were analyzed using positive electrospray ionization and multiple reaction monitoring mode for quantitation of the parent compound and its metabolites (m/z 327/270 and 327/192 for clozapine; m/z 313/270 and 313/192 for norclozapine; m/z 343/256 and 343/192 for clozapine N-oxide). Normalized clozapine concentrations were calculated by dividing the concentration values by the corresponding 24-hour dose on a mg/kg basis.
Data Analysis
The statistical significance of demographic data, CYP1A2 and CYP3A4 expressions, CYP2C19, CYP2D6 and CYP3A5 genotypes as covariates of clozapine, norclozapine and clozapine N-oxide concentrations was analyzed by ANOVA using linear model of covariate effects with constant terms. Several mathematical models (nonlinear Artificial Neuronal Network and linear with various input combinations) were tested to identify the key factor(s) in steady-state clozapine concentrations; however, the best models for the optimal prediction with the smallest standard deviation of model prediction and the smallest number of model parameters were all linear. The dose and patients’ CYP3A4 expression were identified as significant parameters for clozapine concentration (P<.0001). The model provided the best prediction when clozapine dose was divided by the patients’ bodyweight.
The statistical model for the clozapine serum concentration and for the effects of dose and bodyweight on the concentration was defined as:
where cclz is clozapine trough concentration expressed as ng/mL, dose is clozapine dose in mg, and bw is bodyweight in kg. Model parameters k0 and k1 were estimated for low and normal/high CYP3A4 expressers individually. To estimate the optimal dose, the model for clozapine concentration was resolved for the dose:
The lower and upper limits for the optimal dose were estimated from the lower and the upper limits of the optimal clozapine concentration range of 200 to 600 ng/mL. Matlab R2009b was used to perform the analysis and to calculate the optimal dosing.
The data of normalized clozapine concentrations, the metabolite/parent drug ratios, and the dose requirements for the optimal therapeutic level in the patients were expressed as the median and range or mean±SD. Between-group differences were calculated by the use of Kruskal–Wallis ANOVA followed by Dunn’s multiple comparisons test. P<.05 was considered to be statistically significant.
RESULTS
Patients’ Drug-Metabolizing Capacity and Clozapine Exposure
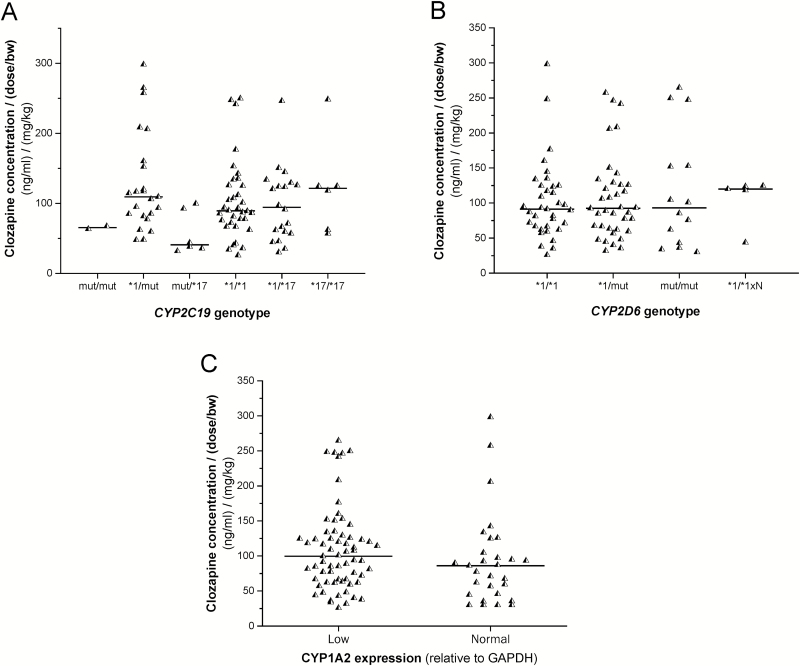
Of 92 patients, 31 carried at least one CYP2C19 loss-of-function allele (CYP2C19*2, CYP2C19*3, or CYP2C19*4), and 32 carried the gain-of-function CYP2C19*17 allele. CYP2C19 genotype seemed to influence neither the clozapine concentrations normalized by the dose nor the concentrations normalized by the dose/bodyweight. No significant differences in clozapine concentrations were observed between the carriers of one or two loss-of-function alleles and the patients with CYP2C19*1/*1 genotype or those with gain-of-function allele (CYP2C19*17) associated with increased CYP2C19 expression (P=.1083) (Figure 1A). Furthermore, no significant association was found between the normalized clozapine concentrations and the patients’ CYP2D6 genotypes (P=.8881) (Figure 1B).
Figure 1.
The influence of the patients’ CYP2C19 (A), CYP2D6 genotypes (B), and CYP1A2 expression (C) on serum clozapine concentrations. GAPDH, glyceraldehide 3-phosphate dehydrogenase; mut, loss-of-function CYP allele; ×N, allele duplication.
