Abstract
Stable isotope assisted metabolomics (SIAM) uses stable isotope tracers to support studies of biochemical mechanisms. We report a suite of data analysis algorithms for automatic analysis of SIAM data acquired on a high resolution mass spectrometer. To increase the accuracy of isotopologue assignment, metabolites detected in the unlabeled samples were used as reference metabolites to generate possible isotopologue candidates for analysis of peaks detected in the labeled samples. An iterative linear regression model was developed to deconvolute the overlapping isotopic peaks of isotopologues present in a full MS spectrum, where the threshold for the weight factor was determined by a simulation study assuming different levels of Gaussian white noise contamination. A normalization method enabling isotope ratio-based normalization was implemented to study the difference of isotopologue abundance distribution between sample groups. The developed method can analyze SIAM data acquired by direct infusion MS and LC-MS, and can handle metabolite tracers containing different tracer elements. Analysis of SIAM data acquired from mixtures of known compounds showed that the developed algorithms accurately identify metabolites and quantify stable isotope enrichment. Application of SIAM data acquired from a biological study further demonstrated the effectiveness and accuracy of the developed method for analysis of complex samples.
TOC Image
Developed a suite of data analysis algorithms for automatic analysis of SIAM data acquired on a high resolution mass spectrometer.
1. Introduction
The metabolome represents all metabolites in a biological sample. These metabolites are commonly derived from enzymatic reactions and form a network where the outputs of preceding enzymatic reactions provide inputs to others. In metabolite profiling metabolomics, the measured concentration of a metabolite is its summed abundance if this metabolite is synthesized in more than one pathways (e.g., lactate). The relative contribution of those pathways to the synthesis of that metabolite remains ambiguous. Therefore, abundance measurements are frequently insufficient for pathway assignment and identification of changes in metabolite fate and flux.
Stable isotope assisted metabolomics (SIAM) uses heavy isotope tracers (e.g., 13C, 18O, 15N) to identify and discern pathways involved in biochemical processes, by measuring the incorporation of heavy atoms into the metabolites produced downstream of the tracer(s) 1–3. While SIAM has been applied in metabolomics, relatively few efforts have been devoted to developing bioinformatics tools to analyze SIAM data. Creek et al. combined multiple software packages to analyze the SIAM data 1, and Huang and colleagues developed the software package X13CMS to track isotopic labels 4. Other developments include geoRge 5, mzMatch-ISO 6, and MIRACLE 7. However, technical challenges persist, especially with respect to automatic and accurate assignment and quantification of isotopologue peaks. These features are crucial to deconvolute SIAM data properly and to resolve biologically relevant pathway information.
The objective of this work was to develop a comprehensive computational platform for analysis of SIAM data acquired by high resolution mass spectrometry (HRMS), in forms of direct infusion mass spectrometry (DI-MS) or liquid chromatography mass spectrometry (LC-MS). We developed a data reduction and analysis strategy capable of high-throughput, simultaneous assignment and quantification of overlapping isotopologue peaks. Several graphical user interfaces (GUIs) were further developed for facile integration and visualization of experimental data. The developed software package was tested by analyzing two sets of SIAM data acquired by DI-MS and LC-MS. One set of data was acquired from mixtures of known compounds, while the other was acquired from two groups of metabolite samples extracted from cardiac-derived cells.
2. Experimental Section
2.1 Mixtures of Known Compounds
A mixture of known metabolites was created by the combination of two commercially-available amino acid mixtures purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA, USA): a mixture of unlabeled amino acids (Cat. No. ULM-2314-1, ALGAL amino acid mixture unlabeled) and a mixture of 13C-labeled amino acids (Cat. No. CNLM-452-0.5, ALGAL amino acid mixture (U-13C, 97–99%)). Each of these two mixtures contains 16 amino acids, including L-alanine, L-arginine, L-aspartic acid, L-glutamic acid, L-glycine, L-histidine, L-isoleucine, L-leucine, L-lysine, L-methionine, L-phenylalanine, L-proline, L-serine, L-threonine, L-tyrosine and L-valine. After dissolving each amino acid mixture into 0.1 M HCl, the combined concentration of 16 amino acids in each mixture was 200 μg/mL. The two amino acid mixtures were then mixed in a ratio of weight Wlabeled : Wunlabeled = 100 μg/mL : 200 μg/mL and 200 μg/mL : 200 μg/mL, respectively. A total of three samples were prepared in parallel for the mixture with a specific weight ratio.
2.2 Cell Culture SIAM Experiment
All animal procedures were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the University of Louisville Institutional Animal Care and Use Committee. Murine cardiac progenitor cells were isolated from adult, male C57BL/6J mouse heart cell outgrowth cultures 8. These cells were cultured in DMEM/F12 medium containing the following constituents: glutamine (2.5 mM), glucose (17.5 mM), pyruvic acid (0.5 mM), leukemia inhibitory factor (1000 U/mL), basic fibroblast growth factor (20 ng/mL), epidermal growth factor (20 ng/mL), 10% embryonic stem cell fetal bovine serum, penicillin/streptomycin (1×), and insulin-transferrin-selenium (1×). The cells were grown in 6-well plates in DMEM media to ~80% confluency and subsequently, replaced with DMEM containing 5 mM [U-13C]-glucose or [U-13C5, 15N2]-glutamine. The cells were then harvested after 3 or 18 h. At the time of harvest, the medium was collected and all plates were washed 3× with ice-cold PBS. To quench cellular metabolism, 0.3 mL of ice-cold acetonitrile was added, followed by addition of 0.225 mL nanopure water.
