Abstract
The nucleosome core particle, the basic repeated structure in chromatin fibers, consists of an octamer of eight core histone molecules, organized as dimers (H2A/H2B) and tetramers [(H3/H4)2] around which DNA wraps tightly in almost two left-handed turns. The nucleosome has to undergo certain conformational changes to allow processes that need access to the DNA template to occur. By single-pair fluorescence resonance energy transfer, we demonstrate fast, long-range, reversible conformational fluctuations in nucleosomes between two states: fully folded (closed), with the DNA wrapped around the histone core, or open, with the DNA significantly unraveled from the histone octamer. The brief excursions into an extended open state may create windows of opportunity for protein factors involved in DNA transactions to bind to or translocate along the DNA.
Keywords: conformational transitions, evanescent field fluorescence microscope, nucleosome dynamics, nucleosome opening
The importance of how nucleosomal DNA can be accessed by protein factors to perform its biological function cannot be overstated, because it concerns all DNA transactions, including transcription, replication, recombination, and repair, that must, by necessity, occur within the context of chromatin. How the nucleosomal DNA is made accessible is not clear. Most recent research in the field focuses on the role of postsynthetic modifications to the histones and the DNA, and on the activity of the so-called nucleosome remodeling factors, hardly ever addressing possible spontaneous mechanisms that may provide such accessibility. The paucity of such research stems from the difficulty in finding the right methodological approach.
The structure of the core particle (1) is now known in atomic detail (2–4); however, we are far from understanding its dynamics. To search for possible fast conformational transitions in nucleosome particles in real time, we used single-pair fluorescence resonance energy transfer (FRET) between two fluorophores, a donor and an acceptor, attached to a molecule of DNA that was reconstituted into nucleosomes by using the salt-jump method (5, 6). The fluorescence of the two dyes was recorded as a function of time in a prism-based evanescent field fluorescence microscope. The time trajectories of the fluorescence intensities of the fluorophores revealed fast conformational transitions between a closed and an open state of the nucleosome. The open state is short-lived and results from a long-range unwrapping of the DNA from around the histone core.
Materials and Methods
Histones. Core histones used for reconstitution were purified from chicken erythrocytes (6).
Oligonucleotide Labeling with Biotin and Fluorescent Dyes. Oligonucleotides modified with 5′ biotin and containing internal aminolink-dC (or aminolink-dT) with six-carbon linker were from MWG Biotech (Ebersberg, Germany). Primary amino groups were labeled with Cy3 and Cy5 N-hydroxysuccimide esters (Amersham Pharmacia). Fluorescently labeled oligonucleotides were separated from unlabeled oligonucleotides by RP-HPLC on a C2/C18 column (Amersham Pharmacia) in a 0–60% acetonitrile (Sigma) gradient (in trimethylammonium acetate; Sigma) (pH 7.0) at 60°C. The final construct (164-bp dsDNA having both Cy3 and Cy5) was prepared by PCR using the labeled oligonucleotides as primers.
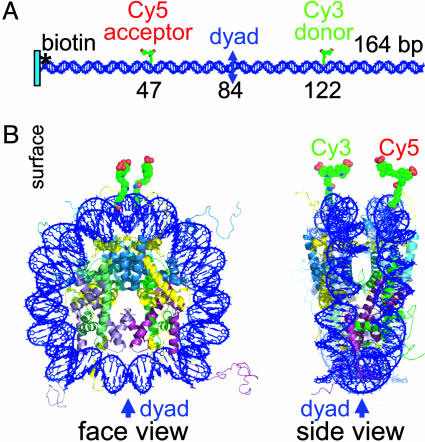
Generation of the 164-bp DNA Sequence with Biotin Labeling at Position 1, Cy5 Labeling at Position 47, and Cy3 Labeling at Position 122 (Position 43 of the Complementary Strand). The 164-bp GUB nucleosome positioning sequence used in these experiments is shown in Fig. 1. In Fig. 1, C is the location of the aminolink-dC in the 50-base oligonucleotide 5′-biotin-ctagaggatc ctctagacgg aggacagtcc tccggttacc ttcgaaCcac that was labeled with the N- hydroxysuccimide ester of Cy5. A is the location of the aminolink-dT on the complementary strand, in the 50-base oligonucleotide atatttcgcg aaggcctccc gggcagctgg atatctttaa acTcgagact that was labeled with Cy3. The two sites for the dyes were chosen by superimposing the GUB sequence (6–8) on the sequence used in the nucleosome crystal structure (2–4) by using the nucleosome pseudodyad (G) that we had determined previously (8). The nucleosome positioning sequence is boxed; the two labeling sites are on the opposite gyres of the nucleosomal DNA in positions that would be close to each other upon nucleosome formation. After purification of the two fluorescently labeled oligonucleotides, the entire 164-bp dsDNA fragment was prepared by PCR using these two fragments as primers with unlabeled GUB as template.