CYP1A2 expression assays revealed that a substantial portion of the patients (70.7%) was low CYP1A2 expressers and less than one-third of the patients expressed CYP1A2 at normal level, whereas no high CYP1A2 expresser was found. Smoking is a potent inducer of CYP1A2, increasing CYP1A2 expression and enzyme activity; however, in the present study, the smoker/nonsmoker ratio was the same in the low and normal CYP1A2 expresser groups (10:17 vs 25:40, P=1.00). Furthermore, the normalized clozapine concentrations were comparable in the patients expressing CYP1A2 at low and normal level (P=.6639) (Figure 1C).
The role of CYP3A4 in the metabolism of clozapine at therapeutic or higher concentrations has been demonstrated; however, the contribution of the highly polymorphic CYP3A5 must be clarified. In the present study, 10 subjects carried the functional CYP3A5*1 allele, whereas most of the patients were CYP3A5 nonexpressers (CYP3A5*3/*3) and were therefore expected to lack the functional CYP3A5 enzyme. Of 92 patients, 18 carried CYP3A4*22 or CYP3A4*1B alleles, resulting in reduced or increased expression of CYP3A4, respectively; however, these alleles cannot explain the inter-individual differences in CYP3A4 mRNA levels. For the categorization of the patients regarding CYP3A4 expression, more useful information can be obtained from CYP3A4 mRNA levels than from CYP3A4 genotyping. CYP3A4 expression assays revealed that about two-thirds of the patients expressed CYP3A4 at normal level, approximately 30% was low CYP3A4 expressers, and 3 patients displayed high CYP3A4 expression (Table 1). One of the high CYP3A4 expressers carried CYP3A5*1, anticipating rapid metabolism of CYP3A substrates. Strong association was found between the patients’ CYP3A4 expression and normalized clozapine serum levels (Figure 2A). In CYP3A5 nonexpressers, the normalized concentrations were significantly higher in those displaying low CYP3A4 mRNA levels than in normal or high CYP3A4 expressers (185.53±56.53 vs 78.05±29.57 or 66.52±0.25 (ng/mL)/(mg/kg), P<.0001). The patients’ CYP3A5 genotype was significantly associated with clozapine concentrations in those patients who expressed CYP3A4 at low levels (P<.0001). Further confirmation of the role of CYP3A5 is needed because of the relatively small number of CYP3A5 expressers.
Figure 2.
The influence of the patients’ CYP3A4 expression and CYP3A5 genotype on serum clozapine concentrations. (A) Clozapine concentrations normalized by the dose and the bodyweight in CYP3A5 expressers (CYP3A5*1/*3) and nonexpresser patients expressing CYP3A4 at low, normal, and high levels, and (B) linear models for low (triangles) and normal/high (points) CYP3A4 expresser patients. The black points (A) indicate low CYP3A4 expresser patients carrying CYP3A5*1. The white point (A) indicates a high CYP3A4 expresser patient carrying CYP3A5*1. *P<.0001. GAPDH, glyceraldehide 3-phosphate dehydrogenase.
According to the present results, the patients’ CYP2C19 or CYP2D6 genotypes and CYP1A2 expression seemed to have no effect on clozapine concentrations, whereas CYP3A4 expression significantly influenced the steady-state levels of clozapine. The patients were divided into 4 groups according to the CYP3A5 genotype (CYP3A5 expressers, nonexpressers) and CYP3A4 expression (low, normal, and high). Significantly higher (about 2-fold) clozapine concentration was observed in CYP3A5 nonexpresser patients displaying low CYP3A4 expression than in all the others (Figure 2A). However, modelling of the association between CYP3A4 expression and clozapine concentrations identified only 2 groups. The first group consisted of the CYP3A5 nonexpressers with low CYP3A4 mRNA levels, whereas the remaining 3 combinations (normal and high CYP3A4 expressers with CYP3A5*3/*3 genotype, and patients with CYP3A5*1/*3 genotype) were grouped, since the separate models for these combinations did not result in significantly different parameters. The same model was applied for the 2 groups; however, the model parameter values were different (ko and k1, Figure 2B). The models were able to predict the normalized clozapine concentrations with the standard deviations of 15.68 and 6.65 ng/mL for the low CYP3A4 expressers with CYP3A5*3/*3 genotype and the rest of the patients, respectively.
