Abstract
We have characterized the interaction of a new class of antibiotics, simocyclinones, with bacterial DNA gyrase. Even though their structures include an aminocoumarin moiety, a key feature of novobiocin, coumermycin A1, and clorobiocin, which also target gyrase, simocyclinones behave strikingly differently from these compounds. Simocyclinone D8 is a potent inhibitor of gyrase supercoiling, with a 50% inhibitory concentration lower than that of novobiocin. However, it does not competitively inhibit the DNA-independent ATPase reaction of GyrB, which is characteristic of other aminocoumarins. Simocyclinone D8 also inhibits DNA relaxation by gyrase but does not stimulate cleavage complex formation, unlike quinolones, the other major class of gyrase inhibitors; instead, it abrogates both Ca2+- and quinolone-induced cleavage complex formation. Binding studies suggest that simocyclinone D8 interacts with the N-terminal domain of GyrA. Taken together, our results demonstrate that simocyclinones inhibit an early step of the gyrase catalytic cycle by preventing binding of the enzyme to DNA. This is a novel mechanism for a gyrase inhibitor and presents new possibilities for antibacterial drug development.
DNA topoisomerases are enzymes that control the topology of DNA in cells (5). The enzymes are divided into two types, topoisomerases I and II, depending on whether their reactions involve breakage of one or both strands of the DNA. Overall, the mechanism of action of topoisomerases involves the passage of one segment of DNA through a break in another. This reaction can achieve all the known reactions of topoisomerases: relaxation and supercoiling, catenation and decatenation, and knotting and unknotting. DNA supercoiling activity is unique to gyrase, and as this enzyme is essential in all bacteria but is not found in humans, it is an ideal target for antibacterials (24). The enzyme consists of an A2B2 heterotetramer. The A subunit (GyrA; 97 kDa in Escherichia coli) comprises an N-terminal domain involved in DNA cleavage and religation and a C-terminal DNA-wrapping domain. The B subunit (GyrB; 90 kDa in E. coli) contains the site of ATP hydrolysis at the N-terminal domain (GyrB43); the C-terminal domain interacts with both GyrA and DNA. To introduce supercoils, the enzyme wraps a double-stranded segment around itself, cleaves this DNA (the gate segment) in both strands, passes the wrapped DNA (transported segment) through the break, and then reseals the DNA.
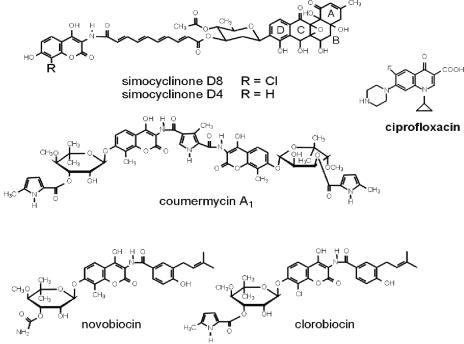
DNA gyrase is a proven target for antibacterial agents (24). Two well-studied classes of drugs target the enzyme: the coumarins and the quinolones. Quinolones are synthetic compounds that are widely used in clinical practice (8). Coumarins are naturally occurring products of Streptomyces species which, despite being more potent inhibitors of gyrase in vitro than quinolones, have had limited clinical use due to their low solubility, poor uptake, and eukaryotic cell toxicity (27). Other gyrase inhibitors include the cyclothialidines and the bacterial toxins CcdB and microcin B17. Quinolone drugs act by stabilizing the gyrase-cleaved DNA complex, and quinolone resistance mutations map predominantly as a cluster in the N-terminal region of GyrA. Coumarins competitively inhibit the ATPase activity of GyrB and exhibit Ki values in the nanomolar range (14). The coumarin drugs all share common structural features: a 3-amino-4,7-dihydroxycoumarin structure, an l-noviosyl sugar, and an aromatic acyl component attached to the amino group of the aminocoumarin moiety (Fig. 1). The interaction of coumarins with gyrase is extremely well characterized, and crystal structures of novobiocin and clorobiocin in complex with the N-terminal subdomain of GyrB (GyrB24) (17, 22, 25, 27, 40) and GyrB43 complexed with novobiocin (21) exist. These structures show that there is overlap of the binding sites of novobiocin and clorobiocin with the binding site of ATP; the noviose moiety of the coumarin overlaps the binding site for the adenine ring of ATP. This explains the competitive nature of inhibition by coumarin drugs. The novobiocin-GyrB24 complex is stabilized by a network of hydrogen bonds and a number of hydrophobic contacts (27). Key hydrogen bonds in the novobiocin-GyrB24 complex include those between Arg136 and the coumarin ring, Asp73 and the 3′-carbamoyl group on noviose, and Asn46 and the 2′-hydroxyl group on noviose. Mutations at these three amino acids have been shown to reduce levels of drug binding, but they also have deleterious effects on enzyme activity (7, 19). The genes involved in the biosynthesis of novobiocin, coumermycin A1, and clorobiocin have been identified as gene clusters in several Streptomyces species (29, 37, 41). The wealth of knowledge that exists on both the interactions of coumarins with gyrase and the genes involved in their synthesis provides an ideal starting point from which to develop new drugs with improved properties by combinatorial biosynthesis, and several such studies are under way (11, 12).
FIG. 1.
Structures of simocyclinones and aminocoumarin drugs; the structure of ciprofloxacin is also shown for comparison.
