Summary
Antibodies to Zika virus (ZIKV) can be protective. To examine the antibody response in individuals that develop high titers of anti-ZIKV antibodies we screened cohorts in Brazil and Mexico for ZIKV envelope domain III (ZEDIII) binding and neutralization. We find that serologic reactivity to dengue 1 virus (DENV1) EDIII before ZIKV exposure is associated with increased ZIKV neutralizing titers after exposure. Antibody cloning shows that donors with high ZIKV neutralizing antibody titers have expanded clones of memory B cells that express the same immunoglobulin VH3-23/VK1-5 genes. These recurring antibodies cross-react with DENV1, but not other flaviviruses, neutralize both DENV1 and ZIKV, and protect mice against ZIKV challenge. Structural analyses reveal the mechanism of recognition of the ZEDIII lateral ridge by VH3-23/VK1-5 antibodies. Serologic testing shows that antibodies to this region correlate with serum neutralizing activity to ZIKV. Thus, high neutralizing responses to ZIKV are associated with preexisting reactivity to DENV1 in humans.
Keywords: Zika virus, dengue virus, flavivirus, antibodies, vaccine, structure
eTOC
Individuals with high neutralizing antibody response against Zika virus have expanded clones of B cells that express the same heavy and light immunoglobulin genes, and that are cross-reactive against Dengue 1 virus.

Introduction
Zika virus (ZIKV) infection typically produces mild symptoms consisting of fever, rash and arthralgia that resolve rapidly, and the infection is also occasionally associated with Guillain-Barré Syndrome (Lessler et al., 2016; Miner and Diamond, 2017; Weaver et al., 2016). However, when infection occurs during pregnancy, vertical transmission can lead to a spectrum of devastating neurodevelopmental aberrations, collectively referred to as Congenital Zika Syndrome. Although the data are still incomplete, infants born to mothers infected with ZIKV during pregnancy carry an up to 42% risk of developing overt clinical or neuroimaging abnormalities (Brasil et al., 2016; Costa et al., 2016; Franca et al., 2016).
ZIKV belongs to the Flavivirus genus, which includes yellow fever (YFV), West Nile (WNV), and the 4 serotypes of dengue virus (DENV1-4). These positive-stranded RNA viruses are responsible for considerable morbidity and mortality in the equatorial and subequatorial regions populated by their mosquito vectors (Kramer et al., 2007; Murray et al., 2013; Weaver and Reisen, 2010). Unlike most other flaviviruses, ZIKV can also be transmitted sexually, and on occasion persists for months (Barzon et al., 2016; Foy et al., 2011; Murray et al., 2017; Suy et al., 2016).
All flaviviruses display a single envelope protein, E, that is highly conserved between different members of this virus family. The E protein ectodomain consists of three structural domains. Domain I (EDI) contains the N-terminus, domain II (EDII) is an extended finger-like structure that includes the dimerization domain and also a pH-sensitive fusion loop that mediates viral fusion in the lysosomes. Finally, domain III (EDIII) is an immunoglobulin-like domain that mediates attachment to target cells (Barba-Spaeth et al., 2016; Dai et al., 2016; Kostyuchenko et al., 2016; Modis et al., 2003; Mukhopadhyay et al., 2005; Rey et al., 1995; Sirohi et al., 2016; Zhang et al., 2004). Several human neutralizing antibodies targeting different E protein epitopes have been described. Antibodies against the EDIII of flaviviruses are among the most potent neutralizers in this group (Beasley and Barrett, 2002; Crill and Roehrig, 2001; Screaton et al., 2015).
Due to the conserved structural features of the E protein, antibodies that develop in response to infection by one flavivirus may also recognize others (Heinz and Stiasny, 2017). Cross-reactivity can lead to cross-protection as first documented by Sabin who showed experimentally in humans that exposure to DENV1 could provide short-term protection from subsequent challenge with DENV2. In contrast, immunity to the autologous strain was long lasting (Sabin, 1950). More recently, human monoclonal antibodies to DENV have been shown to cross-neutralize ZIKV, and vice versa (Barba-Spaeth et al., 2016; Stettler et al., 2016; Swanstrom et al., 2016). However, there is concern that cross-reacting antibodies that fail to neutralize the virus may enhance rather than curb subsequent flavivirus infections (Harrison, 2016; Wahala and Silva, 2011). In vitro and in vivo experiments in mice suggest that this phenomenon, commonly referred to as Antibody Dependent Enhancement (ADE), extends to ZIKV (Bardina et al., 2017; Dejnirattisai et al., 2016; Harrison, 2016; Priyamvada et al., 2016). For this reason, a desirable goal for ZIKV vaccines is to elicit robust and protective antibodies, while avoiding antibodies that bind to the virus but are non-neutralizing and potentially enhancing. Doing so requires a detailed understanding of the neutralizing antibody response to ZIKV.
Several human antibodies to ZIKV have been cloned from convalescent individuals by methods utilizing B cell transformation with Epstein-Barr virus (Sapparapu et al., 2016; Stettler et al., 2016). However, individual donors were not selected for high neutralization titers; whether their antibodies are representative of optimal immune responses, and how these antibodies might relate to previous flavivirus exposure remains unknown.
Here we report on the characteristics of the neutralizing antibody responses that developed in individuals with high levels of serum ZIKV neutralizing activity from two independent cohorts following recent ZIKV outbreaks in Brazil and Mexico, and on their relationship to previous flavivirus exposure.
Results
Serologic Responses to ZIKV in Brazil and Mexico
Individuals infected with pathogens display a spectrum of antibody responses ranging from low levels of non-neutralizing antibodies to high titers of neutralizing antibodies. To determine whether a population infected with ZIKV also displays a range of antibody responses we screened 405 individuals living in ZIKV epidemic areas for serum IgG capable of binding to ZIKV E Domain III (ZEDIII, Fig. 1A).
Figure 1. Identification of individuals with high ZEDIII binding and neutralization capacity.

A- Sera from the Brazilian and Mexican cohorts were screened by ELISA for IgG antibodies binding to ZEDIII. Each dot represents an individual donor. Optical densities are normalized to control serum from a flavivirus naïve individual vaccinated for YFV. In blue are sera selected for neutralization analysis. B- The neutralization capacity of selected sera from Mexico (red), Brazil (light red) and control samples obtained in Brazil in 2010 (grey) was determined by a ZIKV luciferase reporter viral particle (RVP) neutralization assay. The reciprocal of the serum dilution that resulted in 50% inhibition compared to RVP alone is reported as the 50% neutralization titer (NT50). The dotted line indicates the lower limit of dilutions that were examined. The five samples below the dotted line have NT50 values lower than 103. Individuals from whom antibodies were sequenced and cloned are indicated.
Nearly three hundred sera were obtained in November 2015, shortly after ZIKV was introduced in Salvador, Brazil, from participants who were enrolled in a prospective study in 2013 from Pau da Lima, an urban slum community within the city (Cardoso et al., 2015; Felzemburgh et al., 2014; Hagan et al., 2016). An additional 108 sera were from Santa Maria Mixtequilla, a rural town in Oaxaca, Mexico. ZIKV infections were documented by PCR in Santa Maria Mixtequilla at the time of sample collection in April of 2016. Dengue virus (DENV) is endemic at both sites. Sera obtained from the Pau da Lima cohort in 2010 following a DENV outbreak, but before the introduction of ZIKV into Brazil, served as non-ZIKV flavivirus-exposed control for background reactivity against ZIKV (Silva et al., 2016). ZIKV introduction was associated with a broad distribution of serologic reactivity against ZEDIII in both Brazilian and Mexican samples by ELISA (Fig. 1A).
To determine whether serologic reactivity to ZEDIII is associated with ZIKV neutralizing activity, we assessed the top 31 sera (blue symbols in Fig. 1A) for neutralization of luciferase-expressing reporter viral particles (RVP) bearing ZIKV structural proteins (see Methods; Fig. 1B). Neutralizing titers, expressed as the reciprocal of the dilution resulting in a 50% reduction of the luciferase signal achieved in the absence of serum (NT50), varied by over 2 logs indicating a broad range of humoral immune responses to ZIKV (Fig. 1B).
Human Monoclonal Antibodies to ZIKV
To further characterize the antibody response in 6 individuals with high neutralizing titers, 3 from each cohort, we used fluorescently-labeled ZEDIII to identify and purify single memory B cells in the peripheral blood (Fig. 2A). ZEDIII-specific memory B cells were found at frequencies ranging from 0.13 – 1.98% of all circulating IgG+ memory B cells (Fig. 2A, see Methods). Although the sample size is limited, the frequency of ZEDIII-specific memory B cells did not appear to correlate with either ZEDIII binding or neutralizing activity (Fig. 1). We conclude that there is significant variability in the frequency of ZEDIII-specific memory B cells in individuals that show serum ZIKV neutralizing activity.
Figure 2. Discovery of ZEDIII-specific antibodies.

A- Frequency of ZEDIII-specific, IgG+ memory B cells in peripheral blood of 6 donors. Flow cytometry plots display the percentage of all IgG+ memory B cells that bind to a fluorescently tagged ZEDIII bait. Flavivirus naïve peripheral blood samples are shown alongside as negative controls. B- Pie charts show the distribution of antibody clones that share the same IGHV and IGLV; the width of each colored or shaded slice is proportional to the number of clones sharing a distinct combination of IGHV and IGLV sequences. The total number of antibody clones sequenced from each donor is indicated in the center of the pie chart. VH3-23/VK1-5 clones are in red, while other VH3-23 clones are indicated with different shades of blue. Non VH3-23 clones are shown in shades of grey, and singlets are in white. None of the grey clones are recurrent across individuals. C- V(D)J assignments for the VH3-23/VK1-5 clones. IgBLAST was used to assign the germline (GL) reference sequence for IGHV and IGLV. Red highlights differences in D and J usage in the VH3-23 clones between individuals.
Antibody heavy (IGH) and light (IGL) chain genes were amplified from single purified ZEDIII binding B cells by RT-PCR and sequenced (Scheid et al., 2009; von Boehmer et al., 2016). Overall, 290 antibodies were identified from the 6 individuals. As expected based on similar studies on HIV-1 and influenza, nearly one half of all of the antibodies (133) were found in expanded clones that shared the same IGH and IGL variable (IGVH and IGVL) gene segments, and the remaining half were unique (Fig. 2B and Tables S1 and S2).