Fig. 1.
The 164-bp GUB nucleosome positioning DNA sequence. For further details see text.
Flow Cell Preparation. The flow cell was constructed from cleaned quartz slide (3 × 1 inches) (Finkenbeiner, Waltham, MA) with two holes (1-mm diameter) drilled on its diagonal (serving as inlet and outlet ports for solution injections). Two strips of double-stick tape (3M Co.) were placed on the quartz slide, so that a narrow channel a few millimeters wide was formed. The gaps between the two strips of double-stick tape were sealed with epoxy. A coverslip (No. 1, 2 × 1 inches) was placed on top to seal the flow chamber. Then, 30 μl of 0.2 mg/ml streptavidin (Roche or Molecular Probes) in T50 was injected and allowed to passively adsorb onto the quartz surface for 10 min. Later experiments were performed on a polyethylene glycol–streptavidin surface. In this case, the cleaned slide was first treated with 2% aminopropyltriethoxysilane (Sigma) solution in acetone for 1 min. The flow cell was assembled as above, and fresh 20% M-polyethylene glycol-succinimidyl propionate (2,000 molecular weight), 0.2% biotin–polyethylene glycol–succinimidyl propionate (3,400 molecular weight; both Nektar Therapeutics, San Carlos, CA) solution in 0.1 M sodium carbonate buffer (pH 8.5) was injected and allowed to react with the aminopropyltriethoxysilane-treated surface for 2–3 h, followed by a 10-min streptavidin treatment as described above.
Nucleosome Reconstitution. Mononucleosomes were prepared by the salt-jump method (5) using 10 ng of double-labeled PCR product and 5 μg of carrier DNA [plasmid PMSA-3 (8) digested with BamHI]. Then, 4.5 μg of octamers were mixed with 5 μg of DNA in TE (10 mM Tris·HCl/0.5 mM EDTA, pH 7.5) and 2 M NaCl that was stepwise diluted to 1, 0.75, and 0.5 M NaCl with TE, incubating at 37°C for 20 min between each dilution. The reconstituted nucleosomes were dialyzed against TE overnight, and the quality of reconstitution was checked on native 5.5% polyacrylamide gels electrophoresed in 0.5× TBE (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.3). The mononucleosome preparation was injected into the flow cell that had been treated with streptavidin. After 1–3 min of incubation, the flow cell was rinsed with T50. Control experiments showed no fluorescent signal above background when the fluorescently labeled DNA was omitted.
Crosslinking. Crosslinked mononucleosomes were prepared as described in ref. 9. Mononucleosomes were prepared by the salt-jump method as described above. Dimethyl suberimidate (Sigma), freshly dissolved in 0.1 M sodium borate buffer (pH 10.5) at a concentration of 40–50 mg/ml, was added to the dialyzed mononucleosomes to a final concentration of 1 mg/ml. The sample was mixed and incubated at room temperature for 15–20 min; dimethyl suberimidate was repeatedly added in the same way five more times. Histone crosslinking efficiency was checked on 15% SDS/PAGE and/or on 5.5% native PAGE in 0.5× TBE; noncrosslinked material was electrophoresed in parallel.
Wide-Field Prism-Based Evanescent Field Fluorescence Microscope, Data Collection, and Analysis. Samples contained imaging buffer and the oxygen scavenger system [0.4% glucose, 1% mercaptoethanol, glucose oxidase, and catalase (Roche)] (10). The flow cell was placed on the stage of an inverted fluorescent microscope (IX-71, Olympus) with ×60, 1.2-numerical aperture water immersion objective (Olympus). A small pellin broca prism (PLBC-5.0–79.5-ss; CVI Laser, Albuquerque, NM) (11) was placed on top of the quartz slide, using fluorescent-free oil (Cargille type FF, Fisher Scientific) for optical connection. A circularly polarized laser beam was focused into the prism at an angle >65.7° (12), so that an exponentially decaying evanescent field formed inside the flow cell, illuminating only the volume of the buffer closest to the quartz slide. The fluorescence signal traveled through a Dual-view (Optical Insights, Tucson, AZ) filter set system, separating Cy3 and Cy5 signals and forming a two-part image onto an intensified charge-coupled device camera (refs 13–15 and references therein). The filters (Chroma Technology, Rockingham, VT) used with the Dual-view are a dichroic 610DCLP, Cy3 band-pass D580/40 filter for the Cy3 and a long-pass E645 filter for the Cy5. iplab (Scanalytics, Billerica, MA) software was used to operate the camera and collect images and time sequences of images (movies) at the typical speed of 10 frames per second. Photobleaching of Cy5 and Cy3 occurred in seconds; this fast photobleaching likely limited the observation of two-state transitions in every single member of the nucleosomal populations (see Results and Discussion).