Recently, a new antibiotic, simocyclinone D8, was isolated from Streptomyces antibioticus Tü 6040 (18, 33, 38). Like the coumarins, it contains a 3-amino-4,7-dihydroxycoumarin structure with an acyl moiety attached to the amino group (Fig. 1). In contrast to the other coumarin antibiotics, it does not contain an l-noviosyl sugar at the 7-OH of the aminocoumarin ring; but another sugar, d-olivose, is attached to the acyl moiety (which is a tetraene dicarboxylic acid) by an ester bond. This d-olivose is glycosidically linked to an angucyclic polyketide (Fig. 1). Like clorobiocin, simocyclinone D8 contains a chlorine atom at position 8 of the coumarin ring. It is produced together with simocyclinone D4, which lacks the halogen at this position (Fig. 1). Sequence analysis of the biosynthetic gene cluster of simocyclinone revealed that it contains several genes with high degrees of similarity to those involved in the biosynthesis of the coumarin moieties of novobiocin, clorobiocin, and coumermycin (13, 39). However, the simocyclinone gene cluster does not contain an obvious resistance gene that would give a clue as to its target (13, 39); this is in contrast to the biosynthetic gene clusters of novobiocin, clorobiocin, and coumermycin, all of which contain genes encoding aminocoumarin-resistant topoisomerase subunits (35, 36). Simocyclinone D8 has been shown to exhibit antibiotic activity against gram-positive bacteria, as well as cytostatic effects against human tumor cell lines; little or no activity has been found against gram-negative organisms such as E. coli (32, 33). The aminocoumarin moiety is rare in nature and, apart from the simocyclinones and coumarins, is only found in only one other antibiotic, protorubradirin (3). Therefore, the presence of this moiety in the simocyclinones suggested that gyrase is a possible target. Experiments carried out with E. coli by using a reporter gene system sensitive to changes in the degree of DNA supercoiling suggested that simocyclinone D8 reduces supercoiling by inhibiting gyrase in vivo (M. Abu Mraheil and P. Heisig, personal communication). We have studied the interaction of simocyclinone D8 with gyrase and found that it is an even more potent inhibitor than novobiocin and that it inhibits gyrase by a novel mechanism.
MATERIALS AND METHODS
Protein, DNA, and drugs.
GyrA and GyrB from E. coli were purified to homogeneity as described previously (26). Topoisomerase IV from E. coli was purified by a method based on that of Peng and Marians (28). Human type I and IIα topoisomerases were from TopoGEN. Supercoiled and relaxed pBR322 DNAs were prepared as described previously (4, 30) and linearized by cutting with EcoRI. Ciprofloxacin (Sigma Aldrich) was dissolved in water. Novobiocin (Sigma Aldrich), camptothecin (Sigma Aldrich), etoposide (Sigma Aldrich), and simocyclinone D8 were dissolved in dimethyl sulfoxide (DMSO). The 147-bp linear DNA fragment used in this study incorporated the preferred quinolone-mediated gyrase cleavage site of pBR322 (site 990) (9) and was generated by PCR with the following oligonucleotides (Sigma Aldrich): single-stranded 5′-biotinylated 19mer (5′→3′) GCC ATT ATC GCC GGC ATG G and 21mer (5′→3′) CTG CCT GGA CAG CAT GGC CTG. The 119- and 197-bp linear DNA fragments and some of the GyrB protein used in this study were gifts from Sylvain Mitelheiser (John Innes Centre).
Enzyme assays.
Gyrase supercoiling assays were performed as described previously (15, 30). Samples (30 μl) containing 3.4 nM gyrase and 0.5 μg of relaxed pBR322 DNA (6 nM) were incubated at 37°C for 30 min in the presence of 1.26 mM ATP. The DNA was prepared for electrophoresis by the addition of an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), brief vortexing, centrifugation (15,700 × g, 5 min), and the addition of 8 μl of 40% sucrose-0.1 M Tris · HCl (pH 7.5)-0.1 M EDTA-bromophenol blue to the upper phase. The products were analyzed on 0.8% agarose gels.
Gyrase relaxation assays were performed similarly to the supercoiling assays, except that ATP and spermidine were omitted, the enzyme concentration was increased to 60 nM, and the samples were incubated for 1 h at 37°C.
Gyrase ATPase assays (GyrB43 domain) were carried out at 25°C in 96-well plates; and each sample contained 20 μM GyrB43 in 47 mM Tris · HCl (pH 7.5)-1 mM EDTA-9.3% (wt/vol) glycerol-26.6 mM KCl-4 mM dithiothreitol (DTT)-5 mM MgCl2-800 μM phosphoenolpyruvate-400 μM NADH-1% (vol/vol) pyruvate kinase-lactate dehydrogenase mixture (Sigma)-2 mM ATP. For determination of the DNA-dependent ATPase activity, gyrase (70 nM) was incubated with linear pBR322 DNA (2.8 nM) under the same conditions described above, except that the NADH concentration was halved. In both cases a range of drug concentrations was used to test the effect of simocyclinone D8.
DNA cleavage assays contained gyrase (30 to 500 nM), 1 μg of relaxed pBR322 DNA (12 nM), 35 mM Tris · HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 1.8 mM spermidine, 0.1 mg of bovine serum albumin (BSA)/ml, 6.5% (wt/vol) glycerol, and simocyclinone D8 at the desired concentration in a 30-μl reaction volume. Reactions were carried out in the presence or the absence of 1.26 mM ATP. These assays were also carried out with a reduced DTT concentration (0.5 μM), in which drug was added as the final component. Ciprofloxacin was used at 5 μM to observe the ciprofloxacin-induced cleavage complex. For Ca2+ cleavage, MgCl2 was replaced with CaCl2. After incubation for 1 h at 37°C, sodium dodecyl sulfate and proteinase K were added to final concentrations of 0.2% (wt/vol) and 0.1 mg/ml, respectively, and the incubation was continued for 30 min at 37°C. Samples were prepared for electrophoresis as described above. The products were analyzed on 1.1% agarose gels containing 1 μg of ethidium bromide per ml in the gel and the running buffer.
The binding of gyrase to DNA in the absence or the presence of simocyclinone D8 was assayed by a gel shift procedure with 119- and 197-bp DNA fragments derived from pBR322, with the preferred gyrase cleavage site incorporated at position 990 (9). Gyrase (150 nM) was incubated with DNA (160 nM) and increasing concentrations of simocyclinone D8 in 25 mM Tris · HCl (pH 7.5)-45 mM KCl-20 mM MgCl2-1 mM DTT-0.36 mg of BSA/ml-5% (wt/vol) glycerol at 25°C for 2 h. Samples were analyzed on a 6% polyacrylamide gel (37.5:1 [wt/wt] acrylamide to bisacrylamide; Severn Biotech) in 90 mM Tris-90 mM boric acid-5 mM MgCl2 (30 mA, 1 h).