Memory B cells expressing antibodies composed of VH3-23 paired with VK1-5 were found in 5 out of the 6 individuals assayed (red slices in Fig. 2B, and Fig. S1). Moreover, VH3-23/VK1-5 was present as an expanded clone in 4 individuals, and was the largest expanded clone in 3 out of the 6 individuals. The sequence of the VH3-23/VK1-5 antibodies in the expanded clones was further limited in that the VK1-5 gene segment was always recombined with JK1 (Fig. 2C). In addition to VH3-23/VK1-5 clones we also found expanded clones of memory B cells expressing antibodies composed of VH3-23 paired with other IGL genes (VH3-23/VK1-27, VH3-23/VK3-11 and VH3-23/VK3-20; blue slices in Fig. 2B). Of the 6 individuals examined, only BRA 138, who exhibited the lowest level of neutralizing activity, did not have any detectable memory B cells expressing VH3-23/VK1-5 antibodies. We conclude that individuals with high serologic neutralizing titers to ZIKV in geographically distinct outbreak areas frequently show clonally expanded ZEDIII-specific memory B cells that express VH3-23/VK1-5 antibodies.
Cross-reactivity with Other Flaviviruses
Nineteen representative antibodies obtained from expanded memory B cell clones from the 6 individuals were expressed for further testing. This antibody panel included 8 different VH3-23/VK1-5 antibodies from 5 separate volunteers. Antibody binding activity to ZEDIII was measured by ELISA, and found to vary broadly even among the closely related VH3-23/VK1-5 antibodies, with EC50 values ranging from 20 to >4000 ng/ml (Figs. 3A, 3B, and S2A). Like other human antibodies derived from memory B cells, anti-ZEDIII antibodies showed somatic mutations. For example, the number of IGH V gene mutations in the VH3-23/VK1-5 clones ranged from 12–40 nucleotides (average = 27.7, Figure S2B), which is far lower than that seen in antibodies during chronic HIV-1 infection (Escolano et al., 2017). Nevertheless, the mutations in anti-ZEDIII antibodies are essential to the binding activity of the antibodies since reversion of the mutations to the predicted germline sequence impaired binding to the antigen (Fig. 3B).
Figure 3. Binding of cloned antibodies to EDIII from ZIKV and other flaviviruses.

A- Binding of human monoclonal antibodies to ZEDIII. Human anti-HIV antibody 10–1074 was used as a negative control (Mouquet et al., 2012). The average half effective concentration (EC50) from at least two independent experiments is shown. B- Somatic mutations are required for ZEDIII binding. Binding of Z004, its predicted germline (GL), and control antibodies to ZEDIII as assessed by ELISA is shown. C- Human monoclonal antibody cross-reactivity by ELISA. Reactivity to the EDIII of the indicated flaviviruses is shown in blue. The list of antibodies is reported on the left of panel A. D- Z004 binds to the EDIII of DENV1. Binding of Z004, its predicted germline (GL), and control antibodies to DENV1 EDIII as assessed by ELISA is shown.
To determine whether the antibodies cloned from our cohorts cross-react to the EDIII proteins of other flaviviruses, we screened for binding to the four DENV serotypes (DENV1-4), YFV (Asibi and 17D strains), and WNV. We observed 5 different patterns of cross-reactivity with other flaviviruses (Fig. 3C). All 8 of the VH3-23/VK1-5 antibodies tested cross-reacted with DENV1, but not with the other flaviviruses in our panel (Figs. 3C and 3D). Other antibodies showed singular cross-reactivity to WNV, or broader reactivity to DENV1-4, or YF and WNV, and some antibodies were uniquely specific for ZIKV (Fig. 3C). Similar to ZIKV, mutations in the VH3-23/VK1-5 antibodies were required for optimal binding to DENV1 EDIII since reversion to the predicted germline sequence impaired binding to the DENV1 antigen (Fig. 3D). We conclude that anti-ZEDIII VH3-23/VK1-5 antibodies cross-react with DENV1 but not with other flaviviruses.
Neutralizing Activity in vitro and in vivo
To determine whether the anti-ZEDIII antibodies neutralize ZIKV in vitro we measured their neutralizing activity in the ZIKV luciferase RVP assay described above. As expected, neutralizing activity varied among the different antibodies ranging from sub-nanogram 50% inhibitory concentrations (IC50) to non-neutralizing (Figs. 4A, 4B and S3A). The most potent antibody, Z004, a member of one of the VH3-23/VK1-5 clones, displayed an IC50 of 0.7 ng/ml (Figs. 4A, 4B). Similar results were obtained by plaque reduction neutralization test (PRNT) using a Puerto Rican strain of ZIKV (IC50 of 2.2 ng/ml, Fig. S3B). All of the other VH3-23/VK1-5 antibodies tested were also potent neutralizers of ZIKV with IC50 values ranging from 0.7–4.6 ng/ml (Fig. 4A).
Figure 4. VH3-23/VK1-5 antibodies neutralize ZIKV and DENV1.

A- Neutralization potency of human monoclonal antibodies by ZIKV luciferase RVP assay. The human anti-HIV antibody 10–1074 serves as a negative control. Average values of the half maximal inhibitory concentration (IC50) from at least two independent experiments are shown. B and C- Z004 neutralizes ZIKV (B) and DENV1 (C) RVPs. Luciferase activity relative to the no antibody control was determined in the presence of increasing concentrations of Z004 or of its predicted germline antibody as indicated. Control antibody was tested at a single concentration. Data are represented as mean ± SD. D, E, and F- Z004 protects IFNAR−/− mice from ZIKV disease. Mice were infected by footpad (f.p.) injection with the Puerto Rican PRVABC59 ZIKV strain and treated intraperitoneally (i.p.) with Z004 (or 10–1074 control) either before (E) or 1 day after (F) infection. Mice were monitored for symptoms and survival. Survival: p<0.0001 (pre-exposure) and p=0.0027 (post-exposure). Symptoms: p<0.0001 (both pre- and post-exposure, Mantel-Cox test). Three independent experiments, of 4 to 7 mice per group, were combined and displayed. G- Amino acid alignment of a portion of the EDIII lateral ridge region for a panel of flaviviruses. The corresponding accession numbers are indicated in parenthesis. H- The K394 residue in the ZEDIII lateral ridge is required for ZIKV neutralization by Z004. Luciferase activity relative to no antibody control was determined for ZIKV wild type or mutant E393A and K394A RVPs. I- Z004 neutralizes both Asian/American and African strains. RVPs bearing Asian/American ZIKV wild type (E393), mutant (Asian/American with E393D) and African strain (D393) E proteins were neutralized by Z004. In H and I data are represented as mean ± SD.
Z004 is a member of the VH3-23/VK1-5 family that cross-reacts with DENV1. To determine whether Z004 also neutralizes DENV1, we measured its neutralizing activity against DENV1 luciferase RVPs and by flow cytometry using authentic DENV1. We found that Z004 is a potent neutralizer of DENV1 in both assays (IC50=1.6 ng/ml by luciferase assay, and IC50=16.4 ng/ml by flow cytometry; Fig. 4C and S3C). Thus, the VH3-23/VK1-5 antibody Z004 binds and neutralizes both ZIKV and DENV1.
To determine whether VH3-23/VK1-5 antibodies also neutralize ZIKV in vivo, we passively transferred Z004 to IFNRA−/− mice one day before or one day after ZIKV infection (Fig. 4D). In 3 independent pre-exposure experiments, with a total of 14 mice infected with ZIKV in the presence of control antibody, we found that 93% developed clinical symptoms and 79% succumbed to infection. In contrast, pre-exposure prophylaxis with Z004 resulted in a significant reduction in disease symptoms and mortality. Only 12.5% of the Z004 group developed clinical symptoms and none died (p<0.0001 for both disease and survival; Figs. 4E and S3D). Similar results were also obtained when the antibody was administered one day after infection (p<0.0001 for symptoms, p=0.0027 for survival; Figs. 4F and S3D). We conclude that Z004 was protective and significantly reduced both symptoms and mortality when administered either before or after infection. Z004 also displayed a suitable profile of low poly- and auto-reactivity (Fig. S3E). Thus, VH3-23/VK1-5 antibodies have the potential for further pre-clinical evaluation.
VH3-23/VK1-5 Antibodies Recognize the Lateral Ridge of ZEDIII
There are only 2 contiguous amino acids that are uniquely shared between the EDIIIs of ZIKV and DENV1, and not by DENV2, DENV3, DENV4, WNV or YF (E393 and K394 in ZIKV, E384 and K385 in DENV1; Fig. 4G). These 2 amino acids are found in the lateral ridge region of the ZEDIII, which is a region that is associated with virus interaction with cellular receptors (Mukhopadhyay et al., 2005). To determine whether these 2 amino acids are essential for interaction between VH3-23/VK1-5 antibodies and ZIKV, we made alanine substitutions in the context of the ZIKV RVPs and tested the recombinant RVPs for sensitivity to Z004-mediated neutralization. Although ZIKV RVPs bearing the E393A substitution remained sensitive to Z004, K394A mutant RVPs were resistant to the antibody (Fig. 4H). African ZIKV strains differ from others at position 393 carrying aspartic acid at this position. Similar to wild type Asian/American ZIKV RVPs, African ZIKV RVPs carrying D393 instead of E393 were sensitive to Z004, and Asian/American ZIKV RVPs with an E393D substitution were also efficiently neutralized (Figs. 4G and I). Thus a shared epitope in the lateral ridge region could account for the finding that Z004 neutralizes ZIKV and DENV1, but not other flaviviruses.
Structures of ZIKV Antibodies/EDIII Complexes Reveal a Shared Binding Mode
To gain additional insights into the molecular basis of ZEDIII recognition by VH3-23/VK1-5 antibodies, we solved crystal structures of complexes of the antigen-binding fragment (Fab) of two antibodies isolated from different donors, Z006 and Z004, with ZIKV and DENV1 EDIII domains, respectively (Table S5). The Z006 Fab-ZEDIII and Z004 Fab-DENV1 EDIII structures showed a common mode of antigen recognition, as revealed by similar orientations of Fab VH and VL domains when the EDIII domains were superimposed (Fig. 5A). The Z006/Z004 Fab orientation is distinct from orientations in other crystallographically-characterized Fab/ZIKV and Fab/DENV1 EDIII complexes (Fig. 5B). The Z006 epitope extends over much of the EDIII lateral ridge (Fig. 5C): beyond the E393-K394ZIKV region, the next largest parts of the interface consist of the N-terminal region of EDIII (residues 305–311ZIKV), CC’ loop residues 350–352ZIKV, and BC loop residues 333–336ZIKV. The E393-K394ZIKV (E384-K385DENV1) motif is central to the interface (Fig. 5C) and contacts residues within the Fab CDRH3, CDRL3, and CDRL1 loops in both structures (Fig. 5A, D–F). Despite the antibodies originating from different donors and binding to two different flavivirus EDIIIs, a number of contact interactions occur in both complexes (Table S6). Specifically, the side chain of residue K394ZIKV (K385DENV1) occupies a hydrophobic pocket formed by W32LC and Y91LC(Z006)/F91LC(Z004) and forms an H-bond with the latter residue’s backbone oxygen atom (Fig. 5D). The side chain of residue E393ZIKV (E384DENV1) interacts with R96HC, although this interaction differs somewhat in the two structures: in the Z006 structure, E393ZIKV forms an H-bond with the Y91LC hydroxyl and an electrostatic interaction with R96HC, while for Z004, the side chain of residue F91LC lacks a hydroxyl group to form an H-bond, and instead the side chain of E384DENV1 forms a salt bridge with R96HC (Fig. 5E). Other common interactions include the side chain of Y58HC forming an H-bond to the backbone oxygen of L307ZIKV (M301DENV1) (Fig. 5F), and the side chain of T93LC (Z006)/S93LC (Z004) forming an H-bond to the backbone N of T335ZIKV (T329DENV1). Hence the common mode of recognition involves using equivalent pairwise interactions as well as binding with a similar orientation.
