Abstract
Triple negative breast cancer (TNBC) is a subtype of highly aggressive breast cancer with poor prognosis. The main characteristic feature of TNBC is its lack of expression of ER, PR and HER2 receptors that are targets for treatments. Hence, it is imperative to identify novel therapeutic strategies to target TNBC. Our aim was to examine whether GROα is a specific marker for TNBC metastasis. For this we performed qPCR, ELISA, migration/invasion assays, western blotting, and siRNA transfections. Evaluation of baseline GROα expression in different breast cancer (BC) subtypes showed that it is significantly upregulated in breast tumor cells, specifically in TNBC cell line. On further evaluation in additional 17 TNBC cell lines we found that baseline GROα expression was significantly elevated in >50% of the cell lines validating GROα overexpression specifically in TNBC cells. Moreover, GROα-stimulation in MCF7 and SKBR3 cells and GROα-knockdown in MDA-MB-231 and HCC1937 cells elicited dramatic changes in migration and invasion abilities in vitro. Corresponding changes in EMT markers were also observed in phenotypically modified BC cells. Furthermore, mechanistic studies identified GROα regulating EMT markers and migration/invasion via MAPK pathway and specific inhibition using PD98059 resulted in the reversal of effects induced by GROα on BC cells. In conclusion, our study provides strong evidence to suggest that GROα is a critical modulator of TNBC migration/invasion and proposes GROα as a potential therapeutic target for treatment of TNBC metastasis.
Keywords: GROα, triple negative breast cancer, metastasis, epithelial-mesenchymal transition, MAPK
Introduction
Breast cancer (BC) is one of the leading cancers in women accounting for more than one million new cases being diagnosed and >400,000 deaths in the world every year (1,2). Approximately 1.4 million women were diagnosed with BC in 2008 (3). In 2016, US Breast Cancer Statistics estimated the diagnosis of 246,660 new cases of invasive BC in women in the US (4). The probability for a woman to be diagnosed with BC has increased at an alarming rate from 1 to 11 (1975) to 1 to 8 (2013) (5). In 2010, 438,000 deaths were observed globally due to BC (2). Tumor metastasis is one of the primary reasons for its high mortality rate. Despite the 5-year disease-free survival rate for patients diagnosed with and treated for localized BC is an appreciable 89%; the incidence of metastasis always results in poor prognosis (6). Patients diagnosed with distant metastasis have <2-year survival rate; while only 25% of those patients survive longer than five years (7). Such critical statistics urge for immediate action to limit metastasis and drastically improve survivability of BC patients.
Triple negative breast cancer (TNBC) subtype of BC have been defined as cells which lack the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2), have an aggressive phenotype when compared to other BC subtypes and comprises of 10–20% of BCs (8,9). They are generally found in younger women, have a high rate of metastasis, poor prognosis and most importantly, lack clinically beneficial target molecules (10). Also, our previous study identified that the occurrence of TNBC is especially high in the African-American and Hispanic women residing in the South Los Angeles area (11). Treatment options for women with TNBC are very limited. While Trastuzumab and endocrine therapy indicate negative beneficiary effects, cytotoxic chemotherapy and molecular-targeted therapies are a few that have demonstrated affirmative efficacy towards TNBC metastasis treatment (12–14). Thus, it is imperative to identify specific unique markers overexpressed in TNBC that may assist in inhibiting metastasis and also be used as a potential targeted therapy for treatment of women diagnosed with TNBC subtype of breast cancer.
Recent studies have demonstrated that chemokines regulate paracrine survival networks that could be beneficial for cancer cells to thrive, metastasize and cause chemoresistance (6,15–17). GROα/CXCL1 (C-X-C motif ligand 1) is a chemotactic cytokine composed of small peptides that act as a chemoattractant for leukocytes during homeostasis and inflammatory responses (18,19). GROα specifically binds to a seven-transmembrane G protein-coupled CXCR2 chemokine receptor to trigger downstream NF-κB signaling pathway and PI-3K pathway (20–25). Reports also illustrate that GROα plays a vital role in tumor cell transformation, angiogenesis and metastasis in various forms of cancer (15,18,26–29). A study has identified numerous inflammation-related cytokines to be differentially regulated in TNBC cells (30).
Here, we report our findings on the importance of GROα expression in TNBC metastasis. We investigated the GROα expression levels in different BC subtypes along with a panel of additional TNBC cell lines and performed functional and mechanism studies on high-GROα expressing TNBC cell lines versus low-GROα expressing BC cell line in vitro. GROα stimulation or inhibition demonstrated significant differences in migration and invasion abilities indicating the role of GROα in BC metastasis. Also, these results were substantiated by differential alterations in the expression of EMT markers. In this study we also identify that GROα regulates BC migration/invasion via MAPK pathway and that treatment with MAPK inhibitor PD98059 significantly reverses all the effects induced by GROα on BC cells.
Materials and methods
Cell cultures
MDA-MB-231, MCF-7, SKBR3 and BT474 cells were purchased from ATCC (American Type Culture Collection) and grown in DMEM/F12 medium containing 10% FBS, 1% glutamine and 0.5% antibiotics (penicillin/streptomycin) in 5% CO2 incubator at 37°C. Triple-Negative Breast Cancer Panel 3 (TCP 1003) cell lines mentioned below were purchased from ATCC and cultured as per the Company's instructions. BT20 and HCC2157 were grown in DMEM/F12 medium containing 10% FBS, 1% glutamine, 0.5% antibiotics in 5% CO2 incubator at 37°C; HCC2157 was additionally supplemented with 0.02 mg/ml insulin, 0.01 mg/ml transferrin, 25 nM sodium selenite, 50 nM hydrocortisone, 1 ng/ml EGF, 0.01 mM ethanolamine, 0.01 mM O-phosphorylethanolamine, 100 pM 3,3′,5-triiodo-L-thyronine (T3), 0.5% bovine serum albumin, and 0.5 mM sodium pyruvate. DU4475, HCC1187, HCC1937, HCC1143, HCC1599, HCC1806, HCC1395, HCC38 and HCC70 were cultured in RPMI-1640 medium with 10% FBS and 1% glutamine in 5% CO2 incubator at 37°C. BT-549 and HS578T were cultured in DMEM/F12 medium with 10% FBS, 1% glutamine and 0.01 mg/ml Insulin in 5% CO2 incubator at 37°C. MDA-MB-157, MDA-MB-453, MDA-MB-468, MDA-MB-436 were grown in Leibovitz's L-15 medium with 10% FBS, 1% glutamine and 0.5% antibiotics in a free gas exchange incubator with atmospheric air; MDA-MB-436 was additionally supplemented with 16 µg/ml glutathione.