Decatenation assay mixtures containing human topoisomerase IIα (7.8 nM), kinetoplast DNA (200 ng), and DMSO (0.5%) in 50 mM Tris · HCl (pH 8.0)-120 mM KCl-10 mM MgCl2-0.5 mM ATP-0.5 mM DTT-30 μg of BSA/ml were incubated for 1 h at 37°C with etoposide or novobiocin (positive and negative controls, respectively) or simocyclinone D8; and the extracted DNA was analyzed on a 1% agarose gel containing 0.5 μg of ethidium bromide per ml.
Relaxation assays for human topoisomerase I were carried out with 5.5 nM enzyme and 0.5 μg of supercoiled pBR322 DNA in 10 mM Tris · HCl (pH 7.9)-1 mM EDTA-150 mM NaCl-0.1% BSA-0.1 mM spermidine-5% glycerol in the presence of 0.5% DMSO with and without camptothecin and novobiocin (positive and negative controls, respectively) and with or without simocyclinone D8. Samples were incubated for 1 h at 37°C, and DNA was prepared for electrophoresis as described above before it was run on 0.8% agarose gels.
Decatenation assays for topoisomerase IV were carried out by incubating enzyme (0.8 nM ParC dimer, 1.3 nM ParE) and 200 ng of kinetoplast DNA with various concentrations of drug in 40 mM HEPES-KOH (pH 7.6)-10 mM magnesium acetate-100 mM potassium glutamate-10 mM DTT-50 μg of BSA/ml-1 mM ATP-4 μg of tRNA/ml-1% (wt/vol) glycerol-2% DMSO.
SPR.
Surface plasmon resonance (SPR) was performed (at 25°C) with a Biacore X instrument (Biacore AB, Uppsala, Sweden). Proteins were immobilized onto CM5 sensor chips after activation of the surface carboxyl groups by addition of 35 μl of a mixture of N-hydroxysuccinimide (0.05 M in H2O) and 1-ethyl-3-(3-diaminopropyl)carbodiimide (0.2 M in H2O) (Biacore amine coupling kit). Protein (25 nM GyrB43 or 85 to 360 nM GyrA59) diluted in 10 mM sodium acetate (pH 4.5) was injected in 20- to 50-μl aliquots across the surface of the chip until a response of 2,200 to 4,000 resonance units was reached. The active esters were deactivated by injection of 35 μl of ethanolamine, followed by injection of 50 μl of regeneration buffer (2 M KCl). The system was reprimed with running buffer (35 mM Tris · HCl [pH 7.5], 6.5% [wt/vol] glycerol, 24 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.02% Tween 20, 5% DMSO). The flow rate was increased to 50 μl/min, and a range of concentrations of simocyclinone D8 was allowed to flow across the surface. To test whether simocyclinone D8 inhibited the binding of gyrase to DNA, 5′ biotinylated linear DNA (147 bp) was attached in 10 mM Hepes (pH 7.4)-150 mM NaCl-3 mM EDTA-0.005% surfactant P20 (Biacore) to flow cell 2 of a streptavidin-coated sensor chip by flowing 50 μl of 6 nM DNA in 10 mM HEPES (pH 7.4)-3 mM EDTA-0.005% surfactant P20 (Biacore)-160 mM NaCl across the chip at 10 μl/min. Flow cells 1 and 2 were then blocked with 100 μl of 1 mg of biotin per ml. The system was reprimed with running buffer, and the flow rate increased to 50 μl/min. Gyrase (20 nM) was injected alone and in the presence of simocyclinone D8 (50 and 500 nM). To investigate the interaction of DNA with simocyclinone D8, DNA was immobilized onto a streptavidin-coated sensor chip as described above, except that a higher concentration of DNA was used in order to achieve an immobilization level of >1,400 resonance units. Simocyclinone D8 at a range of concentrations was allowed to flow over the chip surface.
Isothermal titration calorimetry.
Enthalpy values were measured by using a VP isothermal titration calorimeter (Microcal, Milton Keynes, United Kingdom). Gyrase subunits and domains (GyrA, GyrA59, GyrB, and GyrB43 at 16 to 22 μM) were dialyzed extensively against binding buffer (50 mM Tris · HCl [pH 7.5], 1 mM EDTA, 100 mM KCl, 5% DMSO). A first dialysis step was carried out prior to addition of the DMSO, and this dialysis buffer was used to dilute simocyclinone D8 and novobiocin (used as a control), which had been solubilized in DMSO. Samples were centrifuged (13,000 rpm, 10 min) and degassed prior to use. During a titration, the protein (2.3 ml) was placed in the sample cell, and 43 successive aliquots of simocyclinone D8 or novobiocin (5-μl volumes except for the first one [2 μl] and volumes 37 to 43 [10 μl]) were injected at 2-min intervals. The upper limit of the injectant concentration was 250 μM owing to the poor solubility of the drug at higher concentrations. The binding data were corrected for the heats of dilution of the drugs. Integrated heat effects were analyzed by nonlinear regression by using a single-site binding model with Origin software (Microcal), which yielded independent values for Kd and the change in enthalpy (44).
RESULTS
Simocyclinones inhibit DNA supercoiling and relaxation by DNA gyrase but not the DNA-independent ATPase activity of DNA gyrase.