Optimization of Clozapine Dose
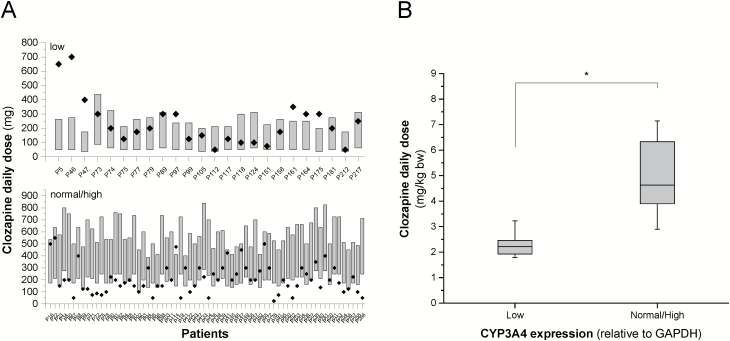
According to the consensus guideline (Hiemke et al., 2011), a clozapine concentration between 350 and 600 ng/mL is recommended for the treatment of schizophrenia; however, lower serum concentrations (200–300 ng/mL) were demonstrated to be as effective as high concentrations (300–450 ng/mL) (Zwaag et al, 1996; Ulrich et al, 2003). Therefore, the concentration of 200 ng/mL was considered as the lower threshold. The 2 models for prediction of clozapine concentration in the patients expressing CYP3A4 at low or normal/high levels had comparable prediction quality (Figure 2B) and were found to be applicable for prediction of optimal dosing. The dose requirement for therapeutic concentration was predicted for each patient setting the minimal dose adjustment to 12.5 mg (the lowest content of clozapine products available is 25 mg, and the pills can be split in half) (Figure 3A). The models proposed significantly lower daily dose for the patients expressing CYP3A4 at a low level than for those with normal/high CYP3A4 mRNA levels or with CYP3A5*1/*3 genotype (the dose range of 51.04–258.33 vs 227.39–586.38 mg/d; P<.0001), whereas the actual dosage applied for low and normal/high CYP3A4 expresser patients or CYP3A5 expressers was identical (245.37±158.88 vs 208.98±120.69 mg/d; P=.4166). In 38% of the patients, the clozapine daily dose was found to be out of the dose range predicted by the models, that is, approximately one-third of the patients might have received inadequate doses. Underdosing occurred more frequently, primarily in the patients expressing CYP3A4 at a normal/high level or in CYP3A5 expressers (Figure 3A). Moreover, the routine dosing regimen appears to be appropriate for the patients with low CYP3A4 mRNA levels rather than for normal/high expressers. Seven of the 24 low expresser patients were overdosed, whereas the clozapine therapy of 17 patients (71%) in the low CYP3A4 expresser group resulted in therapeutic concentrations of clozapine. The mathematical model displayed clozapine dosing to be optimal for 40 patients in the normal/high CYP3A4 expresser and CYP3A5 expresser groups (n=68), whereas 41% were underdosed. An additional approach was also applied for the calculation of clozapine dose requirement for the therapeutic concentrations, and we inferred the dose requirement from the dosing applied in those patients who displayed clozapine concentrations in the range of 200 to 600 ng/mL (Figure 3B). The low CYP3A4 expresser patients required a significantly lower (50%) dose of clozapine for the optimal serum level than normal/high CYP3A4 expressers (2.18±0.64 mg/kg vs 4.98±1.40 mg/kg; P<.0001) (Figure 3B).
Figure 3.
The influence of the patients’ CYP3A4 expression on clozapine dose requirements. (A) Dose requirements for therapeutic concentration of clozapine calculated from the models (bars) and applied clozapine doses (points) in low and normal/high CYP3A4 expresser patients, and (B) dose requirements calculated from applied doses in the patients displaying therapeutic concentrations of clozapine (200–600 ng/mL). *P<.0001. GAPDH, glyceraldehide 3-phosphate dehydrogenase.
Clozapine Metabolite Concentrations in Patients with Various CYP3A4 Expression
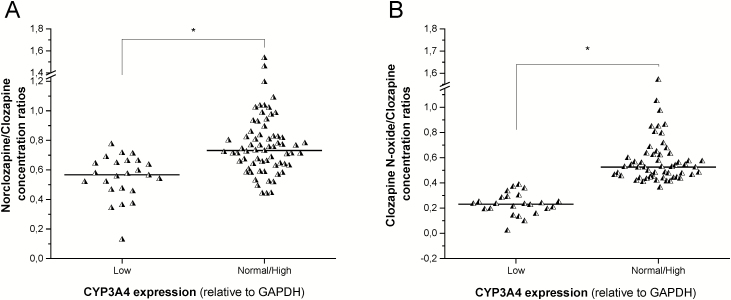
Several CYPs are assumed to be responsible for the formation of norclozapine, while CYP3A4 is likely to catalyze the clozapine N-oxidation. The formation of both principal metabolites widely varied in individuals (Table 1); however, the serum concentrations of clozapine metabolites appeared to be associated merely with the patients’ CYP3A4 expression and not with CYP1A2 mRNA levels. Strong association was observed between the metabolite/clozapine ratios and CYP3A4 mRNA levels. The patients who expressed CYP3A4 at a low level displayed significantly lower norclozapine/clozapine ratios than the CYP3A4 normal/high expressers or those who carry CYP3A5*1 (0.5521±0.1535 vs 0.7655±0.2088; P<.0001) (Figure 4A). Furthermore, significantly lower clozapine N-oxide/clozapine ratios were observed in the patients displaying low CYP3A4 expression than in CYP3A4 normal/high expressers (0.2279±0.0887 vs 0.5659±0.1960, P<.0001) (Figure 4B). The strong association between the patients’ CYP3A status and the serum clozapine concentrations or the metabolite/clozapine ratios demonstrated the potential role of CYP3A enzymes in clozapine clearance. Nevertheless, the norclozapine/clozapine concentration ratios were comparable in patients expressing CYP1A2 at a low or normal level (0.6988±0.2341 vs 0.7070±0.1824; P=0.3966).