The cells were harvested by scraping using a rubber policeman. This process was repeated once more to ensure complete cell harvest. To extract metabolites, a solvent mixture of acetonitrile : water : chloroform (v : v : v = 2 : 1.5 : 1) was used to obtain the polar, non-polar and insoluble proteinaceous fractions. The non-polar (lipid) layer was collected, dried under a stream of nitrogen gas, and then reconstituted in 0.1 mL mixture of chloroform : methanol (v : v = 2 : 1) containing 1 mM BHT. The polar layer of the extract was dried by Speedvac to remove acetonitrile, followed by freeze drying to remove water. The dried sample was reconstituted in 100 μL 20% acetonitrile and used for LC-MS analysis.
For nucleotide analysis, the samples were prepared using a previously published protocol 9, with slight modifications. Briefly, lyophilized polar extracts were first reconstituted in 50 μL of 5 mM aqueous hexylamine (pH 6.3) with acetic acid (Solvent A). Samples were then loaded onto a 100 μL capacity C18 tip (Pierce-Thermo Fisher Scientific, Rockford, IL, USA) via four slow aspirations followed by washing twice with 50 μL of Solvent A. The metabolites were eluted with two 50 μL portions of 70% Solvent A and 30% 1 mM ammonium acetate in 90% methanol, pH 8.5 (Solvent B) by aspirating ten times. For mass spectrometry, the resulting eluates were diluted 3× with methanol and analyzed via DI-FTICR-MS.
2.3 Direct Infusion FTICR-MS Analysis
The direct infusion experiments were performed on an FTICR-MS instrument (LTQ-FT; Thermo Electron Corporation, Bremen, Germany) equipped with a chip-based nanoelectrospray ionization (nESI) ion source (Triversa NanoMate) (Advion Biosciences, Ithaca, NY, US). The TriVersa NanoMate was operated by applying 1.6 kV and 0.7 psi head pressure in the negative mode. High mass accuracy data were collected using the FTICR analyzer over a mass range from 150 to 850 Da (- mode) for 15 min at the target mass resolution of 400,000 at 400 m/z. The LTQ-FT was tuned using the [ATP–H]– peak (m/ztheor = 505.988478) and calibrated according to the manufacturer’s default standard recommendations, to achieve mass accuracy of typically ≤ 2 ppm. The maximum ion accumulation time was set at 1000 ms.
2.4 LC-MS Analysis
All samples were analyzed on a Thermo Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer coupled with a Thermo DIONEX UltiMate 3000 HPLC system (Thermo Fisher Scientific, Inc., Germany). The UltiMate 3000 HPLC system was equipped with a reversed phase (RP) column and a hydrophobic interaction liquid chromatography (HILIC) column. The RP and HILIC columns were configured in parallel mode. The HILIC column was a Thermo Accucore HILIC column (100×3 mm i.d., 2.6 μm, part number: 17526-103030). The RP column was a Waters ACQUITY UPLC HSS T3 column (150×2.1 mm i.d., 1.8 μm, part number: 186003540). The temperature of these two columns was set as 40 °C. The HILIC column was operated as follows: mobile phase A was 10 mM ammonium acetate (pH adjusted to 3.25 with acetate) and mobile phase B was acetonitrile. The gradient was: 0 min, 100% B; 0 to 5 min, 100% B to 35% B; 5 to 12.7 min, 35% B; 12.7 to 12.8 min, 35% B to 95% B; 12.8 to 14.3 min, 95% B. The flow rate was set 0.3 mL/min. For the RP column, the mobile phase A was water with 0.1% formic acid and mobile phase B was 100% acetonitrile. The gradient was as follows: 0 min, 0% B; 0 to 5 min, 0% B; 5 to 6.1 min, 0 to 15% B; 6.1 to 10 min, 15 to 60% B; 10 to 12 min, 60% B; 12 to 14 min, 60% to 100% B; 14 to 14.1 min, 100% to 5% B; 14.1 to 16 min, 5% B. The flow rate was 0.4 mL/min.
The electrospray ionization probe was fixed at level C. The parameters for the probe were set as follows: sheath gas = 55 arbitrary units, auxiliary gas = 15 arbitrary units, sweep gas = 3 arbitrary units, spray voltage = 3.5 kV, capillary temperature = 320 °C, S-lens RF level = 65.0, auxiliary gas heater temperature = 450 °C. The method of mass spectrometer was set as follows: full scan range = 50 to 750 (m/z); resolution = 30,000; maximum injection time = 50 ms; automatic gain control (AGC) = 106 ions for both positive and negative modes.
In order to identify the metabolites in samples, MS/MS analysis was performed with the unlabeled samples. The LC methods and electrospray ionization conditions were the same as those used in analyzing the labeled and unlabeled samples in full MS mode. The method of mass spectrometer was set as follows: for full MS scan, scan range = 50 – 750 (m/z), resolution = 30,000, maximum injection time = 50 ms, automatic gain control (AGC) = 106 ions; for dd-MS2 scan, resolution = 15,000, maximum injection time = 100 ms, automatic gain control (AGC) = 5×104, loop count = 6, isolation window = 1.3 m/z, dynamic exclusion time = 1.2 s, the collision energy was set 10, 20, 40, 60 and 150 eV, respectively.
3. Theoretical Basis
Fig. 1 depicts the experiment design and data analysis workflow of a SIAM project. The unlabeled sample group and the labeled sample group are always generated in parallel under identical experimental conditions except that the labeled precursor metabolites are applied to every sample of the labeled sample groups, while the precursor metabolites without stable isotope tracers are applied to the unlabeled samples. The experimental data of unlabeled sample groups are used to limit the search space of metabolite candidates for identification of isotopologues from the labeled samples. The number of samples in the unlabeled sample groups does not need to be as large as that in the labeled sample groups, depending on the degree of experimental variation. Details of the data analysis modules are explained in the following sections.