To compare the inhibition of gyrase supercoiling caused by simocyclinones D4 and D8 with that caused by other gyrase inhibitors, the simocyclinones were titrated into supercoiling reactions at a range of concentrations in order to estimate the 50% inhibitory concentrations (IC50s); novobiocin and ciprofloxacin were used as control compounds for comparison (Fig. 2, top panel). Simocyclinones D4 and D8 were found to be inhibitors of gyrase supercoiling with potencies similar to that of novobiocin, with IC50s of ∼100 nM for simocyclinone D8 and ∼450 nM for simocyclinone D4, compared with IC50s of ∼250 nM for novobiocin and ∼700 nM for ciprofloxacin under these conditions (Fig. 2, middle panel). These IC50s indicate the relative affinities of these compounds for gyrase. A third simocyclinone, simocyclinone C2 (34), which contains the angucyclic moiety, the d-olivose, and the tetraene dicarboxylic acid but which lacks the aminocoumarin ring, was also tested but showed only weak inhibitory activity towards gyrase (IC50 > 100 μM; data not shown).
FIG. 2.
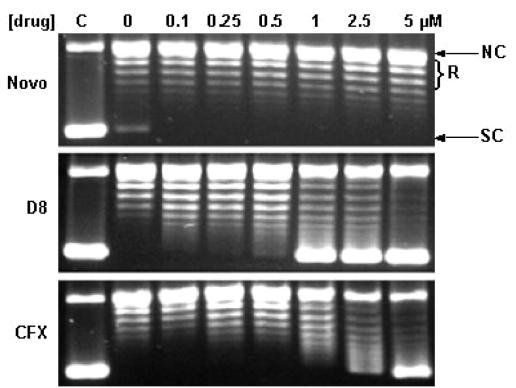
Effects of simocyclinones D4 and D8 on DNA supercoiling by gyrase. (Top panel) Supercoiling in the presence of increasing concentrations of novobiocin (Novo), simocyclinones D4 and D8, and ciprofloxacin (CFX). NC, nicked circle; R, relaxed; SC, supercoiled. (Middle panel) Estimation of IC50s (in parentheses) of simocyclinones D4 and D8 compared with those of novobiocin and ciprofloxacin. (Bottom panel) Effect of ATP concentration on the extent of inhibition. Samples containing the estimated IC50s of the drugs were incubated in the presence of a range of ATP concentrations.
Novobiocin inhibits gyrase supercoiling by competing with ATP (27). In order to determine whether simocyclinones act in a similar way, the effect of the ATP concentration on the level of inhibition of supercoiling was assessed by using the drugs at approximately their IC50s (Fig. 2, bottom panel). Due to its competitive mode of action, novobiocin exerted a greater effect at low ATP concentrations than at high ATP concentrations. In striking contrast, inhibition by simocyclinones D4 and D8, as well as inhibition by the quinolone drug ciprofloxacin, varied little with the ATP concentration, indicating that inhibition was not competitive with ATP.
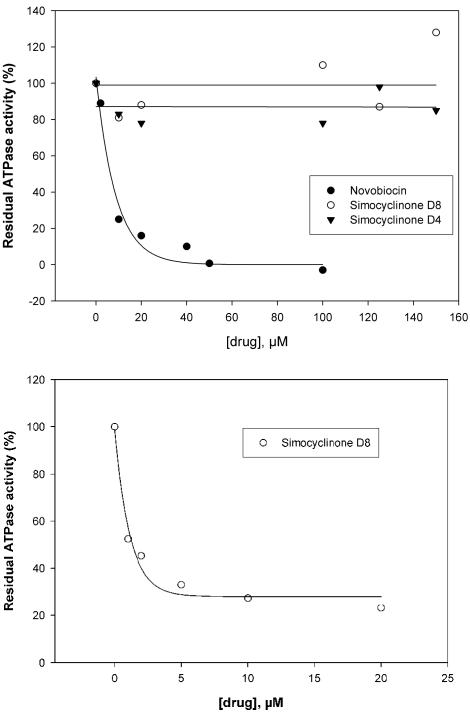
Having found that inhibition by simocyclinone was apparently independent of ATP, we then checked whether ATPase activity was affected by the drug (Fig. 3, top panel). For these experiments we used the N-terminal domain of GyrB, which contains the ATPase active site (2, 42). Whereas novobiocin completely inhibited gyrase ATPase at a 1:1 molar ratio of drug to enzyme, no inhibition was observed with simocyclinones D4 and D8 up to a 7:1 molar ratio of drug to enzyme; it was not possible to achieve a higher molar ratio under the experimental conditions due to the poor solubilities of the drugs. The effect of simocyclinone D8 on the ATPase activity of the full-length enzyme (A2B2) was also tested (Fig. 3, bottom panel). The ATPase activity of gyrase is strongly stimulated in the presence of DNA (25). We found that simocyclinone D8 inhibited the DNA-dependent ATPase activity, whereas the DNA-independent rate was unaffected.
FIG. 3.
Effects of simocyclinones D4 and D8 on gyrase ATPase activity. (Top panel) The N-terminal ATPase domain of GyrB (GyrB43; 20 μM) was incubated with increasing concentrations of novobiocin and simocyclinones D4 and D8, and the rates of hydrolysis were determined and plotted as the residual ATPase activity against the drug concentration. (Bottom panel) Gyrase (A2B2; 70 nM) was incubated with increasing concentrations of simocyclinone D8 in the presence of linear pBR322 DNA (2.8 nM).
To ascertain the effect of simocyclinone on the other topoisomerase activity of gyrase, DNA relaxation, a range of concentrations of the drug were incubated with the enzyme and supercoiled DNA in the absence of ATP (Fig. 4). In parallel, ciprofloxacin and novobiocin were used for comparison. Inhibition by simocyclinone D8 was observed at a concentration of 0.1 μM, with an approximate IC50 of 0.5 to 1 μM. Novobiocin showed no inhibition up to 5 μM, whereas ciprofloxacin showed significant inhibition at concentrations greater than 1 μM. Simocyclinone D4 also inhibited relaxation, but to a lesser extent than simocyclinone D8 (IC50 of simocyclinone D4, approximately five times higher than that of simocyclinone D8).
FIG. 4.