Figure 5. Structures of Fab complexes with ZIKV and DENV1 EDIII domains.

A- Superimposition of Z006 Fab-ZEDIII and Z004 Fab-DENV1 EDIII crystal structures after alignment of the EDIII domains. The VH domain positions differ by a 14° rotation about an axis passing through the center of the interface. Inset: close-up of interactions between the E393ZIKV–K394ZIKV/E384DENV1–K385DENV1 motif (shown as sticks) within the EDIII lateral ridge and the two Fabs. Fab CDRs are highlighted. B- Overlay of the Z006-ZEDIII complex structure (VH-VL in blue and cyan; EDIII in black) with previously solved structures of antibodies in complex with ZIKV and DENV1 EDIII domains. VH-VL domains from ZIKV antibodies are pink; VH-VL from DENV1 antibodies are tan; the E393ZIKV–K394ZIKV side chains in ZEDIII are shown as spheres. Structures were aligned on the EDIII domains; only ZEDIII is shown for clarity. C- ZEDIII epitope: EDIII residues contacted by Z006 Fab are highlighted on a surface representation of the EDIII structure. EDIII residues contacted by VH are blue, residues contacted by VL are cyan, and residues making interactions with both VH and VL are dark blue. The E393ZIKV–K394ZIKV motif is outlined. Contacts between the Z004 Fab and DENV1 EDIII were less extensive than Z006–ZEDIII contacts, in part because of disorder of the CC’ loop in DENV1 EDIII (residues 343–349) (Methods). D to F- Comparison of key antibody-antigen interactions for Z006 Fab-ZEDIII and Z004 Fab-DENV1 EDIII structures. Hydrogen bonds are shown as dotted lines. D- Fab interactions with K394ZIKV/K385DENV1. E- Fab interactions with E393ZIKV/E384DENV1. F- Y58HC (germline-encoded in VH3-23) interactions with antigen. See also Tables S5 and S6.
Pre-existing DENV1 Reactivity is Associated with Enhanced ZEDIII Antibody Responses
The existence of VH3-23/VK1-5 antibodies that neutralize both DENV1 and ZIKV and are recurrently found in expanded clones suggests that prior exposure to DENV1 primes the development of protective ZIKV immunity. To examine this possibility, we tested sera obtained at time points before and after introduction of ZIKV in the Pau da Lima community in Salvador, Brazil. As expected, anti-ZEDIII serum IgG reactivity increased significantly between April and November of 2015 (Fig. 6A). Interestingly, a similar increase was seen for DENV1, although there was no documented DENV1 outbreak in this area at this time, with only 5 DENV1 cases reported between September 2014 and July 2016 (Fig. 6A). In contrast, no significant increase in reactivity was observed for DENV2, DENV3, DENV4, YFV, or WNV EDIIIs (Fig. 6A). Consistent with the hypothesis that DENV1 primes the subsequent response to ZIKV, we observed a significant positive correlation between DENV1 EDIII-reactive IgG levels pre-ZIKV, and ZEDIII-reactive IgG levels post-ZIKV (Pseudo-ρ=0.48, p<0.001, Fig. 6B). Together, these data indicate that the exposure to ZIKV boosted the pre-existing DENV1 antibody response, and that individuals with pre-existing antibodies targeting the DENV1 EDIII are more likely to develop high levels of EDIII antibodies upon ZIKV infection.
Figure 6. EDIII reactivity over time.
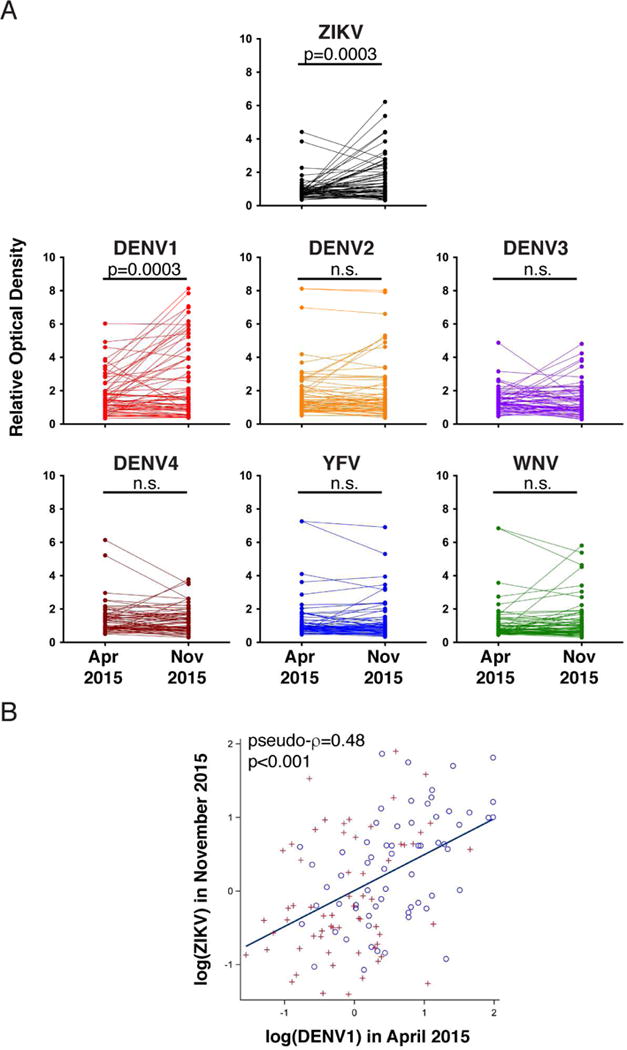
A- A set (n=63) of paired sera from the Brazilian cohort participants were collected in April and November 2015 and assayed for binding to flavivirus EDIII. Optical densities are normalized as described in Fig. 1A. Paired sera from the same individual are connected by a line. Each value represents the average of two independent measurements. P values were determined with the two-tailed paired t test (n.s., not significant). B- Correlation between DENV1 EDIII reactivity in April and ZEDIII reactivity in November 2015. Circles and plus signs distinguish data from two independent experiments. Pseudo-ρ=0.48, p<0.001 by univariate analysis (see statistical methods).
Lateral Ridge Antibodies Are Associated with ZIKV Neutralization
To determine whether antibodies to the lateral ridge region recognized by Z004 contribute to serologic activity against ZIKV in the Pau da Lima cohort, we developed a competition ELISA assay. In this assay we measure inhibition of biotin-Z004 binding to ZEDIII in order to quantify lateral ridge-binding antibodies present in serum (see Methods). Paired samples from April 2015 (before ZIKV) and November 2015 (after ZIKV) showed an increase in lateral ridge reactivity after ZIKV introduction (p=0.0007, Fig. 7A). Levels of antibodies present in the post-ZIKV serum that are capable of blocking Z004 binding to ZEDIII were directly correlated with the overall reactivity of antibodies to ZEDIII (Spearman coefficient, ρ=0.7319, p<0.0001 Fig. 7B), as well as with the increase in reactivity to ZEDIII from prior to after ZIKV (ρ=0.8190, p<0.0001, Fig. 7C). Finally, there was also a significant correlation between ZIKV neutralizing activity and total ZEDIII reactivity (ρ=0.5885, p=0.0012, Fig. 7D), as well as Z004 blocking activity (ρ=0.6585, p=0.0002, Fig. 7E). We conclude that antibodies that block Z004 binding to the lateral ridge make a measurable contribution to the overall serum neutralizing activity to ZIKV in exposed individuals.
Figure 7. EDIII antibodies contribute to the serologic response and ZIKV neutralization capacity.

A- Competition ELISA shows the increase within individuals of serum antibodies that block biotin-Z004 ZEDIII binding after ZIKV exposure. Each dot represents a serum sample (n=62 at each of the indicated time points). A line connects sera from the same individual obtained at different time points. The p value was determined with the two-tailed paired t test. B and C- The estimated quantity (μg/ml) of Z004 blocking antibodies in the serum obtained after ZIKV introduction (X axes) was plotted with the overall serum binding activity to ZEDIII (Y axis, B), and the change in that individual’s serum ZEDIII binding from before to after ZIKV (Y axis, C). Binding activity change was determined by subtracting the pre- from the post-ZIKV ELISA relative optical density value (average of two independent measurements). Each dot represents an individual (n=62, two-tailed Spearman r test). D and E- Serum neutralization potency expressed as NT50 versus the overall serum binding activity to ZEDIII (D), or Z004 blocking antibody concentrations in sera (E) obtained after ZIKV introduction are plotted. Each dot represents a serum sample from a single donor (n=27, two-tailed Spearman r test). Representative of two independent experiments is shown.
Discussion
Previous studies have examined anti-ZIKV antibodies developing in a small number of available individuals (Sapparapu et al., 2016; Stettler et al., 2016; Wang et al., 2016). In contrast, we screened sera from more than 400 donors from ZIKV epidemic areas of Mexico and Brazil to select high responders. As expected serologic reactivity to ZIKV varied greatly among individuals, with neutralization potencies spanning over more than 2 logs. To better understand this activity we isolated 290 memory B cell antibodies from 6 individuals with high serum neutralizing activity. The cloning experiments revealed the existence of expanded clones of memory B cells expressing E protein lateral ridge-specific ZIKV and DENV1 neutralizing VH3-23/VK1-5 antibodies in 4 of the 6 individuals.
Three separate groups have cloned human anti-ZIKV E protein reactive antibodies (Sapparapu et al., 2016; Stettler et al., 2016; Wang et al., 2016). In all they studied 8 individuals and documented 92 antibodies to the E protein. The great majority of these antibodies (79) were obtained by screening supernatants of Epstein-Barr virus transformed B lymphocytes for binding to ZIKV, and only a minority (15) were directed to the ZEDIII. Among all of these antibodies there was only a single expanded clone containing 3 related VH1-46/VK1-39 antibodies specific for a yet to be determined E epitope. These 3 antibodies were relatively poor neutralizers (3-257μg/ml)(Wang et al., 2016). There was also a single VH3-23/VK1-5 antibody that targeted the ZEDIII and neutralized ZIKV (ZIKV-116), but this antibody appears to be somewhat different than those reported here because it failed to neutralize the African ZIKV strain (Sapparapu et al., 2016). Thus, there was no prior indication of a potent dominant neutralizing response to ZIKV.