Fig. 1.
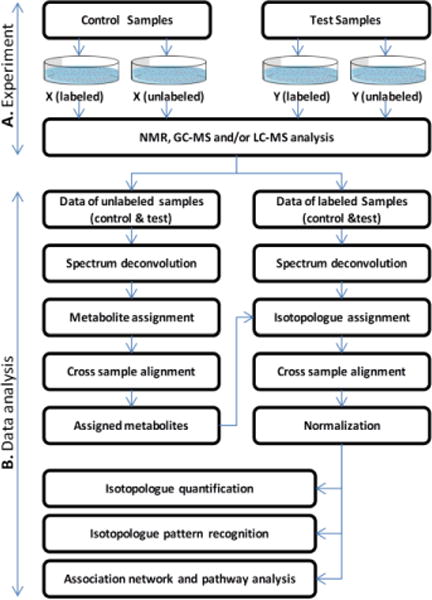
Experimental design and data analysis workflow of a SIAM project. (A) General experimental design of a SIAM study. A total of four sample groups are usually created. The unlabeled sample groups are mainly used to limit the metabolite candidates for identification of isotopologues from the labeled samples. (B) Workflow for analyzing SIAM data. Metabolites assigned for the unlabeled samples are used as the reference library to assign isotopologues in the labeled samples. Assigned isotopologues are then used to assess their abundance alteration between sample groups, pattern recognition, and pathway analysis.
3.1 m/z Value Recalibration
To analyze the SIAM data, the experimental data of both labeled and unlabeled samples are first deconvoluted and reduced into isotopic peak lists using MetSign software 10–12, by setting the minimum time span of a selected ion chromatogram (XIC) as wXIC ≥ 10 scans. To minimize the technical variation of the molecular ion m/z values, users can re-calibrate the m/z values using one or multiple (two or three) internal standards as reference compounds. Given one point calibration, the difference is computed between the theoretical m/z value of the reference and the closest experimental m/z value within the m/z variation window; all other experimental m/z values are corrected based on this difference. For the two and three point calibrations, the linear fitting is applied to the reference m/z values. The other m/z values are adjusted to the new m/z values according to the fitted linear function.
3.2 Fractional Mass Filter
Some compounds that are not originally present in the biological samples may be involved during sample processing, such as contaminates in the solvents and column bleeding. Comparing with metabolites, these compounds usually have different element composition and can be recognized by the fractional mass of their molecular weight. Fig. 2 depicts the fractional mass of all 45,942 metabolites in the Kyoto Encyclopedia of Genes and Genomes (KEGG) 13, LIPID MAPS 14, and the Human Metabolome Database (HMDB) 15. During spectrum deconvolution, any ions with fractional mass not falling into the dotted area in Fig. 2 are considered from compounds that are not in the biological samples, and the XICs of these ions are removed from deconvolution.
Fig. 2.
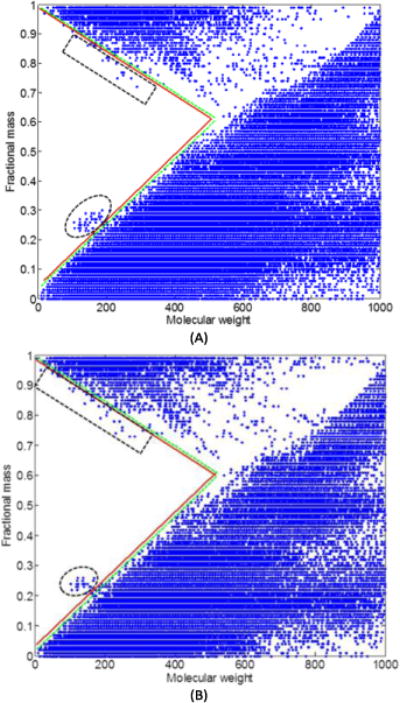
Fractional mass filter. The dotted lines are the boundary of the metabolite fractional mass that covers metabolites from the HMDB, KEGG, LIPID MAPS and our in-house database. The solid lines are the boundary created from the dotted lines using a user defined mass variation, e.g. 2 ppm. The ‘forbidden zone’ the region encompassed by solid lines and axes. The compounds in the ellipse area contain element As, and the compounds in the rectangle area contain element Br. (A) is the fractional mass filter generated for data acquired in positive mode and (B) is for data acquired in negative mode.
3.3 Identification of Metabolites from Unlabeled Samples
After spectrum deconvolution of both the labeled samples and unlabeled samples, the peak lists of unlabeled samples are first aligned based on retention time, parent ion m/z values and isotopic peak profile 10. If MS/MS spectra are available, the spectrum similarity between an experiment MS/MS spectrum X={(x1, m1), (x2, m2), …(xn, mn)} and an MS/MS spectrum of compound standard Y={(y1, m1), (y2, m2), …(yn, mn)} is calculated as follows 16:
| (1) |
where xi and yi are the intensity of the ith fragment ion in the experiment MS/MS spectrum X and the spectrum of a compound standard Y, mi is m/z value of the ith fragment ion, XWand YW are weighted spectra constructed as follows: and , and a, b are the weight factors for peak intensity and m/z value, respectively. The weight factors are set as (a, b) = (0.53, 1.3).
