Abstract
Metronidazole is one of a few antibiotics effective in eliminating Helicobacter pylori infection of the human stomach. Several chromosomal loci have been implicated in resistance to this drug. Saturation transposon mutagenesis of the H. pylori genome revealed inactivation of the rdxA gene as uniquely able to confer metronidazole resistance.
Helicobacter pylori bacteria establish a chronic infection of the stomach in nearly half the human population. Infection is often acquired in childhood and persists for life without specific antimicrobial intervention. While a majority of individuals remain asymptomatic, a significant portion (up to 20%) of those infected develop severe gastric disease (peptic ulcer and/or cancer) requiring eradication of the infection (5). Metronidazole (Mtz) was initially used against a variety of anaerobic microorganisms but was later found to have activity against certain microaerophilic organisms such as H. pylori. Currently, Mtz is a cornerstone of many triple-therapy formulations for the treatment of H. pylori. Unfortunately, resistance approaches 90% in many developing countries and even in Western Europe ranges from 5 to 50% (1). While many triple- and quadruple-therapy formulations use more than one antibiotic, metronidazole resistance has a profound affect on the efficacy of these regimens (13).
The resistance mechanisms in anaerobic organisms and H. pylori differ. Mtz is a prodrug and becomes active when reduced in the cytosol of the microorganism to a toxic metabolite. Unstable Mtz radicals react rapidly with proteins, RNA, and DNA, eventually resulting in cell death. Under the conditions of low-redox potential in anaerobic organisms, drug activation can be catalyzed by nitroreductases such as pyruvate-flavodoxin reductase via a single electron transfer event. H. pylori contains this enzyme, but owing to its microaerophilic nature, molecular oxygen is also present and can compete with the Mtz radical for electrons in a futile cycle that restores the prodrug along with superoxide. Instead, a separate mechanism seems to account for most Mtz sensitivity in H. pylori (6). A non-oxygen-sensitive NADPH nitroreductase encoded by the rdxA gene reduces Mtz by a two-electron transfer step into a toxic metabolite that cannot be retransformed to its parent by molecular oxygen. The vast majority of clinically isolated (12) or experimentally induced (7) Mtz-resistant clones contain a mutation somewhere in the rdxA coding sequence. However, there have been reports that mutation of a second reductase NAD(P)H-flavin oxidoreductase encoded by frxA could also confer low-level Mtz sensitivity in some strains (8) and a role for oxygen sensitive reductases has not been formally excluded. Sequencing candidate genes, such as the reductases described above, in sensitive and resistant isolates has provided support for the idea of these genes playing a role in resistance, but a recent report showed that frameshift mutations in frxA occur with similar frequencies in sensitive and resistant strains (3). We decided to directly test for genes that confer Mtz resistance when inactivated by screening a genome-saturating mutant library.
We recently developed a 10,000-clone transposon-based insertion mutant library targeting all nonessential genes in the genome of the clinical isolate G27 (4, 11). The library was generated using an in vitro mutagenesis system based on the Tn7 transposon with random target site selectivity (2). We screened for Mtz-resistant clones at low and high concentrations to determine whether we could identify transposon insertions within rdxA or any additional genes. The H. pylori GPS-cat mutant library was plated from frozen stock at 10% CO2 and 37°C on horse blood (HB) medium (10) containing 25 μg of chloramphenicol (CHL) ml−1 to give approximately 500 single colonies per plate. Single colonies were transferred using velvet squares to plates containing 36 μg of Mtz ml−1, 9 μg of Mtz ml−1, or no Mtz (in that order). The plates containing Mtz were monitored for the appearance of colonies between 2 and 5 days after plating. Potential Mtz-resistant colonies were picked and patched onto plain HB plates and then retested for Mtz resistance.