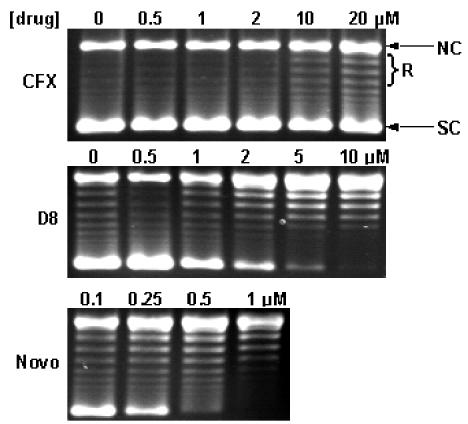
Effect of simocyclinone D8 on gyrase relaxation. Gyrase (60 nM) and supercoiled pBR322 DNA (6 nM) were incubated with increasing concentrations of novobiocin (Novo), simocyclinone D8, and ciprofloxacin (CFX) for 1 h at 37°C. NC, nicked circle; R, relaxed; SC, supercoiled.
Having established that simocyclinones D4 and D8 do not target gyrase at the same site as aminocoumarins, we wanted to determine whether they shared the same site as quinolones. The binding site of quinolones is thought to involve a region adjacent to the active site for DNA cleavage in gyrase that includes GyrA residue Ser83 (16). Using a quinolone-resistant gyrase that contains a Ser83-to-Trp mutation (43), we titrated simocyclinone D8 into a supercoiling assay using ciprofloxacin as a positive control (Fig. 5). The IC50 of simocyclinone D8 increased ∼10-fold with the mutant gyrase, whereas the IC50 of ciprofloxacin increased >30-fold. The extent of novobiocin inhibition was not affected, consistent with the fact that it does not target GyrA. The resistance of the mutant gyrase to simocyclinone D8 may suggest some involvement of this protein region in drug binding.
FIG. 5.
Effect of simocyclinone D8 on DNA supercoiling by quinolone-resistant gyrase. The supercoiling activities of gyrase containing GyrA Trp83 in the presence of increasing concentrations of ciprofloxacin (CFX), simocyclinone D8, and novobiocin (Novo) are indicated. NC, nicked circle; R, relaxed; SC, supercoiled.
To determine whether the actions of simocyclinones are gyrase specific, the effect of simocyclinone D8 on other topoisomerases was tested (data not shown). We found a weak inhibition of topoisomerase IV at 50 μM, whereas a coumermycin A1 control at ∼100 nM abolished the activity. Human topoisomerase I was not inhibited by simocyclinone D8 at 40 μM, whereas a camptothecin control at this concentration caused significant inhibition. We found that simocyclinone D8 inhibited human topoisomerase II decatenation more strongly than etoposide did, with an IC50 of ∼5 μM, suggesting that the drug is also active against eukaryotic topoisomerase II, consistent with its observed effects on human tumor cells (33).
Simocyclinones do not stimulate gyrase-dependent cleavable complex formation but antagonize quinolone-mediated cleavage.
A characteristic feature of quinolones is that they stabilize the covalent complex between gyrase and DNA. To see if this is also true for simocyclinones, simocyclinone D8 was incubated with enzyme and supercoiled DNA both in the presence and in the absence of ATP. To check that the reducing agent DTT did not affect the potential cleavage complex formation, the experiment was also carried out with a low DTT concentration (0.5 μM). No linear DNA was observed on a gel after treatment with sodium dodecyl sulfate and proteinase K, while a linear band was observed in the presence of ciprofloxacin (data not shown).
In order to determine if simocyclinone affects the gyrase cleavage-religation equilibrium, we studied the effect of simocyclinone D8 on ciprofloxacin- and Ca2+-induced cleavage by gyrase. In both cases we found that cleavage was abrogated (Fig. 6). In the case of ciprofloxacin, a less than equimolar concentration of simocyclinone D8 to ciprofloxacin abolished cleavage complex formation, and 1 μM simocyclinone D8 prevented Ca2+-induced cleavage complex formation. Taken together, these results suggest that simocyclinones do not stabilize the cleavage complex but prevent DNA cleavage by gyrase.
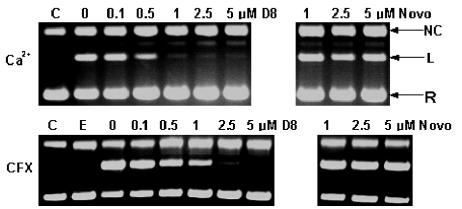
FIG. 6.
Effect of simocyclinone D8 on Ca2+- and ciprofloxacin (CFX)-induced DNA cleavage by gyrase. Increasing concentrations of simocyclinone D8 (left-hand panels) were incubated in the presence of fixed concentrations of Ca2+ (4 mM) or ciprofloxacin (5 μM). Controls with novobiocin (Novo) are shown on the right. Gels were run in the presence of 1 μg of ethidium bromide per ml. C, control (no enzyme or drug); E, enzyme only; NC, nicked circle; L, linear; R, relaxed.
Binding studies.
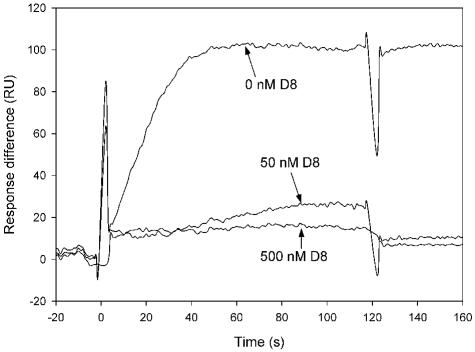
One explanation for the effect of simocyclinone on the gyrase cleavage reaction, as well as the inhibition of DNA-dependent ATPase activity, is that the drug prevents the enzyme from binding to DNA. To see if this was the case, we used SPR to study the interaction of gyrase and DNA in the presence and the absence of simocyclinone D8 (Fig. 7). In the absence of drug, with DNA immobilized on the chip surface, gyrase (20 nM) gave a resonance signal, as expected. In the presence of 50 nM simocyclinone D8, this interaction was virtually abolished, indicating that simocyclinone D8 prevents the binding of gyrase to DNA. The same effect was observed in gel retardation assays (data not shown). We also used SPR to test for the binding of simocyclinone D8 to DNA. We did not observe an interaction, confirming previous observations (32).