Materials
Materials were purchased from various manufacturers: recombinant GROα (R&D Systems), GROα siRNA (Santa Cruz Biotechnology), control siRNA (Santa Cruz Biotechnology), Lipofectamine ltx (Invitrogen), Opti-MEM Reduced Serum medium (Invitrogen), GROα ELISA kit (R&D Systems), GROα primers (IDT), ThermoScript™ RT-PCR systems (Invitrogen), SYBR Green (Qiagen), EMT marker primers (IDT), PD98059 (Cell Signaling), E-cadherin antibody (Santa Cruz Biotechnology), N-cadherin antibody (Santa Cruz Biotechnology), Snail antibody (Abcam), Slug antibody (Abcam), Twist antibody (Santa Cruz Biotechnology), Vimentin antibody (Santa Cruz Biotechnology), VEGF antibody (Santa Cruz Biotechnology), phospho-MAPK antibody (Cell Signaling), MAPK antibody (Cell Signaling), β-actin mouse antibody (Santa Cruz Biotechnology).
Enzyme-linked immunosorbent assay (ELISA)
Medium containing 10% FBS was used for performing all ELISA experiments since preliminary studies to analyze baseline GROα concentration in the complete culture medium used by all the cell lines in this study by ELISA demonstrated no presence of GROα (data not shown). ELISA was performed by following the manufacturer's instructions. Briefly, cells were plated in the 6-well plate until reaching 70–80% confluency. The medium was then replaced with fresh 10% FBS containing medium for 24 h. The culture medium was collected and spun momentarily to remove all particulates; the cells were detached and counted and later used for protein extraction. Cell culture supernatants/lysates were then immediately assayed for GROα expression levels. Briefly, standards or samples were added to each well of the GROα specific ELISA strip and incubated for 1.5 h at room temperature (RT). The wells were washed thrice with wash buffer, and re-incubated with GROα conjugate for 1 h at 4°C. The wells were washed again and incubated with the substrate solution, protected from light for 15 min at RT. The reaction or the color production was ceased by adding stop solution which turned the solution yellow. The absorbance was read colorimetrically (Promega Glomax Multidetection System) at 450 nm. Standard curve was generated to calibrate the concentration of GROα in each supernatant sample and expressed as concentration of GROα (pg/ml) per µg of the protein. The samples were run in duplicates and the experiment repeated thrice for consistent results.
RNA isolation
Cells were washed with cold 1X PBS before lysing them with RNA Bee (Tel-Test) for ~15 min at RT. To the cell lysate, chloroform was added, shaken vigorously and incubated for 15 min on ice. The samples were then centrifuged at 13,000 rpm for 15 min at 4°C resulting in two phases, lower blue chloroform-phenol phase and the color-less upper aqueous phase. The aqueous phase was then transferred to a new tube adding equal volume of isopropanol. The samples were stored on ice for 30 min and then centrifuged at 13,000 rpm for 15 min at 4°C to form a white-yellow pellet of precipitated RNA. The RNA pellet was then washed in 75% ethanol for 10 min at 7,500 rpm at 4°C. The isolated RNA was air-dried and dissolved in DEPC treated water. The concentration and purity of the isolated RNA was calibrated spectrophotometrically using NanoDrop (Thermo Fisher Scientific 2000C).
Quantitative PCR (Q-PCR)
RNA (2 µg) from each sample was used to generate cDNAs using ThermoScript RT-PCR systems kit (Invitrogen). Briefly, RNA along with the first master mix (oligonucleotide, random hexamerase, 10 mM dnTPs) was incubated at 65°C for 5 min. Second master mix (5X buffer, DTT, DEPC water, RNAse Out, Thermoscript) was then added to the solution and the samples were loaded on to the mastercycler (Eppendorf) for converting mRNA to cDNA. The cDNAs were then treated with RNAse H and incubated at 37°C for 15 min. cDNA (1 µl) from each sample was later used along with SYBR Green in a thermocycler (Bio-Rad) to perform Q-PCR. The expression levels of a gene of interest was tested by using specific primers along with an endogenous control 18S by subjecting the cDNA to 58°C primer annealing and 72°C of elongation process for 40 cycles. The Ct values were noted, normalized with an endogenous control (18S) and the results were analyzed by the 2−∆∆Ct method (31). Experiments were performed in triplicates and repeated at least twice.
Transfections
MDA-MB-231 and HCC1937 cells (2×106) were transfected with GROα siRNA using Lipofectamine LTX (Invitrogen) in Opti-MEM (Invitrogen) reduced serum medium following the manufacturer's instructions for 4 h. Scrambled siRNA served as the control. The cells were then incubated at 37°C in a CO2 incubator. Transfected cells were then used for functional studies.
Cell proliferation assay
Cells/well (1×105) of MDA-MB-231 and HCC1937 or serum-starved MCF7 and SKBR3 either transfected with GROα siRNA or induced with recombinant GROα respectively, along with their respective controls were placed in 96-well plates. In another experiment, HCC1937 cells were subjected to increasing concentrations of Olaparib (PARP inhibitor, - 0, 50, 100 and 200 µM) to determine the IC50 value for the drug. After 72 h of treatment, cell proliferation rates were determined using MTT assay (Sigma) as per the manufacturer's protocol. Briefly, the plates were flipped to empty out the media and 50 µl of MTT buffer reagent was added to each well. The setup was incubated for 4 h at 37°C wrapped in an aluminum foil. After incubation, the MTT reagent was replaced with 100 µl of DMSO and incubated for 15 min at RT on a rocker. The absorbance was read at 560 nm. Experiments were performed in triplicates and repeated thrice. The results are presented as percentage for cell proliferation.