We found a total of 69 individual VH3-23/VK1-5 memory B cells antibodies in 5 out of 6 individuals. In addition to recurring V gene segments, VH3-23/VK1-5 antibodies bear the same IGL J gene, and a limited set of IGH D and J genes. These antibodies are closely related and they are potent neutralizers with IC50 values ranging from 0.7–4.6ng/ml. This variation in activity is likely due to somatic mutations, since predicted germline versions of the VH3-23/VK1-5 antibodies bind to ZEDIII and neutralize the virus only weakly. Thus, not unexpectedly, somatic mutations are required for optimal VH3-23/VK1-5 antibody neutralizing activity.
Recurring antibodies that share the same IGV genes and the same molecular interactions with antigen have not been reported for ZIKV or other flaviviruses, but they have been described in other viral infections including HIV-1 and influenza. Broadly neutralizing antibodies targeting the CD4 binding site of HIV-1 frequently utilize VH1-2 or VH1-46 genes (Scheid et al., 2011; West et al., 2012), and broadly neutralizing antibodies to Influenza utilize VH1-69 (Laursen and Wilson, 2013; Pappas et al., 2014; Sui et al., 2009; Throsby et al., 2008; Wrammert et al., 2011). However, in both HIV-1 and influenza, the VH genes can be paired with a collection of different VL genes, a finding that was explained by structural analysis showing that many of the essential contacts made by influenza and HIV-1 antibodies involve variable portions of the IGHV (Ekiert et al., 2009; Pappas et al., 2014; Scheid et al., 2011; Wrammert et al., 2011; Zhou et al., 2010).
Although the Z004 and Z006 antibodies have CDRH3s and CDRL3s of different lengths they share a common mode of EDIII binding. Several shared sequence features may be critical for this binding mode. The most suggestive of these is R96HC. This CDRH3 residue appears to derive from N region addition, thus the different VH3-23/VK1-5 clones do not share this residue due to shared germline genes. Examination of a large collection (n=44,270) of VH3-23-derived antibody sequences indicates that only 13% have R at position 100 (Rubelt et al., 2012). However, 68 of 69 of the sequenced VH3-23/VK1-5 clones contain R96. The clones also tend to conserve the germline residues forming the Z004/Z006/EDIII common interactions: Y58HC, W32LC, Y91LC, and S93LC.
The requirement for conserved IGHV and IGLV genes in VH3-23/VK1-5 antibodies appears to be explained in part by interactions using germline residues. For the light chain, CDRL1 germline residue W32LC interacts with K394ZIKV. Few IGLV genes contain W32; the most common of these is VK1-5 (followed by VK1-12, but this gene is several-fold less common than VK1-5). For the heavy chain, residue Y58HC (present in the VH3-23 germline) makes a contact with EDIII residue 307ZIKV. Y58HC is present in ~50% of VH germlines, potentially explaining a portion of the restriction. Finally, VH3-23 is among the most frequently used VH genes as is VK1-5 (Arnaout et al., 2011; DeKosky et al., 2015). Therefore it is likely that naïve B cell precursors carrying DENV1 and ZIKV reactive VH3-23/VK1-5 antibodies would also be common in the pre-immune repertoire making this epitope a particularly attractive vaccine candidate.
VH3-23/VK1-5 antibodies recognize and neutralize both DENV1 and ZIKV, suggesting that clones of VH3-23/VK1-5 producing B cells originally elicited in response to DENV1 were further expanded in response to ZIKV. This prime-boost, or original antigenic sin hypothesis, is supported by two observations. First, pre-existing antibodies to DENV1 EDIII are associated with a higher antibody response to ZEDIII. Second, at the population level, the introduction of ZIKV correlates with an increase in DENV1 EDIII-reactive antibodies at a time when DENV1 was not circulating. Although DENV1 and ZIKV only share 50% amino acid identity in EDIII, they are structurally very similar, particularly in the lateral ridge region that is recognized by VH3-23/VK1-5 (Fig. 5). Thus, DENV1 EDIII reactive memory B cells have a significant probability of being cross-reactive to ZEDIII. A primary response to DENV1 would increase the frequency of these ZIKV cross-reactive memory B cells and thereby increase their likelihood of undergoing clonal expansion in response to ZIKV. Consistent with this idea, memory B cells with VH3-23/VK1-5 antibodies represent close to half of all ZEDIII-specific B cell clones in 3 of the 6 individuals examined.
Infection by DENV1 confers transient protection to infection by DENV2 (Sabin, 1950, 1952). Whether prior DENV1 infection also protects from ZIKV by cross-priming or in other cases enhances infection is unclear (Castanha et al., 2016; Dejnirattisai et al., 2016; Priyamvada et al., 2016; Swanstrom et al., 2016; Wahala and Silva, 2011). However, the existence of human antibodies to DENV that cross-neutralize or enhance ZIKV in vitro indicates that protection by cross-priming is possible (Barba-Spaeth et al., 2016; Dejnirattisai et al., 2016; Harrison, 2016; Pierson and Graham, 2016; Priyamvada et al., 2016; Stettler et al., 2016; Swanstrom et al., 2016).
ZIKV infection is asymptomatic in most people. Only 20% of ZIKV infected individuals develop symptoms, and in those cases the severity of the disease varies broadly (Miner and Diamond, 2017). Similarly, the spectrum and incidence of developmental sequelae in infants born to women infected with ZIKV during pregnancy differs within and across geographic areas with risk estimates that range from 6–42% (Brasil et al., 2016; Honein et al., 2017). The biological basis for this heterogeneity is not clear. Our experiments point to a cellular and molecular explanation for how a history of DENV1 exposure could alter host responses and susceptibility to ZIKV.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Davide Robbiani (drobbiani@rockefeller.edu). Sharing of antibodies and other reagents with academic researchers may require UBMTA agreements.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Samples of peripheral blood were obtained upon consent from community participants of cohort studies in Pau da Lima (Brazil) and Santa Maria Mixtequilla (Mexico) under protocols approved by the ethical committees of the Rockefeller University (IRB DRO-0898), Yale University (IRB HIC 1603017508), FIOCRUZ (CAAE 63343516.1.0000.5028), Hospital Geral Roberto Santos (1.998.103), and National Institute of Respiratory Diseases (C16-16). Information regarding sex and age of study participants can be obtained upon request. Details on the size of the cohorts and time when samples were obtained is listed in the Results section of the manuscript.
Mice
IFNRA−/− mice were obtained from The Jackson Laboratory and bred and maintained in the AAALAC-certified facility of the Rockefeller University. Mice were specific pathogen free and maintained under a 12 hr light/dark cycle with standard chow diet. Both male and female mice (3–4 week old) were used for all experiments and were equally distributed within experimental and control groups. Animal protocols were in agreement with NIH guidelines and approved by the Rockefeller University Institutional Animal Care and Use Committee (16855-H).
Cell lines
Human embryonic kidney HEK-293-6E suspension cells were cultured at 37°C in 8% CO2, shaking at 120 rpm. All other cell lines described below were cultured at 37°C in 5% CO2, without shaking. Green monkey VERO cells and human hepatocytes Huh-7.5 cells (Blight et al., 2002) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% nonessential amino acids (NEAA) and 5% FBS. Human Lenti-X 293T cells (Clontech) and STAT1−/−, an SV40 large T antigen immortalized skin fibroblast line (Chapgier et al., 2006), were grown in DMEM 10% FBS.
Bacteria
E. coli BL21(DE3) were cultured at 37°C, shaking at 250 rpm. MC1061 cells were cultured in LB medium, with 250 rpm shaking, at 30–37°C depending on the plasmid.
Viruses
Zika virus (ZIKV), 2015 Puerto Rican PRVABC59 strain (Lanciotti et al., 2016), was obtained from the CDC and passaged once in STAT1−/− fibroblasts (STAT1−/− −ZIKV stock, used in mouse experiments) or twice in Huh-7.5 cells (Huh-7.5-ZIKV stock, used in all other experiments). The Thai human isolate of DENV1 PUO-359 (TVP-1140) was obtained from Robert Tesh and amplified by three passages in C6/36 insect cells.
METHOD DETAILS
Collection of human samples
Samples of peripheral blood for serum or mononuclear cells (PBMCs) isolation were obtained from community participants and donors and frozen at the cohorts’ sites. PBMCs were purified using the gradient centrifugation method with Ficoll and cryopreserved in 90% heat-inactivated fetal bovine serum (FBS) supplemented with 10% dimethylsulfoxide (DMSO), prior to shipment to Rockefeller University in liquid nitrogen. Serum aliquots were heat-inactivated at 56°C for 1h and stored at 4°C thereafter.
Production and biotinylation of flavivirus protein
The coding sequences for the EDIII portion of flaviviruses were preceded by sequences encoding the human CD5 signal peptide (MPMGSLQPLATLYLLGMLVASCLG) and followed by a polyhistidine-AviTag (HHHHHH-GLNDIFEAQKIEWHE). The following flavivirus sequences were used.
ZIKV (KJ776791)
5′GTGTCATACTCCTTGTGTACCGCAGCGTTCACATTCACCAAGATCCCGGCTGAAACACTGCACGGGACAGTCACAGTGGAGGTACAGTACGCAGGGACAGATGGACCTTGCAAGGTTCCAGCTCAGATGGCGGTGGACATGCAAACTCTGACCCCAGTTGGGAGGTTGATAACCGCTAACCCCGTAATCACTGAAAGCACTGAGAACTCTAAGATGATGCTGGAACTTGATCCACCATTTGGGGACTCTTACATTGTCATAGGAGTCGGGGAGAAGAAGATCACCCACCACTGGCACAGGAGT.
DENV1 (codon-optimized based on NC_001477)
5′ATGTCATATGTGATGTGTACGGGGTCCTTTAAACTTGAAAAGGAGGTGGCAGAAACACAGCACGGAACAGTACTTGTGCAGGTTAAATATGAGGGAACCGATGCTCCTTGTAAAATACCGTTTTCAAGCCAGGACGAAAAGGGTGTAACACAAAATGGTCGCCTGATTACAGCCAACCCAATAGTCACTGATAAGGAGAAACCTGTGAATATCGAGGCAGAGCCACCATTCGGCGAAAGTTATATCGTAGTTGGTGCTGGAGAAAAGGCCCTGAAACTCTCTTGGTTTAAGAAG.
DENV2 (NC_001474)
5′ATGTCATACTCTATGTGCACAGGAAAGTTTAAAGTTGTGAAGGAAATAGCAGAAACACAACATGGAACAATAGTTATCAGAGTGCAATATGAAGGGGACGGCTCTCCATGCAAGATCCCTTTTGAGATAATGGATTTGGAAAAAAGACATGTCTTAGGTCGCCTGATTACAGTCAACCCAATTGTGACAGAAAAAGATAGCCCAGTCAACATAGAAGCAGAACCTCCATTCGGAGACAGCTACATCATCATAGGAGTAGAGCCGGGACAACTGAAGCTCAACTGGTTTAAGAAA.