FIG. 7.
SPR sensorgram showing the interaction between gyrase and DNA in the presence of simocyclinone D8. A 5′-biotinylated 147-bp linear DNA fragment was immobilized onto the chip, and gyrase (20 nM) was allowed to flow across the chip in the absence and presence of 50 and 500 nM simocyclinone D8. Spikes at the beginning and end of the injection are due to the slight difference in the DMSO concentration between the sample and the running buffer. RU, resonance units
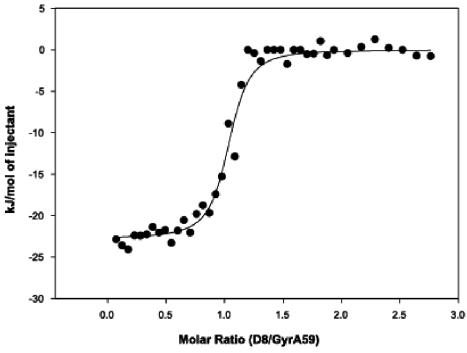
The potential sites of simocyclinone D8 binding on gyrase were investigated by SPR. The N-terminal domains of GyrA and GyrB (GyrA59 and GyrB43, respectively) were immobilized onto separate CM5 sensor chips and were exposed to a range of drug concentrations. These experiments showed no interaction of simocyclinone D8 with GyrB43 (the ATPase domain); the expected binding of novobiocin to GyrB43 was observed (19) (data not shown). Only a weak interaction between simocyclinone D8 and GyrA59 was observed. As this technique involves immobilization of the protein onto a solid surface, this result may not reflect the situation in free solution. Therefore, we used isothermal titration calorimetry as an alternative method to study the binding of simocyclinone D8 to gyrase. An interaction was observed when simocyclinone D8 was injected into a GyrA solution (data not shown). An interaction was also observed with GyrA59, yielding binding constants of 50 to 100 nM and indicating an approximate 1:1 stoichiometry (Fig. 8); in relation to the A2B2 structure of gyrase, this would suggest that two simocyclinone D8 molecules bind per heterotetramer. No binding of simocyclinone D8 to either GyrB or GyrB43 was found by this technique (data not shown); the expected binding of novobiocin was found (14).
FIG. 8.
Binding of simocyclinone D8 to GyrA59 by isothermal titration calorimetry. The integrated data, after correction for the heat of ligand dilution as a function of the molar ratio, is presented as kilojoules per mole of injectant versus the molar ratio of drug/protein. The solid line shows the best fit obtained by least-squares regression by use of a one-site model. This analysis gave an approximate stoichiometry of 1:1.
DISCUSSION
In the present study, we have characterized the interaction of simocyclinone D8 with bacterial DNA gyrase. This compound contains a key structural element present in other gyrase-specific drugs, the aminocoumarin ring. We compared and contrasted the inhibition by simocyclinone D8 with that by the coumarin novobiocin and the quinolone ciprofloxacin.
In terms of supercoiling activity, simocyclinone D8 appears to be a slightly more potent inhibitor than novobiocin (Fig. 2, top and middle panels). However, a striking difference is that inhibition by simocyclinone D8 is ATP independent. Thus, we found that the ATP concentration has no effect on supercoiling inhibition (Fig. 2, bottom panel), and we observed inhibition of gyrase relaxation activity in the absence of ATP (Fig. 4); this reaction is not inhibited by novobiocin, which is a competitive inhibitor of gyrase ATPase activity. Having established that inhibition was ATP independent, we also confirmed that simocyclinone D8 does not inhibit DNA-independent ATPase activity (Fig. 3, top panel). Although the compound shares the aminocoumarin ring structure in common with the coumarins, it does not contain an l-noviosyl sugar, which is implicated in the competitive inhibition of coumarins as the binding sites of the ATP adenine ring and l-noviosyl sugar overlap. Simocyclinone D8 contains a deoxyhexose sugar, but this moiety is situated between the angucyclinone moiety and the tetraene chain and is a d-olivose rather than an l-noviose (Fig. 1). However, although the ATPase activity of the GyrB43 domain was not inhibited, when the DNA-dependent ATPase activity of the full-length enzyme was measured, there was substantial inhibition of this activity by simocyclinone D8 (Fig. 3, bottom panel). Inhibition of the DNA-dependent ATPase activity of gyrase by the quinolones was also observed (20).
Given that the ATPase domain was not the target of simocyclinone D8, we sought to establish what was. Interestingly, simocyclinone D8 was about 10 times less effective an inhibitor of the quinolone-resistant GyrA Trp83 mutant than of the wild-type enzyme. Novobiocin inhibited the mutant enzyme to the same extent as it inhibited the wild-type enzyme, whereas ciprofloxacin, as expected, showed only slight inhibition at concentrations 30 times the IC50 for the wild-type enzyme (Fig. 5). This may suggest that simocyclinone D8 targets GyrA in the same region as the quinolones, although mutation of Ser83 did not have such a drastic effect on simocyclinone D8 inhibition as it did on quinolone inhibition. This similarity both in the inhibition of relaxation and in the reduced inhibition of the mutant enzyme raised the question whether simocyclinone D8 also stimulated cleavage complex formation. We found that it did not either in the presence or in the absence of ATP. However, when the drug was titrated into samples containing ciprofloxacin or Ca2+, both of which induce cleavage complex formation, the amount of cleaved DNA was reduced in a concentration-dependent manner (Fig. 6). This suggests that simocyclinone inhibits the enzyme prior to DNA cleavage. To confirm that this is the case, we used SPR to look at the binding of gyrase to DNA in the presence and the absence of simocyclinone D8 and found that simocyclinone D8 prevented the binding of gyrase to DNA (Fig. 7). Previous work has suggested that simocyclinone D8 does not itself bind to DNA (32), and our SPR experiments confirm this; so it seems likely that simocyclinone D8 binds to gyrase and consequently inhibits DNA binding. To ascertain which domain of gyrase contains the binding site of simocyclinone and to test the affinity and the stoichiometry, we used isothermal titration calorimetry to measure the binding of simocyclinone D8 to GyrA, GyrA59, GyrB, and GyrB43 using novobiocin as a control. As expected, simocyclinone D8 interacted only with GyrA and GyrA59, and the binding constants were comparable to that for novobiocin binding to GyrB43 (∼50 nM for GyrA59-simocyclinone D8 binding [Fig. 8] compared with ∼20 nM for GyrB43-novobiocin binding). The IC50s for inhibition of gyrase supercoiling suggest that simocyclinone D8 has a higher affinity for gyrase than novobiocin. The somewhat higher binding constant measured by isothermal titration calorimetry may be an indication that D8 binding requires more than just the GyrA59 domain.