Scratch assay
MDA-MB-231 and HCC1937 cells along with serum-starved MCF7 and SKBR3 cells were grown up to absolute confluency in a culture-insert (Ibidi) with 500 µM cell-free gap overnight. At 0 hour, the insert was removed and the cell patches were rinsed with 1X PBS to take away all the detached cells. It was then treated with different conditions for 24 h. Images were captured at 0 and 24 h after treatment to mark the changes in migration of phenotypically modified BC cells. The distance remaining in the gap after 24 h was captured again. The experiment was performed in triplicates and repeated at least twice.
Cell invasion assay
The invasion assay was performed in 24-well Boyden chambers comprising of inserts with 8-µM pore membranes which were coated with Matrigel (28 µg/insert; Sigma). Briefly, equal no. of phenotypically modified BC cells in reduced medium along with control (1×105/well) were placed in the upper wells and 10% FBS containing DMEM/F12 culture medium was filled in the lower wells to act as a chemo-attractant. The cells were cultured for 24 h and then fixed with 0.5% glutaraldehyde and stained with 0.5% toluidine blue. The number of invaded cells was counted with 20X objective of a microscope from 3 different fields per membrane. The results are presented as percentage for cell invasion. Each experiment was performed at least twice.
Western blotting
Total protein was extracted using standard RIPA buffer (Thermo Fisher Scientific) with proteinase inhibitors (Thermo Fisher Scientific) and the concentration was measured by BCA protein assay (Thermo Fisher Scientific). Protein (30 µg) from each sample was loaded on to the well and separated using NuPAGE pre-cast gels (Invitrogen). The proteins were then transferred onto the nitrocellulose membrane (0.45 µM, Bio-Rad). The membrane was initially blocked using 5% milk blocking buffer, washed with 1X TBST and later incubated with specific primary antibody overnight at 4°C. The membrane was washed again and then incubated with secondary antibody for 2 h at RT. The results were visualized using SuperSignal® West Pico Chemiluminescence Substrate (Thermo Fisher Scientific) (Bio-Rad ChemiDoc). β-actin was used as the loading control. The densitometric analysis was performed by calculating the volume density ratio of bands of genes of interest over loading control. Experiments were repeated at least twice for consistent results.
Statistical analysis
All the experiments were performed at least in 2 separate sets of experiments with 3 technical replicates in each. Data are presented as mean values ± standard deviation. Differences between 2 groups were analyzed using Student's t-test. p<0.05 was considered statistically significant.
Results
GROα is overexpressed in TNBC cells
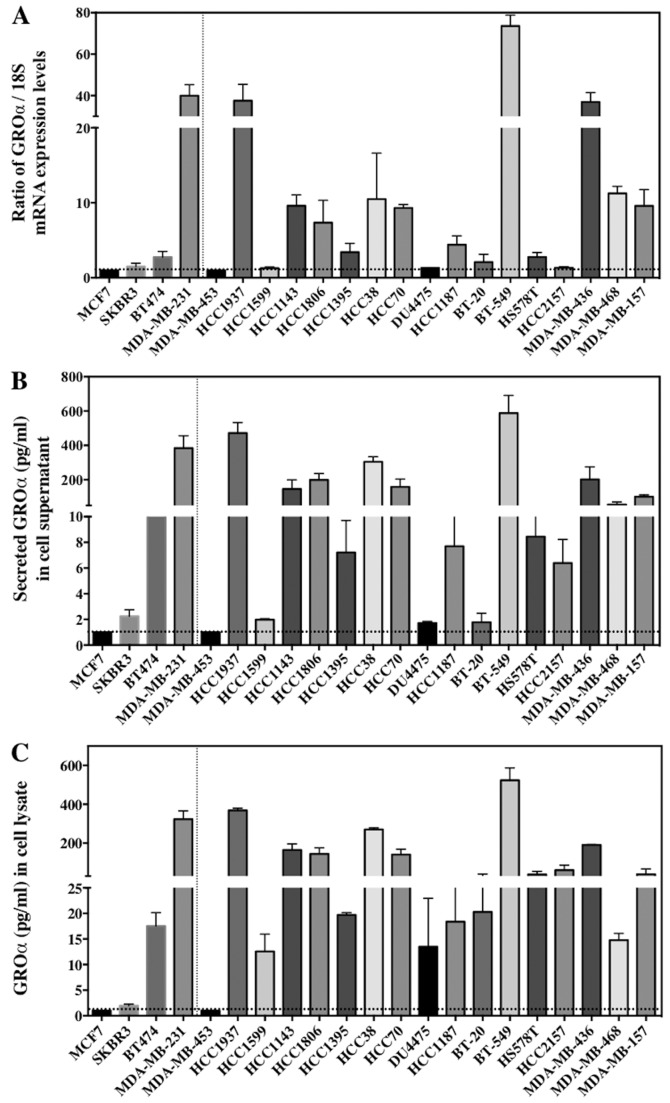
BC comprises of different subtypes. Four BC cell lines were chosen for this study to represent each subtype - SKBR3 (ER−/PR−/HER2+, HER2 subtype), MCF7 (ER+/PR+/HER2−, luminal A subtype), BT474 (ER+/PR+/HER2+, luminal B subtype), and MDA-MB-231 (ER−/PR−/HER2−, basal/TNBC subtype), and cultured for further analysis. Our first approach was to investigate the baseline GROα expression in each subtype. Results from q-PCR and ELISA demonstrated that GROα is significantly overexpressed in the MDA-MB-231 cells with TNBC subtype when compared to other BC cell lines/subtypes. The GROα mRNA levels from q-PCR and protein concentration levels from ELISA were analyzed. Interestingly, expression levels of GROα specifically in MDA-MB-231 cells both in mRNA (30.3, expressed as ratio of GROα/18S) and protein concentration levels (383.92 pg/ml in supernatant) were significantly upregulated when compared to other cell lines expressing <2 and 30 pg/ml (in supernatant) of mRNA and protein respectively (Fig. 1A). Thus, indicating differential GROα expression in BC subtypes.