Software written in iplab allowed us to find fluorescent signals in the sequences of images and obtain intensity–time trajectories. For each dye pair, the Cy5 signal was identified first, and the corresponding Cy3 signal was matched to it. Apparent efficiency of FRET (Eapp) was calculated from the fifth frame, after subtracting the background. After correcting for donor leakage into the acceptor channel and for direct excitation of the acceptor, Eapp values were calculated by using the formula Eapp = Cy5intensity/(Cy3intensity + Cy5intensity). Dwell-time analysis was performed on intensity–time traces. As a criterion for the on/off transition, a 50% difference between low and high states of the Cy5 traces was used. The durations of high and low states between these transitions were measured after smoothing the curve by a two-point moving average that eliminated most noise; however, this averaging also eliminated some of the one-frame transitions.
Despite the huge distance between the two fluorophores in naked DNA, a nonzero Eapp peak was recorded (see Fig. 3A), in accordance with earlier observations (e.g., refs. 14 and 16). The existence of this peak has not yet been satisfactorily explained: distance fluctuations in the DNA double helix due to transient bending, twisting, or local melting are expected to be much faster than the time over which the signal is integrated and thus should cancel each other. The noticeable broadness of Eapp of naked DNA is due to the error in measuring low-intensity fluorescence signals: Cy5 fluorescence is low because of lack of energy transfer, and Cy3 fluorescence is low because it is far away from the surface, where the exponentially decaying evanescent field is weak.
Fig. 3.
Fluorescent behavior of naked DNA and nucleosomes formed on the 164-bp GUB nucleosome positioning sequence labeled with Cy3 and Cy5. Each image is one video frame (inverted contrast) from the intensified charge-coupled device camera and is split into two parts, showing Cy3 fluorescence on the left and Cy5 fluorescence on the right side. Each black dot represents the fluorescence emitted from a single dye. Each Cy5 dye has a one-to-one correspondence with a Cy3 dye (i.e., is located on the same DNA molecule). Increased intensity of Cy5 fluorescence indicates energy transfer from the Cy3 to the Cy5. The Gaussian peak maxima ± SD are listed in each histogram. (A) Naked DNA in 50 mM NaCl and 10 mM Tris·HCl (pH 7.5), in the presence of the oxygen scavenger system (see Materials and Methods). Individual Cy3 signals (black spots) are shown in Left; very few Cy5 signals are in Right. (B) Nucleosomes were reconstituted in solution from DNA and core histones by the salt-jump method. Assembled nucleosomes were attached to the surface, rinsed, and imaged in 5, 50, or 250 mM NaCl. The Gaussian peak parameters and numbers of data points for the different salt concentrations are: 0.87 ± 0.20 (n = 657) for 5 mM NaCl, 0.88 ± 0.22 (n = 636) for 50 mM NaCl, and 0.88 ± 0.21 (n = 701) for 250 mM NaCl. The data for the three salt concentrations are practically indistinguishable and are presented as a single combined peak. (C) Naked DNA mixed with only core histones in 150 mM NaCl, 10 mM Tris·HCl, and 0.5 mM EDTA (pH 7.5); images indistinguishable from those in A indicate no nucleosome formation. (D) Total dissociation of histone octamers in B upon washing with 2 M NaCl to return to naked DNA conformation. The numbers of data points used for the distribution histograms are as follows: A, 461; B, 1,994; C, 326; and D, 472.
Results and Discussion
Nucleosome Formation on DNA Labeled with a Pair of Donor and Acceptor Fluorophores. A 164 bp-long GUB DNA nucleosome positioning sequence was labeled with Cy3 and Cy5 dyes by using amino-linker derivatives of oligonucleotides and N- hydroxysuccimide chemistry. Dye positions were chosen by aligning the crystal structure of the core particle (4) with the GUB sequence (6–8) at the dyad axes, so that the dyes would lie close to the dyad, ≈3 nm apart, when the DNA is wrapped around the histone octamer to form a nucleosome (Fig. 2). Mononucleosomes were reconstituted in bulk solution by mixing the histone octamers and the DNA at high salt (2 M NaCl) and then gradually reducing the salt concentration by stepwise dilution to 0.5 M (or lower) NaCl (5, 17). The particles were attached onto the surface of the flow cell through their biotinylated ends and imaged at three different salt concentrations: 5, 50, or 250 mM NaCl (Fig. 3B).
Fig. 2.
Locations of the donor and acceptor fluorophores on nucleosomal 164-bp DNA. (A) Naked DNA. Marked DNA bases are sites for attachment of acceptor and donor dyes, bases 47 and 122, respectively, on separate strands of DNA. The nucleosomal pseudodyad is at position 84. The distance between the methyl carbons of the two labeled pyrimidines located on opposite strands of the linear DNA is 75 bp (25 nm), but only ≈3 nm (from DNA gyre to DNA gyre) in the particle. This placement of the dyes should result in no single-pair FRET on the linear DNA fragment and high-efficiency energy transfer in the nucleosome. (B) Face and side views of the nucleosome. Modeling of Protein Data Bank structure 1KX5 (4) was performed by using xfit (41), and the model was rendered with pymol (42). We thank J. Harp for help with this figure.