A tier-wise approach is used to assign metabolites to the aligned peaks of unlabeled samples based on the information of each compound contained in each database. Three different compound databases are used in this study. The in-house database contains chemical formula, retention time and MS/MS spectra for each compound standard. The Compound Discoverer (Thermo Fisher Scientific, Inc., Germany) contains chemical formula and MS/MS spectra for each compound. The KEGG, HMDB and LIPID MAPS only contain the chemical formula of each compound. Therefore, every aligned peak of the unlabeled samples is first matched to the compound standards in the in-house database based on the similarity of retention time, parent ion m/z value, isotopic peak profile and MS/MS spectra. Any peaks that do not have a match in the in-house database are then subjected to Compound Discoverer for parent ion m/z values and MS/MS spectrum matching. The rest of peaks are matched to the compounds in KEGG and HMDB databases by parent ion m/z values and isotopic peak profile matching.
In this study, the parameters used for metabolite identification are as follows: retention time variation ≤ 0.1 min, parent ion m/z variation ≤ 3 ppm, Pearson correlation coefficient of the similarity of isotopic peak profile ≥ 0.6, and the threshold of spectrum similarity SWC ≥ 0.6.
3.4 Initial Isotopologue Assignment
The metabolites assigned to the unlabeled samples are then used as reference metabolites to generate possible candidate stable isotope labeled isotopologues. The users can select a combination of up to five types of tracers (i.e., 2H, 13C, 15N, 18O and 34S). For example, given an unlabeled metabolite adenosine triphosphate (ATP, chemical formula C10H16N5O13P3), a total of 33 isotopologues will be generated for this metabolite if the users set a maximum number of stable isotope 13C to eight and 15N to three. After creating all possible isotopologues, each isotopic peak in the peak lists of the labeled samples is assigned to these isotopologues based on similarity of parent ion m/z value, retention time and isotopic peak profile. A correlation score between the experimental isotopic peak profile and the theoretical isotopic peak profile is calculated using Pearson’s correlation for each assignment to assess the confidence of each isotopologue assignment 10. It should be noted that this approach of isotopologue assignment does not require that the unlabeled isotopologue presents in the peak list of a labeled sample.
3.5 Overlapping Isotopic Peak Deconvolution
After the initial isotopologue assignment, the isotopologues of the same metabolite are consolidated together. The theoretical isotopic peak profile of each isotopologue is calculated using its chemical formula by setting the abundance of the monoisotopic peak M0 to 1.0000, and an iterative linear regression model is used to fit the theoretical isotopic peak profiles to the experiment data, i.e., the cluster of overlapping isotopic peaks.
Assuming an experimental isotopic peak cluster contains n isotopic peaks Y={(m/z1, y1), (m/z2, y2), …, (m/zn, yn)} and m isotopologues of the sample metabolite are assigned to these isotopic peaks, the linear regression model without an intercept is defined as follows:
| (2) |
| (3) |
where xij is the theoretical abundance of the jth isotopologue contributed to the ith isotopic peak (i=1,…, n and j=1,…, m)), wj is the weight factor of the jth isotopologue, is fitted peak abundance of the ith isotopic peak and yi is the experimental intensity of the ith isotopic peak in the cluster. The peak abundances of the isotopologues assigned to this isotopic peak cluster W={w1,…,wm } are determined by minimizing the fitting error using equation (3).
To reduce the rate of false-positive isotopologue assignment and increase the accuracy of spectrum deconvolution, a threshold of weight factor is set during regression. The fitting function is then optimized via iterative linear regression. During each iteration, any initially assigned isotopologues with weight factors smaller than a user defined non-negative threshold wmin are considered as false assignment and are removed from the linear combination model. The remaining isotopologues are then used to further fit the experimental data. This iteration process is repeated until all weight factors have values no less than the user defined threshold wmin .
3.6 Determining the Minimum Weight Factor wmin
Owing to experimental variation and the variation introduced during spectrum deconvolution, some isotopologues can be assigned to experiment peaks by error. These isotopologues typically have small weight factor values. To minimize the chance of false isotopologue assignment and increase the accuracy of spectrum deconvolution, the threshold of weight factor wmin is determined by a simulation study described below.
Metabolites recorded in KEGG, LIPID MAPS and HMDB were used for the simulation study. For each metabolite, all isotopic peaks (m/z value and associated abundance) were calculated from its chemical formula, by setting the abundance of the monoisotopic peak M0 to 1.0000 and the minimum abundance of other isotopic peaks not less than 10−5. A training mass spectrum was then formed using the calculated information of this metabolite, where the x-axis is the m/z value of each isotopic peak and the y-axis is its calculated relative abundance. We then added variation to the abundances of all isotopic peaks of this training spectrum by introducing different levels of Gaussian white noise contamination. To add the noise, we implemented two different types of noise, additive noise and multiplicative noise. The mathematical equations are shown as follows:
- Additive noise:
where X is the original intensity, eWGN is the white Gaussian noise, Yadd is the output intensity with additive noise; and(4) - Multiplicative noise:
(5)
where log e is the white Gaussian noise which is added to log X, Ymul is the output intensity with multiplicative noise.(6)
Assuming the simulated abundances of isotopic peaks in the training spectrum was generated by the unlabeled metabolite and all possibly labeled isotopologues of the same metabolite, the weight factor of each isotopologue, wj, was determined using the iterative linear regression model as described in equations (2) and (3), by setting wmin= 0. Because the training spectrum was constructed using the unlabeled metabolite only, any labeled isotopologues with non-zero values of weight factor are false-positive. The maximum of such weight factors was then considered as the simulated threshold of weight factor in equation (2).
3.7 Peak List Alignment and Normalization
After the initial isotopologue assignment and spectrum deconvolution, an isotopologue peak list is generated for each labeled sample. To recognize the abundance alteration of each isotopologue between sample groups, it is necessary to align the isotopologue peak lists of all labeled samples together, i.e., to recognize the same type isotopologue from different labeled samples. All isotopologues detected in the labeled samples are aligned by their retention time and m/z values, where all retention time values are converted to z-score and then aligned using a two-step approach as described in our previous work 11.