Figure 1.
GROα is overexpressed in TNBC cells. Baseline GROα expression levels in breast cancer subtypes and 17 additional TNBC cell lines were evaluated by (A) q-PCR by extracting total RNA and presented as GROα/18S ratio whose average fold was normalized to MCF7 GROα levels (B) ELISA by collecting cell supernatants containing secreted GROα from breast cancer cells (C) ELISA by collecting cell lysates from breast cancer cells. The GROα concentration levels were normalized per microgram (µg) of the total protein. Experiments were performed thrice with triplicates per experimental analysis. Bars correspond to mean ± SD.
To further confirm our observation that GROα is overexpressed specifically in TNBC cells which might suggest the aggressive nature of TNBCs, we examined additional 17 TNBC cell lines (BT20, HCC2157, DU475, HCC1187, HCC1937, HCC1143, HCC1599, HCC1806, HCC1395, HCC38, HCC70, BT-549, HS 578T, MDA-MB-157, MDA-MB-453, MDA-MB-468 and MDA-MB-436), for baseline GROα expression in these cells as a proof-of-principle. All the cell lines were cultured as per the ATCC guidelines. Cells were harvested for total RNA and supernatants were collected for secreted protein analysis by q-PCR and ELISA respectively. Remarkably, >50% of the TNBC cell lines (HCC1937, HCC1143, HCC1806, HCC38, HCC70, BT-549, MDA-MB-436 and MDA-MB-468) expressed significantly elevated levels of GROα mRNA and protein while other cell lines demonstrated lower GROα expression levels (Fig. 1). Cell lines expressing higher levels of GROα such as HCC1937, BT549 and MDA-MB-436 are metastatic in nature and have an aggressive phenotype. Hence, from these results we hypothesized that GROα is an important molecule associated with TNBC metastasis and that upregulation of GROα expression aids BC cell migration and invasion.
GROα effects on BC cell proliferation
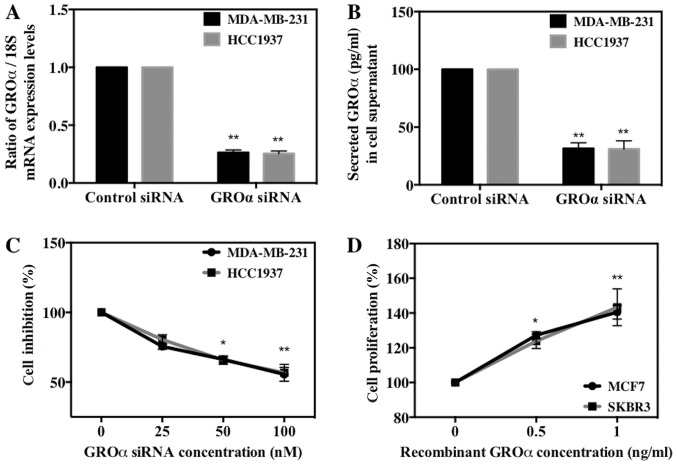
We next investigated the role of GROα in BC cell proliferation in vitro by either knocking down or stimulating BC cells with GROα. To achieve this we used four cell lines, MDA-MB-231 and HCC1937 (high-GROα) and MCF7 and SKBR3 (low-GROα). We then transfected MDA-MB-231 and HCC1937 cells with GROα specific siRNA (100 nM) using scrambled siRNA as control and induced MCF7 and SKBR3 cells with recombinant GROα (1 ng/ml) using water as control for 72 h. The effect of GROα knockdown using GROα specific siRNA on respective cell lines was tested both by qPCR and ELISA (Fig. 2A and B). After 72 h, the treated cells were subjected to MTT assay to assess the effect of GROα on BC cell proliferation. Results obtained demonstrated a gradual decrease (~35–40%) in cell proliferation in GROα-knocked down MDA-MB-231 and HCC1937 cells when compared to control cells (without siRNA treatment) (Fig. 2C). Similarly, a gradual increase (38–42%) in cell proliferation was observed in GROα stimulated MCF7 and SKBR3 cells when compared to control cells (untreated with GROα) (Fig. 2D). These results indicate that GROα induces positive effects on BC cell proliferation over 72-h time period.
Figure 2.
GROα stimulates breast cancer cell proliferation. GROα was silenced using GROα specific siRNA along with scrambled siRNA as control and knockdown was confirmed by (A) qPCR by extracting total RNA and normalizing to 18S expression levels and (B) ELISA by collecting cell supernatants containing secreted GROα and normalizing per µg of the protein. Cell proliferation rates were determined by MTT assay after 72 h of GROα specific siRNA (100 nM) knock down in (C) MDA-MB-231 and HCC1937 cells compared to their control cells treated with scrambled siRNA and after 72-h stimulation with recombinant GROα (1 ng/ml) in (D) MCF7 cells and SKBR3 cells compared to their control cells treated with vehicle (water). Experiments were performed at least twice with triplicates per experimental analysis. Bars correspond to mean ± SD, n=3, *p<0.05, **p<0.01 (paired t-test, sample vs. control).