DENV3 (codon-optimized based on NC_001475.2)
5′ATGTCATACGCAATGTGTACGAACACATTCGTTCTTAAAAAAGAGGTAAGTGAAACCCAACATGGTACTATCCTTATAAAAGTTGAGTACAAGGGCGAGGACGCTCCCTGCAAAATACCGTTTTCCACAGAGGACGGGCAAGGTAAGGCACACAATGGGAGACTTATAACCGCCAATCCAGTAGTGACCAAGAAAGAGGAACCAGTCAACATTGAAGCGGAGCCCCCTTTCGGAGAATCCAACATAGTGATAGGCATTGGGGACAACGCTCTGAAGATCAACTGGTATAAGAAG.
DENV4 (codon-optimized based on NC_002640.1)
5′ATGTCATATACAATGTGTAGCGGTAAATTCAGCATTGATAAAGAAATGGCCGAGACACAGCACGGCACCACCGTGGTAAAAGTGAAATACGAAGGAGCGGGAGCCCCGTGCAAGGTCCCCATCGAAATCAGGGATGTAAACAAAGAGAAGGTCGTTGGTAGAATAATTTCTTCTACACCACTGGCCGAGAACACTAATTCAGTTACGAATATAGAACTTGAGCCCCCCTTTGGTGACAGCTATATAGTTATTGGCGTGGGAAATTCTGCACTGACTCTGCATTGGTTCCGAAAA.
YFV (Asibi strain, KF769016)
5′ACATCCTACAAAATGTGCACTGACAAAATGTCTTTTGTCAAGAACCCAACTGACACTGGCCATGGCACTGTTGTGATGCAGGTGAAAGTGCCAAAAGGAGCCCCCTGCAAGATTCCAGTGATAGTAGCTGATGATCTTACAGCGGCAATCAATAAAGGCATTTTGGTTACAGTTAACCCCATCGCCTCAACCAATGATGATGAAGTGCTGATTGAGGTGAACCCACCTTTTGGAGACAGCTACATTATCGTTGGGACAGGAGATTCACGTCTCACTTACCAGTGGCACAAAGAG.
YFV (17D strain, KF769015)
5′ACATCCTACAAAATATGCACTGACAAAATGTTTTTTGTCAAGAACCCAACTGACACTGGCCATGGCACTGTTGTGATGCAGGTGAAAGTGTCAAAAGGAGCCCCCTGCAGGATTCCAGTGATAGTAGCTGATGATCTTACAGCGGCAATCAATAAAGGCATTTTGGTTACAGTTAACCCCATCGCCTCAACCAATGATGATGAAGTGCTGATTGAGGTGAACCCACCTTTTGGAGACAGCTACATTATCGTTGGGAGAGGAGATTCACGTCTCACTTACCAGTGGCACAAAGAG.
WNV (KX547539.1)
5′ACAACCTATGGCGTCTGTTCAAAGGCTTTCAAGTTTCTTGGGACTCCCGCAGACACAGGTCACGGCACTGTGGTGTTGGAATTGCAGTACACTGGCACGGATGGACCTTGCAAAGTTCCTATCTCGTCAGTGGCTTCATTGAACGACCTAACGCCAGTGGGCAGATTGGTCACTGTCAACCCTTTTGTTTCAGTGGCCACGGCCAACGCTAAGGTCCTGATTGAATTGGAACCACCCTTTGGAGACTCATACATAGTGGTGGGCAGAGGAGAACAACAGATCAATCACCACTGGCACAAGTCT.
Gene synthesis was by Genscript. The proteins were produced by transient transfection into HEK-293-6E cells using PEI (polyethylenimine, branched). After 7 days of incubation, cell supernatants were cleared by centrifugation and histidine-tagged proteins were purified with Ni Sepharose 6 Fast Flow. Purified ZEDIII was biotinylated using the Biotin-Protein Ligase-BIRA kit according to manufacturer’s instructions.
ELISA assays
Serum and recombinant antibody
The binding of serum IgG or recombinant IgG antibodies to the EDIII proteins was measured by standard ELISA. ELISA plates were coated with 250 ng of EDIII protein in PBS per well and stored overnight at room temperature. Plates were then blocked with 1% BSA, 0.1mM EDTA in PBS-T (PBS with 0.05% Tween20) for 1h at 37°C. Plates were washed with PBS-T in between each step above. Serum samples were diluted 1:500 with PBS-T and added for 1h at 37°C. Secondary HRP-conjugated goat anti-human IgG (0.16μg/ml) was added for 1h at 37°C. Plates were then developed using ABTS substrate and read at 405 nm. The relative binding affinity of recombinant monoclonal antibodies was determined similarly, using serially diluted samples. The half effective concentration (EC50) needed for maximal binding was determined by non-linear regression analysis.
Cross-reactivity ELISA
The binding of monoclonal antibodies to the panel of flavivirus EDIII proteins was determined using the standard ELISA setup described above. Antibodies were tested at 10μg/ml alongside a control serum weakly cross-reactive to all flaviviruses. Samples with a relative optical density ratio of >1 compared to control were deemed reactive.
Auto- and poly-reactivity ELISA
To determine the auto- and poly-reactivity of recombinant antibodies, ELISA plates were coated with 50μl PBS containing dsDNA (10μg/ml), ssDNA (10μg/ml, obtained by denaturing dsDNA at 95°C for 30 minutes), LPS (10μg/ml), Insulin (5μg/ml), or keyhole limpet hemocyanin (KLH; 10μg/ml). After washing with PBS-T, plates were blocked with 1% BSA and 0.5mM EDTA in 0.05% PBS-T for 2h at room temperature. Serial dilutions of antibody samples were then incubated for 2h, also at room temperature. Incubation with secondary antibody and ELISA development were performed as described above. Previously reported antibodies ED38 (Wardemann et al., 2003) and mG053 (Yurasov et al., 2005) were used as positive and negative control, respectively.
Competition ELISA
Competition ELISA was performed as described above for serum EDIII binding, with the following modifications. After 1h of incubation with serum (diluted 1:10 in PBS-T) at room temperature, biotinylated antibody Z004 (biotin-Z004) was added at a final concentration of 0.16 μg/ml to compete for an additional 15 min at room temperature. After washing, streptavidin-HRP was used for detection of bound biotin-Z004. The optimal concentration of biotin-Z004 (0.16 μg/ml) was determined by measuring its binding to ZEDIII over a range of concentrations, and corresponds to 50% of the observed maximal binding. The concentration of Z004 blocking antibodies in serum was estimated by interpolation with a standard curve generated by competing biotin-Z004 (0.16 μg/ml) with a range of non-biotinylated Z004 concentrations, and using the stats package nls() function in R 3.3.2.
Antibody discovery and production
Isolation of ZEDIII+ memory B cells
B cell purification, labeling and antibody discovery were performed as previously described in detail (Tiller et al., 2008; von Boehmer et al., 2016), with the following modifications. PBMCs were resuscitated and washed in 37°C RPMI. To enrich for B cells, PBMCs were incubated with CD19 microbeads according to the manufacturer’s instructions. Upon washing, B cells were positively selected using LS magnetic columns, washed with PBS 3%FBS, and incubated with anti-CD20-PECy7, anti IgG-APC, and fluorescently-labeled ZEDIII bait at 4°C for 20 min. The fluorescently-labeled ZEDIII bait was previously prepared by incubating 2–3μg of biotin-ZEDIII with streptavidin-PE for at least 1h at 4°C in the dark. After wash, single CD20+ZEDIII+gG+ memory B cells were sorted into 96-well plates using a FACSAriaII (Becton Dickinson).
Antibody sequencing and production
RNA from single cells was reverse-transcribed using random primers (von Boehmer et al., 2016), followed by nested PCR amplifications and sequencing using the primers listed in (Tiller et al., 2008). V(D)J gene segment assignment and determination of the CDR3 sequences were with IgBlast (Ye et al., 2013). Sequences that were nonproductive, out of frame, or with premature stop codons were excluded. Similarly, sequences for which a matching light or heavy chain sequence was not identifiable were omitted. Cloning for recombinant antibody production was by the Sequence and Ligation-Independent Cloning (SLIC; (Li and Elledge, 2007)) method as detailed in (von Boehmer et al., 2016). Amplicons from the first sequencing PCR reaction were used as template for amplification with the SLIC-adapted primers listed in Table S3, and cloned into IGγ1-, IGκ or IGλ-expression vectors as detailed in (von Boehmer et al., 2016). The recombinantly expressed antibodies correspond to the folIowing antibody sequence IDs (see Tables S1 and S2): Z028 (MEX84_p4–53), Z001 (MEX18_21), Z004 (MEX18_89), Z006 (MEX105_42), Z010 (MEX105_88), Z031 (BRA112_46), Z035 (BRA112_71), Z038 (BRA12_2), Z014 (MEX18_91), Z039 (BRA12_21), Z015 (MEX84_p2–44), Z018 (MEX84_p2–45), Z021 (MEX84_p4–23), Z024 (MEX84_p4–12), Z012 (MEX105_57), Z037 (BRA112_57), Z041 (BRA138_57), Z042 (BRA138_17), Z043 (BRA138_15). The variable portion of the predicted germline antibody Z004-GL was codon-optimized, synthesized by Genscript, and cloned as described above (IGH 5′GAGGTGCAGCTGTTGGAGTCTGGGGGAGGTCTTGTTCAGCCGGGTGGATCATTGAGACTTTCTTGTGCTGCAAGTGGATTTACTTTCTCTTCCTACGCCATGTCTTGGGTTCGACAAGCTCCAGGGAAAGGACTCGAATGGGTTAGTGCGATATCTGGGTCTGGAGGATCTACTTACTACGCAGATTCAGTAAAAGGGCGCTTCACAATATCACGCGATAATTCCAAGAATACGCTCTACCTTCAGATGAACAGTCTTCGGGCAGAGGACACAGCGGTTTATTATTGTGCGAAAGATCGCGGTCCCAGAGGCGTGGGCGAACTGTTCGACTATTGGGGACAAGGCACCCTGGTCACCGTCTCCTCAG and IGK 5′GACATCCAGATGACCCAGTCACCGTCTACCTTGTCAGCGTCAGTTGGTGACCGGGTAACCATTACTTGCCGCGCTAGTCAGAGTATTTCCTCCTGGCTCGCCTGGTATCAACAAAAACCAGGTAAAGCCCCCAAATTGCTGATCTATAAGGCAAGTAGCTTGGAATCAGGAGTTCCCAGCCGCTTCTCTGGCTCAGGGTCCGGTACTGAATTTACATTGACCATCTCTTCTCTCCAGCCAGATGACTTCGCCACGTACTATTGTCAACAGTATAACTCATATCCCTGGACTTTTGGACAGGGGACCAAGGTGGAAATCAAAC). Recombinant antibodies were produced as previously described (Klein et al., 2014). Briefly, HEK-293-6E cells were transiently transfected with equal amounts of immunoglobulin heavy and light chain expression vectors. After 7 days, the supernatant was harvested and antibodies were purified with Protein G Sepharose 4 Fast Flow. For antibody biotinylation, 1.5 mg/ml of Z004 were used with FluoReporter Mini-biotin-XX Protein Labeling Kit as instructed by the manufacturer.