Figure 4.
The influence of the patients’ CYP3A4 expression on the metabolite/clozapine concentration ratios. (A) Norclozapine/clozapine concentrations, and (B) clozapine N-oxide/clozapine concentrations in patients with various CYP3A4 expression. *P<.0001. GAPDH, glyceraldehide 3-phosphate dehydrogenase.
Discussion
Clozapine is extensively metabolized, and approximately 2% of the administered dose is excreted as unchanged parent compound (Dain et al., 1997). The plasma concentration in patients is strongly influenced by their clozapine-metabolizing capacity; therefore, therapeutic drug monitoring for clozapine is strongly recommended to avoid the risk of intolerance/intoxication or the risk of relapse due to subtherapeutic clozapine plasma concentration (Hiemke et al., 2011). Several patient-related factors (gender, age, smoking behavior) have been reported to influence the elimination of clozapine; however, the significance of the associations between these factors and clozapine concentrations varies from cohort to cohort and often depends on data normalization (Haring et al., 1989, 1990; Tang et al., 2007). In addition to gender, age, and smoking, disease and co-medication can also modify patients’ drug-metabolizing activity due to the phenoconversion of CYP enzyme(s) (Shah and Smith, 2015). The role of several CYP enzymes in clozapine metabolism has been assumed; however, the dosing strategy applied in clinical practice does not routinely take the patients’ CYP status into consideration, because the clinical relevance of these CYPs has not been clearly evidenced.
In vitro studies using cDNA-expressed CYP enzymes have demonstrated the contribution of CYP1A2, CYP2C19, CYP3A4, and CYP2D6 to clozapine metabolism (Linnet and Olesen, 1997; Fang et al., 1998; Olesen and Linnet, 2001); however, hepatic microsomal metabolism hardly supported a significant role of CYP2C19 and CYP2D6 (Fang et al., 1998; Tugnait et al., 1999). Our results indicated that CYP2C19 and CYP2D6 were not likely to have a primary role in clozapine clearance. These findings are consistent with the results of previous studies showing that the patients’ response to clozapine treatment did not correlate with their CYP2D6 genotype (Arranz et al., 1995), and the association between clozapine clearance in patients and the polymorphic expression of either CYP2D6 or CYP2C19 have failed to be demonstrated (Dahl et al., 1994). In contrast, Jaquenoud-Sirot et al. (2009) reported higher clozapine concentrations in patients with CYP2C19*2/*2 genotype than in those carrying non-CYP2C19*2/*2 genotypes. For normalization of clozapine concentrations, Jaquenoud-Sirot et al. (2009) did not take the patients’ bodyweight into account, which can influence the interpretation and the comparison of patients’ clozapine concentrations.
CYP1A2 is considered to be one of the major enzymes responsible for clozapine biotransformation (Eiermann et al., 1997; Fang et al., 1998; Olesen and Linnet, 2001), which was confirmed by several clinical data. Induction of CYP1A2 by the components of cigarette smoke seems to increase clozapine dose requirement, primarily of male smokers (Haring et al., 1989; Tang et al., 2007); however, the patients with the polymorphic CYP1A2*1F allele, resulting in an increase in CYP1A2 inducibility, did not require dose adjustment (Weide et al., 2003). On the other hand, Ferrari et al. (2012) demonstrated an association between the frequency of clozapine-induced adverse reactions and the occurrence of the low activity CYP1A2*1C allele in patients. The CYP1A2 substrate caffeine has been reported to inhibit clozapine metabolism, and caffeine consumption in a daily dose of 400 to 1000 mg significantly decreased clozapine clearance (Hagg et al., 2000; Raaska et al., 2004). The adjunctive fluvoxamine to ongoing clozapine therapy was found to increase clozapine plasma concentration, moreover resulting in toxic serum concentration, which was attributed to CYP1A2 inhibition by fluvoxamine (Hiemke et al., 1994; Szegedi et al., 1999; Lu et al., 2004). It should be noted that fluvoxamine is not a CYP1A2-selective inhibitor, but significantly blocks the activities of other CYPs (Kashuba et al., 1998), notably it irreversibly inhibits CYP3A4 by forming a metabolic intermediate complex (Jones et al., 2007).