Taken together, our results demonstrate that simocyclinone D8 has a mode of inhibition that is unlike those of coumarins and quinolones. There are, however, some similarities between simocyclinone D8 and an early series of 5,6-bridged quinolones, which were recently investigated (23). Like simocyclinone D8, these compounds inhibit gyrase supercoiling but do not stimulate gyrase-dependent cleavable complex formation. They also antagonize quinolone-mediated cleavable complex formation, although not at concentrations as low as those at which simocyclinone D8 does. Inhibition by both drugs is reduced approximately 10-fold with the Trp83 mutant enzyme. We propose that simocyclinone D8 acts by preventing binding of DNA to gyrase, and it is possible that the bridged quinolones work in a similar way. Like simocyclinone, the chromosomally encoded inhibitor of gyrase (GyrI) in E. coli has also been found to inhibit the enzyme prior to or at the step of DNA binding (6). Merbarone, an inhibitor of human topoisomerase IIα, did not inhibit DNA binding or ATPase activity but was found to prevent cleavage by the enzyme and impeded the action of etoposide, suggesting that it binds at the same site as cleavage-enhancing agents (10). The molecular basis of the action of simocyclinone D8 on gyrase is not yet known, but the data presented in this paper indicate that it may interact with GyrA such that it blocks DNA binding to the enzyme. Alternatively, it could stabilize a conformation of the enzyme that is unable to bind to DNA. It is likely that structural studies of the gyrase-drug complex will be required to elucidate its molecular mode of action.
The biosynthetic genes for the aminocoumarin moieties of simocyclinone D8 and novobiocin show high degrees of sequence similarity, indicating that these antibiotics have a common evolutionary origin. Both of these compounds share the same target, i.e., gyrase, but their modes of binding to this target are entirely different. The future discovery of further natural aminocoumarins may help to answer the question as to how gyrase-inhibiting antibiotics of the aminocoumarin type have evolved in nature to act on different sites in gyrase. Several antibiotics with an angucycline moiety are known, but none of them have been described to be a potent gyrase inhibitor (1, 31). Our observation that simocyclinone C2, which contains the angucycline moiety but which lacks the aminocoumarin moiety (Fig. 1), is a very inefficient gyrase inhibitor indicates that the aminocoumarin structure is crucial for the activities of the simocyclinones.
In light of the present problems of drug resistance, the production of new drugs is essential if bacterial diseases are to be successfully combated. Gyrase inhibitors, especially fluoroquinolones, are of key importance in antibacterial therapy. Our study has now identified the simocyclinones as a new class of highly potent gyrase inhibitors which may serve as lead compounds for drug development. The sequencing of the biosynthetic gene cluster of simocyclinone (13, 39) offers the prospect of generating novel simocyclinone derivatives by combinatorial biosynthesis. Such studies involving the previously known aminocoumarin drugs are already under way (11, 12, 45). Our biochemical study identifies the targets and modes of action of simocyclinones and increases the possibility of exploiting the simocyclinone biosynthetic pathway in the search for new drugs.
Acknowledgments
We thank David Lawson, Lionel Costenaro, and Olivier Pierrat for helpful comments on the manuscript and Melisa Wall for help with making topoisomerase IV.
This work was supported by grants from the European Commission (CombiGyrase LSHB-CT-2004-503466) and the Wellcome Trust.
REFERENCES
- 1.Abdelfattah, M., R. P. Maskey, R. N. Asolkar, I. Grun-Wollny, and H. Laatsch. 2003. Seitomycin: isolation, structure elucidation and biological activity of a new angucycline antibiotic from a terrestrial streptomycete. J. Antibiot. 56:539-542. [DOI] [PubMed] [Google Scholar]
- 2.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kDa N-terminal fragment of the gyrase B protein hydrolyses ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, B., and B. A. Zapotocky. 1992. Protorubradirin, an antibiotic containing a C-nitroso-sugar fragment, is the true secondary metabolite produced by Streptomyces achromogenes var. rubradiris. Rubradirin, described earlier, is its photo-oxidation product. J. Antibiot. 45:1313-1324. [DOI] [PubMed] [Google Scholar]
- 4.Bates, A. D., and A. Maxwell. 1989. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 8:1861-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 6.Chatterji, M., and V. Nagaraja. 2002. GyrI: a counter-defensive strategy against proteinaceous inhibitors of DNA gyrase. EMBO Rep. 3:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras, A., and A. Maxwell. 1992. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 8.Emmerson, A. M., and A. M. Jones. 2003. The quinolones: decades of development and use. J. Antimicrob. Chemother. 51(Suppl. 1):13-20. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortune, J. M., and N. Osheroff. 1998. Merbarone inhibits the catalytic activity of human topoisomerase II alpha by blocking DNA cleavage. J. Biol. Chem. 273:17643-17650. [DOI] [PubMed] [Google Scholar]
- 11.Freel Meyers, C. L., M. Oberthur, H. Xu, L. Heide, D. Kahne, and C. T. Walsh. 2004. Characterization of NovP and NovN: completion of novobiocin biosynthesis by sequential tailoring of the noviosyl ring. Angew. Chem. Int. Engl. Ed. 43:67-70. [DOI] [PubMed] [Google Scholar]