To make the metabolite abundances between samples comparable, an isotope-ratio based normalization method was developed to study differences in isotopologue abundance distribution between sample groups as follows:
| (7) |
where yi1, yi2, …, yip are the peak abundances of the isotopologues derived from the ith metabolite, and is the normalized abundance of the jth isotopologue.
After normalization, the conventional statistical significance tests such as two-tailed pairwise t-test with sample permutation or false discovery control (FDR), or partial least square discriminant analysis (PLS-DA), can be used to recognize significant differences between sample groups 11,17.
4. Results and Discussion
A significant feature of SIAM data is that one metabolite in an unlabeled sample is represented by multiple isotopologues in a corresponding labeled sample. This is due to the incorporation of different numbers of tracer atoms into the metabolite through different biosynthetic pathways. In order to identify the isotopologues from experimental data, one can directly use all known metabolites recorded in public databases such as KEGG, HMDB and LIPID MAPS to generate all possible isotopologue candidates for each metabolite, and then match the experimental data to each of those isotopologue candidates. Such an approach uses an extremely large search space and results in a high probability of false-positive isotopologue assignments. One method to reduce the rate of false assignment is to assume that the unlabeled isotopologue of each metabolite can be detected in the labeled sample. That is, the monoisotopic peak M0 of an unlabeled metabolite always presents in the cluster of isotopic peaks (generated by the isotopologues of this metabolite) in the labeled samples. However, this is not always valid for two reasons. First, a metabolite in the labeled samples could be fully labeled during biosynthesis, so that the unlabeled isotopologue does not present in the mass spectra of the labeled samples. Second, the abundance of a metabolite in an unlabeled sample is distributed among multiple isotopologues in a corresponding labeled sample. A low abundance metabolite detected in the unlabeled sample may not be detected in the labeled samples (as the unlabeled isotopologue) because its abundance may be less than the lower limit of detection (LOD) of the mass spectrometer.
To resolve these problems, the unlabeled samples are needed for the corresponding labeled samples. The metabolites assigned to each unlabeled sample can be aligned to obtain consensus of the initial metabolite assignment. After aligning assigned metabolites in the unlabeled samples, the metabolites with high confidence can be used as reference metabolites to generate all possible isotopologue candidates, which are then used as the search space for the isotopologues present in the labeled samples (left panel of Fig. 1(B)).
While high resolution mass spectrometry is able to resolve the isotopic peaks generated by the same metabolite, the isotopic peaks of isotopologues overlap with each other in m/z domain in SIAM data. For this reason, isotopic peaks (M0, M1, M2, …) generated by an unlabeled metabolite can always be assigned to a series of its labeled isotopologues based on m/z matching. These initially assigned isotopologues contain a high rate of false assignment, and reduce the accuracy of spectrum deconvolution. To minimize the rate of false isotopologue assignment, we developed an iterative linear regression model to fit the experimental isotopic peaks to the theoretical isotopic peak profiles. These profiles were calculated using the chemical formula of initially assigned isotopologues, where a threshold of weight factor was estimated from a simulation study. After the initial isotopologue assignment, all isotopic peak lists obtained from the labeled samples were aligned and normalized for downstream statistical analyses, including statistical significance tests for isotopologue quantification, pattern recognition, association network analysis and pathway analysis (right panel of Fig. 1(B)). All data analysis algorithms were implemented using a modular design.
4.1 Determining Minimum Weight Factor
To determine the threshold of weight factor wmin, we investigated the effects of noise level, noise type and tracer atoms on spectrum deconvolution by a simulation study. It has been reported that peak intensities in mass spectra are a combination of true signal and several categories of noise, which are generally either additive or multiplicative in nature 18,19. We first filtered out metabolites recorded in the KEGG, LIPID MAPS and HMDB databases that contain elements other than C, H, O, P, S, N and Se, resulting in 27,580 metabolites left for simulation study. After computing an isotopic peak profile for each metabolite, 2%, 5%, 7% and 10% of Gaussian white noise was added to the abundance of all peaks in an isotopic peak profile, respectively. By doing so, each metabolite then has a simulated mass spectrum isotopic peak cluster with a known level of Gaussian white noise at peak intensities. We then assumed that each simulated mass spectrum was acquired from a SIAM experiment, i.e., each simulated mass spectrum may contain isotopic peaks generated by labeled isotopologues of the metabolite. We further generated all possible isotopologues for each metabolite assuming that each metabolite was labeled by 2H, 13C, 15N, 18O, 34S, and 13C-15N, respectively. A linear regression was performed on each simulated mass spectrum as described in Equation (2) and (3) by setting the threshold of weight factor wmin=0.
To evaluate the performance, the simulation was performed three times under each condition. By design, any labeled isotopologues with non-zero weight factors are false assignments. A larger fitted weight factor means that the factor of interest (type of noise, level of noise, or tracer atom) has a significant effect on the accuracy of spectrum deconvolution. Fig. S1 (A) and S1 (C) depict the distribution of simulated maximum weight factors of 13C-labeled isotopologues along with their corresponding m/z values with 7% additive noise and multiplicative noise on the abundance of isotopic peaks, respectively. The correlation coefficient between the m/z value and weight factor in Fig. S1 (A) is 0.0095, while the correlation coefficient is 0.18 in Fig. S1 (C). Such small correlation coefficients demonstrate that the simulated maximum weight factors do not correlate with m/z values of isotopologues. Similar results were also observed for other tracer atoms at different levels of additive and multiplicative noise (data not shown). Therefore, a single value of weight factor threshold can be applied to all isotopologues regardless of their m/z values.