Mouse experiments
All experiments involving mice were performed under protocols approved by the Rockefeller University Institutional Animal Care and Use Committee. 123 μg of monoclonal antibodies in 200 μl of PBS were administered intraperitoneally to 3–4 week old IFNRA−/− mice one day prior or after infection with 1.25 × 105 PFU ZIKV Puerto Rican strain in 50 μl into the footpad. Mice were monitored for symptoms and survival over time.
Virus titration
Viral titers were measured on VERO cells by plaque assay (PA) for ZIKV virus and focus forming assay (FFA) for DENV-1 strains. For PA, 200 μl of serial 10-fold virus dilutions in OPTI-MEM were used to infect 400,000 cells seeded the day prior in a 6-well format. After 90 minutes adsorption, the cells were overlayed with DMEM containing 2% FBS with 1.2% Avicel and Pen/Strep. Four days later the cells were fixed with 3.5% formaldehyde and stained with crystal violet for plaque enumeration. For FFA, 100 μl of serial 10-fold virus dilutions in OPTI-MEM were used to infect 250,000 cells seeded the day prior in a 12-well format. After 90 minutes, the cells were overlayed as described above for PA. Six to 7 days later the cells were fixed and incubated with 2 μg/ml of antibody Z004 in PBS/5% FBS/0.25% Triton X-100 for 1 h at room temperature. Foci were enumerated after reaction with goat anti-human IgG HRP-conjugated antibodies and TrueBlue HRP substrate, followed by addition of a few drops of DAB buffer solution (DAKO). Experiments with infectious ZIKV and DENV strains were performed in a biosafety level 2 laboratory.
Plaque reduction neutralization test
Antibody neutralization activity was measured using a standard plaque reduction neutralization test (PRNT) on VERO cells. Diluent medium consisted of medium 199 (Lonza) supplemented with 1% BSA and Pen/Strep (BA-1 diluent). Briefly, 3- to 10-fold serial human antibody dilutions were added to a constant amount of Huh-7.5-ZIKV stock diluted in BA-1 diluent and incubated for 1 h at 37°C prior to application to VERO cells seeded at 400,000 cells per 6-well the day prior. After 90 min adsorption at 37°C the cells were overlayed as per PA protocol above. After 4 days at 37°C, the wells were fixed and stained to enumerate plaques. PRNT50 values were determined as the antibody concentration that resulted in 50% of the number of plaques obtained with the no antibody control.
RVP plasmid construction
West Nile virus (WNV) subgenomic replicon-expressing plasmid pWNVII-Rep-REN-IB (Pierson et al., 2006) and a ZIKV C-prM-E expression plasmid (pZIKV/HPF/CprME) were obtained from Ted Pierson (NIH). Plasmid pWNVII-Rep-REN-IB encodes a Renilla luciferase-expressing WNV replicon RNA while pZIKV/HPF/CprME encodes the structural proteins (C-prM-E) of the ZIKV French Polynesian strain H/PF/2013, both under the control of a CMV promoter. Co-transfection of the two plasmids into permissive cells allows the WNV replicon RNA to replicate, express luciferase and be packaged by the ZIKV H/PF/2013 structural proteins to generate RVPs that can be used for single round infection studies (Mukherjee et al., 2014; Pierson et al., 2006). To facilitate expression of the envelopes of a wide range of ZIKV strains, plasmid pZIKV/HPF/CprME was engineered to have a unique BspHI restriction enzyme site immediately upstream of the envelope region by PCR-based site-directed mutagenesis of two other BspHI restriction sites located in the plasmid backbone. The resulting plasmid, pZIKV/HPF/CprM*E*, has unique BspHI and SacII restriction sites flanking the envelope region, allowing facile manipulation. PCR-based site-directed mutagenesis was used to introduce E393A or K394A mutations into the envelope of H/PF/2013 in pZIKV/HPF/CprM*E*, resulting in plasmids pZIKV/HPF/CprME(E393A) and pZIKV/HPF/CprME(K394A). We generated pZIKV/HPF-CprM/MR766-E by replacing the H/PF/2013 envelope in pZIKV/HPF/CprM*E* with that of the ZIKV African strain, MR766. Because the African strains contain a BspHI site within the E protein coding region, this site was mutated using assembly PCR prior to swapping in the MR766-based BspHI and SacII fragment. To generate pDENV1/PUO-359/CprME, DENV1 (strain PUO-359) virion RNA was isolated by TRIzol extraction (Thermo Fisher Scientific), cDNA generated using Superscript II reverse transcriptase (Thermo Fisher Scientific), and the C-prM-E region amplified by PCR. After PCR assembly with the upstream promoter and vector sequences, the corresponding region of pZIKV/HPF/CprME was replaced using SnaBI/SacII enzymes. PCR reactions utilized either PfuUltra Hotstart DNA Polymerase (Agilent technologies), Phusion High Fidelity DNA polymerase (NEB) or KOD DNA polymerase (Toyobo). All PCR-derived plasmid regions were verified by sequencing. Primer sequences used for assembly PCR and mutagenesis are listed in Supplemental Table S4.
RVP production
Reporter viral particles (RVPs) were produced in Lenti-X 293T cells, seeded the day before DNA transfection at 1×106 cells/well in collagen coated 6-well plates. One μg of pWNVII-Rep-REN-IB (WNV replicon expression construct) and 3 μg of the appropriate flavivirus CprME expression construct were co-transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Lipid–DNA complexes were removed after 4–5 h incubation at 37°C and replaced with DMEM containing 20 mM HEPES and 10% FBS. After incubation for 48–72 h at 34°C, RVP-containing supernatants were harvested, filtered through a 0.45 micron filter and frozen at −80°C. RVPs were titrated on Huh-7.5 cells to determine the dilution to use in the RVP-based neutralization assay to achieve ~2–5×106 RLU in the absence of serum/antibody.
RVP neutralization assay
The day before infection, 96-well plates were seeded with 15,000 Huh-7.5 cells/well in a volume of 100 μl. RVPs were diluted in BA-diluent (ranging from 1:4 to 1:32 depending on the RVP stock) and 100 μl were added to 100 μl of triplicate samples of 3- or 10-fold serially diluted human serum/antibody. After incubation for 1 h at 37°C, 100 μl of the RVP/antibody mixture was added to the cells. After 24 h incubation at 37°C, the medium was removed and cells were lysed in 75 μl lysis buffer and 20 μl used for Renilla luciferase measurement using the Renilla Luciferase Assay System (Promega) according to the manufacturer’s instructions using a FLUOstar Omega luminometer (BMG LabTech). Neutralization capacity of the serum/antibody was determined by the percentage of luciferase activity obtained relative to activity from RVPs incubated with BA-diluent alone (no serum/antibody). NT50 values represented the reciprocal of the serum dilution or the antibody concentration that resulted in 50% inhibition compared to RVP alone.
Flow cytometry-based neutralization assay
The day prior to infection 5,000 VERO cells/well were seeded in 96-well plates. Serial dilutions of antibody were mixed with ZIKV or DENV-1 virus for 1 h at 37°C and then applied to infect cells, using an MOI of 0.02 for ZIKV and DENV-1. After 3 days, cells were fixed with 2% formaldehyde and permeabilized in PBS containing 1% FBS and 0.5% saponin. Cells were stained with 2 μg/ml of the pan flavivirus anti-E protein 4G2 monoclonal antibody (Henchal et al., 1982). After incubation with Alexa Fluor 488-conjugated anti-mouse IgG antibody (Invitrogen) at 1:1,000 dilution, the number of infected cells was determined by flow cytometry. The percentage of infected cells relative to cells infected with virus in the absence of antibody was calculated for each antibody dilution to estimate the 50% reduction.
Protein production and crystallization
His-tagged Fabs were transiently expressed in HEK-293-6E cells by co-transfecting with appropriate heavy and light chain plasmids. Fabs were purified from the supernatant using Ni-NTA affinity chromatography (GE Healthcare) and size exclusion chromatography (Superdex 200; GE Healthcare) in 20mM Tris pH 8.0, 150mM NaCl, 0.02% NaN3. The Fabs were concentrated to 10–20 mg/mL for crystallography. Untagged constructs of ZEDIII and DENV1 EDIII were expressed in E. coli and refolded from inclusion bodies as previously described (Sapparapu et al., 2016). Briefly, BL21(DE3) E. coli were transformed with an appropriate expression vector encoding ZIKV E protein residues 299–407 (ZIKV EDIII, strain H/PF/2013) or DENV1 E protein residues 297–396 (DENV1 EDIII, strain clone 45AZ5). Cells were grown to mid-log phase and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 hours. The cells were lysed and the insoluble fraction containing inclusion bodies was solubilized in buffer containing 6M guanidine hydrochloride and 20mM β-mercaptoethanol, and then clarified by centrifugation. The solubilized inclusion bodies were refolded using rapid dilution into 400 mM L-arginine, 100 mM Tris-base (pH 8.0), 2 mM EDTA, 0.2 mM phenyl-methylsulfonyl fluoride, and 5 and 0.5 mM reduced and oxidized glutathione at 4° C. The refolded protein was filtered and concentrated, and then purified by size exclusion chromatography (Superdex 75; GE Healthcare) in 20mM Tris pH 8.0, 150mM NaCl, 0.02% NaN3. Antigens were concentrated to 2–15 mg/mL for crystallography. Complexes for crystallization were produced by mixing Fab and antigen at a 1:1 molar ratio and incubating at room temperature for 1–2 hours. Crystals of Z006 Fab–ZEDIII complex (space group H32; a = 385.08 Å, b = 385.08 Å, c = 56.64 Å, α = 90°, β = 90°, γ = 120°; two molecules per asymmetric unit) were obtained by combining 0.2 μL of crystallization sample with 0.2 μL of 10% isopropanol, 0.1M sodium citrate tribasic dihydrate pH 5.0, 26% PEG 400 in sitting drops at 22°C. Crystals of Z004 Fab–DENV1 EDIII complex (space group P43212; a = 74.23 Å, b = 74.23 Å, c = 190.76 Å; one molecule per asymmetric unit) were obtained by combining 0.2 μL of crystallization sample with 0.2 μL of 0.1M sodium acetate trihydrate pH 4.5, 30% w/v PEG 1500 in sitting drops at 22°C.