The clinical observations on the dominant role of CYP1A2 were not confirmed by our findings that the patients’ CYP3A status rather than their CYP1A2 expression was associated with normalized clozapine concentrations. The relative importance of CYP3A4 in clozapine clearance, particularly in patients with low CYP1A2 activity, may be higher than it has been previously considered (Eiermann et al., 1997; Olesen and Linnet, 2001). Chetty and Murray (2007) suggested further research with the potential to demonstrate the critical roles of CYPs other than CYP1A2 for the elucidation of individual differences in clozapine metabolism. The following indirect evidences can predict the potential role of CYP3A4: (1) clozapine N-demethylation to norclozapine significantly correlated with CYP3A4-selective midazolam clearance (Ghassabian et al., 2010); (2) concomitant treatment with CYP3A4 inducers, such as carbamazepine, rifampicin, or phenytoin, was observed to substantially reduce clozapine concentrations (Miller, 1991; Jerling et al., 1994; Tiihonen et al., 1995; Peritogiannis et al., 2007; Parker, 2016); and (3) initiation of the CYP3A4 inhibitor erythromycin therapy increased clozapine levels and evoked toxic symptoms (Cohen et al., 1996). Furthermore, Zhang et al. (2008) provided support for the functional roles of both CYP3A4 and CYP1A2 in clozapine clearance, and suggested that the relative activities of CYP3A4 and CYP1A2 in patients might determine the clearance pathways. Although the majority of the patients with schizophrenia were smokers evoking high CYP1A2 activity (Leon and Diaz, 2005), in the present study, only 38% of the patients were current smokers and 4 subjects (4.3%) were heavy smokers (>20 cigarettes/d). In >70% of the patients, CYP1A2 mRNA was expressed at low concentration, whereas a high proportion of the patients (>70%) were CYP3A4 normal/high expressers, and additionally 3 low CYP3A4 expressers displayed normal clozapine-metabolizing capacity because of the functional CYP3A5*1 allele. Therefore, the relative CYP activities predicted the potential role of CYP3A4/5 in clozapine clearance, which was confirmed by the 2-fold higher normalized clozapine concentrations and by the halved dose-requirement in low CYP3A4 expressers.
Since the majority of the patients were on multi-drug therapy, co-medication as one of the main factors resulting in phenoconversion of drug-metabolizing enzymes must be considered. Haloperidol, risperidone, and propranolol are potent CYP2D6 inhibitors that may influence the minor contribution of CYP2D6 to clozapine clearance (Shin et al., 1999; Obach et al, 2006); however, most of the patients who were co-treated with these drugs carried the CYP2D6 loss-of-function allele. Genetic factors and/or co-medication therefore could result in CYP2D6 poor metabolism. According to the guidance of the Pharmacogenetic Working Group of the Royal Dutch Pharmacists Association, no clozapine dose adjustment is needed for patients with CYP2D6 poor metabolism (Samer et al., 2013). Valproate has been proved to transcriptionally induce CYP3A4 expression (Cerveny et al., 2007); consequently, the patients’ CYP3A4 mRNA levels captured the influence of valproate treatment. The patients’ caffeine consumption data were not available, and the CYP1A2 inhibitory effect on clozapine clearance therefore could not be taken into account.
Approximately 30% of the patients were underdosed, displaying lower clozapine concentration than required for the optimal therapeutic effect because of rapid elimination of the drug. However, it should be noted that none of the patients were in acute phase, and the majority of the subjects were in a relatively stable state, which was a consequence of the inclusion criteria of unmodified antipsychotic doses for at least 2 weeks. Therefore, the clinical relevance of this underdosing may be minor. Additionally, the normal/high CYP3A4 activity was associated with increased metabolite formation, that is, the concentrations of norclozapine and clozapine N-oxide were close to or even exceeded clozapine concentrations. While N-oxidation of clozapine is the inactivation pathway of metabolism, the N-demethylated metabolite exhibits some pharmacological effects and contributes to the overall efficacy of clozapine (Wenthur and Lindsley, 2013). Furthermore, high norclozapine level has been associated with an increased risk of weight gain and seizure, because it is a more potent 5-HT2C antagonist than clozapine (Kuoppamaki et al., 1993; Lu et al., 2004; Polcwiartek and Nielsen, 2016). Therefore, the clinical outcome seemed to be predictable from norclozapine/clozapine ratios rather than from the absolute concentration of clozapine (Weiner et al., 2004; Lameh et al., 2007). Consequently, lower clozapine clearance resulting in lower norclozapine/clozapine ratios has some clinical benefit over rapid metabolism (Légaré et al., 2013).
In conclusion, the patients’ clozapine-metabolizing capacity is influenced by genetic and plenty of nongenetic factors, and the expression/activity of clozapine-metabolizing enzymes can be a useful predictor for the individual clozapine concentration and dose requirement. Although the present work involved a limited number of patients and further investigation enrolling larger cohort is required, it clearly demonstrated that CYP3A status could be the major determinant of normalized clozapine concentration and dose requirement, in particular that the patients expressed CYP1A2 at a relatively low level. For achieving the therapeutic concentration, twice as high a dose was necessary for the normal/high CYP3A4 expressers and CYP3A5*1 carriers than for low CYP3A4 expressers. Tailored medication controlled by patients’ CYP3A4 expression and CYP3A5 genotype may facilitate the improvement of the individual clozapine therapy, leading to the dosage optimization for a more effective therapy.