Fig. 3 depicts the maximum weight factors of the simulation results at different simulation conditions. The numeric numbers and corresponding standard deviations of the simulation results are listed in Supplementary Table S1. The multiplicative noise affects more than the additive noise when the noise level is less than 5%, while this difference is diminished when the noise level is increased to > 5%. The type of tracer atom has a relatively small effect on the maximum of simulated weight factors. While the low level of noise does not affect the maximum weight factors, a level of noise larger than 5% dramatically increases the maximum weight factor, regardless of the type of noise. For example, the average of three weight factors for 2H with 2% additive noise is 2.6×10−14, but increased to 8.8×10−6 at 7% additive noise level (Table S1). Because noise levels in MS data are LC-MS platform-dependent, it appears necessary to assess the noise level by analyzing a set of metabolite standards to determine the threshold of weight factor before analysis of complex samples.
Fig. 3.
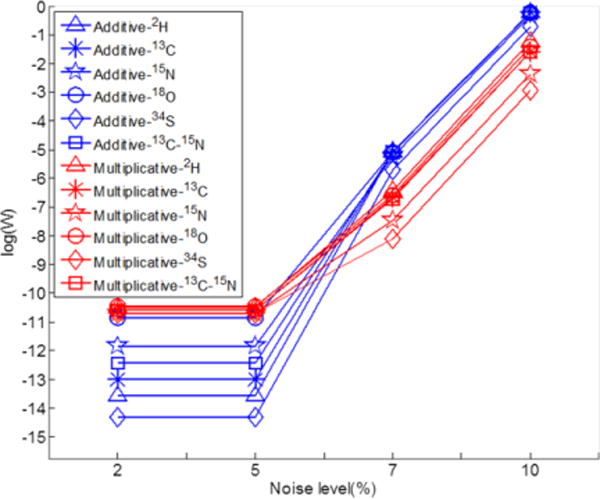
Effects of noise type, noise level and tracer atoms on weight factor.
4.2 Analysis of Mixture of Known Compounds
16 13C-labeled amino acids and 16 unlabeled amino acids were mixed at two different weight ratios (0.5:1 and 1:1), and three samples were prepared in parallel for each mixture. Thus, two sample groups were formed with three samples in each sample group. All six samples were analyzed by DI-FTICR-MS.
Table S2 lists the analysis results, where two tailed pairwise t-test with sample permutation was used for statistical significance test. The amino acids L-isoleucine and L-leucine are isomers. DI-FTICR-MS cannot differentiate these two metabolites based on their parent ion m/z values; therefore, these two metabolites overlapped each other. L-Serine was excluded from further analysis because it can be reliably detected in only one sample due to the very small peak abundance. All 15 13C-labeled amino acids were correctly recognized as the metabolites with significant abundance alterations between two sample groups, with p-values ranging from 2.5×10−3 to 5.1×10−2. While the ideal fold-change is 0.5, L-proline and L-threonine have fold-changes of 0.28 and 0.2, respectively. Fig. S2 depicts that the fold-change and peak abundance have a strong correlation with a correlation coefficient of 0.86 (p < 0.0001). Large fold-change variations of L-proline and L-threonine were mainly induced by their low peak abundance. Such results demonstrate that the developed method can correctly identify and quantify metabolites from mixtures of metabolite standards.
4.3 Analysis of DI-FTICR-MS data of biological samples
To illustrate the applicability of the developed algorithms in the analysis of complex samples, we obtained DI-FTICR-MS metabolomics data from the cardiac-derived murine cells cultured in 5 mM [U-13C]-glucose. Over 200 metabolites were detected and assigned per sample based on parent ion m/z value and isotopic peak profile, each comprising multiple isotopologues.
After isotopologue peak assignment, iterative linear regression was performed to deconvolute the isotopic peak cluster of the isotopologues from the same metabolite and the peak intensities were modified to reflect true isotopic enrichment. It is worth noting, that this process is performed simultaneously and can be applied to data containing any number of isotopic tracers (for example, tracers containing a combination of 13C, 15N, 2H, and 18O can be used). Table S3 shows the original percent distribution of isotopic peaks of 9 nucleotides/nucleotide sugars and the percent distributions of 13C-labeled isotopologues after spectrum deconvolution. Clear differences can be observed for all listed metabolites with many values reduced to zero. In the context of biological data interpretation, the higher the abundance of a particular isotopologue, the higher the utilization of a pathway of interest. Conversely, the lack of a certain type of isotopologue provides direct evidence that the tracer was not metabolized through corresponding pathways.
Once the abundances of all isotopologues were obtained for multiple samples, isotope-ratio based normalization was performed to reduce the concentration effect and experimental variance, which permits better illustration of isotopic fractional enrichment. Fig. 4(A) shows the histogram of isotopologues of UDPHexNAc. A total of seventeen isotopologues were detected for this compound. Using isotope-ratio based normalization, it was recognized that zero, six, eight, eleven, and thirteen 13C-UDPHexNAc had significant change in its abundance level between the Genotype 1 group and Genotype 2 group with fold changes of 2.33 (p = 0.02), 1.84 (p = 0.01), 1.44 (p = 0.02), 0.71 (p = 0.02), and 0.74 (p = 0.03), respectively. Fig. 4(B) is another example showing the histogram of isotopologues of ADP. A total of ten isotopologues were detected for this compound. And it was recognized that five, seven, eight, and nine 13C-ADP had significant change in its abundance level between the Genotype 1 group and Genotype 2 group with fold changes of 1.22 (p = 0.02), 0.21 (p = 0.001), 0.06 (p = 0.01), and 0.09 (p = 0.04), respectively. Details of the biological discovery of this study have been discussed in part in a separate report 3.