Structure Determination and Refinement
X-ray diffraction data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12–2 using a Dectris Pilatus 6M detector. The data were integrated using Mosflm (Battye et al., 2011) and scaled using CCP4 (Winn et al., 2011)(Table S5). The Z006–ZEDIII complex structure (PDB ID 5VIG) was solved by molecular replacement with a Z006 unbound Fab structure (data not shown) and Zika EDIII (PDB ID 5KVG) as the search models using Phaser and Molrep (CCP4) and refined to 3.0 Å using an iterative approach involving (i) refinement in CNS and Phenix applying NCS constraints and (ii) manual rebuilding into electron density maps using O and AntibodyDatabase (Jones, 2004; West et al., 2013). The final model (Rwork = 21.2%; Rfree = 25.7%) contains 7,892 protein atoms and one citrate ion (12 atoms). 90.8, 7.6, and 1.6% of the residues were in the favored, allowed, and disallowed regions, respectively, of the Ramachandran plot (Table S5). Residues 1, 128–133, 214–219, and the 6×-His tag of the Z006 heavy chain; residue 214 of the LC; and residues 299–304 and 405–407 were disordered and are not included in the model. The Z004-DENV1 EDIII complex structure (PDB ID 5VIC) was solved by molecular replacement using the VHVL (CDR residues removed) and CHCL domains from PDB 3SKJ and DENV1 EDIII from PDB 4L5F as search models in Phenix (Adams et al., 2010). The model was refined to 3.0 Å resolution using an iterative approach involving (i) refinement in Phenix and (ii) manual rebuilding into a simulated annealing composite omit map using Coot (Emsley and Cowtan, 2004). The final model (Rwork = 23.6%; Rfree = 28.1%) contains 3,904 protein atoms. 93.0%, 6.8%, and 0.2% of the residues were in the favored, allowed, and disallowed regions, respectively, of the Ramachandran plot (Table S5). Residues that were disordered and not included in the model were Fab HC residues 129–132, 215–219, and the 6×-His tag; LC residues 212–214; and DENV1 EDIII domain residues 315–317, 341–347, 373–374, and 393–396. Structures were superimposed, rmsd calculations done, and figures were generated using PyMOL. Hydrogen bonds were assigned using the following criteria: a distance of < 3.5 Å, and an A-D-H angle of > 90°.
QUANTIFICATION AND STATISTICAL ANALYSIS
Information on the statistical tests used, and the exact values of n and what it represents can be found in the Results and Figure Legends. Unless otherwise noted, statistical analysis was with Prism software. The half effective concentration (EC50) needed for maximal binding by ELISA was determined by non-linear regression analysis (Fig. 3A). Data are the average of at least two independent experiments. Similarly, luciferase- and flow cytometry-based neutralization assays were performed in triplicate wells, and the serum dilution (NT50) or antibody concentration (IC50) that neutralized 50% of the virus or RVP inoculum was calculated by nonlinear, dose-response regression analysis (Fig. 4A). The Mantel-Cox test was applied to analyze disease and survival in mice infection experiments (Fig. 4D–F). The Paired t test was used to analyze changes in seroreactivity over time (Fig. 6A and 7A). Univariate associations were assessed between log-relative optical densities of anti-ZEDIII antibodies at t=2 (November 2015) with log-relative optical densities of anti-DENV1 EDIII antibodies at t=1 (April 2015) using proc mixed in SAS v 9.4 (Fig. 6B). An individual-level random intercept was included to account for non-independence of the two replicated measurements. The two-tailed Spearman r test was used for the correlations in Fig. 7B–E.
DATA AND SOFTWARE AVAILABILITY
The structural data reported in this study were deposited in the Protein Databank (PDB) under ID codes 5VIG (Z006-ZIKV EDIII complex structure) and 5VIC (Z004-DENV1 EDIII complex structure).
Supplementary Material
Figure S1 – Maximum likelihood tree of VH3-23/VK1-5 antibodies. Related to Figure 2 and Table S1.
Antibody amino acid sequences (heavy and light chain combined) are clustered and labeled according to the donor ID (MEX18, MEX105, MEX84, BRA112, BRA12) followed by the clone’s unique sequence ID.
Figure S2 – Features of anti-ZIKV antibodies. Related to Figure 3 and Table S1.
A- Dose-dependent binding of recombinant human monoclonal antibodies to ZEDIII as measured by ELISA. Representative non-linear regression curves are shown. B and C- VH3-23/VK1-5 antibodies have low levels of somatic mutation. The number of V gene nucleotide (B) or amino acid (C) mutations at Heavy (H) and Light (L) chain genes are shown for each donor. Each dot represents one individual antibody V gene (n=69). The average number of nucleotide mutations overall is 27.7 within VH3-23 and 17.5 within VK1-5. The average number of amino acid mutations overall is 14.3 at VH3-23 and 10.6 at VK1-5.
Figure S3 – Neutralization and polyreactivity of anti-ZIKV antibodies. Related to Figure 4.
A- Dose-dependent neutralization of ZIKV RVPs by recombinant human monoclonal antibodies. Luciferase activity relative to no antibody control is determined in the presence of increasing antibody concentrations. Data are represented as mean ± SD. B- ZIKV neutralization by Z004 antibody assessed by PRNT assay (see Methods). C- DENV1 neutralization by Z004 antibody measured by a flow cytometry-based assay. The number of infected cells was determined using the pan-flavivirus monoclonal antibody 4G2 (see Methods). Data are represented as mean ± SD. D- Z004 protects IFNAR−/− mice from ZIKV infection. Three independent experiments were performed as described in Fig. 4D–F; results were pooled and presented in Fig. 4. E- Low auto- and polyreactivity profile of Z004. ELISA measures Z004 binding over a range of concentrations against the following antigens: single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), lipopolysaccharides (LPS), insulin, and keyhole limpet hemocyanin (KLH).
Table S1. V(D)J rearrangements and sequences of anti-ZEDIII antibodies that do cluster in clones. Related to Figure 2.
Table S2. V(D)J rearrangements and CDR3 sequences of anti-ZEDIII antibodies that do not cluster in clones (singlets). Related to Figure 2.
Table S3. List of primers for cloning recombinant antibodies by the SLIC method. Related to Methods.
Table S4. List of primers for the generation of RVP expression constructs. Related to Methods.
Table S5. Data collection and refinement statistics. Related to Figure 5.
Table S6. Antibody-antigen contacts. Related to Figure 5. Contacts are defined as residues in which any atom is within 4 Å of an atom from a residue on the interacting partner using AntibodyDatabase (West et al., 2013). The table is organized by antibody residue, listing all antigen residues contacted by each antibody residue (ordered by contact distance). Antibody residues are highlighted when corresponding interactions occur in both complexes. For the highlighted interactions additional information is listed including the antibody residue’s origin in V(D)J recombination and the residue distribution at that antibody position in the sequenced antibody clones.
Highlights.
-
-
Pre-existing antibodies to DENV1 correlate with high ZIKV neutralizing responses.
-
-
Antibodies to the EDIII lateral ridge correlate with serum neutralization of ZIKV.
-
-
Recurrent VH3-23/VK1-5 antibodies to the EDIII potently neutralize ZIKV and DENV1.
-
-
The structures of VH3-23/VK1-5 antibodies with DENV1 and ZEDIII were solved.
Acknowledgments
We would especially like to thank the members of the Santa Maria Mixtequilla and Pau da Lima communities who agreed to participate in this study, as well as the Fiocruz community research team and staff of Hospital Geral Roberto Santos for their assistance with the clinical protocols. We thank members of the Nussenzweig and Rice labs for helpful discussions and suggestions. A particular thank you to Lotta von Boehmer, Christian Mayer, Yotam Bar-On, and Julio Cetrulo Lorenzi for sharing expertise, protocols, and reagents. We thank Ted Pierson for plasmids pWNVII-Rep-REN-IB and pZIKV/HPF/CprME, Aaron Brault (CDC) for ZIKV PRVABC59, Bob Tesh for DENV1 PUO-359, Matt Evans for ZIKV MR766 cDNA, and Greg Ebel, James Weger and Brian Geiss for ZIKV PRVABC59 cDNA. We are grateful to Laura Kramer (NYSDOH) and Robert Lanciotti (CDC) for PRNT advice and protocols, Kimberly Dowd for RVP advice, and Muhammad Arshad for cloning assistance. We thank Barbara Johnson, Ann Powers, Christin Goodman, Claire Huang, Brandy Russell (CDC) and Claudia Duarte dos Santos for sharing ZIKV information early on. Moreover, we are thankful to Mary Ellen Castillo for facilitating virus work in the cell culture core, Alison Ashbrook for help with PRNT assays, Natalia Frias-Staheli for generation of the DENV1 stock, Corrine Quirk for assistance with mouse colonies, Erica Lee, Alisa Voll, and the Caltech Protein Expression Center for cloning, expression, and purification of proteins for crystallography, and Christopher Barnes and Beth Stadtmueller for help with crystallographic methods. This work was supported by pilot awards from NIH U19AI111825 (to DFR) and UL1TR001866 (to DFR and LB), grants R01AI037526, UM1AI100663, U19AI111825 and UL1TR001866 (to MCN), grants R01AI124690 and U19AI057229 (CCHI pilot project), grants R01AI121207, R01TW009504, R25TW009338 and U01AI088752 (to AIK), The Rockefeller University Development Office and anonymous donors (to CMR), and the Molecular Observatory at Caltech supported by the Gordon and Betty Moore Foundation (to PJB). Operations at the Stanford Synchrotron Radiation Lightsource are supported by the US Department of Energy and the National Institutes of Health. Support was also provided by the Robertson Therapeutic Development Fund (to DFR and MCN). PCO is supported by the Pew Latin American Fellows Program in the Biomedical Sciences, DS-B by Studienstiftung des deutschen Volkes, LU by the Austrian Marshall Plan Foundation, and EES-R is partly supported by Red INMUNOCANEI-Conacyt. MCN is an HHMI Investigator. In connection with this work, DFR and MCN have a provisional patent application with the U.S. Patent and Trademark Office (62/483,001). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies or individuals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DFR conducted the experiments, supervised and designed the experiments, interpreted experimental results and wrote the paper. LB designed and conducted experiments, supervised, interpreted experimental results and edited the manuscript. PCO, AG, DS-B and MS designed and conducted experiments and edited the manuscript. RK, SA-R, and EES-R coordinated cohort studies and blood collection and edited the manuscript. LN, RP, SAA, LFU, MA, KHY, JG, NT, TE and HBG conducted experiments. EAW, GS, NN, CO, FC and MGR identified cohort subjects, coordinated clinical cohorts, and performed clinical work in Brazil. AH and MC coordinated human studies. JP, DW and TO performed statistical and computational analysis. JRK and APW solved and analyzed crystal structures and wrote structure-related parts of the paper together with PJB. GRT and ARPdA coordinated clinical studies in Mexico and Brazil, respectively. AK coordinated clinical studies in Brazil, interpreted results and edited the manuscript. CMR supervised, interpreted experimental results and edited the manuscript. MRM designed experiments, supervised, interpreted experimental results and edited the manuscript. MCN supervised, designed and interpreted experiments and wrote the paper.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-resolution description of antibody heavy-chain repertoires in humans. PloS one. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Loriere E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017 doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L, Pacenti M, Franchin E, Lavezzo E, Trevisan M, Sgarabotto D, Palu G. Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 21. 2016 doi: 10.2807/1560-7917.ES.2016.21.32.30316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallographica Section D, Biological crystallography. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Barrett AD. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. Journal of virology. 2002;76:13097–13100. doi: 10.1128/JVI.76.24.13097-13100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. Journal of virology. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro. The New England journal of medicine. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso CW, Paploski IA, Kikuti M, Rodrigues MS, Silva MM, Campos GS, Sardi SI, Kitron U, Reis MG, Ribeiro GS. Outbreak of Exanthematous Illness Associated with Zika, Chikungunya, and Dengue Viruses, Salvador, Brazil. Emerging infectious diseases. 2015;21:2274–2276. doi: 10.3201/eid2112.151167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanha PM, Nascimento EJ, Cynthia B, Cordeiro MT, de Carvalho OV, de Mendonca LR, Azevedo EA, Franca RF, Rafael D, Marques ET., Jr Dengue virus (DENV)-specific antibodies enhance Brazilian Zika virus (ZIKV) infection. The Journal of infectious diseases. 2016 doi: 10.1093/infdis/jiw638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS genetics. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Sarno M, Khouri R, de Paula Freitas B, Siqueira I, Ribeiro GS, Ribeiro HC, Campos GS, Alcantara LC, Reis MG, et al. Emergence of Congenital Zika Syndrome: Viewpoint From the Front Lines. Annals of internal medicine. 2016;164:689–691. doi: 10.7326/M16-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. Journal of virology. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, Zhang Y, Yuan Y, Song H, Haywood J, et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell host & microbe. 2016;19:696–704. doi: 10.1016/j.chom.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nature immunology. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy-and light-chain antibody repertoire. Nature medicine. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Escolano A, Dosenovic P, Nussenzweig MC. Progress toward active or passive HIV-1 vaccination. The Journal of experimental medicine. 2017;214:3–16. doi: 10.1084/jem.20161765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felzemburgh RD, Ribeiro GS, Costa F, Reis RB, Hagan JE, Melendez AX, Fraga D, Santana FS, Mohr S, dos Santos BL, et al. Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the Leptospira agent. PLoS neglected tropical diseases. 2014;8:e2927. doi: 10.1371/journal.pntd.0002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerging infectious diseases. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca GV, Schuler-Faccini L, Oliveira WK, Henriques CM, Carmo EH, Pedi VD, Nunes ML, Castro MC, Serruya S, Silveira MF, et al. Congenital Zika virus syndrome in Brazil: a case series of the first 1501 livebirths with complete investigation. Lancet. 2016;388:891–897. doi: 10.1016/S0140-6736(16)30902-3. [DOI] [PubMed] [Google Scholar]
- Hagan JE, Moraga P, Costa F, Capian N, Ribeiro GS, Wunder EA, Jr, Felzemburgh RD, Reis RB, Nery N, Santana FS, et al. Spatiotemporal Determinants of Urban Leptospirosis Transmission: Four-Year Prospective Cohort Study of Slum Residents in Brazil. PLoS neglected tropical diseases. 2016;10:e0004275. doi: 10.1371/journal.pntd.0004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Immunogenic cross-talk between dengue and Zika viruses. Nature immunology. 2016;17:1010–1012. doi: 10.1038/ni.3539. [DOI] [PubMed] [Google Scholar]
- Heinz FX, Stiasny K. The Antigenic Structure of Zika Virus and Its Relation to Other Flaviviruses: Implications for Infection and Immunoprophylaxis. Microbiology and molecular biology reviews : MMBR 81. 2017 doi: 10.1128/MMBR.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. The American journal of tropical medicine and hygiene. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, et al. Birth Defects Among Fetuses and Infants of US Women With Evidence of Possible Zika Virus Infection During Pregnancy. Jama. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- Jones TA. Interactive electron-density map interpretation: from INTER to O. Acta crystallographica Section D, Biological crystallography. 2004;60:2115–2125. doi: 10.1107/S0907444904023509. [DOI] [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. The Journal of experimental medicine. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Lim EX, Zhang S, Fibriansah G, Ng TS, Ooi JS, Shi J, Lok SM. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Li J, Shi PY. West Nile virus. The Lancet Neurology. 2007;6:171–181. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor Ldel C. Phylogeny of Zika Virus in Western Hemisphere, 2015. Emerging infectious diseases. 2016;22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral research. 2013;98:476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, Carcelen AC, Ott CT, Sheffield JS, Ferguson NM, et al. Assessing the global threat from Zika virus. Science. 2016;353:aaf8160. doi: 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Miner JJ, Diamond MS. Zika Virus Pathogenesis and Tissue Tropism. Cell host & microbe. 2017;21:134–142. doi: 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Pierson TC, Dowd KA. Pseudo-infectious reporter virus particles for measuring antibody-mediated neutralization and enhancement of dengue virus infection. Methods in molecular biology. 2014;1138:75–97. doi: 10.1007/978-1-4939-0348-1_6. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nature reviews Microbiology. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Murray KO, Gorchakov R, Carlson AR, Berry R, Lai L, Natrajan M, Garcia MN, Correa A, Patel SM, Aagaard K, et al. Prolonged Detection of Zika Virus in Vaginal Secretions and Whole Blood. Emerging infectious diseases. 2017;23:99–101. doi: 10.3201/eid2301.161394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray NE, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clinical epidemiology. 2013;5:299–309. doi: 10.2147/CLEP.S34440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Pierson TC, Graham BS. Zika Virus: Immunity and Vaccine Development. Cell. 2016;167:625–631. doi: 10.1016/j.cell.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 2006;346:53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rubelt F, Sievert V, Knaust F, Diener C, Lim TS, Skriner K, Klipp E, Reinhardt R, Lehrach H, Konthur Z. Onset of immune senescence defined by unbiased pyrosequencing of human immunoglobulin mRNA repertoires. PloS one. 2012;7:e49774. doi: 10.1371/journal.pone.0049774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin AB. The dengue group of viruses and its family relationships. Bacteriological reviews. 1950;14:225–232. [PubMed] [Google Scholar]
- Sabin AB. Research on dengue during World War II. The American journal of tropical medicine and hygiene. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, et al. A method for identification of HIV gp140 binding memory B cells in human blood. Journal of immunological methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nature reviews Immunology. 2015;15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- Silva MM, Rodrigues MS, Paploski IA, Kikuti M, Kasper AM, Cruz JS, Queiroz TL, Tavares AS, Santana PM, Araujo JM, et al. Accuracy of Dengue Reporting by National Surveillance System, Brazil. Emerging infectious diseases. 2016;22:336–339. doi: 10.3201/eid2202.150495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suy A, Sulleiro E, Rodo C, Vazquez E, Bocanegra C, Molina I, Esperalba J, Sanchez-Seco MP, Boix H, Pumarola T, et al. Prolonged Zika Virus Viremia during Pregnancy. The New England journal of medicine. 2016;375:2611–2613. doi: 10.1056/NEJMc1607580. [DOI] [PubMed] [Google Scholar]
- Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, Baric RS. Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. mBio. 2016:7. doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of immunological methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, Nussenzweig MC. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nature protocols. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- Wahala WM, Silva AM. The human antibody response to dengue virus infection. Viruses. 2011;3:2374–2395. doi: 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, Wong G, Peng R, Liu S, Li J, et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Science translational medicine. 2016;8:369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. Zika virus: History, emergence, biology, and prospects for control. Antiviral research. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral research. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic acids research. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC. Defective B cell tolerance checkpoints in systemic lupus erythematosus. The Journal of experimental medicine. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – Maximum likelihood tree of VH3-23/VK1-5 antibodies. Related to Figure 2 and Table S1.
Antibody amino acid sequences (heavy and light chain combined) are clustered and labeled according to the donor ID (MEX18, MEX105, MEX84, BRA112, BRA12) followed by the clone’s unique sequence ID.
Figure S2 – Features of anti-ZIKV antibodies. Related to Figure 3 and Table S1.
A- Dose-dependent binding of recombinant human monoclonal antibodies to ZEDIII as measured by ELISA. Representative non-linear regression curves are shown. B and C- VH3-23/VK1-5 antibodies have low levels of somatic mutation. The number of V gene nucleotide (B) or amino acid (C) mutations at Heavy (H) and Light (L) chain genes are shown for each donor. Each dot represents one individual antibody V gene (n=69). The average number of nucleotide mutations overall is 27.7 within VH3-23 and 17.5 within VK1-5. The average number of amino acid mutations overall is 14.3 at VH3-23 and 10.6 at VK1-5.
Figure S3 – Neutralization and polyreactivity of anti-ZIKV antibodies. Related to Figure 4.
A- Dose-dependent neutralization of ZIKV RVPs by recombinant human monoclonal antibodies. Luciferase activity relative to no antibody control is determined in the presence of increasing antibody concentrations. Data are represented as mean ± SD. B- ZIKV neutralization by Z004 antibody assessed by PRNT assay (see Methods). C- DENV1 neutralization by Z004 antibody measured by a flow cytometry-based assay. The number of infected cells was determined using the pan-flavivirus monoclonal antibody 4G2 (see Methods). Data are represented as mean ± SD. D- Z004 protects IFNAR−/− mice from ZIKV infection. Three independent experiments were performed as described in Fig. 4D–F; results were pooled and presented in Fig. 4. E- Low auto- and polyreactivity profile of Z004. ELISA measures Z004 binding over a range of concentrations against the following antigens: single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), lipopolysaccharides (LPS), insulin, and keyhole limpet hemocyanin (KLH).
Table S1. V(D)J rearrangements and sequences of anti-ZEDIII antibodies that do cluster in clones. Related to Figure 2.
Table S2. V(D)J rearrangements and CDR3 sequences of anti-ZEDIII antibodies that do not cluster in clones (singlets). Related to Figure 2.
Table S3. List of primers for cloning recombinant antibodies by the SLIC method. Related to Methods.
Table S4. List of primers for the generation of RVP expression constructs. Related to Methods.
Table S5. Data collection and refinement statistics. Related to Figure 5.
Table S6. Antibody-antigen contacts. Related to Figure 5. Contacts are defined as residues in which any atom is within 4 Å of an atom from a residue on the interacting partner using AntibodyDatabase (West et al., 2013). The table is organized by antibody residue, listing all antigen residues contacted by each antibody residue (ordered by contact distance). Antibody residues are highlighted when corresponding interactions occur in both complexes. For the highlighted interactions additional information is listed including the antibody residue’s origin in V(D)J recombination and the residue distribution at that antibody position in the sequenced antibody clones.