Statement of Interest
None
Acknowledgments
The authors are indebted to Mária Szabó for her skillful assistance in this study. This work was supported by the grants from the Hungarian Scientific Research Fund (OTKA K104459 and K104738).
References
- Arranz MJ, Dawson E, Shaikh S, Sham P, Sharma T, Aitchison K, Crocq MA, Gill M, Kerwin R, Collier DA. (1995) Cytochrome P4502D6 genotype does not determine response to clozapine. Br J Clin Pharmacol 39:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo Lobos C, Komossa K, Rummel-Kluge C, Hunger H, Schmid F, Schwarz S, Leucht S. (2010) Clozapine versus other atypical antipsychotics for schizophrenia. Cochrane Database Syst Rev 11:CD006633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard A, Taylor DM. (2012) Comparative effectiveness of atypical antipsychotics in schizophrenia: what have real-world trials taught us? CNS Drugs 26:491–508. [DOI] [PubMed] [Google Scholar]
- Cerveny L, Svecova L, Anzenbacherova E, Vrzal R, Staud F, Dvorak Z, Ulrichova J, Anzenbacher P, Pavek P. (2007) Valproic acid induces CYP3A4 and MDR1 gene expression by activation of constitutive androstane receptor and pregnane X receptor pathways. Drug Metab Dispos 35:1032–1041. [DOI] [PubMed] [Google Scholar]
- Chetty M, Murray M. (2007) CYP-mediated clozapine interactions: how predictable are they? Curr Drug Metab 8:307–313. [DOI] [PubMed] [Google Scholar]
- Cohen LG, Chesley S, Eugenio L, Flood JG, Fisch J, Goff DC. (1996) Erythromycin-induced clozapine toxic reaction. Arch Intern Med 6:675–677. [PubMed] [Google Scholar]
- Couchman L, Morgan PE, Spencer EP, Flanagan RJ. (2010) Plasma clozapine, norclozapine, and the clozapine:norclozapine ratio in relation to prescribed dose and other factors: data from a therapeutic drug monitoring service, 1993–2007. Ther Drug Monit 32:438–447. [DOI] [PubMed] [Google Scholar]
- Dahl ML, Llerena A, Bondesson U, Lindström L, Bertilsson L. (1994) Disposition of clozapine in man: lack of association with debrisoquine and S-mephenytoin hydroxylation polymorphisms. Br J Clin Pharmacol 37:71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dain JG, Nicoletti J, Ballard F. (1997) Biotransformation of clozapine in humans. Drug Metab Dispos 25:603–609. [PubMed] [Google Scholar]
- Eiermann B, Engel G, Johansson I, Zanger UM, Bertilsson M. (1997) The involvement of CYP1A2 and CYP3A4 in the metabolism of clozapine. Br J Clin Pharmacol 44:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Coutts RT, McKenna KF, Baker GB. (1998) Elucidation of individual cytochrome P450 enzymes involved in the metabolism of clozapine. Naunyn Schmiedebergs Arch Pharmacol 358:592–599. [DOI] [PubMed] [Google Scholar]
- Ferrari M, Bolla E, Bortolaso P, Callegari C, Poloni N, Lecchini S, Vender S, Marino F, Cosentino M. (2012) Association between CYP1A2 polymorphisms and clozapine-induced adverse reactions in patients with schizophrenia. Psychiatry Res 200:1014–1017. [DOI] [PubMed] [Google Scholar]
- Fischer V, Vogels B, Maurer G, Tynes RE. (1992) The antipsychotic clozapine is metabolized by the polymorphic human microsomal and recombinant cytochrome P450 2D6. J Pharmacol Exp Ther 260:1355–1360. [PubMed] [Google Scholar]
- Ghassabian S, Chetty M, Tattam BN, Glen J, Rahme J, Stankovic Z, Ramzan I, Murray M, McLachlan AJ. (2010) The participation of cytochrome P450 3A4 in clozapine biotransformation is detected in people with schizophrenia by high-throughput in vivo phenotyping. J Clin Psychopharmacol 30:629–631. [DOI] [PubMed] [Google Scholar]
- Hägg S, Spigset O, Mjörndal T, Dahlqvist R. (2000) Effect of caffeine on clozapine pharmacokinetics in healthy volunteers. Br J Clin Pharmacol 49:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring C, Fleischhacker WW, Schett P, Humpel C, Barnas C, Saria A. (1990) Influence of patient-related variables on clozapine plasma levels. Am J Psychiatry 147:1471–1475. [DOI] [PubMed] [Google Scholar]
- Haring C, Meise U, Humpel C, Saria A, Fleischhacker WW, Hinterhuber H. (1989) Dose related plasma levels of clozapine: influence of smoking behaviour, sex and age. Psychopharmacology (Berl) 99:S38–40. [DOI] [PubMed] [Google Scholar]