Fluorescence Patterns of Naked DNA and Nucleosomes Differ. The labeled DNA fragments or reconstituted nucleosomes were attached to streptavidin-coated quartz surface by 5′ biotin linker, to allow observation of individual molecules over time. Samples were illuminated with a 532-nm laser in a prism-based evanescent field fluorescence microscope, which allowed recording fluorescence from the Cy3 and Cy5 dyes in separate windows. With naked DNA, single fluorescent spots were observed on the Cy3 side of the image, whereas practically no signal was detected on the Cy5 side. The lack of measurable energy transfer from Cy3 to Cy5 reflected the large distance between the two dyes, as expected (Fig. 3A; see also Materials and Methods and ref. 18).
Two changes in the fluorescence patterns were observed in the nucleosome population compared with the naked DNA. First, the intensity of Cy3 spots increased. We attribute this increase to the movement of the dye closer to the surface (where the exponentially decaying evanescent field is much stronger) as a result of nucleosome formation. Second, we observed a considerable increase in the number and intensity of fluorescent spots in the Cy5 side of the image, which indicated an increase in the efficiency of FRET from the donor to the acceptor dye. This observation is the expected behavior of the system if the distance between the two dyes decreases as a result of the wrapping of the DNA around the histone octamer, i.e., of the formation of nucleosomes. In all three salt concentrations, high-efficiency FRET was observed, with a single Gaussian peak in the distributions centering on ≈0.87 Eapp (Fig. 3B displays a histogram of data combined for the three salt concentrations). The means of the Gaussian distributions ± SD for this and all other experimental scenarios are given in Table 1. The table also presents the distances between the two dyes calculated according to the Förster equation (19) [EFRET = 1/(1 + (R/R0)6], using 6 nm as R0 for the Cy3/Cy5 pair (18).
Table 1. Eapp and Rapp values and dwell times in the open and closed nucleosomal states.
| Sample | Eapp | Rapp,* nm | τon, s | τoff, s | τon/(τon+τoff),† % |
|---|---|---|---|---|---|
| Salt-jump reconstitution with final salt concentrations of | |||||
| 5 mM NaCl | 0.87 ± 0.20 | 4.4 | 2.10 ± 0.09 | 0.187 ± 0.002 | >92 |
| 50 mM NaCl | 0.88 ± 0.22 | 4.3 | 4.56 ± 0.30 | 0.147 ± 0.002 | >97 |
| 250 mM NaCl | 0.88 ± 0.21 | 4.3 | 4.92 ± 0.30 | 0.137 ± 0.001 | >97 |
| After 2 M NaCl wash | 0.11 ± 0.08 | >8‡ | |||
| Naked DNA | 0.13 ± 0.16 | >8‡ | |||
| Naked DNA plus core histones at 150 mM NaCl | 0.08 ± 0.09 | >8‡ | |||
EFRET = 1/[1 + (R/R0)6], where R0 = 6 nm for Cy3/Cy5 dye pair.
Rapp is calculated based on Eapp, which closely parallels EFRET (18)
These numbers are underestimates because they are based on analysis of only those time trajectories that exhibit the high Eapp to low Eapp (and back) transitions. In many cases, the nucleosomes were in a closed high Eapp state the entire time before one or both of the fluorophores photobleached, and taking these in account was not possible
The useful range of single-pair FRET is ≈2–8 nm (43); >8 nm indicates a lower bound in these measurements
As a control, we performed an experiment where free histones were added to the flow cell at 150 mM NaCl. Whereas the histones will still attach to the DNA nonspecifically, no nucleosomes are expected to form under these conditions (e.g., ref. 20). Indeed, as Fig. 3C demonstrates, the behavior of the system was similar to that of naked DNA, with the center of the Eapp distribution on ≈0.08. To make sure that nucleosomes could form on the immobilized short piece of DNA, we assembled nucleosomes directly in the flow cell by simultaneously adding core histones and nucleosome assembly protein 1 (NAP-1) (21). A high Eapp peak was observed, similar to the one in Fig. 3B, indicating in situ nucleosome assembly (M.T., H.Z., S.H.L., and J.Z., unpublished data).
To prove that the changes in Eapp were caused by nucleosome formation, we made use of the known biochemical properties of the nucleosome particle (22, 23). Treatment of the preformed nucleosomes with 2 M NaCl is expected to completely dissociate the histones from the DNA, resulting in loss of FRET, which is exactly what we observed (Fig. 3D).