GROα promotes BC cell migration and invasion
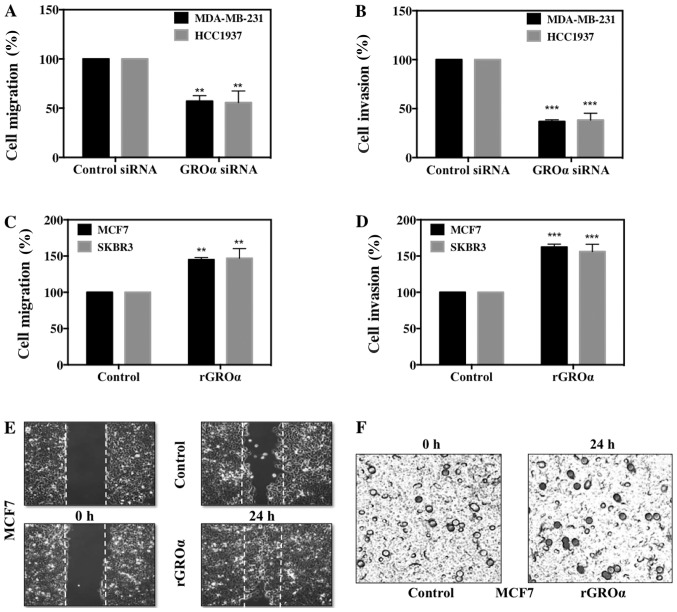
We further evaluated the significance of GROα on BC cell migration and invasion in vitro. For this, we used the phenotypically modified GROα-knocked down MDA-MB-231 and HCC1937 cells and GROα-stimulated MCF7 and SKBR3 cells along with their respective untreated controls and subjected them to scratch assay/wound healing assay and Boyden chamber Matrigel invasion assay. Images were captured at 0 and 24 h for migration and at 24 h for invasion assays to mark the changes in treatment conditions. Results from both migration and invasion assay demonstrated complementary data; while GROα-knocked down MDA-MB-231 and HCC1937 cells displayed significant decrease after 24 h with as low as 43–47% reduced migratory and 59–60% reduced invasive abilities (Fig. 3A and B). In contrast, GROα-stimulated MCF7 and SKBR3 cells showed increase in both migration and invasion, with as high as 45–47% enhanced migratory and 56–69% invasive abilities with reference to their untreated controls (Fig. 3C and D). Representative images of migration and invasion assays in MCF7 cells with GROα-stimulation are shown in Fig. 3E and F. Results from the in vitro functional studies clearly demonstrate that GROα plays a crucial modulatory role in BC cell migration and invasion; suggesting that GROα could be an important target molecule in the treatment for TNBC metastasis.
Figure 3.
GROα promotes breast cancer cell migration and invasion. Cell migration rates were determined by scratch assay in GROα specific siRNA (100 nM) knocked down (A) MDA-MB-231 cells and HCC1937 cells and recombinant GROα-stimulated (1 ng/ml) (C) MCF7 and SKBR3 cells after 24 h by comparing with their respective controls, scrambled siRNA or vehicle (water) respectively. Images were captured at 0 h and 24 h of wound healing process and presented as percentage cell migration. Cell invasion rates were determined by Boyden chamber Matrigel invasion assay by placing treated cells in serum-free medium in the upper chamber and 10% FBS containing medium in the lower chamber. After 24 h, bluish-black cells stained with toluidine blue indicating cell invasion into the Matrigel were counted in three fields of view per chamber and presented as percentage cell invasion in GROα specific siRNA (100 nM) knocked down (B) MDA-MB-231 cells and HCC1937 cells; and recombinant GROα (1 ng/ml) stimulated (D) MCF7 cells and SKBR3 cells after 24 h compared to their respective controls, scrambled siRNA and vehicle (water). Representative images for GROα-stimulated MCF7 cell migration (E) and invasion (F) assays. For invasion assays, the images were captured at 20x where the darkly stained dots represented the invaded cells that passed through the porous membrane matrix. Experiments were performed at least twice with triplicates per experimental analysis. Bars correspond to mean ± SD, n=3, **p<0.01, ***p<0.001 (paired t-test, sample vs. control).
GROα stimulation/knockdown induces phenotypic changes in EMT markers
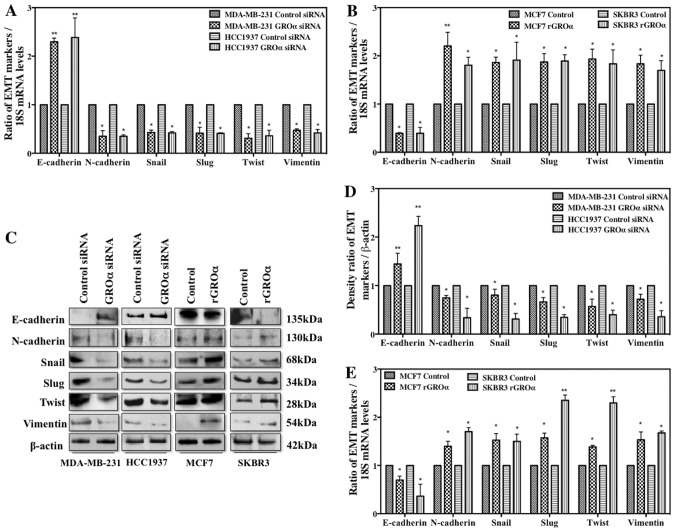
Thus far, our studies imply that GROα is a critical modulator for BC cell metastasis. To further understand the molecular mechanisms that regulate the phenotypically modified MDA-MB 231/HCC1937 and MCF7/SKBR3 cells to inhibit/initiate metastasis process, we evaluated the change in expression of various EMT markers in the presence or absence of GROα in BC cells by q-PCR and western blotting after 48-h treatment. Interestingly, the results from both q-PCR (Fig. 4A and B) and western blotting (Fig. 4C) demonstrated that in the presence of GROα (GROα induced MCF7/SKBR3 cells), a significant downregulation of E-cadherin was accompanied by strong upregulation of all the other mesenchymal markers such as N-cadherin, Snail, Slug, Twist and Vimentin. In contrast, in the absence of GROα (GROα-knocked down MDA-MB-231/HCC1937 cells), an elevated expression of E-cadherin was accompanied with decrease in mesenchymal markers. Densitometric analysis of the bands obtained from western blotting method clearly indicated loss of E-cadherin and gain of other EMT markers in GROα-knocked down cells versus the contrary effects in GROα-stimulated cells (Fig. 4D and E). These results further emphasize the role and the molecular mechanisms regulated by GROα to influence and trigger metastasis process in TNBC cells.