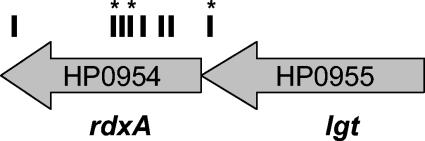
We screened approximately 27,500 clones (18 genome equivalents) and obtained 26 Mtz-resistant mutants. While some originally only grew at the low concentration of drug, upon retesting, all grew at the highest dose. Because Mtz is mutagenic and gives a high spontaneous rate of Mtz resistance, we determined whether resistance was linked to the transposon insertion. Genomic DNA was isolated from each candidate clone by use of a Wizard Genomic preparation according to the instructions of the manufacturer (Promega), and 2 μg was used to transform the parent strain by natural transformation (14). The transformation reactions were plated on HB-CHL medium to select for transposon-containing clones. Two CHL-resistant colonies from each transformation were tested for growth on Mtz-containing plates. Seven clones had resistance unlinked to the transposon, and five clones did not yield transformants after three attempted transformation reactions and were not pursued further. The site of transposon insertion was sequenced for the remaining 14 Mtz-resistant clones by direct sequencing of agarose gel-purified PCR products obtained by semirandom PCR with transposon-specific primers (9). As shown in Fig. 1, the 14 clones represented seven independent transposon insertion events within the rdxA coding sequence and one insertion immediately upstream of the rdxA gene. Six insertions were represented by single clones, while three insertions were each obtained on three occasions from separate plates in the screen. Multiple isolation of some clones indicate that we have performed a saturating screen of our library. Furthermore, mutation of rdxA was uniquely necessary and sufficient to confer Mtz resistance in H. pylori strain G27. The frequency of Mtz-resistant clones in the transposon mutant library was approximately 5 × 10−4.
FIG. 1.
Position of transposon insertion for Mtz-resistant mutants. Hash marks indicate the sites of transposon insertion at positions −25, +146, +150, +235, +249, +252, +258, and +600 of the rdxA coding sequence. Asterisks indicate the isolation of multiple clones with the same insertion position.
In a minority of clinical and experimentally induced Mtz-resistant clones, no mutations within the rdxA coding region could be identified (7, 12). We did not isolate insertions in frxA or any oxygen-sensitive oxidoreductases implicated in Mtz resistance in anaerobic organisms in spite of the fact that mutations in these genes are present in our library (11). We attempted to identify additional modulators of Mtz resistance by repeating the screening using a G27 derivative already lacking the rdxA gene and testing for resistance at even higher levels of drug. These experiments yielded no resistant colonies (unpublished observations). It is possible that the expression of these genes in the human stomach differs from that occurring under our in vitro growth conditions, allowing them to play a role in Mtz sensitivity in vivo that we could not detect. Furthermore, this experiment was done in a single-strain background and cannot rule out the effect of additional genes in other genetic backgrounds.
Interestingly, one of the transposon insertions conferring Mtz resistance mapped to a position 25 bp upstream of the coding sequence, within the coding sequence of a predicted prolipoprotein diacylglyceryl transferase (lgt) (Fig. 1). rdxA is the eighth and last gene in a large predicted operon. Our transposon lacks terminator sequences and is not expected to give polar effects. Furthermore, our library contained several insertions further 5′ of rdxA within this operon (11). Yet this single insertion, just upstream of the ribosomal binding site, likely affects either transcription or translation of this gene. This suggests that mutations outside of the rdxA coding sequence may still mediate their effect through the rdxA gene product and may account for the portion of Mtz resistance for which no mutation in the rdxA coding region was observed. While the experiments described here do not address resistance determinants acquired from other organisms such as pumps or modifying enzymes, our data support a unique role for rdxA in conferring resistance to this drug mediated by mutational inactivation.
Acknowledgments
We acknowledge Benjamin Shepherd for technical assistance.
We acknowledge grants from The Pew Charitable Trusts and a New Development Fund of the Fred Hutchinson Cancer Research Center to N.R.S.
REFERENCES
- 1.Alarcon, T., D. Domingo, and M. Lopez-Brea. 1999. Antibiotic resistance problems with Helicobacter pylori. Int. J. Antimicrob. Agents 12:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisholm, S. A., and R. J. Owen. 2004. Frameshift mutations in frxA occur frequently and do not provide a reliable marker for metronidazole resistance in UK isolates of Helicobacter pylori. J. Med. Microbiol. 53:135-140. [DOI] [PubMed] [Google Scholar]
- 4.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 7.Jenks, P. J., R. L. Ferrero, and A. Labigne. 1999. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 43:753-758. [DOI] [PubMed] [Google Scholar]
- 8.Kwon, D. H., K. Hulten, M. Kato, J. J. Kim, M. Lee, F. A. El-Zaatari, M. S. Osato, and D. Y. Graham. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 45:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manoil, C. 2000. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 326:35-47. [DOI] [PubMed] [Google Scholar]
- 10.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salama, N. R., B. Shepherd, and S. Falkow. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tankovic, J., D. Lamarque, J. C. Delchier, C. J. Soussy, A. Labigne, and P. J. Jenks. 2000. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Wouden, E. J., J. C. Thijs, J. G. Kusters, A. A. van Zwet, and J. H. Kleibeuker. 2001. Mechanism and clinical significance of metronidazole resistance in Helicobacter pylori. Scand. J. Gastroenterol. Suppl. 234:10-14. [DOI] [PubMed]
- 14.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]