From the data presented thus far, we concluded that salt-jump-reconstituted nucleosomes, with the DNA wrapped around the histone octamer, exhibit FRET characteristics predicted from the crystal structure of the core particle. It should be noted that the nucleosomal Eapp peak is unusually broad, with SD of ± 0.20 Eapp units. This observation was rather surprising in view of reported experimental data (obtained with freely diffusing molecules) (16), which demonstrated that the higher the mean of Eapp, the narrower the peak: peaks of ≈0.9 Eapp units exhibit SD of plus or minus ≈0.05 (see figure 2 in ref. 16), whereas the lower Eapp peaks are significantly broader. The fact that our nucleosomal peak is so broad may reflect the presence of a subpopulation of molecules undergoing the conformational transitions reported below during the time of imaging (50-ms exposure time).
We do not favor the alternative formal explanation that the broadness of the peak results from intrinsic heterogeneity of our nucleosome preparation. Indeed, biochemical analyses of such preparations revealed a proper molar ratio of the four core histones and only one translational position (6, 8). Moreover, noncanonical nucleosomes missing one or both H2A/H2B dimers would have the same high-FRET value as intact particles (Fig. 6, which is published as supporting information on the PNAS web site, and data not shown); thus, they cannot contribute to the broadness of the distribution. Conversely, nucleosomes positioned at or extending beyond the ends of the DNA are expected to show a low efficiency of FRET (less than ≈0.35 Eapp units) (Fig. 6); we do not detect a distinct population of such particles in the Eapp histograms (Fig. 3B).
Reversible Conformational Fluctuations in the Nucleosome. We next analyzed the time trajectories of pairs of corresponding Cy3 and Cy5 dyes, i.e., dyes that are situated on the same molecule of DNA within a nucleosome. We found molecules where FRET occurred (Cy5 signal was high) and did not change until photobleaching of one of the dyes. Importantly, we also found a significantly large number of molecules (493 of 1,994 molecules recorded, i.e., ≈25%) that repeatedly switched back and forth between a high-FRET and a low-FRET state. Three examples of such molecules are presented in Fig. 4. We attribute the high Cy5 signal (with concomitant low Cy3 signal) to a “closed” nucleosomal structure, with the DNA wrapped around the octamer core, similar to what is observed in the crystal structure (2–4). The reverse situation with low Cy5 signal (and high Cy3 signal) we interpret as evidence of nucleosome “opening,” i.e., fast conformational transition to states in which the DNA has significantly unraveled from the histone octamer, but the octamer still stays attached. If the octamer were to completely dissociate from the DNA, the process would not have been reversible, because once it had gone into the solution, the octamer would not have been able to find its way back for any of the following reasons: (i) The concentration of the released histones would have been far too small to allow biomolecular interactions to occur to a significant extent; (ii) the dissociated histone octamer would have fallen apart into a H3/H4 tetramer and two H2A/H2B dimers (24); (iii) nonspecific histone binding to DNA would have been favored over nucleosome formation at these salt concentrations (in the absence of histone chaperones, such as NAP-1) (21, 25); or (iv) there are no nearby free DNA or RNA molecules that could have served as a temporary sink for binding of the released proteins (for example, during transcription it has been observed that histones from nucleosomes whose DNA is being transcribed are hosted on the nascent RNA) (25, 26).
Fig. 4.
Fluctuation between closed and open states of a nucleosome. Example time trajectories of the fluorescence intensity of the donor and acceptor dyes from single nucleosomes in Fig. 3B are shown. The time trajectories for the two fluorophores (Cy3, green, and Cy5, red) bound to one single DNA molecule are overlaid. (A–C) Example traces with evidence for nucleosome dynamics. (D) Dwell-time histograms of nucleosomes in the closed state. Each histogram was fitted with a single exponential decay function (solid blue curve) to derive τon in final salt concentration of 5 mM (Top), 50 mM (Middle), and 250 mM (Bottom) NaCl. Numbers of data points are 815, 782, and 613, respectively.
We also must consider the possibility that the observed conformational fluctuations are due to gross rearrangements in the histone octamer itself. For this purpose, we repeated the experiments with salt-jump-assembled nucleosomes in which the histone cores had been crosslinked with dimethyl suberimidate. This treatment does not crosslink the DNA to the histone; Stein et al. (27) have shown they could completely remove the DNA from dimethyl suberimidate-crosslinked nucleosomes with a high salt treatment. The Eapp histograms were indistinguishable from those of the noncrosslinked material (Fig. 3B), and the time trajectories displayed the high-FRET to low-FRET (and back) transitions (Fig. 7, which is published as supporting information on the PNAS web site). Thus, the conformational transitions in the nucleosomal particle that give rise to the FRET transitions cannot result from conformational changes in the histone octamer.
Dynamic Equilibrium Between Open and Closed States of a Nucleosome. Thus, we believe that we are seeing, to our knowledge, the first real-time demonstration of a reversible, fast, long-range, two-state conformational fluctuation between an open and a closed nucleosome state. The open state could be populated by two different conformers, arising by opening the DNA from one (more probable) or both (less probable) ends (Fig. 5 and see below). There are no transition intermediates between the two states detectable at our time-resolution (see example time trajectories in Fig. 4).