Figure 4.
GROα stimulation/knockdown induces phenotypic changes of EMT markers. Effect of GROα on EMT markers were evaluated by (A) q-PCR in GROα specific siRNA (100 nM) knocked down MDA-MB-231 and HCC1937 cells compared to their control cells treated with scrambled siRNA (B) q-PCR in recombinant GROα (1 ng/ml)-stimulated MCF7 and SKBR3 cells compared to their control cells treated with vehicle (water). Total RNA extracted from treated cells was subjected to qPCR and normalized to 18S expression levels and (C) western blot analysis in phenotypically modified MDA-MB-231, HCC1937, MCF7 and SKBR3 BC cells were performed by extracting total proteins. β-actin was used as the loading control. The data obtained from western blotting was analyzed and presented as the density ratio of gene over β-actin in (D) MDA-MB-231 and HCC1937 cells and (E) MCF7 and SKBR3 cells. Experiments were performed at least twice with triplicates per experimental analysis. Bars correspond to mean ± SD, n=2, *p<0.05, **p<0.01 (paired t-test, sample vs. control).
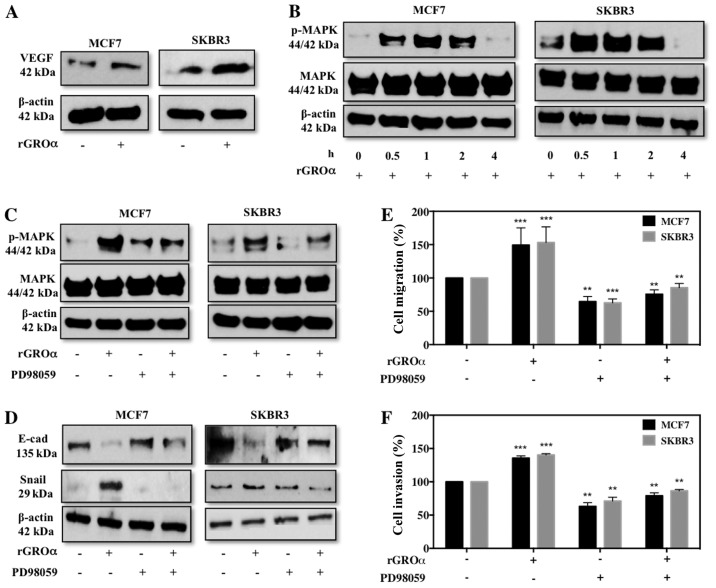
GROα stimulation activates VEGF and MAPK targets
To further investigate the specific pathway through which GROα might induce migration/invasion in BC cells, we evaluated VEGF (angiogenic) and MAPK targets. To our surprise we observed that MCF7 and SKBR3 cells induced with GROα demonstrated an enhanced expression level of VEGF (Fig. 5A). Additionally, GROα induction at different time-points gradually increased p-MAPK with maximum activation at 1 h time-point and then gradually decreased (Fig. 5B). Next we performed experiments to verify specificity of MAPK pathway with regards to GROα. For this, we pre-treated MCF7 and SKBR3 cells with PD98059 (MAPK inhibitor, 50 µM) for 2 h and then induced with GROα (1 ng/ml) to re-confirm whether the effects of GROα were specific through MAPK pathway in BC cells. Interestingly, we observed that while GROα-alone induction activated MAPK, in the presence of PD98059 GROα induction had reduced effect on MAPK activation, clearly demonstrating that GROα induces migration/invasion via MAPK pathway in BC cells (Fig. 5C). To further validate our findings, we performed functional studies in vitro and identified that PD98059 significantly reduces migratory/invasion abilities induced by GROα induction both in MCF7 and SKBR3 cells (Fig. 5D and E). Our immediate follow through experiment was to assess if these changes affected EMT markers; thus we performed western blotting on MCF7 and SKBR3 cells pre-treated with PD98059 and induced with GROα to evaluate changes in EMT markers. As expected, while GROα alone decreased E-cadherin and increased Snail expression, pre-treatment with PD98089 reversed these effects in BC cells (Fig. 5F); suggesting a definitive role of GROα in regulating BC metastasis.
Figure 5.
GROα stimulation activates VEGF and MAPK targets. Effect of recombinant GROα-stimulation (1 ng/ml) on MCF7 and SKBR3 cells were evaluated for change in expression levels of (A) VEGF and (B) pMAPK by western blotting when compared to their respective controls (water). Next, effect of pre-treating MCF7 and SKBR3 cells with MAPK inhibitor PD98059 (50 µM) for 2 h on (C) pMAPK and (D) EMT markers were analyzed by western blotting, (E) migration by wound healing assay and presented as percentage cell migration versus DMSO control and (F) invasion by Boyden chamber assay and presented as percentage cell invasion versus DMSO control in GROα-stimulated MCF7 and SKBR3 cells. β-actin was used as the loading control for western blots. Experiments were performed at least twice. Bars correspond to mean ± SD, n=3, **p<0.01, ***p<0.001 (paired t-test, sample vs. control).
Discussion
Our study mainly focuses on identifying a specific target molecule for the treatment of TNBC metastasis. Numerous studies have reported that chemokines such as IL-6 and IL-8 play a crucial role in tumor metastasis and chemoresistance (6,30). Acharyya et al have put forth a study highlighting a network of paracrine signaling pathway involving inflammatory regulators such as TNF-α, CXCL1/2 and S100A8/9 that assist tumor cells to evade stress and survive severe conditions (6). Literature also signifies the importance of proinflammatory cytokine expression for the growth of TNBC cells (30). Based on literature and our data we hypothesized that GROα is an important inflammatory cytokine regulating TNBC migration/invasion. To prove the hypothesis, we initially performed a baseline GROα expression studies using four BC cell lines - SKBR3, MCF7, BT474 and MDA-MB-231, each representing a subtype of BC. Interestingly, we noted that expression of GROα was upregulated specifically in MDA-MB-231/TNBC cell line while negligible amounts were found in other cell lines from different subtypes (Fig. 1).