- Hiemke C, Weigmann H, Härtter S, Dahmen N, Wetzel H, Müller H. (1994) Elevated levels of clozapine in serum after addition of fluvoxamine. J Clin Psychopharmacol 14:279–281. [PubMed] [Google Scholar]
- Hiemke C, et al. (2011) AGNP consensus guidelines for therapeutic drug monitoring in psychiatry: update 2011. Pharmacopsychiatry 44:195–235. [DOI] [PubMed] [Google Scholar]
- Jaquenoud-Sirot E, Knezevic B, Morena GP, Harenberg S, Oneda B, Crettol S, Ansermot N, Baumann P, Eap CB. (2009) ABCB1 and cytochrome P450 polymorphisms: clinical pharmacogenetics of clozapine. J Clin Psychopharmacol 29:319–326. [DOI] [PubMed] [Google Scholar]
- Jerling M, Lindstrom L, Bondesson U, Bertilsson L. (1994) Fluvoxamine inhibition and carbamazepine induction of the metabolism of clozapine: evidence from a therapeutic drug monitoring service. Ther Drug Monit 16:368–374. [DOI] [PubMed] [Google Scholar]
- Jones DR, Ekins S, Li L, Hall SD. (2007) Computational approaches that predict metabolic intermediate complex formation with CYP3A4 (+b5). Drug Metab Dispos 35:1466–1475. [DOI] [PubMed] [Google Scholar]
- Kashuba AD, Nafziger AN, Kearns GL, Leeder JS, Gotschall R, Rocci ML, Jr, Kulawy RW, Beck DJ, Bertino JS., Jr (1998) Effect of fluvoxamine therapy on the activities of CYP1A2, CYP2D6, and CYP3A as determined by phenotyping. Clin Pharmacol Ther 64:257–268. [DOI] [PubMed] [Google Scholar]
- Kuoppamäki M, Syvälahti E, Hietala J. (1993) Clozapine and N-desmethylclozapine are potent 5-HT1C receptor antagonists. Eur J Pharmacol 245:179–182. [DOI] [PubMed] [Google Scholar]
- Lally J, Gaughran F, Timms P, Curran SR. (2016) Treatment-resistant schizophrenia: current insights on the pharmacogenomics of antipsychotics. Pharmgenomics Pers Med 9:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lameh J, Burstein ES, Taylor E. (2007) Pharmacology of N-desmethylclozapine. Pharmacol Ther 115:223–231. [DOI] [PubMed] [Google Scholar]
- de Leon J, Diaz FJ. (2005) A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr Res 76:135–157. [DOI] [PubMed] [Google Scholar]
- Légaré N, Grégoire CA, De Benedictis L, Dumais A. (2013) Increasing the clozapine: norclozapine ratio with co-administration of fluvoxamine to enhance efficacy and minimize side effects of clozapine therapy. Med Hypotheses 80:689–691. [DOI] [PubMed] [Google Scholar]
- Linnet K, Olesen OV. (1997) Metabolism of clozapine by cDNA-expressed human cytochrome P450 enzymes. Drug Metab Dispos 25:1379–1382. [PubMed] [Google Scholar]
- Lu ML, Lane HY, Chen KP, Jann MW, Su MH, Chang WH. (2000) Fluvoxamine reduces the clozapine dosage needed in refractory schizophrenic patients. Clin Psychiatry 61:594–599. [DOI] [PubMed] [Google Scholar]
- Lu ML, Lane HY, Lin SK, Chen KP, Chang WH. (2004) Adjunctive fluvoxamine inhibits clozapine-related weight gain and metabolic disturbances. J Clin Psychiatry 65:766–771. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Lieberman JA, Stroup TS, Davis SM, Meltzer HY, Rosenheck RA, Swartz MS, Perkins DO, Keefe RS, Davis CE, Severe J, Hsiao JK, CATIE Investigators (2006) Effectiveness of clozapine versus olanzapine, quetiapine, and risperidone in patients with chronic schizophrenia who did not respond to prior atypical antipsychotic treatment. Am J Psychiatry 163:600–610. [DOI] [PubMed] [Google Scholar]
- Miller DD. (1991) Effect of phenytoin on plasma clozapine concentrations in two patients. J Clin Psychiatry 52:23–25. [PubMed] [Google Scholar]
- Ng W, Kennar R, Uetrecht J. (2014) Effect of clozapine and olanzapine on neutrophil kinetics: implications for drug-induced agranulocytosis. Chem Res Toxicol 27:1104–1108. [DOI] [PubMed] [Google Scholar]
- Obach RS, Walsky RL, Venkatakrishnan K, Gaman EA, Houston JB, Tremaine LM. (2006) The utility of in vitro cytochrome P450 inhibition data in the prediction of drug-drug interactions. J Pharmacol Exp Ther 316:336–348. [DOI] [PubMed] [Google Scholar]
- Olesen OV, Linnet K. (2001) Contributions of five human cytochrome P450 isoforms to the N-demethylation of clozapine in vitro at low and high concentrations. J Clin Pharmacol 41:823–832. [DOI] [PubMed] [Google Scholar]