Fig. 5.
Model of the closed and the two open states of the nucleosome (all-or-none conformational fluctuations). The DNA is drawn as a thick line, while the histone octamer is taken from the core particle crystal structure Protein Data Bank ID 1KX5 (4). The nucleosome dwells in the closed state most of the time, with only occasional brief excursions into the open state.
We quantitatively analyzed the “dwell times” of the nucleosome in the two states from individual dye intensity-time traces. Nucleosomal “on” and “off” times (corresponding to the amount of time the nucleosome spends in the closed and open states, respectively) from several hundred molecules were plotted and fitted with a first-order exponential decay function, from which τ, the dwell time for the particular state of nucleosome, can be derived (Fig. 4D and Table 1). Although τoff values were on the order of 100–200 ms (one or two frames of the image sequence), τon values varied between 2.1 and 4.9 s depending on the experimental conditions used (Table 1). Preformed nucleosomes imaged in 5, 50, or 250 mM NaCl concentrations showed increasing τon with increasing salt concentration. The salt effects on the dwell times of both the open (τoff) and the closed (τon) state probably reflect stiffening of the DNA at the lower salt concentrations (see below).
Even more informative is the fraction of time the nucleosome spends in the closed state, which can be calculated as τon/(τon + τoff) (Table 1). The results indicate that the nucleosome is in the closed state ≈95% of the time. It must be noted that the actual time the particle spends in the closed state is even higher (see limitations of the procedure discussed in Table 1). In other words, under all conditions tested, the nucleosome prefers to be in the closed state and makes only brief excursions into the open state, to quickly revert to the closed state again. Nevertheless, even such brief sojourns in the open state (≈100–200 ms) would allow plentiful time for interaction with proteins.
The experiments described here provide, to our knowledge, the first direct evidence for spontaneous, long-range, reversible fluctuations in nucleosome structure. These conformational changes are major: DNA sites situated close to each other in the canonical core particle structure (see Fig. 2) briefly move to a separation of 8 nm or greater. The only plausible explanation for such an observation is a profound unwrapping of the DNA (Fig. 5).
A simple geometrical calculation shows that if the unwrapping is symmetrical, no more than 35 bp can remain attached to the histone core, in the region of the dyad. An examination of the core particle structure in terms of the strength of the protein/DNA contacts along the nucleosomal DNA suggests that the actual length of the DNA remaining bound to the DNA in the symmetrically open state could be as small as only ≈10 bp (28, 29). Brower-Toland et al. (30) observed a force-induced stepwise unraveling of individual nucleosomes in a nucleosomal array and proposed a final predissociation state in which only a very short DNA segment of ≈10 bp near the dyad remains attached to the histone. It is possible that we are seeing a similar conformational state that arises spontaneously, at zero applied force. An additional support for our interpretation comes from theory. Marky and Manning (31, 32) have provided an in-depth theoretical analysis of the possible trajectories of nucleosomal DNA as a function of the length of the fragment that comes into contact with the histone octamer or moves away from it. For lengths between 10 and 60 bp, there are mechanically stable trajectories intermediate between fully wound and fully unwound states. In striking contrast, segments of 70 and 80 bp exhibit a two-state behavior: the DNA is either fully wound or fully unwound, with no stable intermediates. We believe that the long-range conformational fluctuations we are observing experimentally correspond to the all-or-none unpeeling predicted by the modeling for segments of 70 and 80 bp; the unpeeling can be from one side only or, with less probability, from both sides simultaneously. Thus, there are both experimental and theoretical bases for the model we propose (Fig. 5).
Unwrapping of DNA from the histone core has been suggested in a number of contexts (1, 33). Importantly, Polach and Widom (34) have studied restriction-site accessibility along nucleosomal DNA in reconstituted mononucleosomes and found that the level of accessibility decreased in going from the DNA ends to the DNA dyad. It should be emphasized that the results of our experiments do not conflict with the observations of Polach and Widom (34). Exposure of sites near the DNA termini, by partial unwrapping (which we prefer to call “breathing” to distinguish it from the long-range “opening” seen here), would not be detected with the specific construct we are using. However, Polach and Widom (34) also observed a low, but detectable, accessibility of sites close to the dyad, which is to be expected from the small percent of time we observe the nucleosomes to be in the open state. In two recent studies (35, 36), a dynamic equilibrium conformational transition has been deduced from population FRET analysis of reconstituted core particles labeled with one fluorophore at one end of the DNA and the other attached to the unstructured tail of either H3 or H2A. However, data from such constructs cannot be easily interpreted in terms of the extent of the DNA unwrapping from around the histone core, because even a small degree of unwrapping (breathing) would lead to a dramatic decrease in FRET. In contrast, the construct we have used requires major unwrapping to yield significant FRET diminution.