Fig. 4.
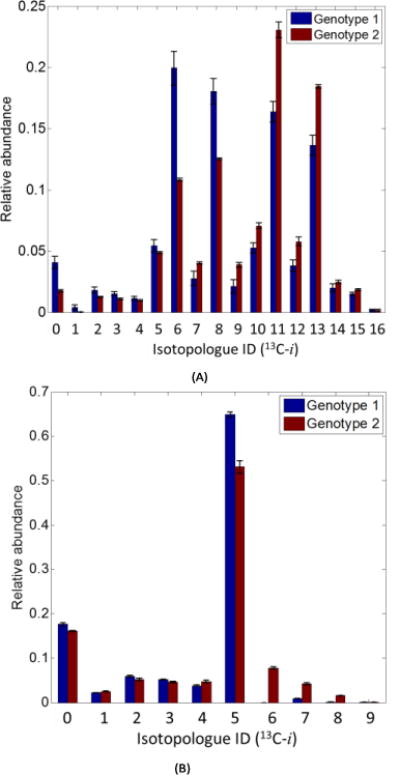
Sample isotopologue abundance distributions. Each plot shows the abundance distribution of all isotopologues of the same metabolite in two sample groups. (A) UDPHexNAc, and (B) ADP.
4.4 Analysis of LC-MS data of biological samples
To further demonstrate the capability of the developed method in analyzing LC-MS data, the polar metabolites of the labeled and unlabeled samples were analyzed by 2DLC-MS, where the RP and HILIC columns were configured in parallel. Fig. S3 shows the TICs of two samples randomly selected from the unlabeled and labeled samples. The high similarity of the two original TICs (Figs. S3 (A) and S3 (C)) demonstrates that incorporating 13C atoms into metabolites did not introduce retention time shift between the labeled and unlabeled samples, and the 2DLC-MS system was very stable during data acquisition. However, the difference can be observed from the two TICs (Figs. S3 (B) and S3 (D)) that were reconstructed after spectrum deconvolution. This difference was introduced by incorporation of 13C atoms into metabolites.
The purpose of using fractional mass filtering during spectrum deconvolution is to detect and remove the non-metabolite peaks. 6443 XICs were constructed from the unlabeled sample, while 6670 XICs from the labeled sample. Using the fractional mass filter, 109 XICs were removed from the unlabeled sample and 103 XICs from the labeled sample.
After fractional mass filtering, a total of 9,917 peaks were picked from the unlabeled sample and 10,422 peaks from the labeled sample. By using retention time, parent ion m/z value and MS/MS spectrum matching, 134 compounds were identified from the unlabeled sample. Using these compounds as reference compounds, a total of 753 possible isotopologues were generated and 378 of these isotopologues were assigned to the peaks detected in the labeled sample.
Fig. 5(A) depicts the mass spectrum of L-glutamic acid detected in a 13C-labeled sample (top) and an unlabeled (bottom) sample. After isotopologue peak deconvolution, six isotopologues of L-glutamic acid were detected from these labeled sample, including zero 13C-, one 13C-, two 13C-, three 13C-, four 13C- and five 13C-glutamic acid. Fig. 5(B) shows the isotopologues distribution of L-alanine eluted at retention time 4.75 min from the RP column. A total of two isotopologues were detected for this compound. Using isotope-ratio based normalization, it was recognized that three 13C-alanine had significant change in its abundance level between the Genotype 1 group and Genotype 2 group with a fold-change of 0.26 (p = 0.02). Fig. S4 is another example, showing the histogram of isotopologues of L-glutathione (reduced). While five isotopologues were quantified for this metabolite, none of the isotopologues were recognized with significant abundance changes. These analysis results demonstrate that the developed algorithms are also able to process SIAM data of biological samples analyzed by LC-MS.
Fig. 5.
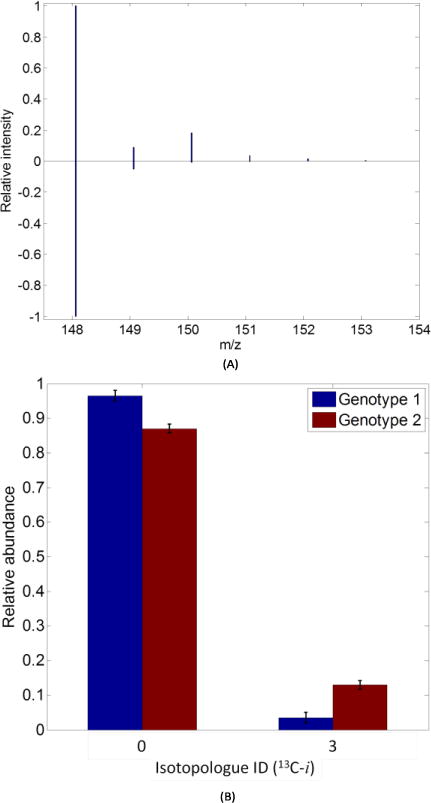
Sample mass spectra and abundance distributions selected from the results of analyzing LC-MS data. (A) Sample mass spectra of 13C-labeled (top) and unlabeled (bottom) mass spectra of L-glutamic acid. (B) Histogram of each isotopologue grouped by sample groups of L-alanine.
5. Conclusions
We developed a suite of algorithms for automatic analysis of stable isotope assisted metabolomics (SIAM) data acquired on a high resolution mass spectrometer. To reduce the rate of false positive isotopologue assignment, metabolites detected in the unlabeled samples were used as the reference metabolites to generate possible isotopologue candidates for assigning an isotopologue to peaks detected in the labeled samples. In this approach, the isotopologue assignment does not require that the unlabeled isotopologue presents in the peak list of a labeled sample.