Intrigued by these results, we analyzed additional 17 TNBC cell lines as a proof-of-principle to further confirm the concept that TNBCs express high levels of GROα thus making them aggressive in nature. Remarkably, 8 cell lines (50%, HCC1937, HCC1143, HCC1806, HCC38, HCC70, BT-549, MDA-MB-436 and MDA-MB-468) of the 17 cell lines tested expressed significantly elevated levels of GROα (Fig. 1). Thus, our results clearly suggest that GROα might play a vital modulatory role in TNBC metastasis.
Furthermore, we investigated the role of GROα in BC cell proliferation, migration and invasion by performing in vitro functional studies such as MTT assay, scratch assay and Boyden chamber Matrigel invasion assay respectively. Numerous cytokine have been demonstrated as an inducer of invasion in cancers such as breast, gastric, bladder and colorectal (6,26–28). Here we report that GROα, an inflammatory cytokine, plays a crucial modulatory role to trigger phenotypic changes in BC cells that induce the cells to acquire aggressive characteristics to regulate BC migration/invasion. For this we either knocked down GROα using GROα specific siRNA in MDA-MB-231 and HCC1937 cells or stimulated MCF7 and SKBR3 cells with recombinant GROα along with their respective controls. These phenotypically modified cells were then tested for the change in proliferation, migration and invasion. Data from Fig. 2C and D demonstrated a reduced proliferation (45%) in GROα-knocked down MDA-MB-231/HCC1937 cells and an enhanced proliferation (40%) in GROα-stimulated MCF7/SKBR3 cells after 72 h. Even more profound results were observed from migration and invasion studies, migration (43%) and invasion (60%) in the GROα siRNA-treated MDA-MB-231/HCC1937 cells (Fig. 3A and B) versus migration (45%) and invasion (69%) in the GROα-stimulated MCF7/SKBR3 cells (Fig. 3C-F) when compared to their respective control cells after 24 h. These results strongly suggest a positive correlation between GROα expression levels and TNBC migration/invasion.
Our next aim was to decipher the molecular mechanisms induced by GROα that trigger the metastasis process in TNBCs. Last decade has evidenced an influx of reports suggesting a phenomenon called epithelial-mesenchymal transition (EMT) to initiate the metastasis process.
EMT is a succession of various events that initially leads to alterations in cell-cell and cell-extracellular matrix (ECM) communications resulting in the conferring of epithelial cells the ability to move from its primary site and later resulting in additional changes for the maintenance of the newly transformed mesenchymal phenotype (32–35). Specific EMT markers have been identified that are unique to either epithelial cells (E-cadherin) or mesenchymal cells (N-cadherin, Vimentin) and others which are EMT regulators (Snail, Slug, Twist) (35–37). Data from our experiments clearly showed an upregulation of EMT regulators such as Snail, Slug, Twist and mesenchymal marker genes such as Vimentin and N-cadherin along with a notable downregulation of the epithelial marker gene E-cadherin in GROα-stimulated MCF7/SKBR3 cells versus contrary results in GROα-knocked down MDA-MB-231/HCC1937 cells (Fig. 4) either increasing or decreasing the motility of BC cells.
Investigation to identify a specific pathway through which GROα influences migration/invasion in BC cells led us to study the effect of GROα stimulation on VEGF and phosphorylation status of MAPK in MCF7 and SKBR3 cells. In the presence of GROα, we observed an increase in both VEGF and pMAPK proteins in BC cells (Fig. 5A and B). To further confirm the specificity of MAPK pathway, we pre-treated cells with PD98059 (MEK inhibitor) and observed for changes in pMAPK levels, effect on EMT markers and eventually migration/invasion of BC cells. With decrease in pMAPK, we detected a significant reversal of EMT marker expression and migration/invasion abilities in MCF7 and SKBR3 GROα-stimulated cells (Fig. 5C-F). These results clearly indicate the effect of GROα-MAPK pathway on BC cell migration/invasion. In conclusion, results from this study for the first time define a strong association between GROα and TNBC subtype while elucidating the modulatory role of GROα in TNBC cell migration/invasion via MAPK pathway and epithelial to mesenchymal transition. We also demonstrated that targeting GROα via targeting MAPK pathway using MAPK inhibitor PD98059 can limit the effects induced by GROα on BC cells. Thus, a promising therapeutic strategy can be postulated by targeting GROα in aggressive breast cancer cells to limit metastasis specifically in TNBC subtype of cancers.
Acknowledgements
This study was supported by grants from NIH/NCI 1U54- CA14393; U56 CA101599-01; CA15083-25S3; R25DK067015-01; Department of Defense Breast Cancer Research Program grant BC043180, NIH-NIMHD U54MD007598, NIH/NCATS CTSI UL1TR000124 to J.V. Vadgama; and NIH/NIMHD CRECD R25 MD007610, U54MD007598-pilot and bridge support, and NIMHD 5S21MD 000103-Faculty Retention Award to Y. Wu. The cell lines used in this study were provided by Integrated Clinical, Tissue, and Biomarker Database Shared Resource Core (ICTBD) funded by NIH/NCI NIH/NCI 1U54CA14393.