- Parker C. (2016) Prescribing clozapine and rifampicin: clinical impact of their interaction. BJPsych Bull 40:153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peritogiannis V, Pappas D, Antoniou K, Hyphantis T, Mavreas V. (2007) Clozapine-rifampicin interaction in a patient with pulmonary tuberculosis. Gen Hosp Psychiatry 29:281–282. [DOI] [PubMed] [Google Scholar]
- Polcwiartek C, Nielsen J. (2016) The clinical potentials of adjunctive fluvoxamine to clozapine treatment: a systematic review. Psychopharmacology (Berl) 233:741–750. [DOI] [PubMed] [Google Scholar]
- Raaska K, Raitasuo V, Laitila J, Neuvonen PJ. (2004) Effect of caffeine-containing versus decaffeinated coffee on serum clozapine concentrations in hospitalised patients. Basic Clin Pharmacol Toxicol 94:13–18. [PubMed] [Google Scholar]
- Samer CF, Lorenzini KI, Rollason V, Daali Y, Desmeules JA. (2013) Applications of CYP450 testing in the clinical setting. Mol Diagn Ther 17:165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RR, Smith RL. (2015) Addressing phenoconversion: the Achilles’ heel of personalized medicine. Br J Clin Pharmacol 79:222–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JG, Soukhova N, Flockhart DA. (1999) Effect of antipsychotic drugs on human liver cytochrome P-450 (CYP) isoforms in vitro: preferential inhibition of CYP2D6. Drug Metab Dispos 27:1078–1084. [PubMed] [Google Scholar]
- Szegedi A, Anghelescu I, Wiesner J, Schlegel S, Weigmann H, Härtter S, Hiemke C, Wetzel H. (1999) Addition of low-dose fluvoxamine to low-dose clozapine monotherapy in schizophrenia: drug monitoring and tolerability data from a prospective clinical trial. Pharmacopsychiatry 32:148–153. [DOI] [PubMed] [Google Scholar]
- Tang YL, Mao P, Li FM, Li W, Chen Q, Jiang F, Cai ZJ, Mitchell PB. (2007) Gender, age, smoking behaviour and plasma clozapine concentrations in 193 Chinese inpatients with schizophrenia. Br J Clin Pharmacol 64:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temesvári M, Kóbori L, Paulik J, Sárváry E, Belic A, Monostory K. (2012) Estimation of drug-metabolizing capacity by cytochrome P450 genotyping and expression. J Pharmacol Exp Ther 341:294–305. [DOI] [PubMed] [Google Scholar]
- Tiihonen J, Vartiainen H, Hakola P. (1995) Carbamazepine-induced changes in plasma levels of neuroleptics. Pharmacopsychiatry 28:26–28. [DOI] [PubMed] [Google Scholar]
- Tugnait M, Hawes EM, McKay G, Eichelbaum M, Midha KK. (1999) Characterization of the human hepatic cytochromes P450 involved in the in vitro oxidation of clozapine. Chem Biol Interact 118:171–189. [DOI] [PubMed] [Google Scholar]
- Ulrich S, Baumann B, Wolf R, Lehmann D, Peters B, Bogerts B, Meyer FP. (2003) Therapeutic drug monitoring of clozapine and relapse - a retrospective study of routine clinical data. Int J Clin Pharmacol Ther 41:3–13. [DOI] [PubMed] [Google Scholar]
- van der Weide J, Steijns LS, van Weelden MJ. (2003) The effect of smoking and cytochrome P450 CYP1A2 genetic polymorphism on clozapine clearance and dose requirement. Pharmacogenetics 13:169–172. [DOI] [PubMed] [Google Scholar]
- Weiner DM, Meltzer HY, Veinbergs I, Donohue EM, Spalding TA, Smith TT, Mohell N, Harvey SC, Lameh J, Nash N, Vanover KE, Olsson R, Jayathilake K, Lee M, Levey AI, Hacksell U, Burstein ES, Davis RE, Brann MR. (2004) The role of M1 muscarinic receptor agonism of N-desmethylclozapine in the unique clinical effects of clozapine. Psychopharmacology (Berl) 177:207–216. [DOI] [PubMed] [Google Scholar]
- Wenthur CJ, Lindsley CW. (2013) Classics in chemical neuroscience: clozapine. ACS Chem Neurosci 4:1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WV, D’Esposito F, Edwards RJ, Ramzan I, Murray M. (2008) Interindividual variation in relative CYP1A2/3A4 phenotype influences susceptibility of clozapine oxidation to cytochrome P450-specific inhibition in human hepatic microsomes. Drug Metab Dispos 36:2547–2555. [DOI] [PubMed] [Google Scholar]
- van der Zwaag C, McGee M, McEvoy JP, Freudenreich O, Wilson WH, Cooper TB. (1996) Response of patients with treatment-resistant schizophrenia to clozapine within three serum level ranges. Am J Psychiatry 153:1579–1584. [DOI] [PubMed] [Google Scholar]