To increase the accuracy of deconvoluting the overlapping isotopologue peaks, an iterative linear regression model was also developed, where the threshold of weight factor was determined by a simulation study assuming different levels of Gaussian white noise contamination. Our study show that the threshold of weight factor depends on the degree of noise level, and there is no significant difference between the effects of additive noise and those of multiplicative noise. To investigate the difference of isotopologue distribution between sample groups, an isotope-ratio based normalization was also developed. These data analysis methods were integrated into a computational package using a modular design.
The developed software package can be applied to data containing any number of isotopic tracers, such as tracers containing a combination of 13C, 15N, 2H, and 18O. The performance of the algorithms was tested by analyzing two sets of SIAM data acquired by DI-MS and LC-MS. The analysis of the DI-MS data acquired from mixtures of known compounds shows that all of the known compounds were correctly identified and quantified. Analysis of DI-MS and LC-MS data acquired from the two groups of metabolite samples extracted from cardiac-derived cells further demonstrated that the developed algorithms can be used to SIAM data acquired from complex samples.
Supplementary Material
Acknowledgments
The authors thank Mrs. Marion McClain for review of this manuscript and Qianhong Li for providing initial vials of cultured cells. This work was supported by NIH grant nos. 1S10OD020106-01 (XZ), 1R01HL130174 (BGH), 1R56HL122580 (BGH), 1P20GM113226 (CJM), 1P50AA024337 (CJM), 1U01AA021893-01 (CJM), 1U01AA021901-01 (CJM), 1U01AA022489-01A1 (CJM) and 1R01AA023681-01 (CJM), the Department of Veterans Affairs 1I01BX002996-01A2 (CJM), and the American Diabetes Association Pathway to Stop Diabetes Grant 1-16-JDF-041 (BGH).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Creek DJ, Chokkathukalam A, Jankevics A, Burgess KE, Breitling R, Barrett MP. Analytical chemistry. 2012;84:8442. doi: 10.1021/ac3018795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Hoene M, Zhao X, Chen S, Wei H, Haring HU, Lin X, Zeng Z, Weigert C, Lehmann R, Xu G. Analytical chemistry. 2013;85:4651. doi: 10.1021/ac400293y. [DOI] [PubMed] [Google Scholar]
- 3.Salabei JK, Lorkiewicz PK, Mehra P, Gibb AA, Haberzettl P, Hong KU, Wei XL, Zhang X, Li QH, Wysoczynski M, Bolli R, Bhatnagar A, Hill BG. Journal of Biological Chemistry. 2016;291:13634. doi: 10.1074/jbc.M116.722496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang XJ, Chen YJ, Cho K, Nikolskiy I, Crawford PA, Patti GJ. Analytical Chemistry. 2014;86:1632. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capellades J, Navarro M, Samino S, Garcia-Ramirez M, Hernandez C, Simo R, Vinaixa M, Yanes O. Analytical Chemistry. 2016;88:621. doi: 10.1021/acs.analchem.5b03628. [DOI] [PubMed] [Google Scholar]
- 6.Chokkathukalam A, Jankevics A, Creek DJ, Achcar F, Barrett MP, Breitling R. Bioinformatics. 2013;29:281. doi: 10.1093/bioinformatics/bts674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mashego MR, Wu L, Van Dam JC, Ras C, Vinke JL, Van Winden WA, Van Gulik WM, Heijnen JJ. Biotechnology and Bioengineering. 2004;85:620. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- 8.Salabei JK, Lorkiewicz PK, Holden CR, Li Q, Hong KU, Bolli R, Bhatnagar A, Hill BG. Stem Cells. 2015;33:2613. doi: 10.1002/stem.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorkiewicz P, Higashi RM, Lane AN, Fan TW. Metabolomics : Official journal of the Metabolomic Society. 2012;8:930. doi: 10.1007/s11306-011-0388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei X, Sun W, Shi X, Koo I, Wang B, Zhang J, Yin X, Tang Y, Bogdanov B, Kim S, Zhou Z, McClain C, Zhang X. Analytical chemistry. 2011;83:7668. doi: 10.1021/ac2017025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X, Shi X, Kim S, Zhang L, Patrick JS, Binkley J, McClain C, Zhang X. Analytical chemistry. 2012;84:7963. doi: 10.1021/ac3016856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X, Shi X, Kim S, Patrick JS, Binkley J, Kong M, McClain C, Zhang X. Analytical chemistry. 2014;86:2156. doi: 10.1021/ac403803a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.KEGG. 2012;2012 [Google Scholar]
- 14.MAPS L. 2012;2012 [Google Scholar]
- 15.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic acids research. 2009;37:D603. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Koo I, Jeong J, Wu S, Shi X, Zhang X. Anal Chem. 2012;84:6477. doi: 10.1021/ac301350n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Shi X, Koo I, Kim S, Schmidt RH, Arteel GE, Watson WH, McClain C, Zhang X. Bioinformatics. 2013;29:1786. doi: 10.1093/bioinformatics/btt275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veselkov KA, Vingara LK, Masson P, Robinette SL, Want E, Li JV, Barton RH, Boursier-Neyret C, Walther B, Ebbels TM, Pelczer I, Holmes E, Lindon JC, Nicholson JK. Analytical chemistry. 2011;83:5864. doi: 10.1021/ac201065j. [DOI] [PubMed] [Google Scholar]
- 19.Anderle M, Roy S, Lin H, Becker C, Joho K. Bioinformatics. 2004;20:3575. doi: 10.1093/bioinformatics/bth446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