References
- 1.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008 GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.National Cancer InstituteAltekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, et al., editors. SEER Cancer Statistics Review, 1975–2007. National Cancer Institute; Bethesda, MD: [Google Scholar]
- 6.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones SE. Metastatic breast cancer: The treatment challenge. Clin Breast Cancer. 2008;8:224–233. doi: 10.3816/CBC.2008.n.025. [DOI] [PubMed] [Google Scholar]
- 8.Suba Z. Triple-negative breast cancer risk in women is defined by the defect of estrogen signaling: Preventive and therapeutic implications. Onco Targets Ther. 2014;7:147–164. doi: 10.2147/OTT.S52600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark O, Botrel TE, Paladini L, Ferreira MB. Targeted therapy in triple-negative metastatic breast cancer: A systematic review and meta-analysis. Core Evid. 2014;9:1–11. doi: 10.2147/CE.S52197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.André F, Zielinski CC. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann Oncol. 2012;23(Suppl 6):vi46–vi51. doi: 10.1093/annonc/mds195. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Sarkissyan M, Elshimali Y, Vadgama JV. Triple negative breast tumors in African-American and Hispanic/Latina women are high in CD44+, low in CD24+, and have loss of PTEN. PLoS One. 2013;8:e78259. doi: 10.1371/journal.pone.0078259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias AD. Triple-negative breast cancer: A short review. Am J Clin Oncol. 2010;33:637–645. doi: 10.1097/COC.0b013e3181b8afcf. [DOI] [PubMed] [Google Scholar]
- 13.Miles DW, Chan A, Dirix LY, Cortés J, Pivot X, Tomczak P, Delozier T, Sohn JH, Provencher L, Puglisi F, et al. Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 2010;28:3239–3247. doi: 10.1200/JCO.2008.21.6457. [DOI] [PubMed] [Google Scholar]
- 14.Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, Shenkier T, Cella D, Davidson NE. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med. 2007;357:2666–2676. doi: 10.1056/NEJMoa072113. [DOI] [PubMed] [Google Scholar]
- 15.Feliciano P. CXCL1 and CXCL2 link metastasis and chemoresistance. Nat Genet. 2012;44:840. doi: 10.1038/ng.2377. [DOI] [Google Scholar]
- 16.Opdenakker G, Van Damme J. The countercurrent principle in invasion and metastasis of cancer cells. Recent insights on the roles of chemokines. Int J Dev Biol. 2004;48:519–527. doi: 10.1387/ijdb.041796go. [DOI] [PubMed] [Google Scholar]
- 17.Van der Cappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 18.Dhawan P, Richmond A. Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol. 2002;72:9–18. [PMC free article] [PubMed] [Google Scholar]
- 19.Strieter RM, Burdick MD, Mestas J, Gomperts B, Keane MP, Belperio JA. Cancer CXC chemokine networks and tumour angiogenesis. Eur J Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Richmond A, Thomas HG. Purification of melanoma growth stimulatory activity. J Cell Physiol. 1986;129:375–384. doi: 10.1002/jcp.1041290316. [DOI] [PubMed] [Google Scholar]
- 21.Luan J, Shattuck-Brandt R, Haghnegahdar H, Owen JD, Strieter R, Burdick M, Nirodi C, Beauchamp D, Johnson KN, Richmond A. Mechanism and biological significance of constitutive expression of MGSA/GRO chemokines in malignant melanoma tumor progression. J Leukoc Biol. 1997;62:588–597. doi: 10.1002/jlb.62.5.588. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Richmond A. Nuclear factor-kappa B activation by the CXC chemokine melanoma growth-stimulatory activity/growth-regulated protein involves the MEKK1/p38 mitogen-activated protein kinase pathway. J Biol Chem. 2001;276:3650–3659. doi: 10.1074/jbc.M006115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson C, Purcell C, Seaton A, Oladipo O, Maxwell PJ, O'Sullivan JM, Wilson RH, Johnston PG, Waugh DJ. Chemotherapy-induced CXC-chemokine/CXC-chemokine receptor signaling in metastatic prostate cancer cells confers resistance to oxaliplatin through potentiation of nuclear factor-kappaB transcription and evasion of apoptosis. J Pharmacol Exp Ther. 2008;327:746–759. doi: 10.1124/jpet.108.143826. [DOI] [PubMed] [Google Scholar]
- 24.Richmond A, Lawson DH, Nixon DW, Chawla RK. Characterization of autostimulatory and transforming growth factors from human melanoma cells. Cancer Res. 1985;45:6390–6394. [PubMed] [Google Scholar]
- 25.Xia M, Hyman BT. GROalpha/KC, a chemokine receptor CXCR2 ligand, can be a potent trigger for neuronal ERK1/2 and PI-3 kinase pathways and for tau hyperphosphorylation - a role in Alzheimer's disease? J Neuroimmunol. 2002;122:55–64. doi: 10.1016/S0165-5728(01)00463-5. [DOI] [PubMed] [Google Scholar]
- 26.Kawanishi H, Matsui Y, Ito M, Watanabe J, Takahashi T, Nishizawa K, Nishiyama H, Kamoto T, Mikami Y, Tanaka Y, et al. Secreted CXCL1 is a potential mediator and marker of the tumor invasion of bladder cancer. Clin Cancer Res. 2008;14:2579–2587. doi: 10.1158/1078-0432.CCR-07-1922. [DOI] [PubMed] [Google Scholar]
- 27.Bandapalli OR, Ehrmann F, Ehemann V, Gaida M, Macher-Goeppinger S, Wente M, Schirmacher P, Brand K. Down-regulation of CXCL1 inhibits tumor growth in colorectal liver metastasis. Cytokine. 2012;57:46–53. doi: 10.1016/j.cyto.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Cheng WL, Wang CS, Huang YH, Tsai MM, Liang Y, Lin KH. Overexpression of CXCL1 and its receptor CXCR2 promote tumor invasion in gastric cancer. Ann Oncol. 2011;22:2267–2276. doi: 10.1093/annonc/mdq739. [DOI] [PubMed] [Google Scholar]
- 29.Keeley EC, Mehrad B, Strieter RM. CXC chemokines in cancer angiogenesis and metastases. Adv Cancer Res. 2010;106:91–111. doi: 10.1016/S0065-230X(10)06003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartman ZC, Poage GM, den Hollander P, Tsimelzon A, Hill J, Panupinthu N, Zhang Y, Mazumdar A, Hilsenbeck SG, Mills GB, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73:3470–3480. doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: A cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radisky DC. Epithelial-mesenchymal transition. J Cell Sci. 2005;118:4325–4326. doi: 10.1242/jcs.02552. [DOI] [PubMed] [Google Scholar]
- 34.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 37.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]