Finally, we note that as salt concentration is decreased, the percent of time the particle resides in the open state increases from 3% to 8%, as does the mean dwell time in the open state. This finding may at first glance seem paradoxical because of the known electrostatic interactions in DNA-histone binding. However, as Manning (31, 37) points out, the additional stiffness of DNA at low salt should increase the probability of a state with extended DNA. Indeed, there has long been evidence for a thermodynamically stable extended form of the nucleosome existing below ≈1 mM salt (38, 39). Furthermore, UV photofootprinting studies indicate that the DNA in that structure is “no longer bent to a significant degree,” although it is still attached to histones (40). We suggest that the fluctuations we observe may represent temporary excursions into a similar open state that also can occur at higher salt concentrations. What is remarkable and important is that we find these opening excursions persisting to salt concentrations in the physiological range. This finding has the implication that most of the DNA on the nucleosome can be sporadically accessible to regulatory proteins and proteins that track the DNA double helix. The possibility that histone modifications or variant replacement may change either the frequency or durations of the open periods is an exciting prospect that should be explored.
Conclusions
Our data were obtained by a method that gives truly dynamic measurements in individual particles in real time. Based on the observed reversible transitions between a low- and a high-state FRET, and the FRET values of these states, we estimated the minimum distance changes that occur during these transitions. Based on the specific placement of the two dyes (deep within the nucleosomal particle, at an area never previously examined by any dynamic approaches), we can assert that we are seeing long-range conformational fluctuations, with at least half of the nucleosomal DNA being unwrapped. Such long-range fluctuations to our knowledge have never been reported previously; they may have important implications concerning the accessibility of nucleosomal DNA to factor binding.
Supplementary Material
Acknowledgments
We thank Dr. J. Harp for Fig. 2 and Drs. T. Ha, I. Rasnik, and S. McKinney for assistance with initial measurements. This work was supported by the National Cancer Institute Career Transition Award K22, the University of Pittsburgh Cancer Institute, University of Pittsburgh School of Medicine startup funds (to S.H.L.), the Pittsburgh Foundation (to S.H.L. and J.Z.), and National Science Foundation Grant MCB-0343583 (to J.Z.). M.T is a recipient of a postdoctoral fellowship from the Eppley Foundation.
References
- 1.van Holde, K. & Zlatanova, J. (1999) BioEssays 21, 776-780. [DOI] [PubMed] [Google Scholar]
- 2.Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251-260. [DOI] [PubMed] [Google Scholar]
- 3.Harp, J. M., Hanson, B. L., Timm, D. E. & Bunick, G. J. (2000) Acta Crystallogr. D 56, 1513-1534. [DOI] [PubMed] [Google Scholar]
- 4.Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. (2002) J. Mol. Biol. 319, 1097-1113. [DOI] [PubMed] [Google Scholar]
- 5.Zivanovic, Y., Duband-Goulet, I., Schultz, P., Stofer, E., Oudet, P. & Prunell, A. (1990) J. Mol. Biol. 214, 479-495. [DOI] [PubMed] [Google Scholar]
- 6.Tomschik, M., Karymov, M. A., Zlatanova, J. & Leuba, S. H. (2001) Struct. Fold. Des. 9, 1201-1211. [DOI] [PubMed] [Google Scholar]
- 7.Adams, C. C. & Workman, J. L. (1995) Mol. Cell. Biol. 15, 1405-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An, W., Leuba, S. H., van Holde, K. & Zlatanova, J. (1998) Proc. Natl. Acad. Sci. USA 95, 3396-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas, J. O. (1989) Methods Enzymol. 170, 549-571. [DOI] [PubMed] [Google Scholar]
- 10.Harada, Y., Sakurada, K., Aoki, T., Thomas, D. D. & Yanagida, T. (1990) J. Mol. Biol. 216, 49-68. [DOI] [PubMed] [Google Scholar]
- 11.Ha, T. (2001) Methods 25, 78-86. [DOI] [PubMed] [Google Scholar]
- 12.Axelrod, D. (1989) Methods Cell Biol. 30, 245-270. [DOI] [PubMed] [Google Scholar]