- 12.Galm, U., M. A. Dessoy, J. Schmidt, L. A. Wessjohann, and L. Heide. 2004. In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic, and synthetic approach. Chem. Biol. 11:173-183. [DOI] [PubMed] [Google Scholar]
- 13.Galm, U., J. Schimana, H. P. Fiedler, J. Schmidt, S. M. Li, and L. Heide. 2002. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tü 6040. Arch. Microbiol. 178:102-114. [DOI] [PubMed] [Google Scholar]
- 14.Gormley, N. A., G. Orphanides, A. Meyer, P. M. Cullis, and A. Maxwell. 1996. The interaction of coumarin antibiotics with fragments of the DNA gyrase B protein. Biochemistry 35:5083-5092. [DOI] [PubMed] [Google Scholar]
- 15.Heddle, J., and A. Maxwell. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249-1264. [DOI] [PubMed] [Google Scholar]
- 17.Holdgate, G. A., A. Tunnicliffe, W. H. Ward, S. A. Weston, G. Rosenbrock, P. T. Barth, I. W. Taylor, R. A. Pauptit, and D. Timms. 1997. The entropic penalty of ordered water accounts for weaker binding of the antibiotic novobiocin to a resistant mutant of DNA gyrase: a thermodynamic and crystallographic study. Biochemistry 36:9663-9673. [DOI] [PubMed] [Google Scholar]
- 18.Holzenkämpfer, M., and A. Zeeck. 2002. Biosynthesis of simocyclinone D8 in an 18O2-rich atmosphere. J. Antibiot. 55:341-342. [DOI] [PubMed] [Google Scholar]
- 19.Kampranis, S. C., N. A. Gormley, R. Tranter, G. Orphanides, and A. Maxwell. 1999. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry 38:1967-1976. [DOI] [PubMed] [Google Scholar]
- 20.Kampranis, S. C., and A. Maxwell. 1998. The DNA gyrase-quinolone complex: ATP hydrolysis and the mechanism of DNA cleavage. J. Biol. Chem. 273:22615-22626. [DOI] [PubMed] [Google Scholar]
- 21.Lamour, V., L. Hoermann, J. M. Jeltsch, P. Oudet, and D. Moras. 2002. Crystallization of the 43 kDa ATPase domain of Thermus thermophilus gyrase B in complex with novobiocin. Acta Crystallogr. D Biol. Crystallogr. 58:1376-1378. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, R. J., O. M. P. Singh, C. V. Smith, T. Skarynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 23.Macinga, D. R., P. J. Renick, K. M. Makin, D. H. Ellis, A. A. Kreiner, M. Li, K. J. Rupnik, E. M. Kincaid, C. D. Wallace, B. Ledoussal, and T. W. Morris. 2003. Unique biological properties and molecular mechanism of 5,6-bridged quinolones. Antimicrob. Agents Chemother. 47:2526-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell, A. 1997. DNA gyrase as a drug target. Trends Microbiol. 5:102-109. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell, A., and M. Gellert. 1984. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 259:14472-14480. [PubMed] [Google Scholar]
- 26.Maxwell, A., and A. J. Howells. 1999. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 94:135-144. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 28.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 29.Pojer, F., S. M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 30.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 31.Rohr, J., and R. Thiericke. 1992. Angucycline group antibiotics. Nat. Prod. Rep. 9:103-137. [DOI] [PubMed] [Google Scholar]
- 32.Schimana, J. 2000. Simocyclinone und weitere neue Metaboliten aus Streptomyces antibioticus Tü 6040. Eberhard Karls University, Tübingen, Germany.
- 33.Schimana, J., H. P. Fiedler, I. Groth, R. Süssmuth, W. Beil, M. Walker, and A. Zeeck. 2000. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 53:779-787. [DOI] [PubMed] [Google Scholar]
- 34.Schimana, J., M. Walker, A. Zeeck, and H. P. Fiedler. 2001. Simocyclinones: diversity of metabolites is dependent on fermentation conditions. J. Ind. Microbiol. Biotechnol. 27:144-148. [DOI] [PubMed] [Google Scholar]
- 35.Schmutz, E., S. Hennig, S. M. Li, and L. Heide. 2004. Identification of a topoisomerase IV in actinobacteria: purification and characterization of ParYR and GyrBR from the coumermycin A1 producer Streptomyces rishiriensis DSM 40489. Microbiology 150:641-647. [DOI] [PubMed] [Google Scholar]
- 36.Schmutz, E., A. Muhlenweg, S. M. Li, and L. Heide. 2003. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob. Agents Chemother. 47:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steffensky, M., A. Muhlenweg, Z. X. Wang, S. M. Li, and L. Heide. 2000. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob. Agents Chemother. 44:1214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theobald, U., J. Schimana, and H. P. Fiedler. 2000. Microbial growth and production kinetics of Streptomyces antibioticus Tü 6040. Antonie Leeuwenhoek 78:307-313. [DOI] [PubMed] [Google Scholar]
- 39.Trefzer, A., S. Pelzer, J. Schimana, S. Stockert, C. Bihlmaier, H. P. Fiedler, K. Welzel, A. Vente, and A. Bechthold. 2002. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob. Agents Chemother. 46:1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, F. T., O. M. Singh, T. Skarzynski, A. J. Wonacott, S. Weston, A. Tucker, R. A. Pauptit, A. L. Breeze, J. P. Poyser, R. O'Brien, J. E. Ladbury, and D. B. Wigley. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41-52. [PubMed] [Google Scholar]
- 41.Wang, Z. X., S. M. Li, and L. Heide. 2000. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 44:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 43.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiseman, T., S. Williston, J. F. Brandts, and L. N. Lin. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179:131-137. [DOI] [PubMed] [Google Scholar]
- 45.Xu, H., L. Heide, and S. M. Li. 2004. New aminocoumarin antibiotics formed by a combined mutational and chemoenzymatic approach utilizing the carbamoyltransferase NovN. Chem. Biol. 11:655-662. [DOI] [PubMed] [Google Scholar]