- 13.Rasnik, I., Myong, S., Cheng, W., Lohman, T. M. & Ha, T. (2004) J. Mol. Biol. 336, 395-408. [DOI] [PubMed] [Google Scholar]
- 14.Hohng, S., Wilson, T. J., Tan, E., Clegg, R. M., Lilley, D. M. & Ha, T. (2004) J. Mol. Biol. 336, 69-79. [DOI] [PubMed] [Google Scholar]
- 15.Zheng, H. C., Tomschik, M., Zlatanova, J. & Leuba, S. H. (2005) in Protein–Protein Interactions: A Molecular Cloning Manual, eds. Golemis, E. & Adams, P. (Cold Spring Harbor Lab. Press, Woodbury, NY), 2nd Ed., Vol. 20, pp. 1-19. [Google Scholar]
- 16.Deniz, A. A., Dahan, M., Grunwell, J. R., Ha, T., Faulhaber, A. E., Chemla, D. S., Weiss, S. & Schultz, P. G. (1999) Proc. Natl. Acad. Sci. USA 96, 3670-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatchell, K. & van Holde, K. E. (1979) Biochemistry 18, 2871-2880. [DOI] [PubMed] [Google Scholar]
- 18.Tan, E., Wilson, T. J., Nahas, M. K., Clegg, R. M., Lilley, D. M. & Ha, T. (2003) Proc. Natl. Acad. Sci. USA 100, 9308-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Förster, T. (1959) Discuss. Faraday Soc. 27, 7-17. [Google Scholar]
- 20.Leuba, S. H., Karymov, M. A., Tomschik, M., Ramjit, R., Smith, P. & Zlatanova, J. (2003) Proc. Natl. Acad. Sci. USA 100, 495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimi, Y., Hirosumi, J., Sato, W., Sugasawa, K., Yokota, S., Hanaoka, F. & Yamada, M. (1984) Eur. J. Biochem. 142, 431-439. [DOI] [PubMed] [Google Scholar]
- 22.Burton, D. R., Butler, M. J., Hyde, J. E., Phillips, D., Skidmore, C. J. & Walker, I. O. (1978) Nucleic Acids Res. 5, 3643-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Holde, K. E. (1988) Chromatin (Springer, New York).
- 24.Godfrey, J. E., Eickbush, T. H. & Moudrianakis, E. N. (1980) Biochemistry 19, 1339-1346. [DOI] [PubMed] [Google Scholar]
- 25.Levchenko, V. & Jackson, V. (2004) Biochemistry 43, 2359-2372. [DOI] [PubMed] [Google Scholar]
- 26.Peng, H. F. & Jackson, V. (1997) Biochemistry 36, 12371-12382. [DOI] [PubMed] [Google Scholar]
- 27.Stein, A., Bina-Stein, M. & Simpson, R. T. (1977) Proc. Natl. Acad. Sci. USA 74, 2780-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luger, K. & Richmond, T. J. (1998) Curr. Opin. Struct. Biol. 8, 33-40. [DOI] [PubMed] [Google Scholar]
- 29.Harp, J. M., Hanson, B. L. & Bunick, G. J. (2004) in Chromatin Structure and Dynamics: State-of-the-Art, eds. Zlatanova, J. & Leuba, S. H. (Elsevier, Amsterdam), Vol. 39, pp. 13-44. [Google Scholar]
- 30.Brower-Toland, B. D., Smith, C. L., Yeh, R. C., Lis, J. T., Peterson, C. L. & Wang, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 1960-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marky, N. L. & Manning, G. S. (1991) Biopolymers 31, 1543-1557. [DOI] [PubMed] [Google Scholar]
- 32.Marky, N. L. & Manning, G. S. (1995) J. Mol. Biol. 254, 50-61. [DOI] [PubMed] [Google Scholar]
- 33.Zlatanova, J., Leuba, S. H. & van Holde, K. (1999) Crit. Rev. Eukaryot. Gene Expr. 9, 245-255. [DOI] [PubMed] [Google Scholar]
- 34.Polach, K. J. & Widom, J. (1995) J. Mol. Biol. 254, 130-149. [DOI] [PubMed] [Google Scholar]
- 35.Li, G. & Widom, J. (2004) Nat. Struct. Mol. Biol. 11, 763-769. [DOI] [PubMed] [Google Scholar]
- 36.Li, G., Levitus, M., Bustamante, C. & Widom, J. (2005) Nat. Struct. Mol. Biol. 12, 46-53. [DOI] [PubMed] [Google Scholar]
- 37.Manning, G. S. (2003) J. Am. Chem. Soc. 125, 15087-15092. [DOI] [PubMed] [Google Scholar]
- 38.Uberbacher, E. C., Ramakrishnan, V., Olins, D. E. & Bunick, G. J. (1983) Biochemistry 22, 4916-4923. [DOI] [PubMed] [Google Scholar]
- 39.Brown, D. W., Libertini, L. J. & Small, E. W. (1991) Biochemistry 30, 5293-5303. [DOI] [PubMed] [Google Scholar]
- 40.Brown, D. W., Libertini, L. J., Suquet, C., Small, E. W. & Smerdon, M. J. (1993) Biochemistry 32, 10527-10531. [DOI] [PubMed] [Google Scholar]
- 41.McRee, D. E. (1993) Practical Protein Crystallography (Academic, San Diego).
- 42.DeLano, W. L. (2002) The pymol Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 43.Deniz, A. A., Laurence, T. A., Dahan, M., Chemla, D. S., Schultz, P. G. & Weiss, S. (2001) Annu. Rev. Phys. Chem. 52, 233-253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.